Abstract
The chaperone homologs RAC (ribosome-associated complex) and Ssb1/2p are anchored to ribosomes; Ssb1/2p directly interacts with nascent polypeptides. The absence of RAC or Ssb1/2p results in a similar set of phenotypes, including hypersensitivity against the aminoglycoside paromomycin, which binds to the small ribosomal subunit and compromises the fidelity of translation. In order to understand this phenomenon we measured the frequency of translation termination and misincorporation in vivo and in vitro with a novel reporter system. Translational fidelity was impaired in the absence of functional RAC or Ssb1/2p, and the effect was further enhanced by paromomycin. The mutant strains suffered primarily from a defect in translation termination, while misincorporation was compromised to a lesser extent. Consistently, a low level of soluble translation termination factor Sup35p enhanced growth defects in the mutant strains. Based on the combined data we conclude that RAC and Ssb1/2p are crucial in maintaining translational fidelity beyond their postulated role as chaperones for nascent polypeptides.
RAC (ribosome-associated complex) is a stable heterodimer that is almost entirely associated with ribosomes of yeast. RAC consists of the Hsp70 homolog Ssz1p and the Hsp40 homolog zuotin (22). A significant fraction of the Hsp70 homolog Ssb1/2p is also bound to the yeast ribosome (41). RAC associates with the ribosome in a salt-sensitive manner (22). Ssb1/2p binding to nontranslating ribosomes is salt sensitive; however, when the ribosome is involved in translation, binding of Ssb1/2p becomes salt resistant (41, 47). RAC and Ssb1/2p functionally interact in vivo and in vitro (23, 30). Based on the idea that they act on nascent polypeptides, these chaperones are thought to localize close to the tunnel exit of the large ribosomal subunit. Although structural data are not available for that point, this view is supported by the finding that Ssb1/2p can form cross-link products with nascent polypeptides as short as 45 amino acids (21). RAC has not been found in contact with nascent polypeptides, but it influences their interaction with Ssb1/2p (23). Mainly based on localization and homology it has been suggested that RAC and Ssb1/2p are involved in folding or maintaining nascent polypeptides in a folding-competent state, facilitating their transit through the ribosomal tunnel and preventing backward movement or clearing aggregated proteins from the ribosome—experimental evidence, however, is scarce (11, 28, 54). Recently it was shown that Ssb1/2p participates in posttranslational completion of protein folding and has partly overlapping function with the chaperonin tailless complex polypeptide 1 (TCP1) ring complex (TRiC) cochaperone GimC (58). However, zuotin was not required for this function, and most likely it is the soluble pool of Ssb1/2p that cooperates with the TRiC/GimC system (58).
In vivo Δssb1 Δssb2, Δssz1, and Δzuo1 strains display a complex set of phenotypes, including hypersensitivity against aminoglycosides, e.g., paromomycin (22, 41, 70). Although the drug is only weakly active against eukaryotic ribosomes (49), paromomycin causes phenotypic suppression of nonsense and missense mutations in yeast, indicating that the frequency of translational errors is increased (46). A number of yeast mutants in proteins or rRNA close to the decoding center display enhanced sensitivity to paromomycin (1, 9, 14, 38, 62). Strains expressing mutant versions of the translation termination factors Sup35p and Sup45p display paromomycin sensitivity (2, 66). The aforementioned examples are readily explained by a direct involvement of the components in the maintenance of translational fidelity; however, mutants exist in which the causes of paromomycin sensitivity are less obvious. For instance, some glycosylation mutants or mutants of the plasma membrane ATPase display sensitivity against aminoglycosides (12, 33, 34).
In an attempt to better define the roles of Ssb1/2p and RAC in vivo, we have examined how the deleterious effects of paromomycin are linked to translational fidelity in strains lacking functional RAC or Ssb1/2p. To this end we have tested translation termination and misincorporation of amino acids in vivo and in vitro. The results indicate that lack of these ribosome-associated chaperones primarily induces readthrough on the ribosome. The data are in agreement with a model in which a combination of paromomycin and the absence of functional RAC or Ssb1/2p cause lethality because of severe defects in translation termination.
MATERIALS AND METHODS
Yeast strains, media, and culture conditions.
Standard yeast genetic techniques were applied (57). MH272-3f a/α (ura3/ura3 leu2/leu2 his3/his3 trp1/trp1 ade2/ade2 HMLa/HMLa) is the parental wild-type strain (22, 29) for the construction of Δssb1 Δssb2 (Ida56A), Δzuo1 Δssz1 (Ida12), Δzuo1 (Ida1), and Δssz1 (Ida2) deletion strains, which has been reported elsewhere (22, 23). In Ida56A, SSB1 is replaced by the kanMX module (65), which causes partial resistance against paromomycin. For this reason a second Δssb1 Δssb2 deletion strain (Ida56C) was generated by replacing the ClaI/AgeI 1,461-bp fragment in SSB1 with the ADE2 marker. Ida56C and derivatives were used in experiments involving paromomycin. During the course of this study the MH272-3f-derived strains turned out to contain the [PSI+] element. For this reason, Δzuo1 (GT-Δzuo1), Δssz1 (GT-Δssz1), and Δzuo1 Δssz1 (GT-Δzuo1 Δssz1) were generated in the [psi−] strain GT197, which is derived from strain GT81 (MATa ade1-14 ura3 leu2 his3 trp1 lys2) (67; M. Boguta, personal communication).
Strains were grown in a solution of 1% yeast extract, 2% peptone, 2% dextrose (YPD) or in minimal medium (0.67% yeast nitrogen base without amino acids, 2% dextrose, supplemented with all amino acids and bases except those needed for selection). Drug effects either were determined by spotting serial 10-fold dilutions of early-log-phase cultures containing the same number of cells on YPD plates with supplements as indicated or were determined in liquid cultures as described previously (13). In order to cure cells from the plasmid overexpressing HSP104, strains expressing the chaperone on a plasmid with a URA3 marker were grown on 5-fluoro-orotic acid-containing media (5). Loss of the plasmid was confirmed by testing the ability of the strains to grow on media lacking uracil. Curing of HSP104 was carried out just before experiments, as MH272-3f and derivatives showed a tendency to revert to [PSI+]. For the same reason, 2 mM GndHCl was included in the medium when the [psi−] status of MH272-3f-derived strains was required. Paromomycin gradient plates were prepared by pouring 20 ml of YPD medium containing the maximum concentration of the drug into plates resting at an incline. After drug-containing medium was solidified, plates were laid flat and 20 ml of YPD medium was poured in (35).
Plasmids.
Vectors used for the expression of ssz1-S295F, zuo1-H128Q, and HSP104 were from the pY series (24) or pRS series (10). ssz1-S295F was generated by exchanging codon TCT (bp 883 to 885) for TTC in SSZ1, resulting in the replacement of Ser-295 with phenylalanine (27). ssz1-S295F plus 300 bp upstream and downstream of the coding region was cloned into pYEPlac112(2μm TRP1) and was expressed in the Ida2 background lacking the wild-type SSZ1 gene. zuo1-H128Q was expressed from either pRS423(2μm HIS3) or pYCPlac33(CEN, URA3) in the Ida1 background lacking wild-type ZUO1 (23). HSP104 plus 500 bp upstream and 300 bp downstream of the open reading frame was cloned into pRS423(2μm HIS3) and pYEPlac112(2μm TRP1), resulting in p423-HSP104 and p112-HSP104. In order to generate strains overexpressing HSP104, p423-HSP104, p112-HSP104, or plasmid, pA1/4(2μm URA3) (7) was transformed into the respective strains according to the requirements for positive selection.
Reporter constructs contained a hybrid lacZ-luc gene under control of the simian virus 40 (SV40) early promoter, which is active in yeast and higher eukaryotes. Construction of the readthrough reporters was based on previously published information (4). All hybrid reporter constructs were expressed from the multicopy plasmid pYEPlac195(2μm URA3). Primers SV40-F1 (GGATCCGCGCAGCACCATGGCCTGAAATAACCTC) and β-Gal-R1 (GTCGACTTAGCTAGCTTTTTGACACCAGACCAACTGGTAATGGTAGCG) were used to amplify the SV40 promoter region plus β-galactosidase with pSV-β-galactosidase (Promega) as a template. The PCR product was cloned into the BamHI/SalI site of pYEPlac195. In the next step primers Luc+-F1 (GCTAGCAGATCTGAAGACGCCAAAAACATAAAGAAAGGCC) and Luc+-R1 (CTGCAGTCAGCTAATTATTACACGGCGATCTTTCCGCCCTTCTTGGCC) were used to amplify luciferase with pSP-luc+ (Promega) as a template. The PCR product was cut with NheI/PstI and was cloned into pYEPlac195 containing β-galactosidase. This procedure generated an in-frame fusion between β-galactosidase and luciferase. The two genes were separated by a short in-frame linker that can be cut with NheI and BglII and was used to insert short oligonucleotides generating lacZ-luc and lacZ-STOP-luc (see Fig. 2A). The basis of the in vivo misincorporation reporters was lacZ-luc. lacZ-luc* was generated by exchanging codon AGA for AGC, resulting in the replacement of R-218 with serine (Fig. 2A). lacZ-luc** was generated by exchanging codon AGA for TCT, also resulting in the replacement of R-218 with serine (Fig. 2A).
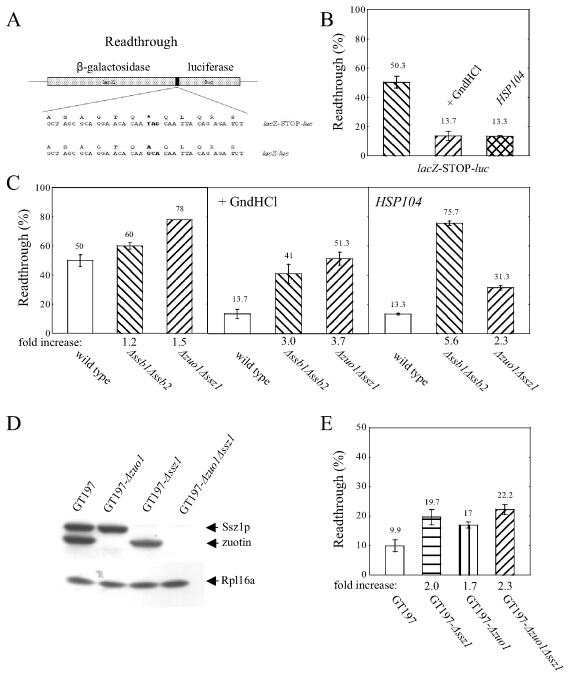
FIG. 2.
Determination of readthrough efficiency in strains lacking Ssb1/2p or the subunits of the RAC complex in vivo. (A) An in-frame fusion between β-galactosidase and luciferase (lacZ-luc) served to determine the relative enzymatic activities upon equimolar expression of the two enzymes. Readthrough efficiency was determined using the lacZ-STOP-luc construct (for details see Results). (B) lacZ-STOP-luc was transformed into wild type or wild type plus HSP104, and readthrough was determined after growth on YPD plus or minus 2 mM GndHCl as indicated. Readthrough frequency is expressed as the ratio of luciferase/β-galactosidase activity (RLU/A420) obtained with the lacZ-STOP-luc construct normalized to the ratio obtained with lacZ-luc. Parts of the results shown in panel B are included in Fig. 4 to facilitate comparison between wild-type and mutant strains. (C) Readthrough efficiency was determined in wild-type, Δssb1 Δssb2, and Δssz1 Δzuo1 strains and in the same set of strains overexpressing HSP104 described above. Strains were grown on YPD or YPD supplemented with 2 mM GndHCl as indicated. Numbers below the columns indicate the relative increase of readthrough frequency compared to that of the wild type. All experiments were performed at least in triplicate; when error bars are missing, numbers were identical. (D) Δzuo, Δssz1, and Δzuo1 Δssz1 were generated in the [psi−] strain GT197. The respective proteins were probed in total extracts derived from GT197 and in the corresponding mutant strains by immunoblotting as described in the legend to Fig. 1. The ribosomal marker Rpl16a was used as a loading control. (E) lacZ-STOP-luc was transformed into GT197, GT197-Δzuo1, GT197-Δssz1, and GT197-Δzuo1 Δssz1. Readthrough was determined after growth on YPD as described in the legend to panel B.
Plasmid pSP-luc+ (Promega) served as a template for the in vitro reporter constructs. pSP-luc+* was generated as described above. In pSP-luc+STOP, leucine 210 was replaced with a stop codon (see Fig. 5A). luc, luc*, and luc-STOP were transferred into pSPUTK (Stratagene). Resulting plasmids pSPUTK-luc+, pSPUTK-luc+*, and pSPUTK-luc+STOP were used to generate mRNA for in vitro translation as described previously (16).
FIG. 5.
Determination of misincorporation efficiency in strains lacking Ssb1/2p or functional RAC in vivo. (A) lacZ-luc* (near cognate) and lacZ-luc** (noncognate) both encode inactive luciferase-R218S. Active luciferase can be regained from lacZ-luc* or lacZ-luc** if serine 218 is mistranslated into arginine (for details see Results). (B) Misincorporation efficiency was determined for the wild type transformed with either lacZ-luc* or lacZ-luc** after growth on YPD or YPD plus 2 mM GndHCl. Misincorporation is expressed as the ratio of luciferase/β-galactosidase activity obtained after expression of the lacZ-luc* or lacZ-luc** constructs normalized to the in-frame control. (C) Misincorporation efficiency was determined in wild-type, Δssb1 Δssb2, Δssz1 Δzuo1, zuo1-H128Q, and ssz1-S295F strains as described above using the lacZ-luc* construct. (D) Misincorporation was determined in wild-type, zuo1-H128Q, and ssz1-S295F strains grown on YPD containing 2 mM GndHCl plus increasing concentrations of paromomycin as indicated. Note that the paromomycin concentration used in this experiment was adjusted according to the inhibitory concentration for each strain (see Fig. 3B).
Ribosome binding assay.
Association of RAC and Ssb1/2p with ribosomes under low-salt (120 mM potassium acetate) or high-salt (800 mM potassium acetate) conditions was determined as described previously (22). The effect of paromomycin was determined by the same approach. In brief, yeast cytosol (40 μl) was layered on top of a 100-μl low-salt sucrose cushion (20 mM HEPES-KOH, 25% sucrose, 120 mM potassium acetate, 2 mM dithiothreitol [DTT], 5 mM magnesium acetate, 1 mM phenylmethylsulfonyl fluoride [PMSF], pH 7.4) or a paromomycin sucrose cushion (20 mM HEPES-KOH [pH 7.4], 25% sucrose, 120 mM potassium acetate, 2 mM DTT, 5 mM magnesium acetate, 100 μg of paromomycin/ml, 1 mM PMSF, pH 7.4). After centrifugation at 200,000 × g in an RC M120 GX ultracentrifuge (Sorvall) for 120 min at 4°C, samples were split into supernatant and pellet and corresponding amounts were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by immunoblotting. A total corresponding to the amount loaded onto the sucrose cushion was analyzed in parallel.
Preparation of cell extracts for the determination of β-galactosidase and luciferase activity.
Twenty-milliliter cultures were grown to an optical density at 600 nm (OD600) of 0.8 to 1.2 on YPD containing 2 mM guanidine hydrochloride (GndHCl). Higher concentrations of GndHCl affected growth of strains lacking functional RAC or Ssb1/2p (32). Cells corresponding to an OD600 of 1 were collected by centrifugation at 10,000 × g, washed once with 1 ml of distilled water, and resuspended in 250 μl of cold TEP-I buffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 1 mM PMSF, 1% dimethyl sulfoxide [DMSO]) containing protease inhibitor mix (1.25 μg of leupeptin/ml, 0.75 μg of antipain/ml, 0.25 μg of chymostatin/ml, 0.25 μg of elastatinal/ml, 5 μg of pepstatin A/ml). Acid-washed glass beads (0.2 g/sample) were added, and cells were disrupted by three cycles of vortexing (1 min) and cooling (1 min) at 4°C. Samples were centrifuged for 1 min at 3,000 × g in order to pellet glass beads and cell debris. Supernatants were transferred to fresh tubes and were spun at 4°C for 10 min at 10,000 × g.
Determination of β-galactosidase activity.
Activity of β-galactosidase was assayed by using o-nitrophenyl-β-d-galactopyranoside (ONPG) as a substrate as described previously (60). In order to ensure that the reaction was in the linear range, each cell extract was diluted 25- and 50-fold into a 1-ml final volume of buffer Z (50 mM NaH2PO4, 45 mM Na2HPO4, 10 mM KCl, 10 mM MgSO4, pH 7). ONPG solution (200 μl; 4 mg/ml in buffer Z) was added, and samples were incubated at 28°C for 30 min with gentle shaking. The reaction was stopped by the addition of 500 μl of 1 M Na2CO3, and the absorbance at 420 nm was measured relative to a blank.
Determination of luciferase activity.
Each cell extract was diluted 25- and 50-fold into a 1-ml final volume of luciferase assay buffer (100 mM KH2PO4, 1 mM EDTA, 1 mM DTT, pH adjusted to 7.8 with KOH). The diluted cell extracts (100 μl) were mixed with 100 μl of luciferase reagent (20 mM Tricine, 5 mM MgCl2, 0.1 mM EDTA, 3.3 mM DTT, 270 μM coenzyme A, 500 μM d-luciferin, 500 μM ATP) using a Lumat LB 9507 device (Berthold Technologies GmbH, Wildbad, Germany). Luciferase activity is given in relative light units (RLU).
Estimation of readthrough and misincorporation efficiencies in vivo and in vitro.
β-Galactosidase activity served as an internal standard for possible variation in translation efficiency, mRNA stability, and folding in vivo. Readthrough and misincorporation was determined based on the relative ratio of luciferase (in RLU) to β-galactosidase (A420) activity. The RLU/A420 obtained was set to 100% when the enzymes were expressed at equimolar quantities from the in-frame lacZ-luc construct. Normalization with lacZ-luc was performed for each strain and growth condition. Efficiency of readthrough and misincorporation was then expressed as the percentage of RLU/A420 obtained when the enzymes were expressed either from the lacZ-STOP-luc, lacZ-luc*, or lacZ-luc** construct (see Fig. 2A and B).
For the determination of in vitro readthrough and misincorporation, translation extracts were primed with mRNA luc, luc*, or luc-STOP (see Fig. 5). After translation, luciferase activity (RLU) was determined as described above. The amount of luciferase synthesized was determined by densitometry of fluorograms and is expressed in relative units (quantum level [QL]) (see Fig. 5B). RLU/QL obtained was set to 100% when luc mRNA was used to prime the translation reaction mixture. Efficiency of readthrough and misincorporation is expressed as the percentage of RLU/QL obtained after translation of luc* or luc-STOP mRNA. Because the 23-kDa luciferase fragment generated in the presence of luc-STOP mRNA contains only 9 methionines (compared to 14 methionine residues in full-length luciferase), relative units in this case were multiplied by 1.55.
Preparation and optimization of yeast translation extracts.
Yeast translation extracts were prepared as described previously (20). Translation reactions were performed in the presence of 35S-labeled methionine as described previously (16). The amount of translation extract for each strain was optimized, and the Mg2+ concentration in each reaction mixture was adjusted precisely to 1 mM. The Mg2+ concentration is a critical parameter for the accuracy of decoding (52). Translation efficiency in the different lysates was similar and did not vary by more than 30%. Translation reaction mixtures (20 μl) were incubated for 55 min at 20°C. Tubes were then placed on ice, and 20 μl of STOP buffer (20 mM HEPES-KOH, 2 mM DTT, 5 mM magnesium acetate, 2 mM EDTA, 50 mM potassium acetate, 5 mM PMSF, protease inhibitor mix, pH 7.4) was added. One aliquot was used for determination of luciferase activity, and another aliquot was boiled in SDS-sample buffer containing 1 μg of ovalbumin/ml. Samples were run on 10% Tris-Tricine gels and were subsequently quantified with an Aida ImageAnalyzer (Raytest; Isotopenmessgeräte GmbH).
Miscellaneous.
Ssb1/2p represents Ssb1p and Ssb2p, two 99% identical proteins with identical function and similar expression levels. The term “mutant strains ” is used collectively for Δzuo1 Δssz1, Δssb1 Δssb2, zuo1-H128Q, and ssz1-S295F strains. Proteins were separated on Tris-Tricine gels (56). 125J-protein A was used to develop the immunoblots (26).
RESULTS
Paromomycin does not affect ribosome association of RAC or Ssb1/2p.
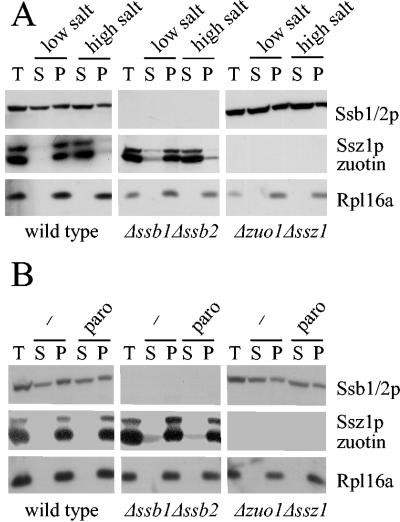
Aminoglycoside binding to the small ribosomal subunit induces structural changes that might affect the entire ribosome (17, 31). Hence, we asked whether paromomycin sensitivity of Δssb1 Δssb2 or Δzuo1 Δssz1 strains resulted from a reduction in ribosomal association of RAC or Ssb1/2p, respectively. The distribution of Ssb1/2p in the Δssz1 Δzuo1 strain and of RAC in the Δssb1 Δssb2 strain was comparable to the distribution in wild-type cytosol under low-salt and high-salt conditions (Fig. 1A). Ribosome association was then tested in the presence of 100 μg of paromomycin/ml, a concentration that completely inhibits growth of Δssb1 Δssb2 and Δzuo1 Δssz1 strains (23). The distribution of RAC and Ssb1/2p in the deletion strains was unaffected, indicating that paromomycin did not directly interfere with ribosome association (Fig. 1B).
FIG. 1.
Paromomycin does not affect binding of RAC and Ssb1/2p to ribosomes. Total yeast cytosol (T) derived from wild-type, Δssb1 Δssb2, or Δssz1 Δzuo1 strains was separated into ribosomal pellet (P) and postribosomal supernatant (S) (A) in the presence of low salt (120 mM potassium acetate) or high salt (800 mM potassium acetate) or (B) in the presence (paro) or absence (−) of 100 μg of paromomycin/ml in a buffer containing 120 mM potassium acetate (see Materials and Methods). Aliquots were separated on 10% Tris-Tricine gels, transferred to nitrocellulose, and decorated with antibodies directed against Ssb1/2p, Ssz1p, zuotin, and Rpl16a (ribosomal marker).
Quantification of translational readthrough reveals that the wild-type strain MH272-3f α contains the [PSI+] element.
Readthrough was determined for the TAG stop codon flanked by CAA, promoting a high basal readthrough level (Fig. 2A, lacZ-STOP-luc) (4, 40). In the wild-type strain MH272-3f α, readthrough efficiency was about 50%, which was significantly higher than that previously reported for a different genetic background (Fig. 2B) (4). Many laboratory strains contain the [PSI+] element, a property that is not readily detectable because no general growth defects are connected to [PSI+] (63). However, [PSI+] strains display a high readthrough frequency, because a large fraction of the translation termination factor Sup35p is trapped in prion aggregates. These aggregates can be dissolved by different means, such as growth on GndHCl, overexpression of HSP104, or deletion of HSP104 (for reviews on [PSI+] see references 37, 45, 64, and 68). We exposed MH272-3f to two conditions that eliminate the prion: growth on GndHCl and overexpression of HSP104. Indeed, both conditions reduced readthrough to about 13%, which is similar to previous results (Fig. 2B) (4). We conclude that readthrough in wild-type MH272-3f can be significantly reduced by conditions that cure [PSI+].
Readthrough is increased in the absence of RAC or Ssb1/2p in vivo.
Readthrough in Δssb1 Δssb2 and Δzuo1 Δssz1 strains approached the limit of the lacZ-STOP-luc reporter assay (Fig. 2C, first panel). To lower readthrough and avoid interference with [PSI+], Δssb1 Δssb2 and Δzuo1 Δssz1 strains were transformed with a plasmid overexpressing HSP104 or were grown on GndHCl. For this, two earlier observations had to be considered. First, Δssb1 Δssb2 (32) and Δzuo1 Δssz1 strains (data not shown) are more sensitive to GndHCl than is wild-type yeast. Second, HSP104 overexpression fails to cure [PSI+] in a Δssb1 Δssb2 background (8, 45). We found that 2 mM GndHCl was sufficient to significantly reduce the readthrough level (Fig. 2C, second panel) while only slightly affecting growth of Δssb1 Δssb2 and Δzuo1 Δssz1 strains (Fig. 3B and data not shown). As predicted, overexpression of HSP104 reduced readthrough in Δzuo1 Δssz1 but not in Δssb1 Δssb2 strains (Fig. 2C, third panel). The combined data indicate that, like the parental strain, Δssb1 Δssb2 and Δzuo1 Δssz1 strains contain the [PSI+] element. Both strains displayed increased readthrough frequency, which was further enhanced when [PSI+] was present (Fig. 2C). In order to determine the effect of the chaperones on translational readthrough without possible interference with [PSI+], we constructed GT197-Δzuo1, GT197-Δssz1, and GT197-Δzuo1 Δssz1 strains (Fig. 2D). These strains were derived from the [psi−] strain GT197 (see Materials and Methods). As expected, GT197 displayed a low level of readthrough, similar to that of MH272-3f cured from [PSI+] (Fig. 2B and E). Growth on GndHCl did not significantly change the efficiency of readthrough in GT197 (data not shown). Readthrough frequency in the mutant strains GT197-Δzuo1, GT197-Δssz1, and GT197-Δzuo1 Δssz1 was approximately twofold increased, confirming that RAC influenced translational readthrough in a [PSI+]-independent manner.
FIG. 3.
zuo1-H128Q and ssz1-S295F strains tolerate higher paromomycin concentrations than Δssb1 Δssb2 or Δssz1 Δzuo1 strains. (A) Serial dilutions of logarithmically growing cultures were spotted on YPD containing the specified supplements and were incubated as indicated. (B) Strains were inoculated at the same OD600 into YPD plus 2 mM GndHCl (control) or into the same medium containing paromomycin as indicated. Cultures were grown until the control without paromomycin had reached an OD600 of 1.2, which was set to 100%.
Readthrough is increased in paromomycin-sensitive strains expressing point mutants of either Ssz1p or zuotin in vivo.
As a possible explanation for defects in translational fidelity, we considered that the complete absence of RAC or Ssb1/2p might have severe consequences on ribosome structure. To address this possibility we made use of point mutants in either RAC subunit which affect paromomycin sensitivity; however, neither affect stability or ribosome association (23; data not shown). We first used zuo1-H128Q, which contains a point mutation in the J domain of zuotin causing growth defects similar to those observed in the deletion strains (23, 30). We next used ssz1-S295F, which contains a single mutation in the ATPase domain of Ssz1p (27). ssz1-S295F was originally identified as a dominant gain-of-function allele increasing pleiotropic drug resistance against, e.g., cycloheximide (27 and Fig. 3A). The ssz1-S295F strain displayed wild-type growth under most conditions; however, the strain was sensitive to elevated concentrations of paromomycin (Fig. 3A). A direct comparison revealed that paromomycin sensitivity increased in the following order: Δzuo1 Δssz1 = Δssb1 Δssb2 > zuo1-H128Q > ssz1-S295F > wild type (Fig. 3B). Both zuo1-H128Q and ssz1-S295F strains displayed an increased level of readthrough that was further increased in the presence of [PSI+] (Fig. 4A). In accord with paromomycin sensitivity, readthrough was more severely affected in zuo1-H128Q (Fig. 4A). The results ruled out purely structural effects caused by the lack of RAC or Ssb1/2p and suggested a requirement for functional RAC to maintain translational fidelity in vivo.
FIG. 4.
Paromomycin strongly affects translational readthrough in zuo1-H128Q and ssz1-S295F strains. (A) Readthrough was determined in wild-type, ssz1-S295F, and zuo1-H128Q strains or the respective strains overexpressing HSP104 grown on YPD or YPD containing 2 mM GndHCl. Numbers below the columns indicate increase of readthrough compared to that of the wild type grown under the same conditions. (B) Readthrough was determined in wild-type, zuo1-H128Q, and ssz1-S295F strains grown on YPD containing 2 mM GndHCl plus increasing concentrations of paromomycin as indicated. Note that the paromomycin concentration used in this experiment was adjusted according to the inhibitory concentration for each strain (see Fig. 3B). Readthrough was quantified as described in the legend to Fig. 2.
Correlation between paromomycin-induced growth inhibition and translational readthrough in zuo1-H128Q and ssz1-S295F strains in vivo.
As Δssb1 Δssb2 and Δzuo1 Δssz1 strains grew only very poorly in the presence of paromomycin, we examined readthrough in the zuo1-H128Q and ssz1-S295F point mutants in the presence of the drug. In the wild-type strain readthrough frequency remained low even at growth-inhibiting concentrations (>500 μg/ml) (Fig. 4B). To the contrary, paromomycin strongly enhanced readthrough in zuo1-H128Q and ssz1-S295F strains (Fig. 4B). Consistent with paromomycin effects on cell growth, the zuo1-H128Q strain responded to lower concentrations than the ssz1-S295F strain. The pronounced difference between wild-type and mutant strains suggests that stop codon recognition became limiting in the presence of paromomycin.
Quantification of misincorporation reveals a paromomycin-induced increase in the mutant strains.
In order to test whether the mutant strains were specifically affected in translation termination or displayed a more general translational defect, we devised an in vivo misincorporation assay. Replacing arginine in position 218 of luciferase with serine inactivates the enzyme (6). To test whether luciferase-R218S was suited as a misincorporation reporter, two different versions were generated. AGA (encoding R218) was either replaced with AGC (encoding S; lacZ-luc*) or TCT (encoding S; lacZ-luc**) (Fig. 5A). Mistranslation of AGC (encoding S) into AGA or AGG (encoding R) requires recognition by a near-cognate tRNA. TCT (encoding S), however, will only be mistranslated into arginine by noncognate tRNAs, a much less frequent event (51). Indeed, the noncognate version produced significantly less active luciferase than the near-cognate version, indicating that the residual activity of luciferase-R218S was lower than activity gained by misincorporation. lacZ-luc* and lacZ-luc** were also tested after curing of [PSI+] (Fig. 5B). In accordance with a specific effect of [PSI+] on translation termination, the misincorporation frequency remained unchanged. The frequency of arginine misincorporation in position 218 of lacZ-luc* was about 1.5 × 10−3. Missense error frequencies vary with codon bias, mRNA template contexts, and the identity of the specific tRNA species involved. The frequency determined for the lacZ-luc* reporter is consistent with previous estimates for prokaryotes and eukaryotes (reference 61 and references therein).
Under normal growth conditions, misincorporation frequencies in the mutant strains were unaltered (Fig. 5C). Paromomycin affected misincorporation in the zuo1-H128Q strain only slightly. In the ssz1-S295F strain, however, which tolerates higher paromomycin concentrations in vivo, misincorporation was raised to significantly higher values than that in the wild-type strain (Fig. 5D). The combined data suggest that the lack of functional RAC primarily affected the fidelity of translation termination and, to a lesser extent, the elongation process. The degree of known translational errors depends on the specific codon and its flanking region; however, the finding suggests that increased misincorporation was unlikely to be the major cause for paromomycin sensitivity.
Readthrough and misincorporation frequencies are increased in the absence of RAC or Ssb1/2p in vitro.
In vivo experiments demonstrated a correlation between translational fidelity and growth defects caused by lack of functional RAC or Ssb1/2p. However, indirect effects, such as increased permeability of the cytoplasmic membrane for paromomycin, cannot readily be excluded based on in vivo data alone. In order to address this possibility and to characterize the mutant strains in more detail, we devised an in vitro system to assess translational fidelity. To this end, luciferase reporter constructs were generated. Readthrough was monitored using a construct in which leucine at position 210 was replaced with a stop codon (luc-STOP) (Fig. 6A). As a result, a truncated, enzymatically inactive version of luciferase was generated (Fig. 6B). Misincorporation was assessed using the active-site mutant R218S (luc*) (Fig. 6A and B). This novel assay closely mimics the in vivo translation process and allowed for the quantification not only of misincorporation but also of readthrough, an advantage over the classical poly(U)-directed system (59).
FIG. 6.
Determination of readthrough and misincorporation efficiency in vitro. (A) luc encodes wild-type luciferase; luc-STOP contains a nonsense mutation in position 210 which, upon translation, generates a 23-kDa fragment; and luc* contains a missense mutation in position 218, generating inactive luciferase. (B) The amount of luciferase synthesized in a wild-type lysate primed with luc, luc-STOP, or luc* was determined via quantification of autoradiography. Activity of an aliquot was determined as described in Materials and Methods. As luc-STOP was generated with higher efficiency, only one-third of the sample was loaded onto the gel and applied to the activity assay. The ratio between luciferase activity and intensity (RLU/QL) obtained for luc mRNA was set to 100%.
In a wild-type translation extract, readthrough frequency for luc-STOP was approximately 10−4 (Fig. 7A). As expected, recognition of the stop codon introduced into the open reading frame of luciferase was much more efficient than the inherently leaky stop codon separating β-galactosidase and luciferase in the lacZ-STOP-luc in vivo construct (Fig. 2). The [PSI+] status of the strains used for generation of yeast translation extracts did not affect readthrough efficiency, indicating that the level of Sup35p was not limiting for translation termination in vitro (data not shown). When luc* mRNA was translated in a wild-type extract, active luciferase was generated with a frequency of 3.6 × 10−4, which is comparable to the frequency determined for the same mistranslation event in vivo (Fig. 5B and 7B). Next, luc, luc-STOP, and luc* mRNAs were used to prime yeast translation extracts derived from wild-type, Δssb1 Δssb2, Δzuo1 Δssz1, zuo1-H128Q, and ssz1-S295F strains. Overall, the in vitro experiments confirmed the results obtained in vivo. Translational fidelity in Δzuo1 Δssz1 and Δssb1 Δssb2 strains was most severely affected, consistent with their growth defects (Fig. 3B and 7A and B). Misincorporation in the mutant strains was more severely affected in vitro than in vivo (Fig. 5C and 7B). Additional factors involved in translation elongation possibly became limiting in the in vitro translation reaction.
FIG. 7.
Paromomycin affects translational fidelity in the absence of functional RAC or Ssb1/2p in vitro. (A to D) Wild-type, Δssb1/2, Δssz1 Δzuo1, zuo1-H128Q, and ssz1-S295F translation extracts were primed with mRNA carrying luc, luc-STOP, or luc*. Paromomycin was added at the indicated concentrations. The ratio of luciferase activity to luciferase intensity (RLU/QL) obtained in the presence of luc mRNA was set to 100%. Experiments depicted in panels A and B were performed in triplicate. (E) Translation reactions performed in wild-type and zuo1-H128Q strains in the presence of increasing concentrations of paromomycin were analyzed on SDS-PAGE followed by autoradiography. Lanes 1, no paromomycin; lanes 2, 1 μg of paromomycin/ml; lanes 3, 5 μg of paromomycin/ml; lanes 4, 7.5 μg of paromomycin/ml; lanes 5, 10 μg of paromomycin/ml; lanes 6, 15 μg of paromomycin/ml; lanes 7, 20 μg of paromomycin/ml. Note that full-length luciferase derived by readthrough is detectable only in zuo1-H128Q.
Effects of paromomycin in vitro correlate with results obtained in vivo.
The overall translation efficiency in vitro was not affected by paromomycin up to a concentration of ∼20 μg/ml. Higher concentrations led to inhibition of translation in all extracts (Fig. 7E and data not shown). The in vitro system enabled us to test the effect of the drug at concentrations that led to growth inhibition in vivo. In agreement with the in vivo results, paromomycin did not significantly affect readthrough in a wild-type translation extract while readthrough in the absence of functional RAC or Ssb1/2p strongly increased (Fig. 7C). The most pronounced effect was obtained in the Δzuo1 Δssz1 extract. The presence of paromomycin led to a more than 10-fold increase in stop codon readthrough (Fig. 7C). Although the basic readthrough level in zuo1-H128Q extract was only slightly higher than that in the wild type, paromomycin caused strong enhancement (Fig. 7A and C). Readthrough in the presence of paromomycin was so efficient in the mutant strains that full-length luciferase became detectable on autoradiograms (Fig. 7E and data not shown). Paromomycin increased misincorporation in wild-type and mutant lysates; however, differences were less pronounced (Fig. 7D). The overall correlation between effects in vivo and in vitro strongly suggests that paromomycin-mediated growth inhibition in the mutant strains is directly linked to the translation process.
[PSI+] enhances the growth defects caused by the lack of functional RAC or Ssb1/2p.
The mutant strains suffered from translational defects, particularly in translation termination. If [PSI+] was present, translation termination was decreased even further (compare Fig. 2C to 4A). For this reason we analyzed the effect of [PSI+] on growth in more detail. Wild-type and mutant strains overexpressing HSP104 were grown on paromomycin gradient plates (Fig. 8A). As expected, growth of the Δssb1 Δssb2 strain was not influenced by overexpression of HSP104 (compare Fig. 2C). The other strains, including the wild type, were able to tolerate significantly higher concentrations of paromomycin when HSP104 was overexpressed (Fig. 8A). In order to cure yeast from [PSI+] it is sufficient to transiently overexpress HSP104 (64). Consistently, even after loss of the HSP104-overexpressing plasmid, higher concentrations of paromomycin were tolerated. Interestingly, growth defects at 30°C and lower temperature in the Δssz1 Δzuo1 strain also were reduced after loss of [PSI+], indicating that defects in translation termination were at least partly also responsible for the general growth defects (Fig. 8B and data not shown).
FIG. 8.
[PSI+] effects growth of Δssb1/2, Δssz1Δzuo1, zuo1-H128Q, and ssz1-S295F strains. (A) Logarithmically growing cultures of wild type, Δssb1/2, Δssz1 Δzuo1, zuo1-H128Q, ssz1-S295F, and the respective isogenic strains overexpressing HSP104 were spotted onto paromomycin gradient plates and were incubated at 30°C for 2 days. (B) Serial dilutions of Δssz1 Δzuo1, Δssz1 Δzuo1 plus HSP104, and Δssz1 Δzuo1* strains were spotted onto YPD and were incubated as indicated. Δssz1 Δzuo1* was cured from the plasmid overexpressing HSP104 before the experiment.
DISCUSSION
A role of RAC and Ssb1/2p in translational fidelity.
We found that the lack of functional RAC or Ssb1/2p caused severe problems in translational fidelity, which were strongly enhanced by paromomycin and correlated with growth inhibition. To the best of our knowledge RAC and Ssb1/2p are the first chaperones implicated in translational fidelity. As they localize far from the decoding center, this effect is at first quite unexpected. RAC and Ssb1/2p may affect translational fidelity via facilitating the folding of one or more proteins involved in translational fidelity. However, increased protein aggregation in the absence of RAC or Ssb1/2p has not been detected so far (data not shown). Given the recently emerging picture of the ribosome as a highly dynamic and flexible machine, the effect of RAC and Ssb1/2p may also be more direct (19).
Paromomycin binds to the rRNA of the small subunit of the eubacterial ribosome and induces conformational changes (15, 31). Kinetic analysis revealed that these changes stabilize the aminoacyl-tRNA (aa-tRNA) in the A site irrespective of whether the codon-anticodon pair is cognate or near cognate (52). It is now understood that aminoglycosides not only increase the binding affinity of near-cognate aa-tRNA but also promote domain closure of the 30S ribosomal subunit—a process otherwise only observed upon binding of cognate aa-tRNA (44). Conformational changes not only provide an explanation for the effect of paromomycin but also explain biochemical and genetic data on translational accuracy (43, 50). Upon a conformational switch in the 16S rRNA, major rearrangements in the entire 70S ribosomal structure have been observed (17). In accord, not only mutants in rRNA of the small subunit but also in the 23S rRNA have been found to affect translational fidelity (42). By using hybrid archaebacterial ribosomes, it was shown that their response to paromomycin depended critically on the large ribosomal subunit (55). A connection to translational fidelity was also suggested for Rpl39p, a protein of the large ribosomal subunit conserved in eukaryotes and archaebacteria. The archaebacterial homolog localizes at the polypeptide tunnel exit and extends into the tunnel (3). Deletion of RPL39 in yeast causes cold sensitivity, slow growth, and paromomycin hypersensitivity, phenotypes very similar to the ones observed in the absence of functional RAC or Ssb1/2p. Using the poly(U)-directed in vitro system it was shown that lack of Rpl39p increases misincorporation in vitro (13). Whether Δrpl39 affects translation termination is presently unknown. Recent experiments suggest that Rpl39 is also involved in the identification of newly synthesized transmembrane segments of proteins targeted to the endoplasmic reticulum membrane and participates in the opening of the seal between ribosome and translocon (53, 69).
Of specific interest is the observation that overexpression of SSB1 results in more efficient stop codon recognition in vivo (I. Hatin and J.-P. Rousset, personal communication). This finding is in excellent agreement with the data presented in this study and supports a model in which Ssb1/2p directly affects the fidelity of translation. Possibly, RAC, Ssb1/2p, and Rpl39p could function in coordinating the movement of the nascent polypeptide with the movement of the mRNA. Irregularities in this coordination obviously may affect the kinetics of polypeptide bond formation and, as a consequence, translational fidelity (25). In fact, evidence has been presented that communication between the two ribosomal subunits regulates gating of the polypeptide tunnel entrance and is required to coordinate the movement of nascent polypeptide and mRNA (18).
Why does yeast lacking functional RAC or Ssb1/2p die in the presence of aminoglycosides?
What causes death of bacteria in the presence of aminoglycosides has been debated for decades and is still not understood. According to the error catastrophe theory, accumulated errors in proteins which themselves function in protein synthesis (such as ribosomal proteins) cause additional errors. This positive feedback loop points toward an inescapable decay of translational accuracy. The issue, however, is still controversial, and it is unclear whether it can account for the bactericidal action of aminoglycosides (48). In the same context, it has been speculated that errors leading to incorrect translation termination (that is, readthrough, frame shifting, premature termination) have a greater effect on cell growth than missense errors (36). The results presented here point in the same direction. Paromomycin sensitivity of the mutant strains correlated well with an effect on readthrough. Reduction of readthrough by eliminating the [PSI+] element significantly improved the fitness of the mutant strains (Fig. 8). Most likely, however, the consequences of [PSI+] (that is, low level of the translation termination factor Sup35p), paromomycin, or the lack of functional RAC or Ssb1/2p overlap but are not identical. [PSI+] or the lack of RAC/Ssb1/2p may well affect different termination events to different degrees. In this context it is interesting that some stop codons are insensitive to the presence of [PSI+] (39). It is not hard to imagine that an increased level of proteins that carry a C-terminal extension can be deleterious. This effect could be specifically relevant for proteins that naturally display a high level of readthrough (39). However, one should keep in mind that RAC and Ssb1/2p, as most chaperone proteins, are likely to serve additional functions in the cell that will contribute to the defects displayed in the absence of these chaperones.
Acknowledgments
This work was supported by the Kultusministerium des Landes Sachsen-Anhalt (to S.R.), the Fonds der Chemischen Industrie (to S.R.), and by SFB 388 and 610 (to S.R.).
We thank Suzanne Ross, Andrej Mun, and Tina Wölfle for excellent technical assistance. We thank Magda Boguta for yeast strain GT197. We also thank Agnieszka Chacinska, Yves Dubaquié, Matthias Gautschi, Klaus Pfanner, and members of the laboratory for discussion and critically reading the manuscript.
REFERENCES
- 1.Alksne, L. E., R. A. Anthony, S. W. Liebman, and J. R. Warner. 1993. An accuracy center in the ribosome conserved over 2 billion years. Proc. Natl. Acad. Sci. USA 90:9538-9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.All-Robyn, J. A., D. Kelley-Geraghty, E. Griffin, N. Brown, and S. W. Liebman. 1990. Isolation of omnipotent suppressors in an [eta+] yeast strain. Genetics 124:505-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289:905-920. [DOI] [PubMed] [Google Scholar]
- 4.Bidou, L., G. Stahl, I. Hatin, O. Namy, J. P. Rousset, and P. J. Farabaugh. 2000. Nonsense-mediated decay mutants do not affect programmed −1 frameshifting. RNA 6:952-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. [DOI] [PubMed] [Google Scholar]
- 6.Branchini, B. R., R. A. Magyar, M. H. Murtiashaw, and N. C. Portier. 2001. The role of active site residue arginine 218 in firefly luciferase bioluminescence. Biochemistry 40:2410-2418. [DOI] [PubMed] [Google Scholar]
- 7.Chacinska, A., M. Boguta, J. Krzewska, and S. Rospert. 2000. Prion-dependent switching between respiratory competence and deficiency in the yeast nam9-1 mutant. Mol. Cell. Biol. 20:7220-7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chernoff, Y. O., G. P. Newnam, J. Kumar, K. Allen, and A. D. Zink. 1999. Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI] prion. Mol. Cell. Biol. 19:8103-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chernoff, Y. O., A. Vincent, and S. W. Liebman. 1994. Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J. 13:906-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 11.Craig, E. A., H. C. Eisenman, and H. A. Hundley. 2003. Ribosome-tethered molecular chaperones: the first line of defense against protein misfolding? Curr. Opin. Microbiol. 6:157-162. [DOI] [PubMed] [Google Scholar]
- 12.Dean, N. 1995. Yeast glycosylation mutants are sensitive to aminoglycosides. Proc. Natl. Acad. Sci. USA 92:1287-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dresios, J., I. L. Derkatch, S. W. Liebman, and D. Synetos. 2000. Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry 39:7236-7244. [DOI] [PubMed] [Google Scholar]
- 14.Eustice, D. C., L. P. Wakem, J. M. Wilhelm, and F. Sherman. 1986. Altered 40 S ribosomal subunits in omnipotent suppressors of yeast. J. Mol. Biol. 188:207-214. [DOI] [PubMed] [Google Scholar]
- 15.Fourmy, D., S. Yoshizawa, and J. D. Puglisi. 1998. Paromomycin binding induces a local conformational change in the A-site of 16 S rRNA. J. Mol. Biol. 277:333-345. [DOI] [PubMed] [Google Scholar]
- 16.Fünfschilling, U., and S. Rospert. 1999. Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol. Biol. Cell. 10:3289-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabashvili, I. S., R. K. Agrawal, R. Grassucci, C. L. Squires, A. E. Dahlberg, and J. Frank. 1999. Major rearrangements in the 70S ribosomal 3D structure caused by a conformational switch in 16S ribosomal RNA. EMBO J. 18:6501-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabashvili, I. S., S. T. Gregory, M. Valle, R. Grassucci, M. Worbs, M. C. Wahl, A. E. Dahlberg, and J. Frank. 2001. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Mol. Cell. 8:181-188. [DOI] [PubMed] [Google Scholar]
- 19.Gabashvili, I. S., M. Whirl-Carrillo, M. Bada, D. R. Banatao, and R. B. Altman. 2003. Ribosomal dynamics inferred from variations in experimental measurements. RNA 9:1301-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia, P. D., W. Hansen, and P. Walter. 1991. In vitro protein translocation across microsomal membranes of Saccharomyces cerevisiae. Methods Enzymol. 194:675-682. [DOI] [PubMed] [Google Scholar]
- 21.Gautschi, M., S. Just, A. Mun, S. Ross, P. Rücknagel, Y. Dubaquié, A. Ehrenhofer-Murray, and S. Rospert. 2003. The yeast Nα-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides. Mol. Cell. Biol. 23:7403-7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gautschi, M., H. Lilie, U. Fünfschilling, A. Mun, S. Ross, T. Lithgow, P. Rücknagel, and S. Rospert. 2001. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc. Natl. Acad. Sci. USA 98:3762-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gautschi, M., A. Mun, S. Ross, and S. Rospert. 2002. A functional chaperone triad on the yeast ribosome. Proc. Natl. Acad. Sci. USA 99:4209-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 25.Gromadski, K. B., and M. V. Rodnina. 2004. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol. Cell. 13:191-200. [DOI] [PubMed] [Google Scholar]
- 26.Haid, A., and M. Suissa. 1983. Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 96:192-205. [DOI] [PubMed] [Google Scholar]
- 27.Hallstrom, T. C., D. J. Katzmann, R. J. Torres, W. J. Sharp, and W. S. Moye-Rowley. 1998. Regulation of transcription factor Pdr1p function by an Hsp70 protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:1147-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartl, F. U., and M. Hayer-Hartl. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852-1858. [DOI] [PubMed] [Google Scholar]
- 29.Heitmann, J., N. R. Movva, P. C. Hiestand, and M. N. Hall. 1991. FK 506-binding protein proline rotamase is a target for the immunosuppressive agent FK 506 in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 88:1948-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hundley, H., H. Eisenman, W. Walter, T. Evans, Y. Hotokezaka, M. Wiedmann, and E. Craig. 2002. The in vivo function of the ribosome-associated Hsp70, Ssz1, does not require its putative peptide-binding domain. Proc. Natl. Acad. Sci. USA 99:4203-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jerinic, O., and S. Joseph. 2000. Conformational changes in the ribosome induced by translational miscoding agents. J. Mol. Biol. 304:707-713. [DOI] [PubMed] [Google Scholar]
- 32.Jones, G. W., Y. Song, and D. C. Masison. 2003. Deletion of the Hsp70 chaperone gene SSB causes hypersensitivity to guanidine toxicity and curing of the [PSI+] prion by increasing guanidine uptake in yeast. Mol. Genet. Genomics 269:304-311. [DOI] [PubMed] [Google Scholar]
- 33.Kaminska, J., A. Tobiasz, M. Gniewosz, and T. Zoladek. 2000. The growth of mdp1/rsp5 mutants of Saccharomyces cerevisiae is affected by mutations in the ATP-binding domain of the plasma membrane H+-ATPase. Gene 242:133-140. [DOI] [PubMed] [Google Scholar]
- 34.Kandl, K. A., R. Munshi, P. A. Ortiz, G. R. Andersen, T. G. Kinzy, and A. E. Adams. 2002. Identification of a role for actin in translational fidelity in yeast. Mol. Genet. Genomics 268:10-18. [DOI] [PubMed] [Google Scholar]
- 35.Katzmann, D. J., E. A. Epping, and W. S. Moye-Rowley. 1999. Mutational disruption of plasma membrane trafficking of Saccharomyces cerevisiae Yor1p, a homologue of mammalian multidrug resistance protein. Mol. Cell. Biol. 19:2998-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurland, C. G. 1992. Translational accuracy and the fitness of bacteria. Annu. Rev. Genet. 26:29-50. [DOI] [PubMed] [Google Scholar]
- 37.Kushnirov, V. V., and M. D. Ter-Avanesyan. 1998. Structure and replication of yeast prions. Cell 94:13-16. [DOI] [PubMed] [Google Scholar]
- 38.Masurekar, M., E. Palmer, B. I. Ono, J. M. Wilhelm, and F. Sherman. 1981. Misreading of the ribosomal suppressor SUP46 due to an altered 40 S subunit in yeast. J. Mol. Biol. 147:381-390. [DOI] [PubMed] [Google Scholar]
- 39.Namy, O., G. Duchateau-Nguyen, I. Hatin, S. Hermann-Le Denmat, M. Termier, and J. P. Rousset. 2003. Identification of stop codon readthrough genes in Saccharomyces cerevisiae. Nucleic Acids Res. 31:2289-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Namy, O., I. Hatin, and J. P. Rousset. 2001. Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep. 2:787-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelson, R. J., T. Ziegelhoffer, C. Nicolet, M. Werner-Washburne, and E. A. Craig. 1992. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71:97-105. [DOI] [PubMed] [Google Scholar]
- 42.O'Connor, M., and A. E. Dahlberg. 1995. The involvement of two distinct regions of 23 S ribosomal RNA in tRNA selection. J. Mol. Biol. 254:838-847. [DOI] [PubMed] [Google Scholar]
- 43.Ogle, J. M., A. P. Carter, and V. Ramakrishnan. 2003. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 28:259-266. [DOI] [PubMed] [Google Scholar]
- 44.Ogle, J. M., F. V. Murphy, M. J. Tarry, and V. Ramakrishnan. 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111:721-732. [DOI] [PubMed] [Google Scholar]
- 45.Osherovich, L. Z., and J. S. Weissman. 2002. The utility of prions. Dev. Cell. 2:143-151. [DOI] [PubMed] [Google Scholar]
- 46.Palmer, E., J. M. Wilhelm, and F. Sherman. 1979. Phenotypic suppression of nonsense mutants in yeast by aminoglycoside antibiotics. Nature 277:148-150. [DOI] [PubMed] [Google Scholar]
- 47.Pfund, C., N. Lopez-Hoyo, T. Ziegelhoffer, B. A. Schilke, P. Lopez-Buesa, W. A. Walter, M. Wiedmann, and E. A. Craig. 1998. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 17:3981-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puglisi, J. D., K. D. Blanchard, R. G. E. Dahlquist, R. G. Eason, D. Fourmy, S. R. Lynch, M. I. Recht, and S. Yoshizawa. 2000. Aminoglycoside antibiotics and decoding, p. 419-429. In R. A. Garrett, S. R. Douthwaite, A. Liljas, P. B. Matheson, P. B. Moore, and H. F. Noller (ed.), The ribosome: structure, function, antibiotics, and cellular interactions. ASM Press, Washington, D.C.
- 49.Recht, M. I., S. Douthwaite, and J. D. Puglisi. 1999. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J. 18:3133-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodnina, M. V., T. Daviter, K. Gromadski, and W. Wintermeyer. 2002. Structural dynamics of ribosomal RNA during decoding on the ribosome. Biochimie 84:745-754. [DOI] [PubMed] [Google Scholar]
- 51.Rodnina, M. V., T. Pape, A. Savelsbergh, D. Mohr, N. B. Matassova, and W. Wintermeyer. 2000. Mechanisms of partial reactions of the elongation cycle catalyzed by elongation factors Tu and G, p. 301-317. In R. A. Garrett, S. R. Douthwaite, A. Liljas, P. B. Matheson, P. B. Moore, and H. F. Noller (ed.), The ribosome: structure, function, antibiotics, and cellular interactions. ASM Press, Washington, D.C.
- 52.Rodnina, M. V., and W. Wintermeyer. 2001. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu. Rev. Biochem. 70:415-435. [DOI] [PubMed] [Google Scholar]
- 53.Rospert, S. 2004. Ribosome function: how to govern the fate of a nascent polypeptide. Curr. Biol. 14:R386-R388. [DOI] [PubMed] [Google Scholar]
- 54.Rospert, S., M. Gautschi, M. Rakwalska, and U. Raue. 2004. Ribosome-bound proteins acting on newly synthesized polypeptide chains. In J. Buchner and T. Kiefhaber (ed.), Handbook in protein folding, vol. II, in press. Wiley-VCH Verlag GmbH & Co., Weinheim, Germany.
- 55.Ruggero, D., and P. Londei. 1996. Differential antibiotic sensitivity determined by the large ribosomal subunit in thermophilic archaea. J. Bacteriol. 178:3396-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 57.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 58.Siegers, K., B. Bolter, J. P. Schwarz, U. M. Bottcher, S. Guha, and F. U. Hartl. 2003. TRiC/CCT cooperates with different upstream chaperones in the folding of distinct protein classes. EMBO J. 22:5230-5240. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Singh, A., D. Ursic, and J. Davies. 1979. Phenotypic suppression and misreading Saccharomyces cerevisiae. Nature 277:146-148. [DOI] [PubMed] [Google Scholar]
- 60.Stahl, G., L. Bidou, J. P. Rousset, and M. Cassan. 1995. Versatile vectors to study recoding: conservation of rules between yeast and mammalian cells. Nucleic Acids Res. 23:1557-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stansfield, I., K. M. Jones, P. Herbert, A. Lewendon, W. V. Shaw, and M. F. Tuite. 1998. Missense translation errors in Saccharomyces cerevisiae. J. Mol. Biol. 282:13-24. [DOI] [PubMed] [Google Scholar]
- 62.Synetos, D., C. P. Frantziou, and L. E. Alksne. 1996. Mutations in yeast ribosomal proteins S28 and S4 affect the accuracy of translation and alter the sensitivity of the ribosomes to paromomycin. Biochim. Biophys. Acta 1309:156-166. [DOI] [PubMed] [Google Scholar]
- 63.True, H. L., and S. L. Lindquist. 2000. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407:477-483. [DOI] [PubMed] [Google Scholar]
- 64.Tuite, M. F., and S. L. Lindquist. 1996. Maintenance and inheritance of yeast prions. Trends Genet. 12:467-471. [DOI] [PubMed] [Google Scholar]
- 65.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 66.Wakem, L. P., and F. Sherman. 1990. Isolation and characterization of omnipotent suppressors in the yeast Saccharomyces cerevisiae. Genetics 124:515-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wegrzyn, R. D., K. Bapat, G. P. Newnam, A. D. Zink, and Y. O. Chernoff. 2001. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol. Cell. Biol. 21:4656-4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wickner, R. B., K. L. Taylor, H. K. Edskes, M. L. Maddelein, H. Moriyama, and B. T. Roberts. 2000. Prions of yeast as heritable amyloidoses. J. Struct. Biol. 130:310-322. [DOI] [PubMed] [Google Scholar]
- 69.Woolhead, C. A., P. J. McCormick, and A. E. Johnson. 2004. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell 116:725-736. [DOI] [PubMed] [Google Scholar]
- 70.Yan, W., B. Schilke, C. Pfund, W. Walter, S. Kim, and E. A. Craig. 1998. Zuotin, a ribosome-associated DnaJ molecular chaperone. EMBO J. 17:4809-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]