Abstract
Epiregulin, an epidermal growth factor family member, acts as a local signal mediator and shows dual biological activity, stimulating the proliferation of fibroblasts, hepatocytes, smooth muscle cells, and keratinocytes while inhibiting the growth of several tumor-derived epithelial cell lines. The epiregulin gene (Ereg) is located on mouse chromosome 5 adjacent to three other epidermal growth factor family members, epigen, amphiregulin, and betacellulin. Gene targeting was used to insert a lacZ reporter into the mouse Ereg locus and to ablate its function. Although epiregulin is broadly expressed and regulated both spatially and temporally, Ereg null mice show no overt developmental defects, reproductive abnormalities, or altered liver regeneration. Additionally, in contrast to previous hypotheses, Ereg deficiency does not alter intestinal cancer susceptibility, as assayed in the ApcMin model, despite showing robust expression in developing tumors. However, Ereg null mice are highly susceptible to cancer-predisposing intestinal damage caused by oral administration of dextran sulfate sodium.
Epiregulin, a novel epidermal growth factor (EGF)-related growth factor first identified in NIH 3T3 cell conditioned medium (55), is one of seven known ligands for the epidermal growth factor receptor (EGFR) (59). Epiregulin is synthesized as a transmembrane precursor before being proteolytically cleaved to release a 46-amino-acid active protein. Unlike other EGFR ligands, epiregulin shows dual biological activity; it stimulates proliferation of fibroblasts, hepatocytes, smooth muscle cells, and keratinocytes but inhibits growth of several tumor-derived epithelial cell lines (43, 51, 55). Epiregulin also shows more potent bioactivity in vitro than other EGF-like growth factors (36, 42, 55). Part of this potent activity can be attributed to the fact that, like two other EGFR ligands, betacellulin and diphtheria toxin receptor (formerly called heparin-binding EGF) (16, 32, 34), epiregulin is promiscuous, binding and activating the EGFR family member ERBB4 via heterodimeric interactions with ERBB2 (18, 35, 42).
Mutations generated in Egfr and the other Erbb receptor genes by gene targeting have been used to demonstrate the requirement for these receptors during embryonic development (7, 11, 21, 30, 45, 53). Unlike the other ERBB receptors that are involved in cardiac and neuronal development, EGFR is primarily required for normal placental development and shows a strong genetic background-dependent variation in embryonic survival, with some genetic backgrounds supporting survival to term before manifesting perinatal lethality caused by multiorgan defects (44, 45, 53). Egfrwa2, a hypomorphic allele of Egfr (10, 24), has been used to demonstrate the importance of this receptor in various disease states. Previous studies have shown the importance of the EGFR signal axis in the establishment of intestinal tumors (37) and in the maintenance of intestinal homeostasis in response to chemically induced ulcerative colitis (4). Phenotypic analysis of targeted mutations in genes coding for the EGFR ligands have been less informative, probably due to functional redundancy or overlap between different ligands. Although mutations in members of the EGFR ligand gene family have manifested only minor phenotypic abnormalities, they appear to be important for normal gastrointestinal and cardiac homeostasis, lactation, and hair follicle morphogenesis (14, 25, 26).
Functional characterization of Ereg has predominantly been based upon Northern blot and in situ expression analysis, with results suggesting that epiregulin is a local cell-signaling mediator involved in many biological processes. The func tions proposed for epiregulin have included roles in reproduction, where Ereg expression is highly localized in the ovary, uterus of early pregnancy, placenta, and macrophages (3, 33, 41, 56, 57), mediation of the mitogenic effects of vasoactive antagonists in smooth muscle cells (51), and liver regeneration (19, 57). Although Ereg expression is highly correlated with survival for those with bladder cancer (52), reports have suggested that epiregulin is also important for pancreatic and prostate cancer development, based upon its upregulation and bioactivity (54, 60). The strongest evidence for a role of epiregulin during tumorigenesis is in Ki-ras-mediated signaling in colon cancer cells (1); Ereg transcripts are also the most frequently detected of the EGFR ligands in serial analysis of gene expression libraries from colorectal cancers (2). More recently, epiregulin has been shown to be required for immortalization of human fibroblasts by telomerase, further suggesting a role for epiregulin during tumorigenesis (23).
In this study, we cloned and determined the genomic structure of the mouse Ereg gene and inserted a lacZ cassette into the locus by homologous recombination in embryonic stem (ES) cells to create an Ereg null allele, Eregtm1Dwt. These mice were used to show that Ereg is widely expressed in many tissues where the expression profile is restricted to specific subsets of cells, displaying highly specific spatial regulation. Homozygous Eregtm1Dwt mice are morphologically normal on two genetic backgrounds and show normal reproductive characteristics. Likewise, the Eregtm1Dwt null mice show normal liver regeneration after partial hepatectomy and unaltered susceptibility to intestinal tumorigenesis when crossed to the ApcMin model despite dramatic pathology-associated alterations in expression. However, Ereg null mice exhibit dramatically enhanced susceptibility to dextran sulfate sodium (DSS)-induced intestinal damage, an intestinal tumor-predisposing condition, similar to that reported previously for the Egfrwa2 hypomorphic allele.
MATERIALS AND METHODS
Cloning of Ereg.
Genomic DNA from the Ereg promoter region was amplified from a DraI GenomeWalker library (Clontech) with the following primers derived from Ereg cDNA sequence (55): Ereg-AS-GSP1, 5′-AAGGTGCCTGTGGCTGAGTTGGAACCGAG-3′, and Ereg-AS-GSP2, 5′-TGATCCAGGTACCAGAGCCCACCAAGTCGC-3′. The PCR-amplified fragment from the kb −2.4 to −2.0 promoter region was used as a probe to screen a 129S6/SvEvTAC bacterial artificial chromosome (BAC) library (RPCI-22; Roswell Park Cancer Institute). Genomic DNA from BAC clone 96A12 was amplified with the TaqPlus long PCR system (Stratagene) with a series of primers based on the mouse Ereg cDNA sequence to resolve the exon-intron boundaries: Ereg-S3 (5′-CTCAGAGTCACAGCGACTTG-3′) and Ereg-AS2 (5′-CTAAGGCGGTACAGTTATCC-3′) were used for amplification of exons 1 to 2, and Ereg-S2 (5′-CAGGCAGTTATCAGCACAAC-3′) and Ereg-AS1 (5-CCTTGTCCGTAACTTGATGG-3′) were used for amplification of exons 2 to 5. The PCR products were subcloned into pCR2.1-TOPO (Invitrogen) and sequenced with BigDye terminator cycle sequencing (Perkin-Elmer).
Radiation hybrid mapping.
The T31 mouse-hamster radiation hybrid (RH) panel (Research Genetics) was screened with PCR primers designed from both Btc and Ereg. PCR mixtures consisted of 25 ng of radiation hybrid DNA, 1 μl of 10× PCR buffer, 0.3 μl of 50 mM MgCl2, 0.5 μl of 10 mM dNTPs, 1 μl of each primer at 2 μM, and 0.2 μl of Taq polymerase (2.5 U/μl) in a 10-μl final volume. PCR was performed with 1 min of denaturation at 95°C, 1 min of annealing at 60°C, and 1 min of extension at 72°C for 35 cycles. Both primer pairs yield PCR products of approximately 300 bp from genomic DNA of 129S6/SvEvTAC mice. PCR products were separated on a 1.5% agarose gel and visualized with ethidium bromide staining. The primer pairs did not amplify hamster genomic DNA and therefore, PCRs were performed in duplicate to ensure the accuracy of a negative result. The strain distribution pattern of the presence or absence of product was entered into MapManager QT (28), and a chromosomal location was determined by comparison to the latest release for the radiation hybrid panel data (Jackson Laboratory).
Generation of Ereg null mice.
Genomic DNA from BAC clone 96A12 was digested with EcoRI, and an approximately 9-kb fragment containing exon 1 of Ereg was subcloned into the pBSII vector (Stratagene) as follows. The sequence encoding the 3′ NdeI-EcoRI fragment was deleted, and the start codon was replaced with a unique ClaI site by site-directed mutagenesis (Promega). A PmeI-XbaI fragment from pHM4 (17) containing the nuclear localization signal (nls), lacZ, simian virus 40 polyadenylation signal, and pgk-neo cassettes was inserted into the ClaI site. Four base pairs (+1 to +4 from the translation start) were deleted in the generation of the final targeting construct. All junctions were verified by DNA sequencing.
Homologous recombination in TL-1 ES cells was performed as described previously (13). Clones were screened for accurate homologous recombination by Southern blot hybridization as follows. DNA isolated from ES cell clones was digested with EcoRV, electrophoretically separated on a 0.8% agarose gel, and transferred to a nylon membrane (Schleicher & Schuell). The membrane was screened with a 0.45-kb probe derived from an NsiI-EcoRI genomic DNA fragment 5′ to the homologous arm of the targeting construct. Chimeric mice were generated by blastocyst injection into C57BL/6J-derived blastocysts (13) and implanting into a surrogate mother. Chimeric pups were born from two independently targeted ES cell clones.
Selected chimeras were bred to identify germ line transmission of the targeted allele. Subsequent generations of mice carrying the Eregtm1Dwt targeted allele were maintained coisogenic on 129S6/SvEvTAC and congenic on C57BL/6J genetic backgrounds. Genotypes were determined by PCR with three primers: Ereg-S3, 5′-CTCAGAGTCACAGCGACTTG-3′ (sense primer from the 5′ noncoding region of exon 1); Ereg-AS1, 5′-CAGCGTCAAGACCCAAGAGG-3′ (antisense primer from the coding region of exon 1); and lacZ-AS2, 5′-GCTGCAAGGCGATTAAGTTGG-3′ (antisense primer from the nls-lacZ region). Primers Ereg-S3 and Ereg-AS1 amplify a 159-bp product specific for the wild-type Ereg allele, and primers Ereg-S3 and lacZ-AS2 amplify a 253-bp product specific for the targeted allele.
RT-PCR.
Total RNA was prepared from tissues isolated from two wild-type 3-month-old 129S6/SvEvTAC mice with Trizol (Invitrogen). One microgram of total RNA was reverse transcribed with Superscript II reverse transcriptase (Invitrogen) with random hexamers. One twentieth of the reverse transcription product was amplified by PCR with epiregulin primers Ereg-S2, 5′-CAGGCAGTTATCAGCACAAC-3′ (sense primer from exon 2) and Ereg-AS4, 5′-CCTTGTCCGTAACTTGATGG-3′ (antisense primer from exon 5) or with a β-actin (Actb) primer set (sense strand, 5′-CTACAATGAGCTGCGTGTGG-3′; antisense strand, 5′-CACAGGATTCCATACCCAAG-3′). The reverse transcription (RT)-PCR products were resolved with 1.2% agarose gel electrophoresis and visualized with ethidium bromide staining. Total RNA from the colons of wild-type, heterozygous, and Ereg null mice was also analyzed by RT-PCR with Ereg-S3 and Ereg-AS4 to verify the absence of Ereg transcripts in null mice. The Ereg-S3 and Ereg-AS4 primers produce a 635-bp PCR product.
Western blots.
An extract of freshly collected jejunum from wild-type, heterozygous, and Ereg null mice was prepared by homogenization in 10 volumes (10 ml/g) of homogenization buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin per ml, 1 μg of aprotinin per ml). The cleared lysate was filtered through a Microcon-30 filter column (Amicon), and eluted solution was further concentrated with a Microcon-3 filter column. Equal amounts of protein lysate were loaded onto a 20% acrylamide gel, electrophoresed, and transferred to a polyvinylidene difluoride membrane (Bio-Rad). The membrane was incubated in blocking solution containing 5% nonfat dried milk in TBST (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.05% Tween 20) for 1 h at room temperature and subsequently incubated with 2 μg of polyclonal rabbit anti-EREG antibody per ml in TBS (10 mM Tris-HCl [pH 7.4], 150 mM NaCl) at 4°C overnight. Following incubation with primary antibody, the membrane was washed four times in TBST and then incubated in blocking solution containing goat anti-rabbit immunoglobulin conjugated with horseradish peroxidase for 1 h at room temperature. The membrane was further washed four times in TBST, and specific protein complexes were visualized with the enhanced chemiluminescence system (Amersham-Pharmacia).
LacZ staining.
Tissues collected from Eregtm1Dwt mice were fixed in 0.2% paraformaldehyde-2 mM MgCl2-5 mM EGTA in 1× phosphate-buffered saline (PBS), pH 7.8, at 4°C for 10 h before equilibrating in a sucrose solution (30% sucrose and 2 mM MgCl2 in 1× PBS, pH 7.8) at 4°C overnight. The sucrose-infused tissues were embedded in OCT compound (Fisher) and frozen in isopentane at −80°C. Cryosections (10 μm) were prepared from the tissues, postfixed in fixative for 10 min at 4°C, and incubated with a detergent rinse (2 mM MgCl2, 0.01% sodium deoxycholate, 0.02% Igepal CA-630 in 1× PBS, pH 7.8) for 10 min. The sections were incubated from 4 h to overnight at 37°C with LacZ staining solution containing 0.04% 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), 2 mM MgCl2, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 0.02% NP-40, and 0.01% sodium deoxycholate in PBS, pH 7.8. The X-Gal-stained tissues were counterstained with either eosin Y or hematoxylin and eosin Y.
Macroadenoma counts.
129S6/SvEvTAC-Eregtm1Dwt mice were bred to C57BL/6J ApcMin/+ mice, and the heterozygous Eregtm1Dwt and ApcMin F1 mice were intercrossed to produce ApcMin/+ mice on an Ereg wild-type, heterozygous, or Ereg null background. The small intestines from the pylorus to the cecum and the colon were removed from 3-month-old mice and processed as described previously (37). Polyp number and diameter were also determined and analyzed as previously described (37).
Partial hepatectomies.
Three-month-old mice were anesthetized with Isoflurane (Halocarbon Laboratories), and the left and median lobes of the liver were resected corresponding to a 70% partial hepatectomy. The regenerated right lobe was collected for RNA isolation and LacZ staining. Ereg RNA was detected by RT-PCR at 0, 1, 2, 4, and 8 h and 1, 2, 3, and 4 days after resection. LacZ staining for epiregulin localization was performed at 0, 2, and 4 h and 1, 2, 3, and 4 days after resection.
DSS treatment and histological scoring.
DSS (ICN, 36 to 50-kDa) was dissolved in deionized water at 1.5% (wt/vol) and administered to mice ad libitum in drinking water. The body weight of each mouse was measured daily. Mice were fed Purina Mills Lab Diet 5058 under specific-pathogen-free conditions in an American Association for the Accreditation of Lab Animal Care-approved facility. Mice were euthanized by CO2 asphyxiation. The colon was excised at 9 days after treatment, and gross blood content was determined by measuring the blood clot region through the entire colon. The colon was then fixed in 10% buffered formalin and embedded with paraffin.
Paraffin sections (7 μm) were stained with hematoxylin and eosin for histological analysis. The severity of mucosal injury was graded on a scale of 0 to 3 as previously described with minor modifications: grade 0, normal; grade 1, partial destruction of crypts; grade 2, complete loss of crypts; and grade 3, complete loss of crypts and epithelial cells (6). To produce a histological score for any given section the histological score was determined by adding the portion of injured surface multiplied by the grade; for example, if 50% of the region was grade 1 and 50% of the region was grade 2, the final score was (0.5 × 1) + (0.5 × 2) = 1.5. Two sections of each proximal, middle, and distal region were used for histological scoring. The histological scores from individual mice were determined by adding all six values. Histological scoring was performed in a blinded fashion.
Statistical analysis.
An unpaired t test was used to analyze associations between genotype with polyp number and size and with histological scores. DSS-induced body weight loss was analyzed with a Mann-Whitney test. Statistical analysis was performed with StatView (SAS Institute).
RESULTS
Structure and mapping of Ereg.
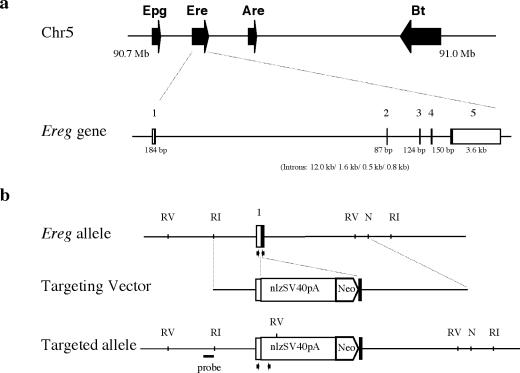
A PCR-amplified 2.4-kb genomic DNA fragment from kb −2.4 to −2.0 of the Ereg promoter region was used as a probe to isolate five positive BAC clones from a 1296/SvEvTAC BAC library (RPCI-22; Roswell Park Cancer Institute). A series of primers were designed based on mouse Ereg cDNA sequences to amplify sequences from one of the BAC clones (55). The amplified PCR products were subcloned, sequenced, and compared with mouse Ereg cDNA sequences to determine the genomic structure. Ereg was found to consist of five exons that spanned approximately 19 kb of genomic DNA (Fig. 1a). The mature form of epiregulin is encoded by exons 3 and 4.
FIG. 1.
Structure and targeting strategy for the mouse Ereg gene. Four genes for the EGF family of ligands, Epgn, Ereg, Areg, and Btc, are located in chromosome 5 from Mb 90.7 to 91.0. (a) The exons of Ereg are labeled 1 to 5. The sizes of exons are given under each exon, and the sizes of introns (introns 1 to 4) are given in parentheses. The empty square at exons 1 and 5 indicates 5′ and 3′ nontranslated region, respectively. The Ereg wild-type allele, targeting vector, and targeted Eregtm1Dwt allele are shown in b. RI, RV, and N denote EcoRI, EcoRV, and NdeI sites, respectively. The vector region flanking the 5′ homology arm of the targeting construct is not included in this schematic, and nls, SV40pA, and neo represent the nuclear localization signal-tagged lacZ reporter gene, a simian virus 40 polyadenylation signal, and a neomycin transphosphorylase expression cassette, respectively. The genomic DNA fragment between NsiI and EcoRI, represented by a solid line, was used as a probe for Southern blot hybridizations. Arrows indicate primer sets used for genotyping wild-type and targeted alleles.
The genomic location of Ereg was determined by screening 100 clones from the T31 mouse-hamster radiation hybrid panel (Research Genetics) with PCR primers specific to mouse Ereg and Btc. Analysis of the strain distribution pattern revealed that the Btc and Ereg primers gave identical patterns, indicating that they are closely linked (logarithm of the odds [LOD], 25.6). Both primer sets showed linkage to markers from mouse chromosome 5 (D5Mit20, LOD 10.7; and D5Mit155, LOD 20.3) that corresponds to approximately 51 centimorgans (cM) on the consensus mouse genetic map (www.informatics.jax.org). Subsequently, a more detailed location of Ereg was determined by comparison of Ereg cDNA sequences with a mouse draft genome assembly (genome.ucsc.edu, February 2002); this localized Ereg at chr5:90,717,211-90,736,235. Interestingly, genes coding for three other EGF family ligands, Epgn, Areg, and Btc, are tightly clustered within a 376-kb region of chromosome 5 (Fig. 1a).
Normal development and reproduction of Ereg null mice.
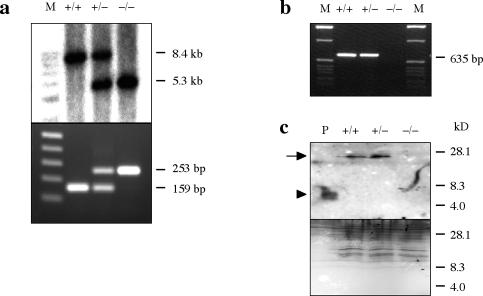
To identify in vivo biological roles for epiregulin, a null mutation, Eregtm1Dwt, was generated by homologous recombination in ES cells (Fig. 1b). An nls-lacZ reporter was integrated behind the 5′ noncoding region of Ereg in the targeting construct to facilitate histological analysis of endogenous Ereg expression. Correct homologous recombination events were identified by Southern blot hybridization of EcoRV-digested genomic DNA with an external probe, resulting in an 8.4-kb fragment from the endogenous Ereg allele and a 5.3-kb fragment from the targeted allele. Two of eight correctly targeted ES clones were used to generate chimeric mice. The Eregtm1Dwt allele was successfully transmitted through the germ line, and heterozygous male and female offspring were intercrossed to produce Ereg null mice. The genotype of each mouse was determined initially by Southern blot hybridization and then by PCR (Fig. 2a).
FIG. 2.
Characterization of the Eregtm1Dwt allele. Genotypes of offspring were done by Southern blot (top, a) and PCR (bottom, a). The wild-type allele results in an 8.4-kb EcoRV fragment, and the targeted allele results in a 5.3-kb EcoRV fragment. M, 1-kb DNA ladder (Promega); +/+, wild-type; ±, heterozygous; −/−, Ereg null mice. PCR amplification of DNA extracted from the tail gives rise to a 159-bp product specific for the wild-type Ereg allele and a 253-bp product specific for the targeted allele. Lane M, 1-kb DNA ladder (Invitrogen). (b) RT-PCR analysis of Ereg mRNA levels in the colon of wild-type (+/+), heterozygous (±), and Ereg null (−/−) mice. Total RNA (1 μg) prepared from the colons of each genotype was analyzed by RT-PCR for 35 cycles with the Ereg-S3 and Ereg-AS4 primers. (c) Equal amounts (33 μg) of small intestine tissue extracts were analyzed by Western blot with rabbit anti-Ereg antibody (top). The membrane was stained with Coomassie blue to confirm equal loading and transfer after Western blot (bottom). Lane P, 100 ng of in vitro-synthesized Ereg peptide, marked by an arrowhead, was used as a positive control. The arrow indicates the precursor form of epiregulin.
Total RNA was extracted from the tissues of mice representing each genotype and analyzed by RT-PCR with Ereg-S3 and Ereg-AS4 to demonstrate the absence of Ereg transcripts in Ereg null mice. An RT-PCR product was only observed in wild-type and heterozygous mice (Fig. 2b), while an RT-PCR product from Actb was detected equally from each genotype (data not shown). Interestingly, a PCR product from Eregtm1Dwt homozygous mice was detected with a sense primer from the 5′ coding region of exon1 and Ereg-AS4 that was transcribed from the pgk promoter in the targeting construct. However, this transcript contained a frameshift mutation and produced no functional epiregulin protein, as detected by Western blot analysis (Fig. 2c); the mature 27-kDa epiregulin protein, which is posttranslationally modified (1), was detected only in the small intestine of wild-type and heterozygous mice.
Eregtm1Dwt homozygous mice were produced at Mendelian ratios from heterozygous intercrosses and were without any overt abnormal phenotype. Both male and female null mice were fertile. The pregnancy rate and litter size were similar between wild-type and Eregtm1Dwt homozygous mice, with an average litter size of 3.88 ± 0.32 (n = 17 litters) and 3.44 ± 0.34 (n = 16 litters), respectively.
To further elucidate the in vivo function of Ereg, the Eregtm1Dwt homozygous mice were crossed to mice carrying the Egfrwa2 mutation, a hypomorphic allele of Egfr (24). Eregtm1Dwt Egfrwa2 double mutant mice were also developmentally normal and fertile. Furthermore, triple knockout mice with null mutations in Egf, Ereg, and Tgfa were produced on a 129S6/SvEvTAC and C57BL/6J mixed background by breeding Ereg null mice with Areg, Egf, and Tgfa triple null mice (25). Egf, Ereg, and Tgfa triple null mice were developmentally normal and fertile, manifesting only wavy coats and incompletely penetrant open eyes, caused by Tgfa deficiency; since Areg is tightly linked to Ereg, the compound effect of these two mutations could not be determined.
Ereg null mice have normal liver regeneration.
We used semiquantitative RT-PCR analysis to show that Ereg mRNA levels increase rapidly after partial hepatectomy in wild-type CD-1 mice, supporting previous reports of elevated epiregulin in liver extracts 24 h after partial hepatectomy (data not shown). However, liver regeneration, as measured by the weight of regenerated liver, was normal in both Eregtm1Dwt homozygous and Eregtm1Dwt, Egfrwa2 double mutant mice (data not shown). Despite an increase in Ereg mRNA levels detected by RT-PCR, LacZ staining for expression of β-galactosidase encoded by the Egfrtm1Dwt allele revealed that the ligand was produced in only a subpopulation of the hepatocytes (data not shown).
Broad cellular expression of Ereg.
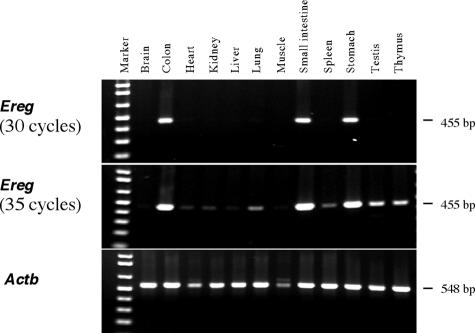
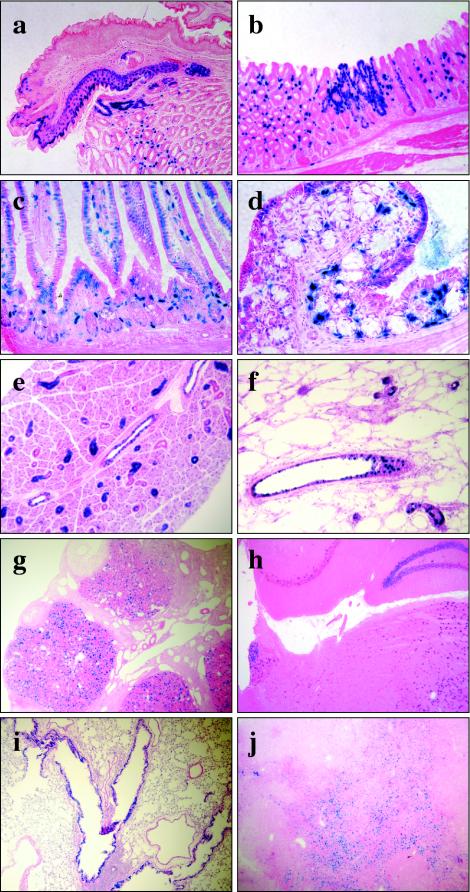
Semiquantitative RT-PCR analysis, used to compare the relative Ereg mRNA levels across various tissues, revealed that relatively high levels of Ereg transcripts were present throughout the gastrointestinal tract, including the stomach, small intestine, and colon (Fig. 3); lower levels of Ereg expression were observed in most other tissues. Ereg expression was further analyzed by histological examination of β-galactosidase activity encoded by the lacZ reporter integrated into Eregtm1Dwt (Fig. 4). There were no spatial staining differences between heterozygous and Eregtm1Dwt homozygous mice. High levels of LacZ activity were detected in punctate patches throughout the digestive tract. Intensive staining was observed in the squamous epithelium of the limiting ridge in the stomach (Fig. 4a), the lamina propria, and some epithelial cells of the glandular stomach (Fig. 4b) as well as in the tunica muscularis of the esophagus (data not shown). Similar intensive staining was also observed in a subpopulation of cells within the lamina propria of the small intestine, with weak expression in the small intestinal epithelial cells (Fig. 4c). Relatively moderate expression was observed in the epithelial cells of the proximal colon (Fig. 4d) but was rare in the distal colon (see Fig. 6f). Compared to the strong expression of a subpopulation of cells within the lamina propria of the proximal colon, only a few sporadic LacZ-positive cells were observed in the lamina propria of the distal colon, and no expression was observed in gut-associated lymphoid tissues (Fig. 6d to f).
FIG. 3.
Semiquantitative RT-PCR analysis of relative Ereg mRNA levels in various tissues. Total RNA (1 μg) was prepared from the tissues of 3-month-old 129S6/SvEvTAC mice and reverse transcribed. One twentieth of the reverse transcription product was amplified by PCR with 30 or 35 cycles for Ereg (455-bp product) and 25 cycles for Actb (548-bp product). The RT-PCR product was resolved by 1.2% agarose gel electrophoresis. Lane Marker, 1-kb DNA ladder (Invitrogen).
FIG. 4.
X-Gal staining of tissues from Ereg null mice. Samples were fixed, infused with sucrose, and sectioned at 10 μm with a cryostat. The sectioned tissue was further fixed and incubated at 37°C with X-Gal staining solution for 4 h (a to g) or overnight (h to j). The X-Gal-stained tissues were counterstained with eosin Y (a, b, g, h, and j) or hematoxylin and eosin (c to f and i). (a and b) Stomach; (c) jejunum; (d) proximal colon; (e) salivary gland; (f) mammary gland; (g) ovary; (h) brain; (i) lung; (j) thymus.
FIG. 6.
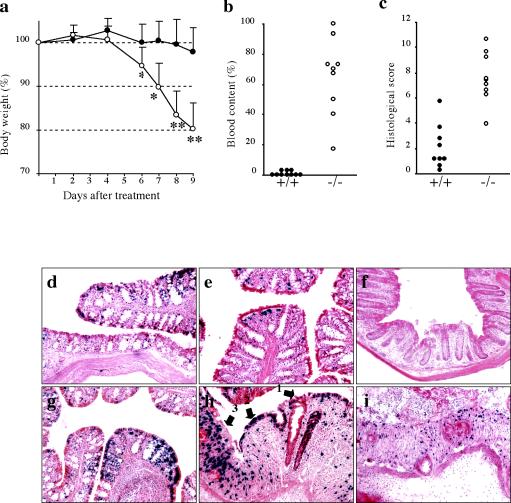
Effect of Eregtm1Dwt on response to DSS treatment. (a) Adult male mice (n = 10 for wild-type and n = 9 for Ereg null mice) were fed 1.5% (wt/vol) DSS in drinking water. Body weight was measured at the indicated time after treatment and expressed as a percentage of the weight on the day of first exposure to DSS. Solid circles, wild-type; open circles, Ereg null mice. Data are means ± standard deviation. *, P < 0.05; and **, P < 0.005, based on a Mann-Whitney test. The colons were collected 9 days after treatment was initiated. (b) Blood content was determined by measuring the portion of the area including blood from the entire colon. (c) Histological damage was scored from cross sections of the colon as described in Materials and Methods. Each dot represents an individual mouse of the wild-type (+/+, solid circles) and Ereg null (−/−, open circles) mice. Blood content and histological score were significantly different (P < 0.0001 with an unpaired t test) between wild-type and Ereg null mice. The colons of Ereg null mice were collected either after no treatment (d to f) or after 9 days of DSS treatment (g to i) and stained with X-Gal. (d and g) Proximal colon; (e and h) mid-colon; (f and i) proximal colon. Arrows numbered 1 to 3 in h indicate histological damage scores as described in Materials and Methods.
Intensive LacZ staining was also detected in the ductal epithelial cells of the salivary (Fig. 4e) and mammary glands (Fig. 4f) and in the corpus luteum of the ovary (Fig. 4g). Weak expression was detected in specific subregions of the brain (Fig. 4h), such as the hippocampus, thalamus, pons, medulla, and a few Purkinje cells in the cerebellum, in the epithelial cells of bronchioles (Fig. 4i), and in the thymus (Fig. 4j). Staining in the thymus was confined to the subcapsular regions in the cortex while being evenly distributed in the medulla. There was only an occasional LacZ-positive cell in the heart, liver, kidney, pancreas, spleen, testis, skeletal muscle, skin, peripheral blood leukocytes, bone marrow, day post coitum (dpc) 12 and dpc 19 embryo proper, and placenta. Analysis of cell proliferation by immunohistochemical detection of bromodeoxyuridine incorporation did not reveal any differences between wild-type and Eregtm1Dwt mice in the stomach, small intestine, and colon (data not shown).
Unaltered intestinal tumorigenesis in Ereg null mice.
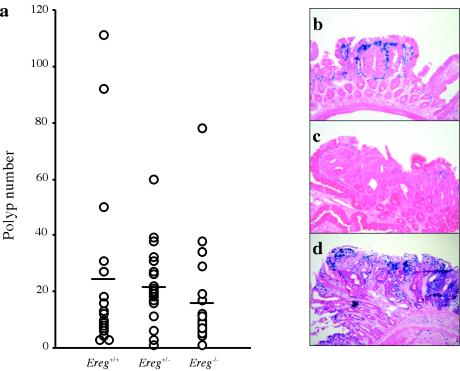
To analyze the potential involvement of Ereg in tumor formation, we introduced the ApcMin allele into wild-type, heterozygous and Eregtm1Dwt homozygous mice. The F2 mice used in this study were generated from 129.B6 F1 intercrosses. At 3 months of age, the total numbers of polyps were counted and their sizes were measured. All ApcMin/+ mice examined (n = 60) had tumors (>0.3 mm) regardless of Ereg genotype, and the mean tumor multiplicities were 24.7 ± 7.6 for wild-type, 21.5 ± 3.0 for heterozygous, and 16.3 ± 4.2 for Eregtm1Dwt homozygous mice (Fig. 5a). Although slightly lower in Ereg null mice, suggesting that epiregulin may contribute to the previously reported role of EGFR signaling in intestinal tumorigenesis (37), tumor multiplicity was not statistically different between the groups, implying that additional EGFR ligands are probably participating to promote tumorigenesis. The distribution of tumors along the small intestine was also similar among the groups.
FIG. 5.
Effect of the Ereg null allele on ApcMin tumor development. (a) Polyp number in the small intestine of ApcMin/+ mice with each Ereg genotype. Each dot represents the total polyp (>0.3 mm) number from a single 3-month-old mouse, with the horizontal lines representing the mean for each genotype (n = 17 Ereg+/+, 22 Ereg+/−, and 19 Ereg−/−). (b) X-Gal staining of a polyp from an ApcMin/+ Eregtm1Dwt/tm1Dwt mouse. The X-Gal-stained tissues were counterstained with eosin Y (b and c) or hematoxylin and eosin (d). Panels b and c represent microadenoma and macroadenoma in the jejunum, respectively. Panel d represents macroadenoma of the colon.
Despite variable tumor size within individual mice, the mean size was equivalent among the groups (data not shown). Tumors from ApcMin/+ Eregtm1Dwt/tm1Dwt mice showed intensive LacZ staining restricted to cells of the lamina propria surrounding nascent microadenomas (Fig. 5b), while sporadic LacZ staining was frequently observed in the lamina propria and occasional epithelium of macroadenomas (Fig. 5c); LacZ-positive cells were concentrated just beneath the epithelium at the tips of villi. Only five colon tumors from four mice were observed out of 60 ApcMin/+ mice regardless of Ereg genotype. Two colon tumors from one ApcMin/+ Eregtm1Dw/+ mouse showed intensive LacZ staining within the tumor cells, in fibroblasts in the lamina propria, and rare interstitial cells within the subjacent tunica muscularis (Fig. 5d).
Increased susceptibility of Ereg null mice to DSS-induced intestinal damage.
Susceptibility to intestinal injury was measured in the DSS model of acute colitis through continuous exposure to 1.5% DSS in the drinking water. Analysis of body weight loss over 9 days of DSS treatment revealed a statistically significant difference between wild-type and 129S6/SvEvTAC-Eregtm1Dw homozygous mice (Fig. 6a). Female Eregtm1Dw homozygous mice showed less severe body weight loss than the male mice; heterozygous mice of either gender and their matched wild-type controls showed similar susceptibilities to DSS-induced colitis (data not shown). A comparison with 2.5% DSS in drinking water showed severe weight loss for both wild-type and Eregtm1Dw homozygous mice (data not shown).
Following 9 days of treatment with 1.5% DSS, colon histology and the pattern of LacZ expression were examined. Statistically significant differences in overall blood content, as measured by the blood clot area through the entire length of the colon (Fig. 6b), and histological score (Fig. 6c) were detected between wild-type and 129S6/SvEvTAC-Eregtm1Dw homozygous mice. The proximal colon was the most resistant to DSS-induced damage, with the middle and distal colon showing significantly more severe lesions (Fig. 6g to i). Mild lesions revealed loss of mucus cells, glandular hyperplasia, and increased leukocytes in the lamina propria. Severe lesions had a complete loss of glandular epithelium, accompanied by collapse of the denuded lamina propria, submucosal edema, fibrinoid necrosis of lamina propria vessels, and infiltration of neutrophils and plasma cells.
DSS treatment induced Ereg expression, as detected by LacZ activity, in the epithelial cells of the colon (Fig. 6g and h) and small intestine (data not shown). Though elevated lacZ expression was observed as early as 2 days after initiating treatment with DSS in the proximal colon, stronger induction of lacZ expression was observed depending on the progression of ulceration in the middle and distal colon (data not shown). No spatial difference in lacZ expression was observed between heterozygous and null mice after DSS treatment. Intense staining was observed in subpopulations of epithelial cells and in lamina propria fibroblasts in the ulcerative lesions, but no staining was observed in the infiltrating leukocytes of the submucosal layer.
DISCUSSION
The ERBB receptor tyrosine kinase family and their ligands play important roles in cell proliferation, differentiation, migration, and apoptosis (59). Targeted mutagenesis shows that all four receptor family members, Egfr, Erbb2, Erbb3, and Erbb4, are essential during embryogenesis (7, 11, 21, 30, 45, 53). Contrastingly, analysis of null mutations in the Erbb ligand genes has been less instructive as to unique functions, with most exhibiting functional redundancy. Although EGFR ligands are often coexpressed, each ligand also potentially has its own specific function in addition to having functionally redundant activities. For example, Areg appears to be essential for normal mammary duct outgrowth during puberty, although Tgfa, Btc, and Dtr show overlapping expression in the developing mammary gland (25, 40). Likewise, mice triply null for Egf, Tgfa, and Areg show only minor defects in mammary duct morphogenesis and gastrointestinal development, where at least six EGFR ligands are expressed, but are otherwise phenotypically normal (25, 58).
Like mice lacking other EGFR ligands, Ereg null mice do not manifest any overtly abnormal phenotype, suggesting that loss of epiregulin activity can be compensated for by other ligands. Interestingly, three other EGFR ligands, Epgn, Areg, and Btc, are tightly linked to Ereg; all four genes lie within a 376-kb region on mouse chromosome 5. We have found that the genomic structures of these genes are very similar; each contains five conserved exons, suggesting that they arose from a common ancestral gene through repeated duplication. Exceptions to the absence of reported unique ligand functions are neuregulin 1, which is essential for normal heart and neuronal cell development (29), and diphtheria toxin receptor, which is required postnatally to maintain normal cardiac function (14, 15). Interestingly, the phenotypes resulting from elimination of Nrg or Dtr are reminiscent of those previously reported for Erbb receptors, suggesting that these phenotypes may be due to perturbation of non-EGFR or EGFR-ERBB heterodimer interactions.
Epiregulin expression has been reported to be highly restricted, including in the uterus during early pregnancy, placenta, ovary, and macrophages (3, 33, 55, 56). Using RT-PCR analysis and LacZ staining, we detected Ereg expression in a wider variety of cell types and in additional organs, suggesting that epiregulin may play a broader role in tissue homeostasis rather than having a specific developmental function. Although Ereg transcripts were previously detected in 7-day-old embryos (55), our results suggest that these analyses were probably due to contamination with decidual cells surrounding the embryo proper. We have not detected any LacZ-positive cells in the embryo during embryogenesis, further suggesting that epiregulin is not essential for normal embryonic development.
Contrary to previous findings of high Ereg expression in human placenta and macrophages (56), we also did not detect lacZ expression in the mouse placenta or in peripheral blood leukocytes. However, it is conceivable that the regulatory factors involved in tissue-specific expression may differ between the two species. Ereg is also one of the gene transcripts activated by gonadotropins, such as pregnant serum gonadotropin (41) and human chorinoic gonadotropin (8) in immature rat ovaries. Recently, it was also reported that Ereg is expressed in ovarian granulosa cells of mice after treatment with luteinizing hormone (33). Contrastingly, we mainly detected LacZ staining in the corpus luteum in the cycling adult mouse, although we did detect high levels of epiregulin, marked by LacZ staining, in the granulosa cells after hormone induction (data not shown). One source of this discrepancy is that the previous report did not investigate normal cycling females, suggesting that the dramatic upregulation of EGFR ligands in response to hormones may not represent normal physiology.
Aberrant expression of ERBB receptors and ligands is commonly observed in tumors and thought to play an important role in their development and growth (39). Of the EGFR ligands, Tgfa is most frequently overexpressed in human cancer (59). However, Ereg-specific SAGE tags are the most frequently observed EGFR ligand detected in colorectal cancers (2). Furthermore, overexpression of Ereg is observed in several tumor cell lines (56), including pancreatic ductal carcinoma (60) and bladder cancer of muscle-invasive stage T2 to T4 tumors (52).
We used the ApcMin mouse model of intestinal cancer, widely used to investigate the in vivo effects of specific mutations on epithelial cancer development (31, 50), to test the role of epiregulin during intestinal cancer development. Although we previously demonstrated ApcMin tumors are highly dependent upon the EGFR signaling axis (37), we did not detect a statistically significant reduction in tumor multiplicity or size in Eregtm1Dwt homozygous mice with a population of F2 hybrids between C57BL/6J and 129S6/SvEvTAC, strains both carrying Mom1s, the sensitive allele of the major ApcMin modifier (12). Our studies revealed that although epiregulin does not individually have a major influence on intestinal tumor development, it may nonetheless be acting in concert with other EGFR ligands to support tumor growth; mutations in no single EGFR ligand have been shown to have an equivalent effect on tumorigenesis as mutations in Egfr (37).
Unlike other EGFR ligands, epiregulin appears to be primarily expressed in tumor stroma during early tumorigenesis rather than by the transformed cells. Additionally, a striking difference in Ereg expression pattern correlates with the size of tumors. Stromal expression of Ereg is more robust in nascent microadenomas, indicating activation of Ereg gene expression during early tumor development, possibly as a host response to the transformed cells. Interestingly, localization of Ereg expression in macroadenomas is very similar to that of cyclooxygenase 2 (Ptgs2) expression, which is primarily detected in fibroblasts and endothelial cells (47). Ereg is one of the immediate-early genes highly induced by growth factor activation of fibroblasts (9) and a major mitogenic protein secreted from vascular smooth muscle cells (51). In larger adenomas, particularly those in the colon, Ereg expression is also dramatically upregulated in tumor epithelial cells, suggesting that epiregulin may also contribute to the expansion of tumors.
Since epiregulin may be playing a role during host response to tissue pathology, we tested Ereg null mice for their response to severe intestinal damage and colitis, a contributing factor to intestinal cancer susceptibility (22). Oral administration of DSS to mice causes intestinal mucosa damage and both acute and chronic ulcerative colitis with features similar to the symptomatic and histological findings in human inflammatory bowel disease (49). Previous reports suggested that mice deficient for Tgfa (6) or with impaired EGFR activity (4) showed heightened susceptibility to DSS-induced damage, while ectopic activation of EGFR in the intestines of transgenic mice is protective against DSS (5). In contrast to a Tgfa single mutation, when Tgfa, Egf, and Areg triple null mice were tested, they did not show an altered DSS response (58). Since these previous reports used mutant mice on mixed genetic backgrounds, the discrepancy in attributing a role for Tgfa in DSS response may be related to genetic background, which is known to strongly influence DSS susceptibility (27). Our results with isogenic mice conclusively demonstrate a requirement for epiregulin in protection from DSS-induced intestinal damage.
An alternative interpretation of why Tgfa- and Ereg-deficient mice manifest similar responses to DSS treatment even though both activate EGFR is that they may have nonoverlapping activities that are required to provide optimal protection from intestinal injury. Epiregulin, like other EGFR ligands, is an autocrine growth factor that induces the expression of other EGF ligand family members (19, 43). However, unlike TGFA, epiregulin can activate ERBB4 in addition to EGFR (36). Furthermore, despite binding various ERBB receptor combinations with relatively low affinity, epiregulin shows more potent bioactivity than other ERBB ligands (42, 57). Thus, increased expression of epiregulin in stromal and epithelial cells after DSS treatment may induce activation of other ERBB receptors in addition to providing an autocrine induction of TGFA, allowing amplification of the EGFR signal cascade.
Other studies with mouse models of mucosal inflammation have provided support that inflammatory bowel disease is related to a dysfunctional mucosal immune response to environmental factors (49). For example, spontaneous induction of colitis was observed in Il2 (38) and Il10 (20) knockout mice, and, in contrast, decreased susceptibility to DSS-induced colitis was observed in Casp1 (interleukin-1β converting enzyme) (46) and Il4 (48) knockout mice. However, the exact mechanism of DSS-induced colitis is not fully understood; it is probably related to a direct toxic effect on epithelial cells and a change in epithelial cell barrier function (49). Finally, the high expression of epiregulin in epithelial cells and lamina propria fibroblasts in the damaged area suggests that epiregulin may also be providing support for reconstitution of the crypt structure after inflammation-mediated tissue injury.
Acknowledgments
We thank Toshi Komurasaki (Taisho Pharmaceutical Co., Ltd, Japan) for providing antiepiregulin antibody, David Lee (University of North Carolina at Chapel Hill) for providing Areg Tgfa Egf triple null mice, and Brigid Hogan (Duke University) for providing the TL-1 ES cell line.
This work was supported by National Institutes of Health (NIH) grants CA092479, CA084239, and HD039896 (D.W.T.) and HD12304 and HD33994 (S.K.D.) and NIH Postdoctoral Training Grant CA009385 (D.L.).
REFERENCES
- 1.Baba, I., S. Shirasawa, R. Iwamoto, K. Okumura, T. Tsunoda, M. Nishioka, K. Fukuyama, K. Yamamoto, E. Mekada, and T. Sasazuki. 2000. Involvement of deregulated epiregulin expression in tumorigenesis in vivo through activated Ki-Ras signaling pathway in human colon cancer cells. Cancer Res. 60:6886-6889. [PubMed] [Google Scholar]
- 2.Boon, K., E. C. Osorio, S. F. Greenhut, C. F. Schaefer, J. Shoemaker, K. Polyak, P. J. Morin, K. H. Buetow, R. L. Strausberg, S. J. De Souza, and G. J. Riggins. 2002. An anatomy of normal and malignant gene expression. Proc. Natl. Acad. Sci. USA 99:11287-11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Das, S. K., N. Das, J. Wang, H. Lim, B. Schryver, G. D. Plowman, and S. K. Dey. 1997. Expression of betacellulin and epiregulin genes in the mosue uterus temporally by the blastocyst solely at the site of its apposition is coincident with the “window” of implantation. Dev. Biol. 190:178-190. [DOI] [PubMed] [Google Scholar]
- 4.Egger, B., M. W. Buchler, J. Lakshmanan, P. Moore, and V. E. Eysselein. 2000. Mice harboring a defective epidermal growth factor receptor (waved-2) have an increased susceptibility to acute dextran sulfate-induced colitis. Scand. J. Gastroenterol. 35:1181-1187. [DOI] [PubMed] [Google Scholar]
- 5.Egger, B., H. V. Carey, F. Procaccino, N. N. Chai, E. P. Sandgren, J. Lakshmanan, V. S. Buslon, S. W. French, M. W. Buchler, and V. E. Eysselein. 1998. Reduced susceptibility of mice overexpressing transforming growth factor alpha to dextran sodium sulphate induced colitis. Gut 43:64-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egger, B., F. Procaccino, J. Lakshman, M. Reinshagen, P. Hoffmann, A. Patel, W. Reuben, S. Gnanakkan, L. Liu, L. Barajas, and V. E. Eysselein. 1997. Mice lacking transforming growth factor alpha have an increases susceptibility to dextran sulfate-induced colitis. Gastroenterology 113:825-832. [DOI] [PubMed] [Google Scholar]
- 7.Erickson, S. L., K. S. O'Shea, N. Ghaboosi, L. Loverro, G. Frantz, M. Bauer, L. H. Lu, and M. W. Moore. 1997. ErbB3 is required for normal cerebellar and cardiac development: a comparison with ErbB2-and heregulin-deficient mice. Development 124:4999-5011. [DOI] [PubMed] [Google Scholar]
- 8.Espey, L. L., and J. S. Richards. 2002. Temporal and spatial patterns of ovarian gene transcription following an ovulatory dose of gonadotropin in the rat. Biol. Reprod. 67:1662-1670. [DOI] [PubMed] [Google Scholar]
- 9.Fambrough, D., K. McClure, A. Kazlauskas, and E. S. Lander. 1999. Diverse signaling pathways activated by growth factor receptors induce broadly overlapping, rather than independent, sets of genes. Cell 97:727-741. [DOI] [PubMed] [Google Scholar]
- 10.Fowler, K. J., F. Walker, W. Alexander, M. L. Hibbs, E. C. Nice, R. M. Bohmer, G. B. Mann, C. Thumbwood, R. Maglitto, J. A. Danks, R. Chetty, A. W. Burgess, and A. R. Dunn. 1995. A mutation in the epidermal growth factor receptor in waved-2 mice has a profound effect on the receptor biochemistry that results in impaired lactation. Proc. Natl. Acad. Sci. USA 92:1465-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gassmann, M., F. Casagranda, D. Orioli, H. Simon, C. Lai, R. Klein, and G. Lemke. 1995. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature 378:390-394. [DOI] [PubMed] [Google Scholar]
- 12.Gould, K. A., W. F. Dietrich, N. Borenstein, E. S. Lander, and W. F. Dove. 1996. Mom1 is a semi-dominant modifier of intestinal adenoma size and multiplicity in Min/+ mice. Genetics 144:1769-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hogan, B., R. Beddington, F. Costantini, and E. Lacy. 1994. Manipulating the mouse embryo, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Iwamoto, R., S. Yamazaki, M. Asakura, S. Takashima, H. Hasuwa, K. Miyado, S. Adachi, M. Kitakaze, K. Hashimoto, G. Raab, D. Nanba, S. Higashiyama, M. Hori, M. Klagsbrun, and E. Mekada. 2003. Heparin-binding EGF-like growth factor and ErbB signaling is essential for heart function. Proc. Natl. Acad. Sci. USA 100:3221-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson, L. F., T. H. Qiu, S. W. Sunnarborg, A. Chang, C. Zhang, C. Patterson, and D. C. Lee. 2003. Defective valvulogenesis in HB-EGF and TACE-null mice is associated with aberrant BMP signaling. EMBO J. 22:2704-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, J. T., R. W. Akita, and M. X. Sliwkowski. 1999. Binding specificities and affinities of egf domains for ErbB receptors. FEBS Lett. 447:227-231. [DOI] [PubMed] [Google Scholar]
- 17.Kaestner, K. H., L. Montoliu, H. Kern, M. Thulke, and G. Schutz. 1994. Universal beta-galactosidase cloning vectors for promoter analysis and gene targeting. Gene 148:67-70. [DOI] [PubMed] [Google Scholar]
- 18.Komurasaki, T., H. Toyoda, D. Uchida, and S. Morimoto. 1997. Epiregulin binds to epidermal growth factor receptor and ErbB-4 and induces tyrosine phosphorylation of epidermal growth factor receptor, ErbB-2, ErbB-3 and ErbB-4. Oncogene 15:2841-2848. [DOI] [PubMed] [Google Scholar]
- 19.Komurasaki, T., H. Toyoda, D. Uchida, and N. Nemoto. 2002. Mechanism of growth promoting activity of epiregulin in primary cultures of rat hepatocytes. Growth Factors 20:61-69. [DOI] [PubMed] [Google Scholar]
- 20.Kuhn, R., J. Lohler, D. Rennick, K. Rajewsky, and W. Muller. 1993. Interleukin-10-deficient mice develop chronic entercolitis. Cell 75:263-274. [DOI] [PubMed] [Google Scholar]
- 21.Lee, K. F., H. Simon, H. Chen, B. Bates, M. C. Hung, and C. Hauser. 1995. Requirement for neuregulin receptor Erbb2 in neural and cardiac development. Nature 378:394-398. [DOI] [PubMed] [Google Scholar]
- 22.Lewis, J. D., J. J. Deren, and G. R. Lichtenstein. 1999. Cancer risk in patients with inflammatory bowel disease. Gastroenterol. Clin. North Am. 28:459-477. [DOI] [PubMed] [Google Scholar]
- 23.Lindvall, C., M. Hou, T. Komurasaki, C. Zhang, M. Henricksson, J. M. Sedivy, M. Bjorkholm, B. T. Teh, M. Nordenskjold, and D. Xu. 2003. Molecular characterization of human telomerase reverse transcriptase-immortalized human fibroblasts by gene expresion profiling: activation of the Epiregulin gene. Cancer Res. 63:1743-1747. [PubMed] [Google Scholar]
- 24.Luetteke, N. C., H. K. Phillips, T. h. Qiu, N. G. Copeland, H. S. Earp, N. A. Jenkins, and D. C. Lee. 1994. The mouse waved-2 phenotype results from a point mutation in the EGF receptor tyrosine kinase. Genes Dev. 8:399-413. [DOI] [PubMed] [Google Scholar]
- 25.Luetteke, N. C., T. H. Qiu, S. E. Fenton, K. L. Troyer, R. F. Riedel, A. Chang, and D. C. Lee. 1999. Targeted inactivation of the EGF and amphiregulin genes reveals distinct roles for EGF receptor ligands in mouse mammary gland development. Development 126:2739-2750. [DOI] [PubMed] [Google Scholar]
- 26.Luetteke, N. C., T. H. Qiu, R. L. Peiffer, P. Oliver, O. Smithies, and D. C. Lee. 1993. TGF alpha deficiency results in hair follicle and eye abnormalities in targeted and waved-1 mice. Cell 73:263-278. [DOI] [PubMed] [Google Scholar]
- 27.Mahler, M., I. J. Bristol, E. H. Leiter, A. E. Workman, E. H. Birkenmeier, C. O. Elson, and J. P. Sundberg. 1998. Differential susceptibility of inbred mouse strains to dextran sulfate-induced colitis. Am. J. Physiol. 274:G544-G551. [DOI] [PubMed] [Google Scholar]
- 28.Manly, K. F., and J. M. Olson. 1999. Overview of QTL mapping software and introduction to Map Manager QT. Mammalian Genome 10:327-334. [DOI] [PubMed] [Google Scholar]
- 29.Meyer, D., and C. Birchmeier. 1995. Multiple essential functions of neuregulin in development. Nature 378:386-390. [DOI] [PubMed] [Google Scholar]
- 30.Miettinen, P. J., J. E. Berger, J. Meneses, Y. Phung, R. A. Pedersen, Z. Werb, and R. Derynck. 1995. Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature. 376:337-341. [DOI] [PubMed] [Google Scholar]
- 31.Moser, A. R., W. F. Dove, K. A. Roth, and J. I. Gordon. 1992. The Min (multiple intestinal neoplasia) mutation: its effect on gut epithelial cell differentiation and interaction with a modifier system. J. Cell Biol. 116:1517-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paria, B. C., K. Elenius, M. Klagsbrun, and S. K. Dey. 1999. Heparin-binding EGF-like growth factor interacts with mouse blastocysts independently of ErbB1: a possible role for heparan sulfate proteoglycans and ErbB4 in blastocyst implantation. Development 126:1997-2005. [DOI] [PubMed] [Google Scholar]
- 33.Park, J. Y., Y. Q. Su, M. Ariga, E. Law, S. L. Jin, and M. Conti. 2004. EGF-like growth factors as mediators of LH action in the ovulatory follicle. Science 303:682-684. [DOI] [PubMed] [Google Scholar]
- 34.Plowman, G. D., J.-M. Culouscou, G. S. Whitney, J. M. Green, G. W. Carlton, L. Foy, M. G. Neubauer, and M. Shoyab. 1993. Ligand-specific activation of HER4/p180erbB4, a fourth member of the epidermal growth factor receptor family. Proc. Natl. Acad. Sci. USA 90:1746-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riese, D. J., 2nd, and D. F. Stern. 1998. Specificity within the EGF family/ErbB receptor family signaling network. Bioessays 20:41-48. [DOI] [PubMed] [Google Scholar]
- 36.Riese, D. J., T. Komurasaki, G. D. Plowman, and D. F. Stern. 1998. Activation of ErbB4 by the bifunctional epidermal growth factor family hormone epiregulin is regulated by ErbB2. J. Biol. Chem. 273:11288-11294. [DOI] [PubMed] [Google Scholar]
- 37.Roberts, R. B., L. Min, M. K. Washington, S. J. Olsen, S. H. Settle, R. J. Coffey, and D. W. Threadgill. 2001. Importance of epidermal growth factor receptor signaling in establishment of adenomas and maintenance of carcinomas during intestinal tumorigenesis. Proc. Natl. Acad. Sci. USA 99:1521-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sadlack, B., H. Merz, H. Schorle, S. Schimpl, A. C. Feller, and I. Horak. 1993. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell 75:253-261. [DOI] [PubMed] [Google Scholar]
- 39.Salomon, D. S., R. Brandt, F. Ciardiello, and N. Normanno. 1995. Epidermal growth factor-related peptides and thier receptors in human malignancies. Crit. Rev. Oncol. Hematol. 19:183-232. [DOI] [PubMed] [Google Scholar]
- 40.Schroeder, J. A., and D. C. Lee. 1998. Dynamic expression and activation of Erbb receptors in the developing mouse mammary gland. Cell Growth Differ. 9:451-464. [PubMed] [Google Scholar]
- 41.Sekiguchi, T., T. Mizutani, K. Yamada, T. Yazawa, H. Kawata, M. Yoshino, T. Kajitani, T. Kameda, T. Minegishi, and K. Miyamoto. 2002. Transcriptional regulation of the epiregulin gene in the rat ovary. Endocrinology 143:4718-4729. [DOI] [PubMed] [Google Scholar]
- 42.Shelly, M., R. Pinkas-Kramarski, B. C. Guarino, H. Waterman, L. M. Wang, L. Lyass, M. Alimandi, A. Kuo, S. S. Bacus, J. H. Pierce, G. C. Andrews, and Y. Yarden. 1998. Epiregulin is a potent pan-ErbB ligand that preferentially activates heterodimeric receptor complexes. J. Biol. Chem. 273:10496-10505. [DOI] [PubMed] [Google Scholar]
- 43.Shirakata, Y., T. Komurasaki, H. Toyoda, Y. Hanakawa, K. Yamasaki, S. Tokumaru, K. Sayama, and K. Hashimoto. 2000. Epiregulin, a novel member of the epidermal growth factor family, is an autocrine growth factor in normal human keratinocytes. J. Biol. Chem. 275:5748-5753. [DOI] [PubMed] [Google Scholar]
- 44.Sibilia, M., J. P. Steinbach, L. Stingl, A. Aguzzi, and E. F. Wagner. 1998. A strain-independent postnatal neurodegeneration in mice lacking the EGF receptor. EMBO J. 17:719-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sibilia, M., and E. F. Wagner. 1995. Strain-dependent epithelial defects in mice lacking the EGF receptor. Science 269:234-238. [DOI] [PubMed] [Google Scholar]
- 46.Siegmund, B., H. A. Lehr, G. Fantuzzi, and C. A. Dinarello. 2001. IL-1 beta-converting enzyme (caspase-1) in intestinal inflammation. Proc. Natl. Acad. Sci. USA 98:13249-13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sonoshita, M., K. Takaku, M. Oshima, K. Sugihara, and M. M. Taketo. 2002. Cyclooxygenase-2 expression in fibroblasts and endothelial cells of intestinal polyps. Cancer Res. 62:6846-6849. [PubMed] [Google Scholar]
- 48.Stevceva, L., P. Pavlidis, A. Husband, A. Ramsay, and W. F. Doe. 2001. Dextran sulphate sodium-induced colitis is ameliorated in interleukin 4 deficient mice. Genes Immun. 2:309-316. [DOI] [PubMed] [Google Scholar]
- 49.Strober, W., I. J. Fuss, and R. S. Blumber. 2002. The immunology of mucosal models of inflammation. Annu. Rev. Immunol. 20:495-549. [DOI] [PubMed] [Google Scholar]
- 50.Su, L.-K., K. W. Kinzler, B. Vogelstein, A. C. Preisinger, A. R. Moser, C. Luongo, K. A. Gould, and W. F. Dove. 1992. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science 256:668-670. [DOI] [PubMed] [Google Scholar]
- 51.Taylor, D. S., X. Cheng, J. E. Pawlowski, A. R. Wallace, P. Ferrer, and C. J. Molloy. 1999. Epiregulin is a potent vascular smooth muscle cell-derived mitogen induced by angiotensin II, endothelin-1, and thrombin. Proc. Natl. Acad. Sci. USA 96:1633-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thogersen, V. B., B. S. Sorensen, S. S. Poulsen, T. F. Orntoft, H. Wolf, and E. Nexo. 2001. A subclass of HER1 ligands are prognostic markers for survival in bladder cancer patients. Cancer Res. 61:6227-6233. [PubMed] [Google Scholar]
- 53.Threadgill, D. W., A. A. Dlugosz, L. A. Hansen, T. Tennenbaum, U. Lichti, D. Yee, C. LaMantia, T. Mourton, K. Herrup, R. C. Harris, J. A. Barnard, S. H. Yuspa, R. J. Coffey, and T. Magnuson. 1995. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science 269:230-234. [DOI] [PubMed] [Google Scholar]
- 54.Torring, N., P. E. Jorgensen, B. S. Sorensen, and E. Nexo. 2000. Increased expression of heparin binding EGF (HB-EGF), amphiregulin, TGF alpha and epiregulin in androgen-independent prostate cancer cell lines. Anticancer Res. 20:91-95. [PubMed] [Google Scholar]
- 55.Toyoda, H., T. Komurasaki, Y. Ikeda, M. Yoshimoto, and S. Morimoto. 1995. Molecular cloning of mouse epiregulin, a novel epidermal growth factor-related protein, expressed in the early stage of development. FEBS Lett. 377:403-407. [DOI] [PubMed] [Google Scholar]
- 56.Toyoda, H., T. Komurasaki, D. Uchida, and S. Morimoto. 1997. Distribution of mRNA for human epiregulin, a differentially expressed member of the epidermal growth factor family. Biochem. J. 326:69-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Toyoda, H., T. Komurasaki, D. Uchida, Y. Takayama, T. Isobe, T. Okuyama, and K. Hanada. 1995. Epiregulin: a novel epidermal growth factor with mitogenic activity for rat primary hepatocytes. J. Biol. Chem. 270:7495-7500. [DOI] [PubMed] [Google Scholar]
- 58.Troyer, K. L., N. C. Luetteke, M. L. Saxon, T. H. Qiu, C. J. Xian, and D. C. Lee. 2001. Growth retardatoin, duadenal lesions, and aberrant ileum architecture in triple null mice lacking Egf, amphiregulin, and Tgf-alpha. Gastroenterology 121:68-78. [DOI] [PubMed] [Google Scholar]
- 59.Yarden, Y., and M. X. Sliwkowski. 2001. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2:127-137. [DOI] [PubMed] [Google Scholar]
- 60.Zhu, Z., J. Kleeff, H. Friess, L. Wang, A. Zimmermann, Y. Yarden, M. W. Buchler, and M. Korc. 2000. Epiregulin is upregulated in pancreatic cancer and stimulates pancreatic cancer cell growth. Biochem. Biophys. Res. Commun. 273:1019-1024. [DOI] [PubMed] [Google Scholar]