Conspectus
Surface-enhanced Raman spectroscopy (SERS) fingerprinting is highly promising for identifying disease markers from complex mixtures of clinical sample, which has the capability to take medical diagnoses to the next level. Although vibrational frequency in Raman spectra is unique for each biomolecule, which can be used as fingerprint identification, it has not been considered to be used routinely for biosensing due to the fact that the Raman signal is very weak. Contemporary SERS has been demonstrated to be an excellent analytical tool for practical label-free sensing applications due its ability to enhance Raman signals by factors of up to 108–1014 orders of magnitude. Although SERS was discovered more than 40 years ago, its applications are still rare outside the spectroscopy community and it is mainly due to the fact that how to control, manipulate and amplify light on the “hot spots” near the metal surface is in the infancy stage.
In this Account, we describe our contribution to develop nanoachitecture based highly reproducible and ultrasensitive detection capability SERS platform via low-cost synthetic routes. Using one-dimensional (1D) carbon nanotube (CNT), two-dimensional (2D) graphene oxide (GO), and zero-dimensional (0D) plasmonic nanoparticle, 0D to 3D SERS substrates have been designed, which represent highly powerful platform for biological diagnosis. We discuss the major design criteria we have used to develop robust SERS substrate to possess high density “hot spots” with very good reproducibility. SERS enhancement factor for 3D SERS substrate is about 5 orders of magnitude higher than only plasmonic nanoparticle and more than 9 orders of magnitude higher than 2D GO. Theoretical finite-difference time-domain (FDTD) stimulation data show that the electric field enhancement |E|2 can be more than 2 orders of magnitude in “hot spots”, which suggests that SERS enhancement factors can be greater than 104 due to the formation of high density “hot spots” in 3D substrate. Next, we discuss the utilization of nanoachitecture based SERS substrate for ultrasensitive and selective diagnosis of infectious disease organisms such as drug resistance bacteria and mosquito-borne flavi-viruses that cause significant health problems worldwide. SERS based “whole-organism fingerprints” has been used to identify infectious disease organisms even when they are so closely related that they are difficult to distinguish. The detection capability can be as low as 10 CFU/mL for methicillin-resistant Staphylococcus aureus (MRSA) and 10 PFU/mL for Dengue virus (DENV) and West Nile virus (WNV). After that, we introduce exciting research findings by our group on the applications of nanoachitecture based SERS substrate for the capture and fingerprint detection of rotavirus from water and Alzheimer’s disease biomarkers from whole blood sample. The SERS detection limit for β-amyloid (Aβ proteins) and tau protein using 3D SERS platform is several orders of magnitude higher than the currently used technology in clinics. Finally, we highlight the promises, major challenges and prospect of nanoachitecture based SERS in biomedical diagnosis field.
1. Introduction
Accurate diagnosis of superbug organisms and disease markers from clinical fluids is highly challenging task for clinical practitioner.1−8 Since in the early stage of disease the amount of superbug or biomarkers for cancer and other diseases are too low in abundance, traditional diagnostic tools used in clinics are not capable of finding them.1−8 As a result, there is a huge demand in clinics to find ultrasensitive probe for disease marker detection from clinical sample, which can be used to move forward the medical diagnoses to the next level.1−8 Since past decade, scientists are trying to develop different nanophotonics based techniques which have the capability to identify different biomarkers for diagnosing dementia disease, superbug organisms for infectious disease, and disease markers for cancer.6−20
Surface-enhanced Raman spectroscopy (SERS),8−16 where a weak Raman scattering signal is enhanced by 6–11 orders of magnitude by adsorbing the bio-organisms onto a roughened plasmonic surface, became an ultrasensitive vibrational fingerprinting technique for identifying biological and chemical analytes.17−27 Since the SERS based bio-organisms identification offers several distinct advantages such as Raman intensities ∼50–100 times narrower than emission bandwidth and very stable against photodegradation or photobleaching due to the instantaneous nature of the process, it will be much superior compared to other biomedical sensing technique used in clinics.28−40
SERS substrate which exhibits strong chemical enhancement and extremely high electromagnetic enhancement has the capability to achieve excellent sensitivity for real life applications.40−50 Chemical enhancement occurs due to the charge-transfer interactions between the SERS substrate with bio-organisms. On the other hand, electromagnetic enhancement occurs due to the formation of plasmonic “hot spot”.15−30 In SERS, due to the squeezing of incident light into extremely small regions in the interface of two plasmon couple metallic nanoparticles, it generates thousands- to million-fold local electromagnetic field enhancements in the “hot spot”, which allows SERS to be used as ultrasensitive technique with the detection of only a few molecules.15−25 Although the first paper on SERS was published in 1974,8 which is more than 40 years ago, its applications are still rare outside the spectroscopy community and it is mainly due to the fact that until now scientists are facing enormous challenges on how to design reproducible plasmonic SERS substrate with large number of “hot spots” and also how to manipulate light on the “hot spots”.20−30
The current Account highlights our recent progress in the development of one-dimensional (1D) to three-dimensional (3D) plasmonic SERS substrate, which can be used as highly powerful platform for biological diagnosis,22,32−42 as shown in Figure 1. We outline the major design criteria one needs use to develop robust SERS substrate to possess high density “hot spots” with very good reproducibility. We examine how the plasmonic coupling enhances SERS intensity via theoretical finite-difference time-domain (FDTD) stimulation modeling, as shown in Figure 1. Then we highlight the exciting research findings by our group on the applications of SERS substrate for the capture and fingerprint identification of disease biomarkers, mosquito-borne flavi-viruses, and drug resistance superbugs.32−42 At the end, we conclude with the promises and challenges for future evolution on nanoachitecture based SERS in biomedical diagnosis field.
Figure 1.
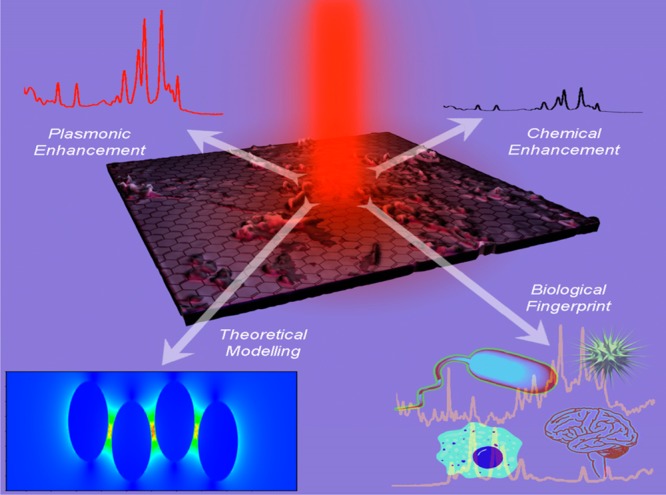
Schematic representation shows the SERS substrate with high plasmonic and chemical enhancement capability can be used for biological fingerprint.
2. Zero Dimensional Gold Nanoparticle Based SERS for Fingerprinting Infectious Disease Agents
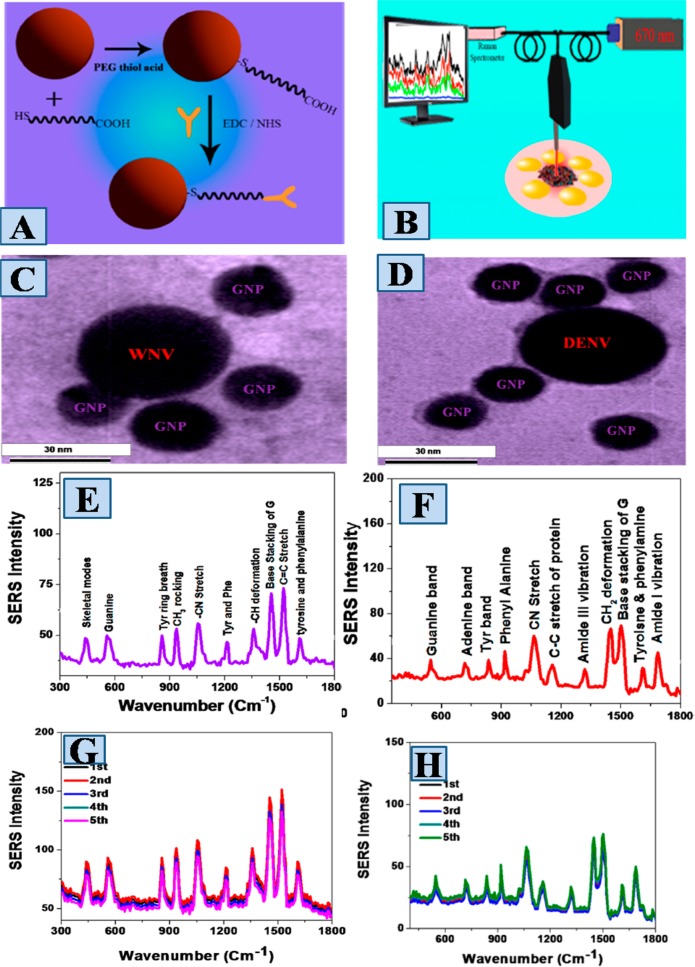
Unique fingerprint spectrum is essential for personal identification of any chemicals.5−9 Recently we have reported the development of nanoparticle based SERS, which has the capability to be used as fingerprint spectra for viruses responsible for infectious disease.32Figure 2 shows the design criteria we have used for the development of antiflaviviral antibody conjugated plasmonic nanoplatform based SERS which has the capability for the fingerprint identification of viruses responsible for mosquito-borne diseases. For this purpose, initial plasmonic nanoparticles were coated with polyethylene glycol (PEG) thiol acid via Au–S linkage.32 In the next step, antiflaviviral 4G2 antibodies were conjugated with PEG coated plasmonic nanoparticles via 1-ethyl-3-[3-(dimethylamino)-propyl] carbodiimide hydrochloride (EDC) coupling method, as shown Figure 2A. transmission electron microscopy (TEM) image data, as shown in Figure 2C and D, indicate that due to the antigen–antibody interaction, 4G2 antibody conjugated plasmonic nanoparticles form “hot-spot” on the surface of Dengue virus (DENV) and West Nile virus (WNV), and as a result, we are able to use antiflaviviral antibody conjugated plasmonic nanoplatform for specific identification of DENV and WNV.32Figure 2B shows the portable Raman design we have used for all SERS measurements, where the QE6500 portable spectrometer has been used for Raman data acquisition and a portable NIR laser has been used as excitation source. Figure 2E and F shows the Raman spectra from DENV and WNV attached plasmonic nanoparticle, where the amide I Raman band due to the protein backbone and the −CH2 deformation Raman band due to the lipid are unique for WNV.32 On the other hand, the skeleton mode Raman band and the CH3 rocking Raman modes are unique for DENV-2.
Figure 2.
(A) Synthetic path for the development of antiflaviviral antibody attached plasmonic nanoparticle based SERS. (B) Portable Raman probe we have designed for SERS measurement. (C) TEM image of WNV attached plasmonic nanoparticle assembly. (D) TEM image of DENV attached plasmonic nanoparticle assembly. (E) Raman spectra from DENV conjugated nanoarchitecture. (F) Raman spectra from WNV conjugated nanoarchitecture. (G) Reproducibility of Raman spectra from DENV conjugated nanoarchitecture produce in different batches. (H) Reproducibility of Raman spectra from WNV conjugated nanoarchitecture produce in different batches. Reproduced with permission from ref (32). Copyright 2015 American Chemical Society.
Those unique Raman bands can be used for identification of DENV and WNV selectively.32Figure 2G and H shows that the SERS intensity stability is excellent and the fluctuation is less than 8% for nanoparticle based SERS substrate developed in different batches. The fluctuation is mainly due to the fact that the spatial distribution of “hot spots” can be different for different batches of experiments.32 We have reported that the detection capability can be as low as 10 PFU/mL for Dengue virus (DENV) and West Nile virus (WNV) respectively.32 Although we have shown SERS application for infection diseases viruses, until now there is a lack of good understanding on the molecular mechanism of interaction between antibody conjugated plasmonic platforms and viruses, which is very important for better performance of the proposed SERS sensor.
3. Developing Hybrid CNT Based 1D SERS Substrate for Fingerprint Sensing of Explosives
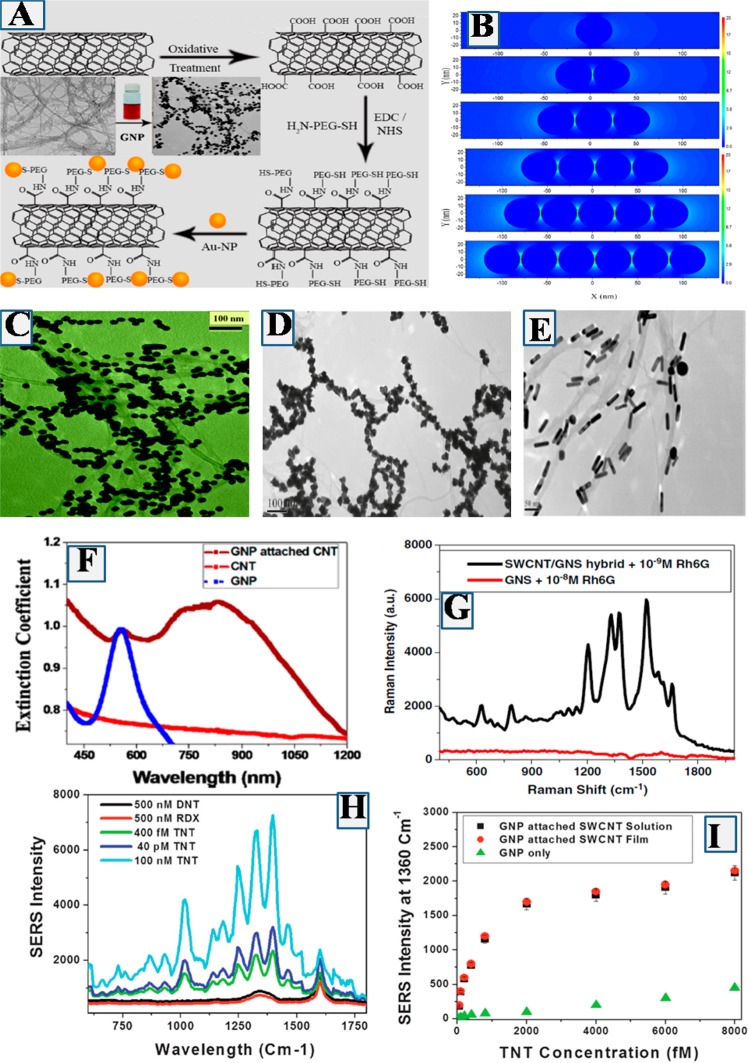
To develop a 1D plasmonic SERS substrate with a large number of “hot spots”, we have designed a hybrid carbon nanotube (CNT) based substrate, where one-dimensional CNTs are conjugated with zero-dimensional plasmonic gold nanoparticles in different size and shapes.37,39,41,42 In our design, plasmonic gold nanoparticle with different shapes have been used as reactive sites for the binding with chemicals in interest.37,39,41,42 Since high aspect ratio CNTs possess a huge surface area, we have used CNT templates to attach plasmonic nanoparticles of different shapes,37,39,41,42 as shown in Figure 3C–E. In our design, zero-dimensional plasmonic gold nanoparticles attached 1D CNTs were synthesized from carboxy functionalized SWCNT, as shown in Figure 3A. For this purpose, initially we have developed water-soluble carboxy functionalized single wall CNTs (SWCNTs) from solid SWCNT using strong oxidizing agent like nitric acid and sulfuric acid oxidizing agent.37,39,41,42 After that, thiol poly(ethylene glycol) (PEG) functionalized SWCNTs were developed by treating HS-PEG-NH2 with carboxy functionalized SWCNTs in the presence of as a cross-linking agent,37,39,41,42 as shown in Figure 3A. Figure 3C–E shows that, in the SERS substrate, different shaped nanoparticles generate a huge number of “‘hot spot’” sites which allow to enhance the confinement of local electromagnetic fields by several orders of magnitude. Finite difference time domain (FDTD) simulation calculation, as shown in Figure 3B, indicates that field enhancement for spherical nanoparticle based assembly containing five “‘hot sites’” is around 2 orders of magnitude higher than that of the individual spherical nanoparticle.37 It is now well documented that SERS enhancement is proportional to the square of the plasmon coupling confinement field. As a result, FDTD simulation data indicate that one can achieve about 4 orders of magnitude SERS enhancement by just forming several “‘hot sites’” on SWCNT surfaces.37Figure 3F shows that when nanoparticles generate huge number of “‘hot sites’” on SWCNT surface, the plamonic band becomes very strong and broad, which allows the incident NIR light to be resonance with the SERS substrate.37 Due to the resonance phenomena between incident light and SERS substrate extinction, 2–3 orders of magnitude SERS enhancement are expected.37,39,41,42
Figure 3.
(A) Synthetic path for the development of plasmonic nanoparticle attached hybrid CNT. (B) FDTD simulated data show the electric field enhancement profiles for 40 nm gold nanoparticle assembly structure. (C) TEM image shows plasmonic spherical gold nanoparticles are attached on SWCNT and formed large number of “hot spots” on CNT (Reproduced with permission from ref (37). Copyright 2015 American Chemical Society). (D) TEM image shows plasmonic popcorn shape gold nanoparticles are attached on SWCNT via the formation of “hot spots” (reproduced with permission from ref (39). Copyright 2011 American Chemical Society). (E) TEM image showing plasmonic rod shaped gold nanoparticles are attached on SWCNT via the formation of “hot spot” (Reproduced with permission from ref (42). Copyright 2011 Elsevier). (F) Extinction spectra for GNP, GNP attached SWCNT, and only SWCNT (Reproduced with permission from ref (37). Copyright 2015 American Chemical Society). (G) Raman intensity from Rh6G on nanoparticle and nanoparticle attached SWCNT. (H) Raman intensity from TNT on popcorn shape nanoparticle and nanoparticle attached SWCNT. (I) TNT Raman intensity enhancement on nanoparticle and nanoparticle attached SWCNT (Reproduced with permission from ref (41). Copyright 2012 Royal Society of Chemistry).
As shown in Figure 3G, due to the simultaneous electric field and resonance enhancement, the different shape nanoparticle attached 1D SWCNT substrate can exhibit around 9 orders of magnitude higher Raman signal from Rhodamine 6G (Rh-6G).41 The SERS enhancement factor is about 2 orders of magnitude higher than only nanoparticle.41 Due to the huge SERS enhancement via high density “hot site” formation, we have used 1D hybrid CNT substrate based SERS for explosive detection.41Figure 3H shows the SERS spectra from explosive 2,4,6-trinitrotoluene on 1D hybrid CNT surface, which exhibits several strong Raman modes due to the NO2 symmetric and asymmetric stretching vibrations,41 as shown in Table 1. Similarly we have also observed strong Raman bands for C6H2–C vibration, C–H out-of-plane mode vibration and CH3 deformation modes, which can be used as fingerprint Raman bands for TNT.41Figure 3I shows that due to the huge number of “‘hot sites’” formation, 1D SERS substrate can be used for the fingerprint detection of TNT even at 100 fM concentration level.41 Although we have demonstrated that 1D hybrid CNT based SERS substrate can be used for ultrasensitive detection of explosives, we need to have better understanding on how to control the distance between two particles on the CNT surface.
Table 1. Raman Bands Observed in SERS Spectra from TNT, RDX, and MRSA.
| peak position for TNT (cm–1) | peak position for RDX (cm–1) | peak position for MRSA (cm–1) | vibration mode |
|---|---|---|---|
| 1560 | 1560 | NO2 asymmetric stretch | |
| 1460 | δ(CH2) saturated lipids | ||
| 1303 | ν(NH2) stretch for adenine | ||
| 1258 | CH2 scissoring and N–N stretch | ||
| 1210 | C6H2–C vibration | ||
| 1026 | CH3 deformation | ||
| 930 | 930 | N–O deformation band | |
| 855 | ring breathing Tyr protein | ||
| 580 | C–O–C glycosidic ring deformation | ||
| 420 | skeletal modes CC |
4. Developing Hybrid Graphene Oxide Based 2D SERS Substrate with Excellent SERS Enhancement Capability via Plasmonic and Chemical Enhancement
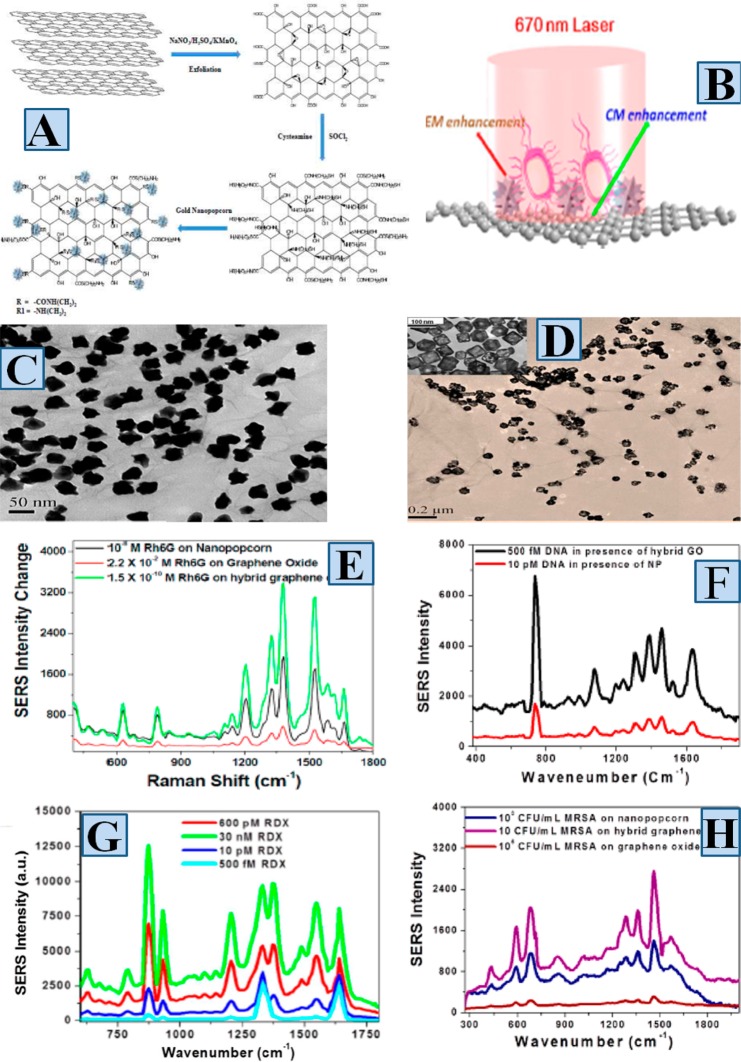
To develop 2D SERS substrate which possesses strong chemical enhancement and extremely high electromagnetic enhancement, we have designed hybrid graphene oxide (GO) based substrate, where two-dimensional GOs are conjugated with zero-dimensional plasmonic gold nanoparticles in different sizes and shapes,36,38 as shown in Figure 4B and D. In our design, the two-dimensional chemically treated graphene based material has been used to enhance Raman signal via chemical enhancement mechanism,36,38 as shown in Figure 4B. Also, we have used 2D GO as strong fluorescence quencher and as a result, it suppressed the background emission signal from biological media.36,38 The above two factors allowed us to use 2D GO as excellent SERS substrate which has very good biocompatibility.36,38 To develop 2D substrate which exhibits strong plasmonic and chemical enhancement, in our design, different size and shaped plasmonic nanoparticles have been used for the best enhancement of SERS signal via the formation of large amount “‘hot spot’” sites on 2D GO surface,36,38 as shown in Figure 4C and D. Zero-dimensional plasmonic gold nanoparticles attached 2D-GOs were synthesized from carboxy functionalized 2D GO,36,38 as shown in Figure 4A. For this purpose, initially strong oxidizing agents were used for graphite exfoliation to produce 2D graphene oxide using modified Hummers reported method.36,38 In the next step, acyl chloride functionalized 2D GO was developed from carboxy functionalized 2D GO by treating with thionyl chloride.36,38 After that, amine functionalized different shape plasmonic nanoparticles were attached on 2D GO surface via the acyl chloride group.36,38Figure 4C and D shows that in the developed hybrid 2D GO based SERS substrate, different shape nanoparticles generates huge number of “‘hot spot’” sites which enhance the Raman signal by several orders of magnitude.36,38Figure 4E shows that we have observed strong Raman bands from few picomolar Rh6G adsorbed on hybrid 2D surface, whereas the Raman band from same Rh6G molecule at 100 mili molar concentration on graphene oxide are almost negligible.38 Using 1511 cm–1 vibrational mode which is due to the skeleton stretching frequency, we found out that only GO can enhance the Raman signal by 2 orders of magnitude via chemical enhancement mechanism. On the other hand, the enhancement factor is around 1011 in the case of gold nanopopcorn attached 2D GO based SERS substrate.38 The SERS enhancement factor is about 4 orders of magnitude higher for hybrid 2D GO based SERS substrate than only nanoparticle. It is mainly due to the cooperative SERS enhancement mechanism like electromagnetic enhancement, chemical enhancement and lighting rod effect enhancement, works simultaneously in hybrid 2D GO based SERS substrate.38
Figure 4.
(A) Synthetic path for the development of plasmonic nanoparticle attached hybrid 2D GO. (B) Scheme shows 2D hybrid SERS substrate using plasmonic nanoparticle attached GO has capability to tune electromagnetic and chemical enhancement simultaneously. (C) TEM image shows that plasmonic popcorn shape gold nanoparticles are attached on 2D GO and formed large number of “hot spots” on GO (Reproduced with permission from ref (38). Copyright 2013 American Chemical Society). (D) TEM image shows plasmonic gold-nanocages are attached on 2D GO via the formation of “hot spots” (Reproduced with permission from ref (36). Copyright 2014 American Chemical Society). (E) Raman intensity from Rh6G on nanoparticle and nanoparticle attached 2D GO (reproduced with permission from ref (38)., Copyright 2013, American Chemical Society). (F) Raman intensity from partial sequence of the HIV-1 gag gene on nanoparticle and nanoparticle attached 2D GO. (G) Raman intensity from different concentration of RDX on plasmonic gold nanoparticle attached 2D GO (Reproduced with permission from ref (36). Copyright 2014 American Chemical Society). (H) MRSA Raman intensity enhancement on GO, nanoparticle, and nanoparticle attached 2D GO (Reproduced with permission from ref (38). Copyright 2013 American Chemical Society).
Since the SERS enhancement factor is in the order of 11 for hybrid GO based 2D SERS substrate, we have used these 2D substrates for fingerprint identification of DNA, drug resistance superbugs MRSA, and RDX explosives.36,38Figure 4F shows the SERS spectra of a partial sequence of the HIV (human immunodeficiency virus) gag-gene with a sequence of 5′-AGAAGATATTTGGAATAACAT-3′, which was attached on 2D hybrid SERS substrate via Au–S bond. Observed Raman vibrational modes are the adenine ring stretching modes, the phosphate backbone vibration modes and the ring breathing modes for thymine and cytosine.38 We have also noted that the SERS bands from gag-gene are combined with the D and G bands of 2D hybrid GO. Figure 4F shows that due to the huge number of “‘hot sites’” formation on 2D SERS substrate and the availability of electromagnetic and chemical enhancement effects simultaneously, hybrid 2D substrate can be used for the fingerprint detection of DNA even at 500 fM concentration level.38Figure 4H shows the fingerprint SERS spectra from methicillin-resistant Staphylococcus aureus (MRSA), when MRSA was captured by aptamer APTSEB1- modified hybrid graphene oxide based 2D SERS substrate.38 SERS spectra from MRSA, as shown in Figure 4H and Table 1, show several prominent Raman bands, and these are δ(CH2) saturated lipids, ring breathing mode of protein, C–O–C glycosidic ring deformation, and skeletal modes.38 Our result shows that the APTSEB1- modified 2D hybrid SERS substrate can be used for the label-free detection of MRSA at the concentration of 10 CFU/mL.38Figure 4G shows the fingerprint SERS spectra of RDX, when RDX were adsorbed on 2D SERS substrate.36 In SERS spectra, the main Raman bands are the symmetric ring-breathing mode, the N–O deformation band and CH2 scissoring stretch vibration bands.36 Our reported data show that the 2D hybrid SERS substrate can be used for the label-free detection of RDX at the concentration of 500 fM level,36 as shown in Figure 4G. Although reported experimental data indicate that hybrid GO based 2D SERS substrate can have huge capability for chemical and biological sensing, until now there is a lack of good understanding on how to maximize the chemical and plasmonic enhancement simultaneously, where theoretical modeling will play an important role. Until now we are in infancy on the development of theoretical model which can predict how to enhance the chemical enhancement mechanism via charge transfer as well as the dipole and multipolar interaction, which can be an excellent future research direction for nanoscience researchers.
5. Developing Hybrid 3D Graphene Oxide Based SERS Substrate for “Hotspot” Formation in Third Dimensions
To enhance the “‘hot spot’” formation in third dimension, in past few years we and others have been focusing to develop 3D SERS substrate, where “‘hot spot’” formation occurs in all the three x, y and z-direction.33,35,43−45 Since the analytes can be adsorbed in all the three dimensions for 3D SERS substrate, the overall surface area available for probe molecules adsorption is much higher for 3D substrate than that for 2D SERS substrate.33,35,43−45 Due to the increase of number of analytes adsorption, as well as number of “hot sites” in 3D SERS substrate, it is a better choice for real life applications. Here we will discuss the design criteria we have used to develop 3D plasmonic “hot spots” SERS substrate in 3D space, which has the capability for capturing, separating and label-free ultrasensitive detection of pathogenic virus and brain disease markers.33,35 To develop SERS substrate with the extension of the “‘hot spot’” formation in third dimension, we have designed hybrid 3D graphene oxide (GO) based substrate, where two-dimensional hybrid GOs are conjugated using a cross-linker.33,35 As shown in Figure 5A, at first, magnetic core-plasmonic gold shell nanoparticle decorated hybrid 2D GO was developed by mixing 2D GO with amine functionalized core–shell nanoparticle in the presence of thionyl chloride.33,35
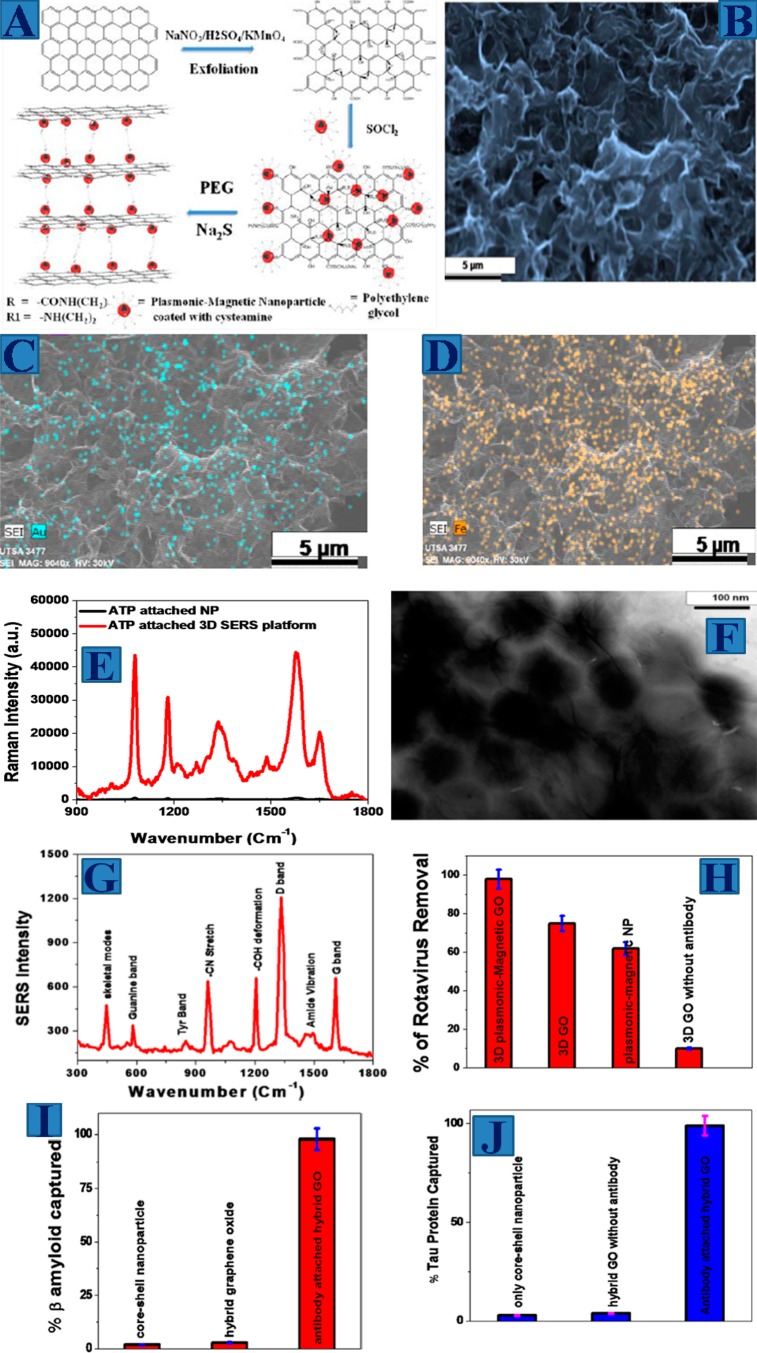
Figure 5.
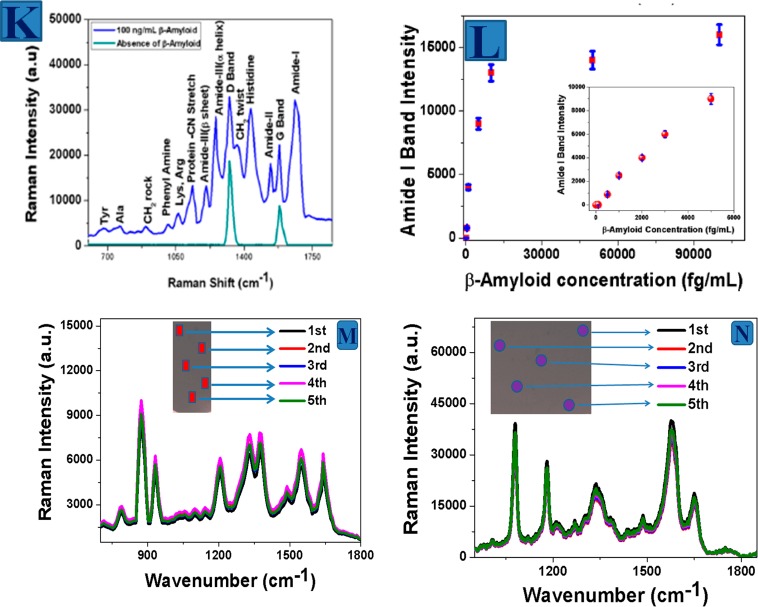
(A) Synthetic path for the development of magnetic core-plsmonic shell nanoparticle attached hybrid 3D GO based 3D SERS substrate. (B) SEM image of hybrid 3D graphene oxide based SERS substarte. (C) EDX mapping shows the presence of Au in hybrid 3D GO. (D) EDX mapping shows the presence of Fe in hybrid 3D GO (Reproduced with permission from ref (35). Copyright 2014 American Chemical Society). (E) Raman intensity from p-aminothiophenol on nanoparticle attached 3D GO based SERS substrate (Reproduced with permission from ref (30). Copyright 2016 Royal Society of Chemistry). (F) TEM image shows rotaviruses are captured by 3D SERS substrate. (G) Raman spectra from rotavirus captured by 3D SERS substrate. (H) Rotavirus captured efficiency using SERS substrate (Reproduced with permission from ref (35). Copyright 2014 American Chemical Society). (I) β-amyloid capture efficiency using SERS substrate (Reproduced with permission from ref (35). Copyright 2014 American Chemical Society). (J) Tau protein captured efficiency using SERS substrate. (K) Raman spectra from β amyloid captured by the plasmonic-magnetic hybrid GO substrate. (L) SERS detection efficiency for β amyloid (Reproduced with permission from ref (33). Copyright 2015 American Chemical Society). (M) Distribution of the SERS intensities from p-aminothiophenol over randomly chosen portions on 3D substrate. (N) Distribution of the SERS intensities from RDX over randomly chosen portions on 3D substrate.
After that, we have designed core–shell nanoparticle decorated 3D GO based SERS substrate from hybrid 2D GO using amine-functionalized PEG as a cross-linking agent.33,35Figure 5B–D shows the SEM characterization of the 3D SERS substrate, which indicate “hot spot” formations are in 3D space. EDX mapping data,33 as shown in Figure 5C and D, indicate the presence of Au and Fe in the 3D substrate. Figure 5E shows the Raman spectra from p-aminothiophenol on core–shell nanoparticle and 3D SERS substrate, where p-aminothiophenol is attached with nanosurface via Au–S linkage. In the SERS spectra, the observed Raman bands are dominated by A1 and B2 vibrational mode peaks for p-aminothiophenol. Using 1590 cm–1 band due to the ν(CC+ NH2 bend) mode, we have found the SERS enhancement factor is around 3.9 × 1012 for 3D SERS substrate whereas the enhancement factor is 1.1 × 107 for p-aminothiophenol attached core–shell nanoparticle. Our finding of 5 orders of magnitude SERS enhancement for 3D SERS substrate with respect to the 0D plasmonic magnetic substrate is mainly due to the fact that in 3D SERS substrate, GO enhances the SERS intensity from p-aminothiophenol via charge transfer chemical enhancement mechanism.33,35 On the other hand, plasmonic-magnetic nanoparticle enhances SERS intensity by several orders of magnitude via the hotspot in first, second and third dimension of interior and exterior surfaces.33,35
Since the 3D substrate has porous structure, we have used the porous 3D SERS substrate for capturing, removal and identification of rotavirus from water.35 For this purpose, we have designed antirotavirus antibody attached 3D SERS substrate. Figure 5H shows that the 3D SERS substrate is able to remove more than 99% of rotavirus from water.35 TEM image shown in Figure 5F indicates that the viruses are attached on the surface of the 3D SRS substrate. Figure 5G shows the SERS spectra from rotavirus captured by 3D SERS substrate, where the Raman bands consist of amide I, −COH deformation and the skeletal vibrations bands.35 We have also designed core–shell nanoparticle attached hybrid graphene oxide platform based SERS for capturing and fingerprint identification of trace levels of Alzheimer’s disease (AD) biomarkers such as β-amyloid and tau protein selectively from whole blood sample.33 For this purpose, we have developed anti-β amyloid and antitau antibody conjugated SERS substrate. Figure 5I and J shows that the capturing efficiency is more than 98% for β-amyloid and tau protein from whole blood sample.35Figure 5K shows the Raman bands from β-amyloid after separation from blood sample using anti-β amyloid antibody conjugated nanoarchitecture. In the reported SERS spectra the strongest Raman bands are mainly amide I, II and III bands due to the α-helical and β-sheet conformation of β-amyloid. Other bands are associated with histidine residue bands, phenylalanine, tyrosine bands and D, G bands for GO.35Figure 5L shows that the SERS detection limit is 500 fg/mL β amyloid. Similarly, we have also shown that the detection limit for tau protein is 0.15 ng/mL, using SERS substrate.35
Since good uniformity is one of the main criteria for SERS signal reproducibility, we have also determined the uniformity of the SERS substrate by measuring the SERS intensity over several randomly chosen portions on the surface of SERS substrate. As shown in Figure 5M and N, the variation of SERS intensity from p-aminothiophenol and RDX is excellent and the fluctuation over five randomly chosen portions is less than 10% for 3D substrate. Although developing 3D SERS substrate is started very recently, we and other have shown that there are several advantages like sensitivity, uniformity, and stability for using 3D material as SERS substrate.33,35,43−45
6. Summary
In this Account, we discussed our recent development on zero- to three-dimensional SERS substrates which have the capability of fingerprint identification and ultrasensitive detection of infectious disease organisms, disease biomarkers and explosives. We discussed the systematic design criteria we have used for the development of SERS substrates of different dimensions with high density “hot spots”, to enhance the SERS sensitivity tremendously. We have shown that in case of 2D and 3D GO based SERS substrate, due to the presence of both chemical and plasmonic enhancement capability, the SERS enhancement factor can be 5 orders of magnitude higher than only zero dimensional plasmonic nanoparticle. Theoretical FDTD stimulation data on electric field enhancement |E|2 agreed with experimental observation that high density “hot spots” in SERS substrate is the main key for achieving huge SERS enhancement. Although until SERS is infancy as routinely used analytical technique, we demenstrated the utilization of nanoachitecture based SERS substrate for fingerprint diagnosis of drug resistance bacteria at 10 CFU/mL level, mosquito-borne flavi-viruses at 10 PFU/mL level and explosive in femto-gram levels. We have also reported the applications of nanoachitecture based SERS substrate for the capture and fingerprint detection of Alzheimer’s disease biomarkers at 500 fg/mL level which is much higher sensitivity than current technology used in clinics.
In the future, robust synthetic techniques needs to be improved to design handy SERS substrate, before it can be used in clinics. To develop cost-effective and high scientific merit SERS substrate, huge amount of challenges need to be addressed, where interdisciplinary collaboration is the key for the success. Substantial fundamental study using experimental and theoretical models are necessary to find how to achieve maximum chemical and electromagnetic enhancement simultaneously for fingerprint identification. Animal model study for finding the biocompatibility of SERS substrates, as well as possible cytoxicity studies need to be performed thoroughly before it can be used in medicine.
Acknowledgments
P.C.R. thanks NSF-PREM Grant # DMR-1205194, NSF CREST Grant # 1547754, NSF RISE Grant # 1547836, and RCMI NIH Grant # G12MD007581 for their generous funding.
Biographies
Sudarson Sekhar Sinha is currently a postdoctoral fellow at Jackson State University. He obtained his Ph.D. from IACS India. His current research involves the design of the plasmonic nanoparticle for SERS.
Stacy Jones is currently a graduate student in department of Chemistry at Jackson State University. His current research involves the design of the hybrid graphene oxide for SERS.
Avijit Pramanik is currently a postdoctoral fellow at Jackson State University. He obtained his Ph.D. from IIT Guwahati, India. His current research involves the design of different 2D and 3D materials for SERS.
Paresh C. Ray is Professor of Chemistry at Jackson State University, Jackson, MS. He obtained his Ph.D. from the IISC, India. He currently serves as Editor for several book series and associate Editor for three international nanoscience journals.
The authors declare no competing financial interest.
References
- World Health Organization. Worldwide Country Situation Analysis: Response to Antimicrobial Resistance. 2015.
- Tackling Drug-Resistant Infections Globally: Final Report and Recommendations (http://amr-review.org/) (accessed March. 17, 2016).
- Courtney C. M.; Goodman S. M.; McDaniel J. A.; Madinger N. E.; Chatterjee A.; Nagpal P. Photoexcited quantum dots for killing multidrug-resistant bacteria. Nat. Mater. 2016, 15, 529–534. 10.1038/nmat4542. [DOI] [PubMed] [Google Scholar]
- Blair J. M. A.; Webber M. A.; Baylay A. J.; Ogbolu D. O.; Piddock L. J. V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- Hazarika P.; Russell D. A. Advances in Fingerprint Analysis. Angew. Chem., Int. Ed. 2012, 51, 3524–3531. 10.1002/anie.201104313. [DOI] [PubMed] [Google Scholar]
- Ray P. C.; Khan S. A.; Singh A. K.; Senapati D.; Fan Z. Nanomaterials for targeted detection and photothermal killing of bacteria. Chem. Soc. Rev. 2012, 41, 3193–3209. 10.1039/c2cs15340h. [DOI] [PubMed] [Google Scholar]
- Henry A. I.; Sharma B.; Cardinal M. F.; Kurouski D.; Van Duyne R. P. Surface-Enhanced Raman Spectroscopy Biosensing: In Vivo Diagnostics and Multimodal Imaging. Anal. Chem. 2016, 88, 6638–6647. 10.1021/acs.analchem.6b01597. [DOI] [PubMed] [Google Scholar]
- Fleischmann M.; Hendra P. J.; McQuillan A. J. Raman Spectra of Pyridine Adsorbed at a Silver Electrode. Chem. Phys. Lett. 1974, 26, 163–166. 10.1016/0009-2614(74)85388-1. [DOI] [Google Scholar]
- Albrecht M. G.; Creighton J. A. Anomalously Intense Raman Spectra of Pyridine at a Silver Electrode. J. Am. Chem. Soc. 1977, 99, 5215–5217. 10.1021/ja00457a071. [DOI] [Google Scholar]
- Jeanmaire D. L.; Van Duyne R. P. Surface Raman Spectroelectrochemistry. J. Electroanal. Chem. Interfacial Electrochem. 1977, 84, 1–20. 10.1016/S0022-0728(77)80224-6. [DOI] [Google Scholar]
- Moskovits M. Surface-enhanced spectroscopy. Rev. Mod. Phys. 1985, 57, 783–826. 10.1103/RevModPhys.57.783. [DOI] [Google Scholar]
- Kneipp K.; Wang Y.; Kneipp H.; Perelman L. T.; Itzkan I.; Dasari R. R.; Feld M. S. Single molecule detection using surface-enhanced Raman scattering (SERS). Phys. Rev. Lett. 1997, 78, 1667–1670. 10.1103/PhysRevLett.78.1667. [DOI] [Google Scholar]
- Nie S.; Emory S. R. Probing single molecules and single nanoparticles by surface-enhanced Raman scattering. Science 1997, 275, 1102–1106. 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- Michaels A. M.; Nirmal M.; Brus L. E. Surface-Enhanced Raman Spectroscopy of Individual Rhodamine 6G Molecules on Large Ag Nanocrystals. J. Am. Chem. Soc. 1999, 121, 9932–9939. 10.1021/ja992128q. [DOI] [Google Scholar]
- Moskovits M. Surface-enhanced Raman spectroscopy: a brief retrospective. J. Raman Spectrosc. 2005, 36, 485–496. 10.1002/jrs.1362. [DOI] [Google Scholar]
- Kneipp K.; Kneipp H.; Itzkan I.; Dasari R. R.; Feld M. S. Ultrasensitive Chemical Analysis by Raman Spectroscopy. Chem. Rev. 1999, 99, 2957–2976. 10.1021/cr980133r. [DOI] [PubMed] [Google Scholar]
- Xu H.; Bjerneld E. J.; Käll M.; Börjesson L. Spectroscopy of Single Hemoglobin Molecules by Surface Enhanced Raman Scattering. Phys. Rev. Lett. 1999, 83, 4357–4360. 10.1103/PhysRevLett.83.4357. [DOI] [Google Scholar]
- Stiles P. L.; Dieringer J. A.; Shah N. C.; Van Duyne R. P. Surface-Enhanced Raman Spectroscopy Annu. Annu. Rev. Anal. Chem. 2008, 1, 601–626. 10.1146/annurev.anchem.1.031207.112814. [DOI] [PubMed] [Google Scholar]
- Haran G. Single-Molecule Raman Spectroscopy: A Probe of Surface Dynamics and Plasmonic Fields. Acc. Chem. Res. 2010, 43, 1135–1143. 10.1021/ar100031v. [DOI] [PubMed] [Google Scholar]
- Lane L. A.; Qian X.; Nie S. SERS Nanoparticles in Medicine: From Label-Free Detection to Spectroscopic Tagging. Chem. Rev. 2015, 115, 10489–10529. 10.1021/acs.chemrev.5b00265. [DOI] [PubMed] [Google Scholar]
- Chen H. Y.; Lin M. H.; Wang C. Y.; Chang Y. M.; Gwo S. Large-Scale Hot Spot Engineering for Quantitative SERS at the Single-Molecule Scale. J. Am. Chem. Soc. 2015, 137, 13698–13705. 10.1021/jacs.5b09111. [DOI] [PubMed] [Google Scholar]
- Singh A. K.; Khan S. A.; Fan Z.; Demeritte T.; Senapati D.; Kanchanapally R.; Ray P. C. Development of a Long-Range Surface-Enhanced Raman Spectroscopy Ruler. J. Am. Chem. Soc. 2012, 134, 8662–8669. 10.1021/ja301921k. [DOI] [PubMed] [Google Scholar]
- Frank O.; Dresselhaus M. S.; Kalbac M. Raman Spectroscopy and in Situ Raman Spectroelectrochemistry of Isotopically Engineered Graphene Systems. Acc. Chem. Res. 2015, 48, 111–118. 10.1021/ar500384p. [DOI] [PubMed] [Google Scholar]
- Zhao J.; Zhang K.; Li Y.; Ji J.; Liu B. High-Resolution and Universal Visualization of Latent Fingerprints Based on Aptamer-Functionalized Core–Shell Nanoparticles with Embedded SERS Reporters. ACS Appl. Mater. Interfaces 2016, 8, 14389–14395. 10.1021/acsami.6b03352. [DOI] [PubMed] [Google Scholar]
- Granger J. H.; Schlotter N. E.; Crawford A. C.; Porter M. D. Prospects for point-of-care pathogen diagnostics using surface-enhanced Raman scattering (SERS). Chem. Soc. Rev. 2016, 45, 3865–3882. 10.1039/C5CS00828J. [DOI] [PubMed] [Google Scholar]
- Ling X.; Huang S.; Deng S.; Mao N.; Kong J.; Dresselhaus M. S.; Zhang J. Lighting Up the Raman Signal of Molecules in the Vicinity of Graphene Related Materials. Acc. Chem. Res. 2015, 48, 1862–1870. 10.1021/ar500466u. [DOI] [PubMed] [Google Scholar]
- Morton S. M.; Jensen L. Understanding the Molecule-Surface Chemical Coupling in SERS. J. Am. Chem. Soc. 2009, 131, 4090–4098. 10.1021/ja809143c. [DOI] [PubMed] [Google Scholar]
- Payton J. L.; Morton S. M.; Moore J. E.; Jensen L. A Hybrid Atomistic Electrodynamics-Quantum Mechanical Approach for Simulating Surface-Enhanced Raman Scattering. Acc. Chem. Res. 2014, 47, 88–99. 10.1021/ar400075r. [DOI] [PubMed] [Google Scholar]
- Premasiri W. R.; Moir D. T.; Klempner M. S.; Krieger N.; Jones G.; Ziegler L. D. Characterization of the Surface Enhanced Raman Scattering (SERS) of Bacteria. J. Phys. Chem. B 2005, 109, 312–320. 10.1021/jp040442n. [DOI] [PubMed] [Google Scholar]
- Jones S.; Sinha S. S.; Pramanik A.; Ray P. C. Three-dimensional (3D) plasmonic hot spots for label-free sensing and effective photothermal killing of multiple drug resistant superbugs. Nanoscale 2016, 8, 18301–18308. 10.1039/C6NR05888D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W.; Crozier K. B. Quantum Mechanical Limit to Plasmonic Enhancement as Observed by Surface-Enhanced Raman Scattering. Nat. Commun. 2014, 5, 5228. 10.1038/ncomms6228. [DOI] [PubMed] [Google Scholar]
- Paul A. M.; Fan Z.; Sinha S. S.; Shi Y.; Le L.; Bai F.; Ray P. C. Bioconjugated Gold Nanoparticle Based SERS Probe for Ultrasensitive Identification of Mosquito-Borne Viruses Using Raman Fingerprinting. J. Phys. Chem. C 2015, 119, 23669–23675. 10.1021/acs.jpcc.5b07387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeritte T.; Viraka Nellore B. P.; Kanchanapally R.; Sinha S. S.; Pramanik A.; Chavva S. R.; Ray P. C. Hybrid Graphene Oxide Based Plasmonic-Magnetic Multifunctional Nanoplatform for Selective Separation and Label-Free Identification of Alzheimer’s Disease Biomarkers, T. ACS Appl. Mater. Interfaces 2015, 7, 13693–13700. 10.1021/acsami.5b03619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S. S.; Paul D. K.; Kanchanapally R.; Pramanik A.; Chavva R. S.; Viraka Nellore B. P.; Jones S. J.; Ray P. C. Long-range Two-Photon Scattering Spectroscopy Ruler for Screening Prostate Cancer Cells. Chem. Sci. 2015, 6, 2411–2418. 10.1039/C4SC03843F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z.; Yust B.; Nellore B. O. V; Sinha S. S.; Kanchanapally R.; Crouch R. A.; Pramanik A.; Chavva S. R.; Sardar D.; Ray P. C. Accurate identification and selective removal of rotavirus using a plasmonic–magnetic 3D graphene oxide architecture. J. Phys. Chem. Lett. 2014, 5, 3216–3221. 10.1021/jz501402b. [DOI] [PubMed] [Google Scholar]
- Kanchanapally R.; Sinha S. S.; Fan Z.; Dubey M.; Zakar E.; Ray P. C. Graphene Oxide-Gold Nanocage Hybrid for Trace Level Identification of Nitro Explosives Using Raman Fingerprint. J. Phys. Chem. C 2014, 118, 7070–7075. 10.1021/jp5015548. [DOI] [Google Scholar]
- Tchounwou C.; Sinha S. S.; Nellore B. P. V.; Pramanik A.; Kanchanapally R.; Jones S.; Chavva S. R.; Ray P. C. Hybrid Theranostic Platform for Second Near-IR Window Light Triggered Selective Two-Photon Imaging and Photothermal Killing of Targeted Melanoma Cells. ACS Appl. Mater. Interfaces 2015, 7, 20649–20656. 10.1021/acsami.5b05225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z.; Kanchanapally R.; Ray P. C. Hybrid Graphene Oxide Based Ultrasensitive SERS Probe for Label-Free Biosensing. J. Phys. Chem. Lett. 2013, 4, 3813–3818. 10.1021/jz4020597. [DOI] [Google Scholar]
- Beqa L.; Fan Z.; Singh A. K.; Senapati D.; Ray P. C. Gold Nano Popcorn Attached SWCNT Hybrid Nanomaterial for Targeted Diagnosis and Photothermal Therapy of Human Breast Cancer Cells. ACS Appl. Mater. Interfaces 2011, 3, 3316–3324. 10.1021/am2004366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W.; Singh A. K.; Khan S. A.; Senapati D.; Yu H.; Ray P. C. Gold nano-popcorn-based targeted diagnosis, nanotherapy treatment, and In Situ monitoring of photothermal destruction response of prostate cancer cells using surface-enhanced Raman spectroscopy. J. Am. Chem. Soc. 2010, 132, 18103–18114. 10.1021/ja104924b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeritte T.; Kanchanapally R.; Fan Z.; Singh A. K.; Senapati D.; Dubey M.; Zakar E.; Ray P. C. Highly Efficient SERS Substrate for Direct Detection of Explosive TNT Using Popcorn-Shaped Gold Nanoparticle-Functionalized SWCNT Hybrid. Analyst 2012, 137, 5041–5045. 10.1039/c2an35984g. [DOI] [PubMed] [Google Scholar]
- Beqa L.; Singh A. K.; Fan Z.; Senapati D.; Ray P. C. Chemically Attached Gold Nanoparticle–Carbon Nanotube Hybrids for Highly Sensitive SERS Substrate. Chem. Phys. Lett. 2011, 512, 237–242. 10.1016/j.cplett.2011.07.037. [DOI] [Google Scholar]
- Zhang Q.; Lee Y. H.; Phang I. Y.; Lee C. K.; Ling X. Y. Hierarchical 3D SERS substrates fabricated by integrating photolithographic microstructures and self-assembly of silver nanoparticles. Small 2014, 10, 2703–2711. 10.1002/smll.201303773. [DOI] [PubMed] [Google Scholar]
- Wang P.; Liang O.; Zhang W.; Schroeder T.; Xie Y.-H. Ultra-Sensitive Graphene-Plasmonic Hybrid Platform for Label-Free Detection. Adv. Mater. 2013, 25, 4918–4924. 10.1002/adma.201300635. [DOI] [PubMed] [Google Scholar]
- Liu H.; Yang Z.; Meng L.; Sun Y.; Wang J.; Yang L.; Liu J.; Tian Z. Three-dimensional and Time-ordered Surface-enhanced Raman Scattering Hotspot Matrix. J. Am. Chem. Soc. 2014, 136, 5332–5341. 10.1021/ja501951v. [DOI] [PubMed] [Google Scholar]
- Thrall E. S.; Crowther A. C.; Yu Z.; Brus L. E. R6G on Graphene: High Raman Detection Sensitivity, Yet Decreased Raman Cross-Section. Nano Lett. 2012, 12, 1571–1577. 10.1021/nl204446h. [DOI] [PubMed] [Google Scholar]
- Ling X.; Xie L. M.; Fang Y.; Xu H.; Zhang H. L.; Kong J.; Dresselhaus M. S.; Zhang J.; Liu Z. F. Can Graphene Be Used as a Substrate for Raman Enhancement?. Nano Lett. 2010, 10, 553–561. 10.1021/nl903414x. [DOI] [PubMed] [Google Scholar]
- Leem J.; Wang M. C.; Kang P.; Nam W. S. Mechanically Self-Assembled, Three-Dimensional Graphene–Gold Hybrid Nanostructures for Advanced Nanoplasmonic Sensors. Nano Lett. 2015, 15, 7684–7690. 10.1021/acs.nanolett.5b03672. [DOI] [PubMed] [Google Scholar]
- Zhao J.; Pinchuk A. O.; McMahon J. M.; Li S.; Ausman L. K.; Atkinson A. L.; Schatz G. C. Methods for Describing the Electromagnetic Properties of Silver and Gold Nanoparticles. Acc. Chem. Res. 2008, 41, 1710–1720. 10.1021/ar800028j. [DOI] [PubMed] [Google Scholar]
- Solís D. M.; Taboada J. M.; Obelleiro F.; Liz-Marzán L. M.; Garcia de Abajo F. J. Toward Ultimate Nanoplasmonics Modeling. ACS Nano 2014, 8, 7559–7570. 10.1021/nn5037703. [DOI] [PubMed] [Google Scholar]