Abstract
E2A transcription factors, E12 and E47, are important regulators of lymphocyte development. Notch signaling pathways have been shown to regulate E2A function by accelerating the degradation of E2A proteins through a mitogen-activated protein kinase-dependent and ubiquitin-mediated pathway. To further understand the mechanism underlying E2A ubiquitination and degradation, we conducted a yeast two-hybrid screen and identified the carboxyl terminus of Hsc70-interacting protein (CHIP) as an E47 binding protein. Here, we show that CHIP associates with E2A proteins in vivo and that overexpression of CHIP induces E47 degradation in a phosphorylation-dependent manner. Conversely, knocking down CHIP with small interfering RNA alleviates Notch-induced E47 degradation. CHIP binds E47 through the E protein homology domains 2 and 3 (EHD2 and EHD3). This interaction between CHIP and E47 is independent of the U-box domain with E3 ubiquitin ligase activity but requires the chaperone binding tetratricopeptide repeats domain. The ability of CHIP to induce E47 ubiquitination and degradation correlates with its ability to bind E47. We propose that CHIP, together with its partner Hsc70, forms a preubiquitination complex (PUC) with E47 and Skp2, thus facilitating the interaction between E47 and Skp2. CHIP also associates with Cul1, which introduces PUC to the SCF E3 ligase complex, responsible for E47 ubiquitination. Therefore, CHIP plays a crucial role in the ubiquitination and degradation of E2A proteins.
The E2A gene encodes two transcription factors, E12 and E47, which differ only in their basic helix-loop-helix domain due to alternative splicing (29, 43). Together with products of the HEB and E2-2 genes, E12 and E47 form a family of closely related transcription factors called E proteins (25). Disruption of E protein genes or inhibition of E protein function arrests B- and T-cell development at early stages, demonstrating a crucial role for these proteins in lymphocyte development (3, 4, 6, 14, 21, 27, 42, 46, 47). Therefore, controlling the function of these E proteins becomes an effective means for regulating lymphopoiesis. Indeed, the Notch signaling pathway, known to be important for lymphoid lineage commitments (2, 37), has been shown to augment the function of E2A proteins (30, 32). Activation of Notch receptors accelerates the degradation of E2A proteins through a ubiquitin-mediated and proteasome-dependent pathway (30). In addition, phosphorylation of E2A proteins by mitogen-activated protein (MAP) kinases is essential for their ubiquitination and subsequent degradation. Mutations in MAP kinase phosphorylation sites found in the conserved EHD3 region shared by all E proteins render E47 resistant to Notch-induced degradation. The ability of Notch signals to trigger E2A degradation is influenced by levels of active Erk1/2 present in a given cell, thus providing additional means for regulating E2A function. However, activation of Erk1/2 alone is not sufficient to cause E2A degradation.
The molecular mechanism underlying Notch-induced E2A ubiquitination is not known. To obtain a better understanding of the mechanism, a more comprehensive knowledge about the machinery responsible for E2A ubiquitination is necessary. Our laboratory has previously shown that the SCFSkp2 E3 ubiquitin ligase complex, including Cul1, Skp1, Skp2 and Roc1, is involved in E2A ubiquitination (17, 30). Interaction between Skp2 and E2A proteins depends on E2A phosphorylation. To search for possible additional components involved in E2A degradation, we used a fragment of E2A known to be crucial for its degradation as bait to conduct a Saccharomyces cerevisiae two-hybrid screen. We identified CHIP as an interacting partner of E47 not only in yeast but also in mammalian cells.
CHIP is a cochaperone protein identified through its interaction with Hsc70 (5). In addition to its well-characterized tetratricopeptide repeats (TPR) that mediate interaction with chaperones (15, 40), it also has a U-box, representing a subclass of ubiquitin ligases (13, 23). Diverse functions have been ascribed to CHIP. CHIP itself has been shown to possess E3 ubiquitin ligase activity by using luciferase, ErbB2, Hsc70, and CHIP as substrates (7, 18, 28, 45). However, CHIP has also been shown to facilitate the ubiquitination of Pael receptor and cystic fibrosis transmembrane-conductance regulator by other E3 ligases (16, 26). In addition, CHIP is postulated to be a cochaperone that facilitates the degradation of misfolded proteins (1, 8, 10, 44). Moreover, CHIP also performs functions unrelated to protein ubiquitination and degradation, such as inducing the trimerization of HSF transcription factor and partitioning soluble endothelial nitric oxide synthase into an insoluble and inactive cellular compartment (9, 19). Here we provide evidence suggesting that CHIP acts as a carrier of the ubiquitination substrate, E47, thus promoting its association with the F box protein Skp2, through the formation of a preubiquitination complex (PUC). CHIP also binds to Cul1, ultimately facilitating the assembly of the entire SCFSkp2-E3 ligase complex and the ubiquitination of E47. This function of CHIP is essential for Notch-induced E2A degradation.
MATERIALS AND METHODS
Yeast two-hybrid screen.
To construct the bait, a PCR product encoding the E protein homology domains 2 and 3 (amino acids 319 to 382 of both E12 and E47) was subcloned into the pEG202 plasmid and fused with the LexA DNA binding domain (11). A unidirectionally synthesized LyD9 (pro-B-cell line) cDNA library was inserted into the pJG45 galactose-inducible expression vector and fused to the B42 activation domain (11). The bait plasmid exhibited no basal trans-activating activity in the absence of any interacting protein encoded by cDNAs in the pJG4-5 vector. The bait and cDNA library plasmids were transfected into the yeast strain EGY48 along with a LacZ reporter driven by multiple LexA binding sites. A total of 5 × 106 clones were screened, and positive clones were confirmed by secondary screening. Plasmids containing cDNA inserts were recovered and sequenced by the DNA sequencing facility at Oklahoma Medical Research Foundation.
Cell lines and transfection.
NIH 3T3 and 293T cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. The BaF3 cell line was grown in modified RPMI 1640 supplemented with 10% fetal bovine serum, 50 μM β-mercaptoethanol, and 20% WEHI-3-conditioned medium as a source of interleukin-3. Jagged-1-expressing and control NIH 3T3 cell lines were generous gifts from T. Kadesch, University of Pennsylvania. Transient transfection with desired constructs was performed using the calcium phosphate precipitation method.
Plasmids and retroviral constructs.
Full-length CHIP cDNA with a 3′ hemagglutinin (HA) tag was cloned by PCR amplification using total mouse cDNA as templates and the following primers: 5′ primer, ATTGGTACCATGAAGGGCAAGGAGGAAAAGGAG; 3′ primer, TTCAGGCGTAGTCTGGGACGTCCTAAGGGTAATAGTCCTCTACCCAGCCGTTCTCAG. The PCR product was first cloned into the pGEM-T easy vector (Promega, Madison, Wis.). The insert was then excised out with Asp718 and EcoR1 and cloned into the same sites of pcDNA3 (Invitrogen, Carlsbad, Calif.). HPC4-tagged CHIP-pcDNA3 was generated by inserting oligonucleotides containing sequences encoding the HPC4 tag (38) into the Asp718 site at the 5′ end of CHIP in pcDNA3. The CHIP K31A point mutant was generated by replacing the wild-type SacI-Bpu11021 fragment within the coding sequence with a mutant fragment generated by PCR amplification with the following oligonucleotides: 5′ primer, AGAGCTCGCGGAGCAGGGAAAC (mutated codon is underlined); 3′ primer, TGAATTCAATCATCTTCATGACCCTCGTGG. The CHIP H261A/P270A point mutant construct (13) was obtained from S. Hatakeyama, University of Kyushu, Fukuoka, Japan, and modified by swapping the 5′ portion of the wild-type CHIP with the HPC4 tag. To produce TPR and U-box domain-containing constructs, we performed PCR using CHIP-pcDNA3 as a template with the following oligonucleotides: for TPR, TGGATCCATGAAGGGCAAGGAGGAAAAGG and TGAATTCAATCATCTTCATGACCCTCGTGG; for U-box, TGGATCCAAGAAGCGCTGGAACAGTATCGAGG and TGAATTCACTAGTGATTTCAGGCGTAG. The PCR products were cloned into the BamHI and EcoRI sites of the pcDNA3 vector carrying the HPC4 tag at the Asp718 site. To generate the RNA interference construct targeting CHIP, double-stranded oligonucleotides containing a hairpin structure were cloned into the XhoI and HindIII sites of the pBS/U6 vector (41). The top-strand sequence of the oligonucleotides for small interfering RNA (siRNA) against CHIP (CHIPi) was TCGAGGAAGCGAGATATCCCTGACGAGTACTGGTCAGGGATATCTCGCTTCCTTTT, and that for siRNA against green fluorescent protein (GFPi) was as described elsewhere (30). Retroviral constructs for CHIPi and GFPi were generated by inserting the BamHI-EcoRI fragments containing the entire U6 transcription units from the above pBS/U6 constructs into the BglII and EcoRI sites of the MiGR1 vector (36). Retrovirus production was carried out as described previously (34).
E47 point and deletion mutants were made by two-step PCR-assisted mutagenesis using the parental plasmid E47-pcDNA3 as a template (33). T7-tagged Skp2 construct was a gift from H. Zhang, Yale University School of Medicine. The Skp2 construct used for in vitro translation was generated by PCR amplification of the coding sequence of Skp2 using the T7-tagged Skp2 construct as a template. The PCR product was cloned into the Asp718 and EcoRI sites of pcDNA3. The constructs, Cul1-pcDNA3 and HA-Skp1-pHR, were kindly provided by W. Harper, Harvard Medical School.
Immunoblotting and immunoprecipitation.
Cells were collected and lysed in NP-40 lysis buffer containing 15 mM Tris-HCl (pH 7.6), 150 mM NaCl, 0.1 mM EDTA, 10% glycerol, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin/ml, 4 μg of leupeptin/ml, 2 μg of pepstain/ml, and 5 μM MG132. Supernatants were obtained after centrifugation at 20,000 × g in a microcentrifuge for 15 min at 4°C. Protein concentrations were determined by BCA protein assay kit (Pierce, Rockford, Ill.). For immunoblotting, cells were collected 36 h after transfection. For in vivo ubiquitination assays, cells were treated with 25 μM MG132 for 5 h prior to harvest. The immunoreactive proteins were detected using enhanced chemiluminescence reagents (Amersham, Piscataway, N.J.) and a LumiImager (FUJIFILM Medical Systems USA Inc., Stamford, Conn.). Loading controls were carried out by probing for β-actin. Antibodies against E47, Hsc70, HA tag, and β-actin were from Santa Cruz Biotechnology (Santa Cruz, Calif.), while those against Skp1 and Cul1 were from NeoMarkers (Fremont, Calif.). Anti-Skp2 was purchased from Zymed (San Francisco, Calif.). The antiserum against CHIP was generated by immunizing rabbits with full-length recombinant CHIP (carried out by Pocono Rabbit Farm & Laboratory Inc., Canadensis, Pa.). For immunoprecipitation assays, cell lysates were precleared with 60 μl of protein A or protein A/G-Sepharose (50% slurry equilibrated in phosphate-buffered saline) for 2 h at 4°C and then incubated with 5 μg of specific antibodies for 4 to 5 h at 4°C. Protein A or protein A/G-Sepharose was added and incubated for an additional 1 h at 4°C to capture primary immune complexes. The beads were washed three times in NP-40 lysis buffer and heated at 100°C in sodium dodecyl sulfate (SDS) sample buffer before loading on 10 or 12% polyacrylamide gels. For immunoprecipitation with the anti-HPC4 antibody (a generous gift from C. Esmon, Oklahoma Medical Research Foundation), which binds to the HPC4 tag in a calcium-dependent manner (38), 1 mM CaCl2 was included in all buffers used. The immunoprecipitates were eluted with 10 mM EDTA by shaking for 30 min at room temperature. The SDS loading buffer was then added to the supernatants, and the samples were boiled before electrophoresis.
In vitro translation and binding assays.
35S-labeled Skp1, Skp2, Cul1, and CHIP were produced by programming the TNT-coupled reticulocyte lysate in vitro translation system (Promega) with 1 μg of each expression plasmid DNA, according to the manufacturer's instructions. A 50-μl aliquot of in vitro-translated reaction mixtures diluted with 300 μl of NP-40 lysis buffer was used for immunoprecipitation with the antibody against the HPC4 tag on CHIP. The resulting immunoprecipitates were electrophoresed on SDS-10% polyacrylamide gels and analyzed by autoradiography using a PhosphorImager.
RESULTS
Interaction of CHIP with E47.
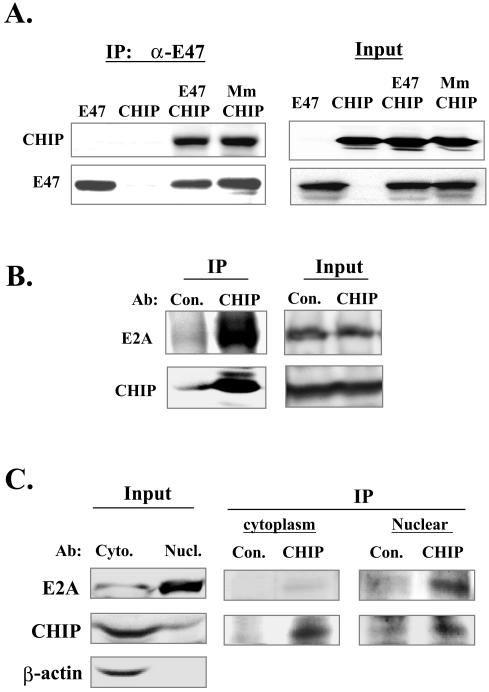
We have previously shown that E47 is ubiquitinated and subsequently degraded by proteasomes (30). To search for additional proteins involved in the degradation of E47, we conducted a yeast two-hybrid screen using bait containing a 64-amino-acid polypeptide sequence including EHD2 and EHD3, which are present in both E12 and E47. We obtained several independent cDNA clones encoding either full-length CHIP or truncated proteins lacking 28 or 32 amino acids at the N terminus. To confirm the interaction in mammalian cells, constructs expressing E47 and CHIP were transfected into 293T cells, individually or together. Whole-cell lysates of transfected cells were immunoprecipitated with polyclonal antibodies against E47 (Fig. 1A). Immunoblotting revealed that both E47 and CHIP were present in the immunoprecipitates, suggesting that E47 and CHIP interact with each other in mammalian cells. We have previously shown that mutation of the MAP kinase phosphorylation sites in E47 (Mm mutation) abolishes E47 ubiquitination and degradation (30). We tested if CHIP was able to bind to the Mm mutant in the same coimmunoprecipitation assays. The Mm mutant bound to CHIP as avidly as wild-type E47 (Fig. 1A), suggesting that the association of CHIP with E47 is independent of phosphorylation at these sites.
FIG. 1.
Interaction between E47 and CHIP in mammalian cells. (A) Coimmunoprecipitation assays. Wild-type E47 or the Mm mutant construct was cotransfected with HA-tagged CHIP into 293 T cells. Whole-cell lysates were used for immunoprecipitation with anti-E47 antibodies. The resulting immunoprecipitates and 5% of the lysates (input) were analyzed by immunoblotting the same membrane with anti-HA and anti-E47 antibodies sequentially. (B) Association of endogenous CHIP and E47 in whole-cell lysates of BaF3 cells. Immunoprecipitations were performed with whole-cell lysates of BaF3 cells with an antiserum from a rabbit immunized with recombinant CHIP or a preimmune serum (Con.). The precipitates were analyzed by immunoblotting with antibodies against E47 (which also recognize E12) and CHIP. (C) Distribution of E2A and CHIP in nuclear and cytoplasmic fractions of BaF3 cells. Nuclear and cytoplasmic extracts were prepared using BaF3 cells and analyzed by immunoblotting for the indicated proteins (input). These extracts were used in coimmunoprecipitation assays as described for panel B.
To demonstrate interaction between endogenous E2A proteins and CHIP in vivo, a hematopoietic progenitor cell line, BaF3, was used in a coimmunoprecipitation assay. Immunoprecipitation against endogenous CHIP was carried out using antibodies raised in rabbits by immunizing with full-length recombinant CHIP. The immunoprecipitates obtained with the anti-CHIP serum but not preimmune serum contained E2A proteins as detected by immunoblotting with anti-E47 antibodies which recognize both E12 and E47 (Fig. 1B). This result confirmed that E2A proteins indeed bind to CHIP under physiological conditions.
Furthermore, because CHIP was previously reported to be localized to the cytoplasm and E2A proteins are in the nucleus (5, 26), an issue arises as to whether the two proteins have any opportunity to interact with each other. To clarify this issue, we prepared cytoplasmic and nuclear fractions from BaF3 cells and examined the relative levels of these proteins (Fig. 1C). While the majority of E2A proteins were present in the nucleus and CHIP was in the cytoplasm, a small fraction of CHIP was found in the nucleus. This was not due to contamination of the nuclear fraction with the cytoplasmic fraction, because actin was detected only in the cytoplasmic fraction when the same membrane was reprobed for actin. However, the small amount of E2A proteins in the cytoplasmic fraction might be due to contamination from broken nuclei or proteins transiting the cytoplasm as they are newly synthesized or degraded. To examine the interaction between E2A and CHIP in cytoplasmic and nuclear fractions, we carried out coimmunoprecipitation experiments (Fig. 1C). In the cytoplasmic fraction, very low levels of E2A proteins were detected in the anti-CHIP precipitates despite a large amount of CHIP brought down by the antiserum against CHIP. This is because E2A proteins are rarely present in the cytoplasm. In contrast, the anti-CHIP immunoprecipitates from the nuclear fraction brought down a substantial amount of E2A proteins. A preimmune serum was also used in parallel as a negative control. These results suggest that a small portion of CHIP is present in the nucleus and capable of interacting with E2A proteins in vivo.
CHIP is important for E47 degradation.
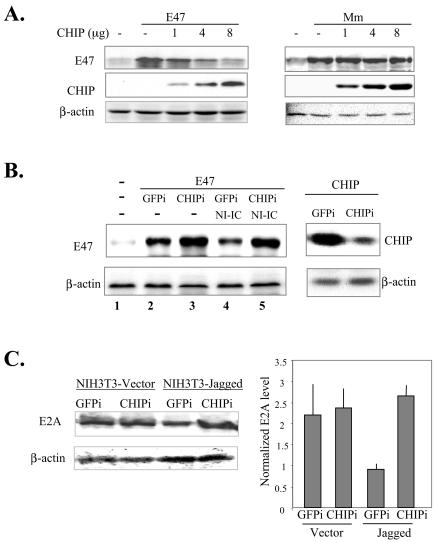
Next, we examined the effect of CHIP on E47 degradation. E47 was cotransfected with increasing amounts of the CHIP expression construct into NIH 3T3 cells. The amount of E47 present in transfected cells was measured by immunoblotting with anti-E47 antibodies. With an increasing amount of CHIP expressed, the amount of wild-type E47 decreased, but that of the E47 Mm mutant remained unchanged (Fig. 2A). This result suggested that CHIP facilitated the degradation of E47 in a phosphorylation-dependent manner. It should be noted that induction of E47 degradation by CHIP is not apparent in 293T cells. This is probably because E47 is expressed at much higher levels in these cells, thus overwhelming endogenous degradation machineries. We also found that, compared to NIH 3T3 cells, 293T cells have much lower MAP kinase activities, which are required for E47 phosphorylation prior to its degradation. Therefore, the 293T cell line is ideal for examining protein interaction but not for E47 degradation.
FIG. 2.
CHIP enhances E47 degradation. (A) CHIP induces degradation of wild-type but not Mm mutant E47. One microgram of E47 or Mm construct was cotransfected with the indicated amounts of HPC4-tagged CHIP into NIH 3T3 cells. The whole-cell lysates were used for immunoblotting for the indicated proteins. The anti-HPC4 antibody was used to detect CHIP. The first lane of each panel contains the lysate from cells transfected with vector control. The amounts of β-actin served as loading controls. (B) Inhibition of Notch-induced E47 degradation by CHIPi. (Left) Constructs expressing CHIPi or GFPi were cotransfected into NIH 3T3 cells with E47 plus or minus NI-IC. Thirty-six hours after transfection, whole-cell lysates were immunoblotted for the indicated proteins. (Right) To control for the efficacy of CHIPi, CHIP was cotransfected into NIH 3T3 cells with constructs expressing CHIPi or GFPi. The amount of CHIP present in transfected cells was determined by immunoblotting with the anti-HPC4 antibody. The amount of β-actin served as a loading control. (C) Inhibition of endogenous E2A degradation by CHIPi. BaF3 cells were infected with retroviruses expressing CHIPi or GFPi. Infected cells were sorted by flow cytometry and propagated before coculturing with NIH 3T3 cells expressing a Notch ligand, Jagged1, or vector control cells for 4 h. BaF3 cells were then harvested, and whole-cell lysates were used in immunoblotting with anti-E47 and anti-actin antibodies. The amounts of detected proteins were quantified with a LumiImager, and the amount of E2A was normalized with that of β-actin as shown on the right.
We have previously shown that Notch signaling induces E47 degradation by stimulating E47 ubiquitination in a phosphorylation-dependent manner (30). To test if CHIP is involved in E47 degradation induced by activated Notch1 receptor, constructs expressing siRNA against CHIP or GFP (as a control) were cotransfected with E47 in the absence or presence of the construct expressing the intracellular domain of Notch1 (N1-IC) (Fig. 2B). In the presence of GFPi, the amount of E47 was lower in N1-IC-transfected cells than in vector-transfected cells (lanes 2 and 4). This suggested that N1-IC accelerated the degradation of E47, and GFPi could not rescue the degradation. In contrast, introduction of CHIPi inhibited E47 degradation caused by N1-IC (lanes 4 and 5). In addition, the basal level of E47 in the absence of N1-IC overexpression was also increased slightly by CHIPi (lanes 2 and 3). Thus, CHIP plays an important role not only in N1-IC-induced E47 degradation but also in the steady-state turnover of E47. Furthermore, to determine the role of CHIP in E2A degradation under physiological conditions, we introduced the same siRNAs into BaF3 cells by infecting the cells with retroviral expression vectors for GFPi and CHIPi. To activate Notch signaling, BaF3 cells were cocultured with NIH 3T3 cells producing a Notch ligand, Jagged1. Coculturing GFPi-expressing BaF3 cells with NIH 3T3-Jagged1 cells led to an over-twofold reduction in E2A protein level compared to coculturing the same cells with control NIH 3T3 cells (Fig. 2C). In contrast, CHIPi-expressing BaF3 cells had similar levels of E2A proteins after cocultivation with either NIH 3T3 control or Jagged1-expressing cells (Fig. 2C), suggesting that CHIP is important for Notch-induced E2A degradation in B cells.
Mutational analysis of the CHIP-binding domain in E47.
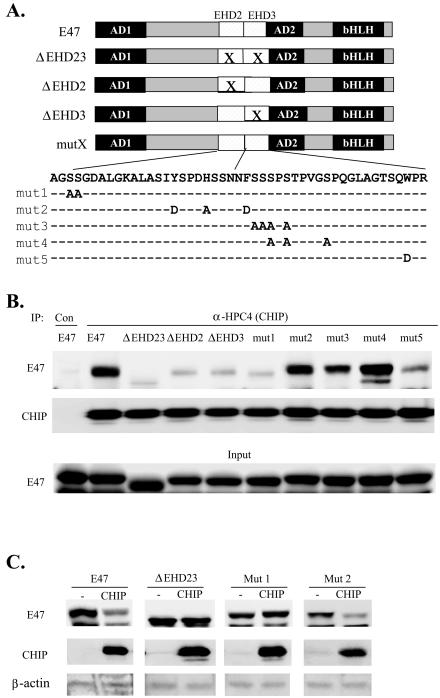
To map the CHIP-interacting domain in E47, we first focused on the region originally used for screening the yeast two-hybrid library and identifying CHIP as an interacting protein. This region contains two domains highly conserved in all E proteins, called EHD2 and EHD3 (Fig. 3A). EHD3 carries the MAP kinase sites shown to be important for Skp2 binding and E47 ubiquitination. CHIP was cotransfected into 293T cells with various E47 mutants, and whole-cell lysates were prepared 36 h after transfection. Coimmunoprecipitation experiments were carried out with the monoclonal antibody against the HPC4 tag on CHIP, and the precipitates were analyzed by immunoblotting with anti-E47 antibodies (Fig. 3B). The amount of CHIP brought down in each reaction was determined by anti-HPC4 immunoblotting, while the expression level of each E47 construct was measured by immunoblotting the whole-cell extract with anti-E47 antibodies. Deletion of EHD2 and -3 individually or together dramatically decreased the interaction of E47 with CHIP in coimmunoprecipitation assays, suggesting that both domains are involved in the interaction (Fig. 3B). The extremely low level of interaction remaining in these constructs might result from interaction with the junction sequences on either side of EHD2 plus -3 or from a nonspecific interaction of the mutants with the anti-HPC4 antibody. Furthermore, point mutations in EHD2 and EHD3 were also examined in the same manner (Fig. 3B). Surprisingly, mut1 and mut5, located at the N terminus of EHD2 and the C terminus of EHD3, respectively, lost most of their ability to bind to CHIP. In contrast, clusters of mutations near the junction of EHD2 and -3 in mut2, mut3, and mut4 (also known as Mm) constructs had little or no effect on CHIP interaction (Fig. 3B). Taken together, it appears that EHD2 and -3 together form a CHIP binding domain and CHIP binds to both ends of the domain, leaving the internal region available for Skp2 interaction.
FIG. 3.
Mapping the CHIP interacting domain in E47. (A) Diagram of E47 structure. The two activation domains and the DNA binding-dimerization domain are marked in dark boxes and labeled as AD1, AD2, and bHLH, respectively. Two of the E protein homology domains (EHD2 and -3) are shown in hatched boxes, and their deletion is symbolized by white boxes with an X. Wild-type sequences of EHD2 and -3 were presented with amino acid substitutions in each mutant listed below. (B) Interaction between CHIP and E47. E47 and its mutant constructs were cotransfected with or without CHIP into 293T cells, and whole-cell lysates were used in immunoprecipitations with the anti-HPC4 antibody. The precipitates were analyzed by immunoblotting with anti-E47 and anti-HPC4 antibodies. The expression levels of E47 and its mutants in transfected cells were determined by probing with anti-E47 antibodies (input). (C) CHIP-induced degradation of E47 mutants. One microgram of CHIP was cotransfected into NIH 3T3 cells with 0.5 μg of E47 construct. Whole-cell lysates were analyzed as described for Fig. 2A.
Furthermore, we tested the correlation between the abilities of E47 mutants to interact with CHIP and their susceptibilities to CHIP-stimulated degradation. We selected several mutants representing those that bind to CHIP and those that do not (Fig. 3C). As expected, cotransfection of wild-type E47 with CHIP significantly reduced the amount of E47 present in the cells. However, the ΔEHD23 mutant lacking the entire CHIP interacting domain was resistant to CHIP-induced degradation. Similarly, the mut1 point mutant was neither bound by CHIP nor induced to degrade by CHIP. In contrast, the level of mut2 was sensitive to CHIP, because mut2 was capable of interaction with CHIP. Therefore, these results further confirm the involvement of CHIP in E47 degradation.
Mutational analysis of the E47 binding domain in CHIP and the functional consequences of the mutations.
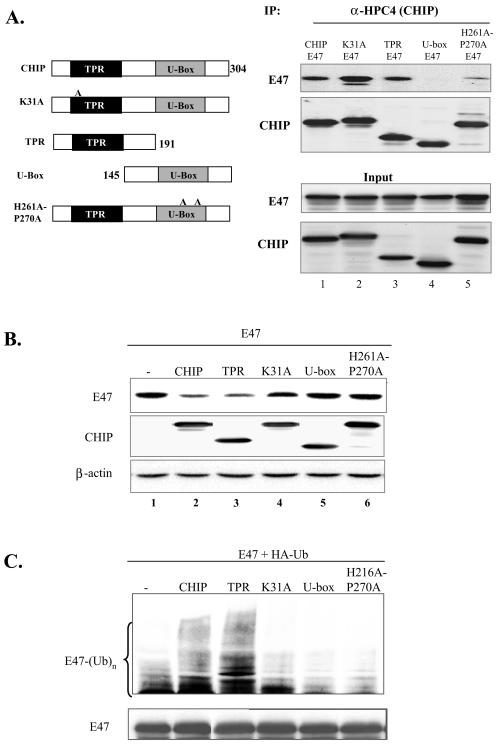
CHIP contains two well-characterized domains: the TPR domain responsible for chaperone binding, and the U-box domain important for its E3 ubiquitin ligase activity (8). Coimmunoprecipitation assays were performed by incubating antibodies against E47 with whole-cell lysates prepared from 293T cells transfected with E47 and wild-type or mutant CHIP. The precipitates were analyzed by immunoblotting with the anti-HPC4 antibody to detect CHIP (Fig. 4A). The TPR domain, located 28 amino acids downstream from the N terminus, was necessary and sufficient for E47 binding, but the K31A point mutation within the TPR domain, known to disrupt interaction with chaperones (45), did not affect E47 binding. This suggests that CHIP associates with E47 independently of chaperones. However, the U-box domain did not associate with E47 (Fig. 4A). Intriguingly, the H261A and P270A mutations within the U-box, thought to inhibit the ubiquitin ligase activity (13), also impaired the interaction between E47 and CHIP. This is in contrast to the finding with the TPR construct, that complete deletion of the U-box domain in the TPR construct had minimal effect on E47 interaction. We hypothesize that the integrity of the U-box domain or the ubiquitin ligase activity is necessary for maintaining a proper conformation to allow the TPR domain to bind to E47. A charged region lies between the TPR domain and the U-box which has recently been shown to mediate dimerization of CHIP (31). Since this domain is present in both the TRP and U-box constructs, it is unlikely involved in binding to E47.
FIG. 4.
Mapping the domain in CHIP responsible for E47 interaction, degradation, and ubiquitination. (A) Domain involved in E47 interaction. CHIP structure is diagrammed on the right, with the two major functional domains, TPR and the U-box, marked in dark and shaded boxes. The end points of deletion mutations are labeled according to the position of the amino acids. Positions of the amino acid substitutions in point mutants are indicated by the numbers in the names of the mutants. E47 was transfected into 293T cells with or without wild-type or mutant CHIP as indicated above each lane. Immunoprecipitation was carried out with the anti-HPC4 monoclonal antibody to pull down CHIP. The precipitates and 5% of cell lysates used for immunoprecipitation (input) were analyzed with antibodies against E47 or the HPC4 tag. (B) Domain involved in E47 degradation. NIH 3T3 cells were cotransfected with 1 μg each of E47 and wild-type or mutant CHIP constructs as indicated above each lane. Whole-cell lysates were used for immunoblot analysis with antibodies against E47. The same membrane was stripped and reprobed with anti-HPC4 (for CHIP) and anti-β actin. (C) Domain involved in E47 ubiquitination. E47 and HA-ubiquitin were cotransfected with wild-type or mutant CHIP into 293T cells. Twenty-four hours after transfection, cells were treated with 25 μM MG132 for 5 h. Whole-cell lysates were immunoprecipitated with antibodies against E47. Precipitates were analyzed by immunoblotting with the anti-HA antibody for ubiquitinated proteins (upper panel) or with anti-E47 antibodies for the amount of E47 precipitated (lower panel).
Next, we examined the correlation between the ability of CHIP mutants to bind to E47 and their effects on E47 degradation. E47 was cotransfected with wild-type or mutant CHIP constructs, and levels of E47 and CHIP were measured using immunoblotting (Fig. 4B). The TPR domain construct not only bound to E47 but also induced E47 degradation. Conversely, the U-box domain construct and the H261A-P270A mutant failed to either bind to E47 or cause E47 degradation. Interestingly, the K31A mutation bound to E47 avidly but did not promote E47 degradation.
Since we had previously shown that E47 degradation is ubiquitin mediated, we examined the effect of CHIP on E47 ubiquitination (Fig. 4C). E47 was cotransfected into 293T cells with an HA-tagged ubiquitin construct along with wild-type or mutant CHIP constructs. Immunoprecipitation was carried out with antibodies against E47, and the precipitates were analyzed by immunoblotting with anti-HA and anti-E47 antibodies. Coexpression of wild-type CHIP with E47 dramatically enhanced the ubiquitination of E47, suggesting that CHIP is involved in the process of E47 ubiquitination. Consistent with the effects of CHIP mutants on E47 degradation, the TPR domain stimulated E47 ubiquitination, while the U-box domain as well as the K31A and H261A-P270A mutants did not alter the level of E47 ubiquitination. Therefore, we conclude that the TPR domain of CHIP is responsible for facilitating E47 degradation by enhancing its ubiquitination. Furthermore, the inability of the K31A mutant to promote E47 degradation indicates that although the ability of CHIP to bind to chaperones is not necessary for binding to E47, it is required for promoting E47 ubiquitination and degradation.
CHIP binds to SCFskp2 through interaction with Cul1.
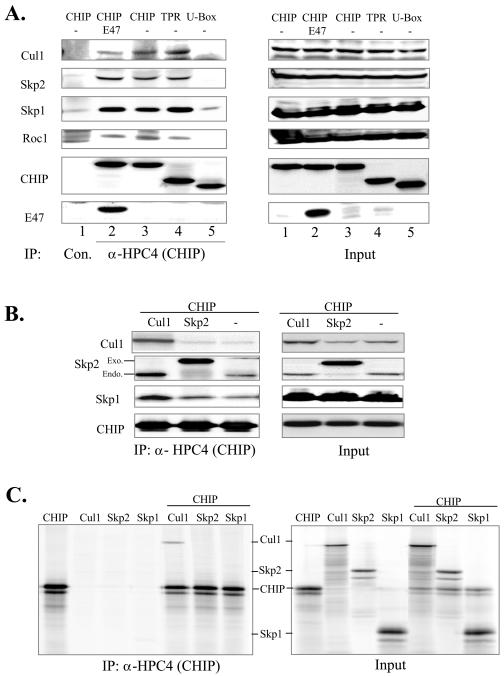
To understand the mechanism by which CHIP enhances E47 ubiquitination, we examined the association between CHIP and the SCFskp2 complex, which we have previously shown to be involved in E47 ubiquitination (30). CHIP or its deletion mutants were transfected with or without E47 into 293T cells. Immunoprecipitation was carried out using the antibody against the HPC4 tag on CHIP or a control antibody. The immunoprecipitates were analyzed by blotting with antibodies against components of the SCFskp2 complex (Fig. 5A). CHIP or the mutant containing the TPR domain (lanes 2 to 4) coimmunoprecipitated with components of the endogenous SCFSkp2 complexes, including Cul1, Skp2, Skp1, and Roc1 (12). However, expression of the mutant containing the U-box domain did not lead to coprecipitation with SCFSkp2, as shown by the comparable amounts of each component brought down with the anti-HPC4 and control antibodies (lanes 1 and 5). These results suggested that the TPR domain mediates the interaction between CHIP and SCFSkp2, which is consistent with its role in stimulating E47 ubiquitination and degradation (Fig. 4). The interaction between CHIP and SCFskp2 was independent of E47, since the amounts of SCFskp2 components bound to CHIP were similar in cells transfected with and without E47 (Fig. 5A).
FIG. 5.
CHIP associates with the SCFSkp2 complex through Cul1. (A) CHIP associates with the SCFSkp2 complex. Wild-type or mutant CHIP was cotransfected with or without E47 into 293T cells. Immunoprecipitation was performed with the anti-HPC4 antibody or control antibodies. The precipitates were analyzed by immunoblotting for endogenous components of the SCFSkp2 complex, transfected HPC4-CHIP, or E47 as indicated at left. Input controls were analyzed similarly. (B) CHIP was cotransfected with or without Cul1 or T7-tagged Skp2 into 293T cells. Whole-cell lysates were analyzed as described for panel A. The endogenous (endo) and exogenous (exo) Skp2 are indicated. (C) Interaction between CHIP and Cul1 in vitro. Skp1, Skp2, or Cul1 was synthesized alone or together with HPC4-tagged CHIP by in vitro translation in the presence of [35S]methionine. The resulting proteins were immunoprecipitated with the anti-HPC4 antibody. The precipitates were analyzed by electrophoresis followed by autoradiography on a PhosphorImager.
Interestingly, cotransfection of a construct expressing Cul1 with CHIP not only increased the amount of Cul1 coprecipitated with CHIP but also those of endogenous Skp2 and Skp1 (Fig. 5B). In contrast, cotransfection of a construct expressing Skp2 did not increase the amounts of endogenous Cul1 and Skp1 brought down by the anti-HPC4 antibody. These results suggested that Cul1 was a limiting factor important for the assembly of the SCF complex and CHIP directly bound to Cul1. Indeed, a coimmunoprecipitation assay using in vitro-translated proteins showed that when cosynthesized, the HPC4-tagged CHIP interacted with Cul1 in rabbit reticulocyte lysates but not with Skp2 or Skp1 (Fig. 5C). As negative controls, Cul1, Skp2, and Skp1 synthesized individually without CHIP were not brought down by the anti-HPC4 antibody (Fig. 5C). These data thus indicate that CHIP binds to Cul1 but not Skp2 or Skp1 in reticulocyte lysates, most likely in the absence of SCF complexes. However, coexpression of Skp2 with CHIP did increase the amount of Skp2 coprecipitated with CHIP (Fig. 5B). This was probably due to an indirect interaction between Skp2 and CHIP in a complex described in Fig. 6.
FIG. 6.
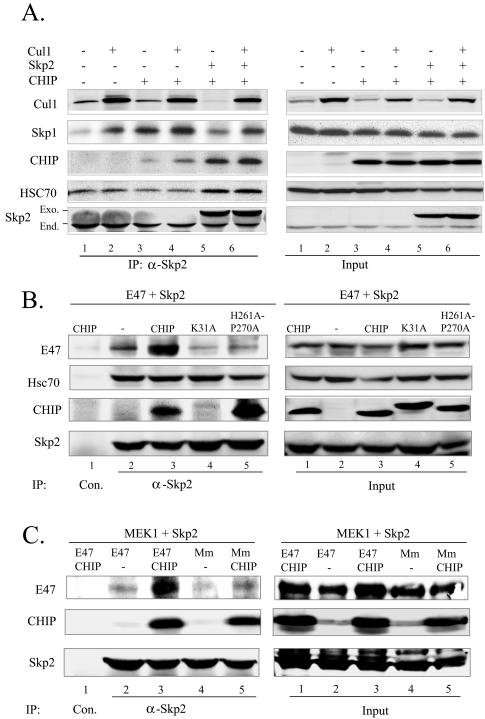
Characterization of a PUC. In 293T cells, CHIP, Cul1, and Skp2 were transfected individually or in combinations as indicated above each lane (A); E47 and Skp2 were cotransfected with wild-type and mutant CHIP as indicated (B); or Skp2 and a constitutively active form of MEK1 were cotransfected with E47 or the Mm mutant plus or minus CHIP (C). Whole-cell lysates from these transfected cells were used in immunoprecipitation assays with anti-Skp2 or control antibodies. The precipitates and input were analyzed by immunoblotting for the indicated proteins.
Characterization of a preubiquitination complex (PUC).
As shown in Fig. 5B, cotransfection of Skp2 with CHIP led to an increased association between Skp2 and CHIP independent of other components in the SCF complex, even though Skp2 did not bind to CHIP directly (Fig. 5C). This result suggested that Skp2 and CHIP might be present in an additional intermediary complex different from the SCF complex. To explore this possibility, coimmunoprecipitation experiments were carried out with antibodies against Skp2 in 293T cells transfected with Cul1, Skp2, and CHIP individually or in combinations (Fig. 6A). Coprecipitation between Skp2 and Cul1 was dramatically increased by overexpression of Cul1, further supporting the notion that the amount of endogenous Cul1 is limiting in 293T cells (lanes 1 and 2). Although Skp1 has been shown to interact with Skp2 directly (39), overexpression of Cul1 apparently enhanced the association between Skp1 and Skp2, probably by facilitating the assembly of the SCF complex (lanes 1 and 2). Cotransfection of CHIP also greatly increased the amount of Skp1 coprecipitated with Skp2, consistent with the notion that CHIP helps in recruiting Skp2 to the SCF complex (lanes 1 and 3). Cul1 overexpression also increased the amount of cotransfected CHIP brought down by anti-Skp2 antibodies (lanes 3 and 4), which confirmed that CHIP is present in the SCF complex, as demonstrated in Fig. 5.
Significantly, expression of Skp2 drastically increased the amount of CHIP coprecipitated without increasing the amounts of Cul1 and Skp1, suggesting that Skp2 and CHIP form an additional non-SCF complex, which we named the PUC (Fig. 6A, compare lanes 3 and 4 to lanes 5 and 6). In fact, amounts of Cul1 and Skp1 in the anti-Skp2 precipitates in lane 5 were lower than that in lane 3. This may be explained by the fact that overexpressed Skp2 was able to increase the amount of PUC, but the amount of SCF complex was limited by the quantity of Cul1 available in the cells. Therefore, PUC or free Skp2 outcompeted the SCF complex for anti-Skp2 antibodies during immunoprecipitation. When Cul1 was also overexpressed (lane 6), the amounts of Cul1 and Skp1 coprecipitated were increased. Moreover, the amount of endogenous Hsc70 coprecipitated was significantly elevated by overexpression of Skp2 but not Cul1 (compare lanes 4 and 5), thus raising the possibility that PUC also contains Hsc70.
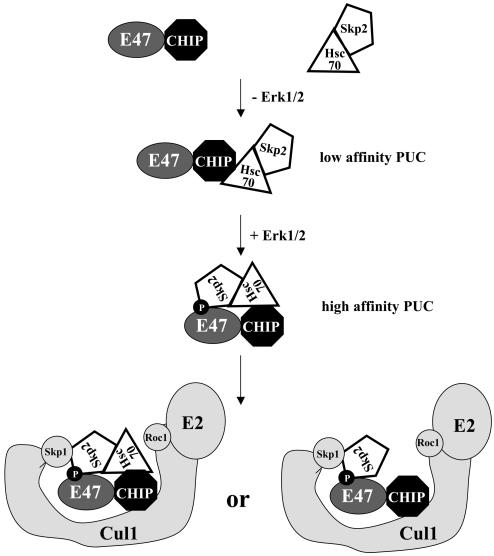
Data shown in Fig. 6A led us to postulate that CHIP and its interacting partner, Hsc70, provide a scaffold for the formation of PUC that promotes the interaction between the F-box protein of the SCF complex, Skp2, and its substrate, E47. Therefore, CHIP binds to E47 on one hand and Hsc70 on the other. Similarly, Hsc70 interacts with both CHIP and Skp2. Together, this tetrameric complex brings E47 and Skp2 together and allows Skp2 to recognize and bind with high affinity to phosphorylated E47. To test this hypothesis, E47 and Skp2 were cotransfected with or without wild-type CHIP or with CHIP mutants defective in binding to either Hsc70 or E47. Immunoprecipitation was carried out with anti-Skp2 or control antibodies. Amounts of coprecipitated E47, Hsc70, and CHIP were assayed by immunoblotting (Fig. 6B). In the absence of CHIP, anti-Skp2 antibodies brought down a small amount of E47, likely via interaction with endogenous CHIP (lane 2). However, coexpression with CHIP dramatically increased the amount of E47 coprecipitated, suggesting that CHIP indeed facilitated the interaction between E47 and Skp2 (lane 3). In contrast, the K31A and H261A-P270A mutants of CHIP failed to facilitate coimmunoprecipitation of E47 and Skp2 (lanes 4 and 5). Since the K31A mutant is defective in binding to Hsc70 (45) and the H261A-P270A mutant does not bind to E47 (Fig. 4A), these results thus suggest that the ability of CHIP to promote E47 and Skp2 interaction depends on its activities in interacting with both E47 and Hsc70. Hsc70, however, was found to bind to Skp2 in the presence or absence of wild type and mutant CHIP, indicating its direct interaction with Skp2 (Fig. 6B, lanes 2 to 5). Moreover, CHIP could associate with Skp2 only if it was able to interact with Hsc70 (compare lane 3 to lane 4), which is consistent with the finding that CHIP did not bind to Skp2 directly (Fig. 5C). The H261A-P270A mutant was pulled down readily by anti-Skp2 antibodies but failed to assist in recruiting of E47 due to its poor affinity to E47 (lane 5). Taken together, these results suggest that components of PUC connect linearly in the order of E47-CHIP-Hsc70-Skp2 (Fig. 7).
FIG. 7.
Mechanism of action for CHIP-facilitated E47 ubiquitination. Interaction between E47 and Skp2 is facilitated by their respective affinities to CHIP and Hsc70. In the absence of phosphorylation of E47 by Erk1/2, a low-affinity tetrameric complex (PUC) is formed but is stabilized by phosphorylation of E47, which leads to a high-affinity PUC. This complex can then join the SCF complex to initiate ubiquitination of E47.
Further experiments suggested that phosphorylation of E47 apparently led to its high-affinity binding to Skp2, probably by stabilization of PUC. As shown in Fig. 6C, Skp2 and a constitutively active form of MEK1 (which activates Erk1/2) were cotransfected with wild-type E47 and the Mm mutant lacking the phosphorylation sites, in the presence or absence of CHIP. Immunoprecipitation was carried out with antibodies against Skp2, and the precipitates were analyzed by immunoblotting for E47, CHIP, and Skp2. In the absence of CHIP, a low level of interaction between E47 and Skp2 was detected, which was probably facilitated by endogenous CHIP and Hsc70. However, wild-type E47 associated with Skp2 more efficiently than Mm, suggesting that phosphorylation of E47 is important. Coexpression of CHIP dramatically increased the interaction between Skp2 and wild-type E47 but not the Mm mutant, indicating that phosphorylation of E47 probably allows Skp2 to recognize and bind E47, thus reorganizing PUC into a high-affinity complex. CHIP caused a slight enhancement of the association between the Mm mutant and Skp2. This was likely due to a low-affinity interaction between components of PUC, because binding of CHIP to E47 per se does not require its phosphorylation (Fig. 1A). These data further emphasize the important role of CHIP in facilitating the association of E47 with Skp2 and, consequently, the ubiquitination of E47.
DISCUSSION
E2A proteins, E12 and E47, have previously been shown to be degraded through a MAP kinase-dependent and ubiquitin-mediated pathway (30). Ubiquitination of E2A proteins involves the SCFSkp2-E3 ligase complex (12, 17). Here, we have shown that the chaperone binding protein, CHIP, is also crucial for E2A protein degradation. CHIP interacts with E2A proteins not only in vitro but also in vivo. Overexpression of CHIP enhances E2A degradation, while reducing the level of CHIP protein by siRNA alleviates Notch-induced E47 degradation. However, Notch signaling does not appear to induce E2A degradation by up-regulation of CHIP gene expression, as cellular levels of CHIP mRNA did not change in the presence or absence of Notch signaling (data not shown). It remains to be determined if Notch induces the expression of other factors involved in E2A ubiquitination and degradation. Nevertheless, it is evident that CHIP is intimately involved in the ubiquitination process of E2A proteins, and knowledge about its mechanism of action will contribute to an understanding of Notch-induced E2A degradation.
Ubiquitination of proteins destined for degradation is mostly accomplished through cooperation of three enzymes with ubiquitin-activating, -conjugating, and -ligating activities, namely E1, E2, and E3 (35). It is believed that in mammalian cells a single E1 enzyme exists to serve the activating function for several E2 enzymes. The large number and diversity of E3 ligase complexes are thought to be responsible for substrate specificity, for example, by specific interaction between the F-box-containing component of the SCF complex and the substrate (12, 20). Even so, it would not seem economical to assign thousands of E3 ligase complexes to the same number of proteins to be ubiquitinated so that each substrate would have its own E3 ligase. Additional layers of complexity could be provided by forming PUCs, which may augment substrate specificity or affinity between substrates and E3 ligases. We propose the presence of one such complex for the E47 substrate which consists of E47, CHIP, Hsc70, and Skp2 (Fig. 7).
The purpose of forming PUC for E12/E47 is probably to facilitate their interaction with the substrate-recognizing subunit of the SCF complex, Skp2. Because of the relatively low abundance of E12/E47 and Skp2, the opportunity for them to interact with each other would be significantly increased if they independently bound to the scaffold provided by CHIP and Hsc70. CHIP mutants that fail to mediate the formation of PUC, such as the K31A and H261A-P270A mutants, are incapable of stimulating E47 degradation (Fig. 4). However, PUC would be rather indiscriminate, because CHIP is able to bind to E47 with or without its phosphorylation by MAP kinases, which is known to be required for high-affinity binding of E47 to Skp2 and ubiquitination of E47. Therefore, phosphorylation of E47 might induce a conformational change in PUC to allow a better fit between E47 and Skp2, preparing them to enter the E3 ligase complex. Furthermore, the affinity of CHIP to Cul1 may also enable CHIP to introduce the E47-containing PUC to the SCF complex and promote the ubiquitination of E47. Cul1 appears to play an important role in assembling the SCF complex, since overexpression of Cul1 not only increased the amount of Cul1 coprecipitated with CHIP but also those of other components of the SCF complex. Taken together, CHIP, through its multiple interfaces with Hsc70, E47, and Cul1, plays a crucial role in the ubiquitination process of the E47 transcription factor. We propose that CHIP mediates the formation of PUC and SCFskp2 based on results from overexpression and coimmunoprecipitation studies. The confirmation for the existence of these complexes awaits elaborate biochemical fractionation.
A similar cast of characters as those described here has also been shown to be involved in the ubiquitination of the Pael receptor by Parkin (16). Parkin is a ring-finger-containing subunit of the E3 ubiquitin ligase, whose mutation is responsible for juvenile Parkinson's disease (22). In this case, CHIP and Hsp70/Hsc70 interact with Parkin and its substrate, the membrane-bound Pael-R. Similar to its role in E47 ubiquitination, CHIP enhances the ubiquitin ligase function of Parkin. However, there are several important differences. First, Parkin interacts with either CHIP or Hsp70/Hsc70 mutually exclusively, while Skp2 only binds to Hsc70 but not CHIP. Second, CHIP associates with E47 but not Pael-R. Finally, the U-box domain is necessary for Parkin interaction, while the TPR domain of CHIP is required for binding to E47. Consequently, these differences lead to different mechanisms of action proposed for CHIP: CHIP displaces Hsp70/Hsc70 to take misfolded Pael-R from the endoplasmic reticulum (ER) to ubiquitination and degradation. A similar mechanism has also been ascribed to the degradation of the immature cystic fibrosis transmembrane conductance regulator (26). Therefore, it has been established that CHIP plays an important role in quality control for proteins synthesized and matured through the ER (8). In contrast, CHIP, together with Hsc70, facilitates the ubiquitination of the nuclear protein E47 by forming a PUC before joining the SCF-E3 ligase complex. Production of E47 does not involve ER, and E47 does not directly bind to Hsc70 or Hsp90 (data not shown). It is likely that the E47-containing PUC forms in the nucleus (Fig. 1C). E47 is the first nuclear protein shown to associate with CHIP, even though other transcriptional regulators, such as Smads, also bind to CHIP (24). However, these proteins stay in the cytoplasm and only translocate to the nucleus upon activation.
Despite the ubiquitin ligase activity associated with its U-box domain, CHIP did not act as an E3 ligase for E47 because the U-box domain is dispensable for its function in stimulating E47 ubiquitination and degradation. This is an unusual feature of CHIP, which appears to act more as an adaptor than as an enzyme to enhance the efficiency of E2A ubiquitination. Intriguingly, although deletion of the U-box domain did not alter its effect on E47, point mutations in the U-box domain which abolished the ubiquitin ligase activity disrupted the interaction between CHIP and E47. This raises an interesting question as to whether the ubiquitin ligase activity of CHIP, through autoubiquitination, augments its own conformation and consequently controls its ability to interact with different proteins. The answer to this question awaits further investigation.
Acknowledgments
We acknowledge Stuart Robinson for participating in the yeast two-hybrid library screen as a summer research student. We are grateful to S. Scott Perry and Darryll Dudley for critical reading of the manuscript.
The work was supported by grants from the National Institutes of Health to X.-H.S. (AI33597, CA77553, AI056129, and RR15577 [COBRE award]). Z.H. was funded by training grant 1T32-AI07633-01A1 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Alberti, S., J. Demand, C. Esser, N. Emmerich, H. Schild, and J. Hohfeld. 2002. Ubiquitylation of BAG-1 suggests a novel regulatory mechanism during the sorting of chaperone substrates to the proteasome. J. Biol. Chem. 277:45920-45927. [DOI] [PubMed] [Google Scholar]
- 2.Allman, D., J. A. Punt, D. J. Izon, J. C. Aster, and W. S. Pear. 2002. An invitation to T and more: notch signaling in lymphopoiesis. Cell 109(Suppl.):S1-S11. [DOI] [PubMed] [Google Scholar]
- 3.Bain, G., I. Engel, E. C. Robanus Maandag, H. P. te Riele, J. R. Voland, L. L. Sharp, J. Chun, B. Huey, D. Pinkel, and C. Murre. 1997. E2A deficiency leads to abnormalities in αβ T-cell development and to rapid development of T-cell lymphomas. Mol. Cell. Biol. 17:4782-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bain, G., E. C. Robanus Maandag, D. J. Izon, D. Amsen, A. M. Kruisbeek, B. Weintraub, I. Krop, M. S. Schlissel, A. J. Feeney, M. van Roon, M. van der Valk, H. P. J. te Riele, A. Berns, and C. Murre. 1994. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell 79:885-892. [DOI] [PubMed] [Google Scholar]
- 5.Ballinger, C. A., P. Connell, Y. Wu, Z. Hu, L. J. Thompson, L. Y. Yin, and C. Patterson. 1999. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 19:4535-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barndt, R. J., M. Dai, and Y. Zhuang. 2000. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol. Cell. Biol. 20:6677-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connell, P., C. A. Ballinger, J. Jiang, Y. Wu, L. J. Thompson, J. Hohfeld, and C. Patterson. 2001. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 3:93-96. [DOI] [PubMed] [Google Scholar]
- 8.Cyr, D. M., J. Hohfeld, and C. Patterson. 2002. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem. Sci. 27:368-375. [DOI] [PubMed] [Google Scholar]
- 9.Dai, Q., C. Zhang, Y. Wu, H. McDonough, R. A. Whaley, V. Godfrey, H. H. Li, N. Madamanchi, W. Xu, L. Neckers, D. Cyr, and C. Patterson. 2003. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 22:5446-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demand, J., S. Alberti, C. Patterson, and J. Hohfeld. 2001. Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr. Biol. 11:1569-1577. [DOI] [PubMed] [Google Scholar]
- 11.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791-803. [DOI] [PubMed] [Google Scholar]
- 12.Harper, J. W. 2001. Protein destruction: adapting roles for Cks proteins. Curr. Biol. 11:R431-R435. [DOI] [PubMed] [Google Scholar]
- 13.Hatakeyama, S., M. Yada, M. Matsumoto, N. Ishida, and K. I. Nakayama. 2001. U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 276:33111-33120. [DOI] [PubMed] [Google Scholar]
- 14.Heemskerk, M. H., B. Blom, K. Oda, A. P. Stegmann, A. Q. Bakker, K. Weijer, P. C. Res, and H. Spits. 1997. Inhibition of T cell and promotion of natural killer cell development by the dominant negative helix loop helix factor Id3. J. Exp. Med. 186:1597-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirano, T., N. Kinoshita, K. Morikawa, and M. Yanagida. 1990. Snap helix with knob and hole: essential repeats in S. pombe nuclear protein nuc2+. Cell 60:319-328. [DOI] [PubMed] [Google Scholar]
- 16.Imai, Y., M. Soda, S. Hatakeyama, T. Akagi, T. Hashikawa, K. I. Nakayama, and R. Takahashi. 2002. CHIP is associated with Parkin, a gene responsible for familial Parkinson's disease, and enhances its ubiquitin ligase activity. Mol. Cell 10:55-67. [DOI] [PubMed] [Google Scholar]
- 17.Jackson, P. K., and A. G. Eldridge. 2002. The SCF ubiquitin ligase: an extended look. Mol. Cell 9:923-925. [DOI] [PubMed] [Google Scholar]
- 18.Jiang, J., C. A. Ballinger, Y. Wu, Q. Dai, D. M. Cyr, J. Hohfeld, and C. Patterson. 2001. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem. 276:42938-42944. [DOI] [PubMed] [Google Scholar]
- 19.Jiang, J., D. Cyr, R. W. Babbitt, W. C. Sessa, and C. Patterson. 2003. Chaperone-dependent regulation of endothelial nitric-oxide synthase intracellular trafficking by the co-chaperone/ubiquitin ligase CHIP. J. Biol. Chem. 278:49332-49341. [DOI] [PubMed] [Google Scholar]
- 20.Karin, M., and Y. Ben Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 21.Kim, D., X. C. Peng, and X. H. Sun. 1999. Massive apoptosis of thymocytes in T-cell-deficient Id1 transgenic mice. Mol. Cell. Biol. 19:8240-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitada, T., S. Asakawa, N. Hattori, H. Matsumine, Y. Yamamura, S. Minoshima, M. Yokochi, Y. Mizuno, and N. Shimizu. 1998. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392:605-608. [DOI] [PubMed] [Google Scholar]
- 23.Koegl, M., T. Hoppe, S. Schlenker, H. D. Ulrich, T. U. Mayer, and S. Jentsch. 1999. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96:635-644. [DOI] [PubMed] [Google Scholar]
- 24.Li, L., H. Xin, X. Xu, M. Huang, X. Zhang, Y. Chen, S. Zhang, X. Y. Fu, and Z. Chang. 2004. CHIP mediates degradation of Smad proteins and potentially regulates Smad-induced transcription. Mol. Cell. Biol. 24:856-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massari, M. E., and C. Murre. 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20:429-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meacham, G. C., C. Patterson, W. Zhang, J. M. Younger, and D. M. Cyr. 2001. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 3:100-105. [DOI] [PubMed] [Google Scholar]
- 27.Morrow, M. A., E. W. Mayer, C. A. Perez, M. Adlam, and G. Siu. 1999. Overexpression of the helix-loop-helix protein Id2 blocks T cell development at multiple stages. Mol. Immunol. 36:491-503. [DOI] [PubMed] [Google Scholar]
- 28.Murata, S., Y. Minami, M. Minami, T. Chiba, and K. Tanaka. 2001. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2:1133-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murre, C., P. S. McCaw, and D. Baltimore. 1989. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD and myc proteins. Cell 56:777-783. [DOI] [PubMed] [Google Scholar]
- 30.Nie, L., M. Xu, A. Vladimirova, and X. H. Sun. 2003. Notch-induced E2A ubiquitination and degradation are controlled by MAP kinase activities. EMBO J. 22:5780-5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nikolay, R., T. Wiederkehr, W. Rist, G. Kramer, M. P. Mayer, and B. Bukau. 2004. Dimerization of the human E3 ligase CHIP via a coiled-coil domain is essential for its activity. J. Biol. Chem. 279:2673-2678. [DOI] [PubMed] [Google Scholar]
- 32.Ordentlich, P., A. Lin, C. P. Shen, C. Blaumueller, K. Matsuno, S. Artavanis-Tsakonas, and T. Kadesch. 1998. Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol. Cell. Biol. 18:2230-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park, S. T., and X. H. Sun. 1998. The Tal1 oncoprotein inhibits E47-mediated transcription. Mechanism of inhibition. J. Biol. Chem. 273:7030-7037. [DOI] [PubMed] [Google Scholar]
- 34.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 36.Pui, J. C., D. Allman, L. Xu, S. DeRocco, F. G. Karnell, S. Bakkour, J. Y. Lee, T. Kadesch, R. R. Hardy, J. C. Aster, and W. S. Pear. 1999. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 11:299-308. [DOI] [PubMed] [Google Scholar]
- 37.Radtke, F., A. Wilson, and H. R. MacDonald. 2004. Notch signaling in T- and B-cell development. Curr. Opin. Immunol. 16:174-179. [DOI] [PubMed] [Google Scholar]
- 38.Rezaie, A. R., M. M. Fiore, P. F. Neuenschwander, C. T. Esmon, and J. H. Morrissey. 1992. Expression and purification of a soluble tissue factor fusion protein with an epitope for an unusual calcium-dependent antibody. Protein Expr. Purif. 3:453-460. [DOI] [PubMed] [Google Scholar]
- 39.Schulman, B. A., A. C. Carrano, P. D. Jeffrey, Z. Bowen, E. R. Kinnucan, M. S. Finnin, S. J. Elledge, J. W. Harper, M. Pagano, and N. P. Pavletich. 2000. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408:381-386. [DOI] [PubMed] [Google Scholar]
- 40.Sikorski, R. S., M. S. Boguski, M. Goebl, and P. Hieter. 1990. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell 60:307-317. [DOI] [PubMed] [Google Scholar]
- 41.Sui, G., C. Soohoo, E. B. Affar, F. Gay, Y. Shi, W. C. Forrester, and Y. Shi. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 99:5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun, X.-H. 1994. Constitutive expression of the Id1 gene impairs mouse B cell development. Cell 79:893-900. [DOI] [PubMed] [Google Scholar]
- 43.Sun, X.-H., and D. Baltimore. 1991. An inhibitory domain of E12 prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell 64:459-470. [DOI] [PubMed] [Google Scholar]
- 44.Wiederkehr, T., B. Bukau, and A. Buchberger. 2002. Protein turnover: a CHIP programmed for proteolysis. Curr. Biol. 12:R26-R28. [DOI] [PubMed] [Google Scholar]
- 45.Xu, W., M. Marcu, X. Yuan, E. Mimnaugh, C. Patterson, and L. Neckers. 2002. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc. Natl. Acad. Sci. USA 99:12847-12852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhuang, Y., P. Cheng, and H. Weintraub. 1996. B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes, E2A, E2-2, and HEB. Mol. Cell. Biol. 16:2898-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhuang, Y., P. Soriano, and H. Weintraub. 1994. The helix-loop-helix gene E2A is required for B cell differentiation. Cell 79:875-884. [DOI] [PubMed] [Google Scholar]