Abstract
Previously, we identified PHR1 as an abundantly expressed gene in photoreceptors and showed that it encodes four isoforms, each with N-terminal pleckstrin homology (PH) and C-terminal transmembrane domains. To better understand PHR1 function and expression, we made a Phr1 null mouse by inserting a β-galactosidase/neor cassette into exon 3. In addition to photoreceptors, we found abundant expression of specific Phr1 splice forms in olfactory receptor neurons and vestibular and cochlear hair cells. We also found Phr1 expression in cells with a possible sensory function, including peripheral retinal ganglion cells, cochlear interdental cells, and neurons of the circumventricular organ. Despite this discrete expression in known and putative sensory neurons, mice lacking PHR1 do not have overt sensory deficits.
To clone genes exclusively or preferentially expressed in retina, our investigators performed a differential hybridization screen of an arrayed human retinal cDNA library (38, 39). Among the several genes identified, we focused on one, PHR1, which we found to be highly expressed in retina and brain. Our group showed that in humans and mice, PHR1 uses two promoters and alternative splicing of an internal exon to produce four transcripts in a tissue-specific manner (46). The PHR1 5′ promoter appeared to be photoreceptor specific, controlling the expression of transcripts 1 and 2, which differ only by the presence (transcript 1) or absence (transcript 2) of the alternatively spliced exon 7. A second PHR1 promoter in intron 2 directs expression of transcripts 3 and 4, both encoding proteins lacking the N-terminal 19 amino acids of the products of transcripts 1 and 2 and containing or lacking the sequence encoded by exon 7, respectively. All four PHR1 isoforms have a pleckstrin homology (PH) domain at their N terminus and a transmembrane domain at their C terminus. In the retina, PHR1 is an abundant integral membrane protein in the outer segments of the photoreceptors. Using in vitro binding assays, our investigators showed that the PH domain of PHR1 does not bind any of several inositol phosphates (IP), including 1-IP, 1,4-IP2, 1,4,5-IP3, and 1,3,4,5-IP4, nor several phosphatidylinositol phosphates (PIPs), but it does exhibit specific binding to transducin βγ subunits (46). On the basis of these results, our group suggested that PHR1 might play a role in modifying signal transduction in the photoreceptors.
PH domains are compact protein modules formed by sequences of 100 to 120 amino acids (17, 24, 29, 32, 36). Despite only 10 to 30% sequence identity, they have a common tertiary structure (8, 23-25). The N-terminal sequence comprises seven antiparallel β-strands, with the first four and the last three arranged in two β-sheets, oriented with respect to each other at an angle of about 60°, forming a β-sandwich structure. The C-terminal sequence of the PH domain forms an α-helix that folds over and “closes off” the wider end of the β-sandwich (8, 36). Proteins containing PH domains participate in cellular signaling, cytoskeleton organization, and other processes (23, 25, 28) and are found in a wide variety of species, from yeast to mammals. In Saccharomyces cerevisiae, there are 27 PH domain-containing proteins, while the draft human genome sequence has 252 different human proteins containing at least one PH domain, making it the 11th most common motif in the human proteome (19, 25). Although the function of most PH domains is uncertain, several have been shown to mediate reversible association of their host protein to cellular membranes binding PIPs and/or Gβγ subunits on the membrane (21, 24, 28, 35). In this regard, PHR1 is atypical in two ways. First, with its C-terminal transmembrane domain, PHR1 appears to be integrated into the membrane by a mechanism independent of its PH domain. As far as we know, PHR1 is the only integral membrane protein containing a PH domain. Second, despite extensive in vitro binding studies, neither we (46; S. Xu and D. Valle, unpublished data) nor others (22) have been able to identify binding of the PH domain of PHR1 to PIPs or IPs.
Other groups using different nomenclature have also reported studies of PHR1 (3, 22). Krappa et al. identified PHR1 and a homolog with 40% amino acid identity, PHR2, which they designated evectin 1 and 2, respectively (22). RNA blotting showed high expression of Phr1 in retina and brain, with more general expression of Phr2. In situ hybridization suggested that Phr1 expression was prominent in photoreceptors, oligodendrocytes, and Schwann cells. Based on this expression pattern and the results of a pulse-labeling experiment in isolated frog photoreceptors, Krappa et al. suggested that PHR1 was a mediator of post-Golgi protein trafficking in cells that produce large amounts of membrane. Andrews et al. identified PHR1, which they designated KPL1, as a gene whose expression increased dramatically when rat tracheal epithelial cells were grown under conditions that stimulate ciliogenesis (3). RNA blotting showed the highest expression in the brain, with lower but detectable levels in liver, spleen, trachea, and lung (the retina was not tested).
To extend our studies of PHR1 expression and function, we produced a Phr1 knockout-knockin mouse, disrupting expression of Phr1 and introducing a β-galactosidase (β-Gal) gene downstream of the Phr1 promoters. Here, we describe the cellular expression pattern of Phr1 by using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) histochemistry and phenotypic studies on Phr1β-Gal/β-Gal mice.
MATERIALS AND METHODS
Phr1 targeting construct.
We cloned and sequenced a 10.6-kb segment (AF071001) of the murine Phr1 structural gene extending from 1.3 kb 5′ of the transcription start site to intron 6 containing two HindIII fragments, a 5′ 7.5-kb and a 3′ 3.1-kb fragment (subclones pH7.5 and pH3.1 in pBluescript [BioCrest/Stratagene, Cedar Creek, Tex.]). To create the 5′ arm of the construct, we made a primer (ClaI F) by creating a ClaI site, 5′-TTGAGACTCATCGATAGACTGAGACA-3′, corresponding to sequence about 3.1 kb 5′ of exon 3 and amplified with a reverse primer imperfectly complementary to exon 3, and by creating a XhoI site (KO-4, 5′-GCCCCTCTCGAGGGCCATTTCT). We made a forward primer, KO-3, complementary to primer KO-4, containing the XhoI site (5′-GAAATGGCCCTCGAGAGGGGCG-3′). Using KO-3 and the T7 primer of the vector of H7.5, we amplified a modified fragment containing exon 3 with an XhoI site on the 5′ end and the 3′ HindIII site of the clone pH7.5 at its 3′ end. Using the introduced restriction sites, we ligated and cloned the two amplified fragments as a single fragment with an engineered XhoI site at the targeting site in exon 3 in a pBK-CMV (ClaI/HindIII) vector (BioCrest/Stratagene). To develop the 3′ arm of the construct, we designed a reverse primer, KO-2 (5′-AGTTATCCGAATTCAGCTGGGGAG-3′), creating an EcoRI site, about 700 bp into intron 5. Using KO-2 and the vector T3 primer of pH 3.1, we amplified a modified fragment, extending from the 3′ end of intron 3 to intron 5, with an EcoRI site corresponding to sequence at the 3′ end. We cloned this HindIII/EcoRI fragment into pBK-CMV. Using a ClaI/HindIII double digestion, we released the left arm and ligated it to the right arm to produce the construct pKO-10. We obtained a β-Gal/neor cassette plasmid, ploxpSAβgalneo (modified from pSAβgal, a gift from S. Cole and R. Reeves, Johns Hopkins University School of Medicine, Baltimore, Md.) (11, 12). We released the β-Gal/neor cassette with XhoI digestion and ligated it into the XhoI site of KO-10 to produce the finished targeting construct, pKO-12.
ES cells, homologous recombination, and blastocyst injections.
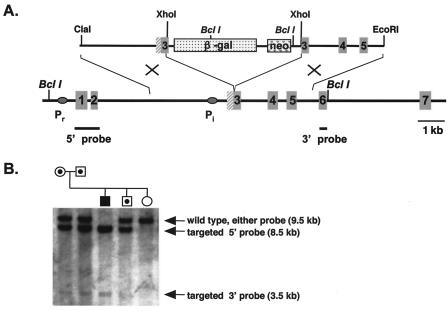
We transfected R1 cells (from J. Rossant, Mount Sinai Hospital, Toronto, Ontario, Canada) and J-1 cells (from A. Lawler, Johns Hopkins University School of Medicine) with ∼30 μg of the AsuII-linearized pPhr1KO-12-β-gal by electroporation (Gene Pulser; Bio-Rad, Melville, N.Y.) in two separate experiments. We selected G418-resistant embryonic stem (ES) cell colonies as described elsewhere (44). We picked 302 colonies and screened them for homologous recombination by digesting the DNA with BclI and hybridizing Southern blots with the indicated 5′ and 3′ probes. The 5′ probe was a 757-bp genomic fragment covering Phr1 exon 1 and exon 2 amplified with the forward primer H7.5-6 (5′CAGAGAATGAGTTAAAGGCAC-3′) and reverse primer H7.5-8 (5′-GTCGCTACTTTGACTAGCTC-3′). This probe detects an 8.0-kb fragment in the targeted allele and a 9.5-kb fragment in the wild-type allele. The 3′ probe was a 258-bp genomic fragment complementary to exon 6 amplified with a forward primer, KO-1 (5′-CTCCCCAGCTGAATTCGGATAACT-3′), and reverse primer, M-R-14 (5′-GTGGAATTTGCCTCCATCAG-3′). This probe detects a 3.5-kb fragment in targeted alleles and a 9.5-kb fragment in the wild-type allele. Two ES cell clones, one from the R1 cells (1E7) and one from J-1 cells (II-C-12), demonstrated homologous recombination. We injected cells from each clone into C57BL/6J blastocysts to produce two lines of chimeric mice in the Transgenic Facility of the Johns Hopkins University School of Medicine. For the studies reported here, we used mice derived from the 1E7 clone. We obtained similar expression and phenotypic results in mice derived from the II-C-12 clone (data not shown).
Breeding and manipulation of knockout mice.
We identified male chimeric mice by coat color and bred them to female C57BL/6J mice (Jackson Labs, Bar Harbor, Maine) to generate mice heterozygous for the targeted Phr1 gene (F1) and bred these to produce homozygotes. All animal breeding and manipulations were carried out according to National Institutes of Health guidelines at the Johns Hopkins University School of Medicine.
DNA preparation and genotyping.
We followed the protocol described by Wang et al. (42) for mouse tail genomic DNA extraction. Genotyping was performed either by Southern blot hybridization as described above or by PCR. For the PCR assay, the wild-type allele was amplified with H7.5-23 and H7.5-2 (H7.5-23, 5′-GCAGGAGCAGAGCCTTAGG-3′; H7.5-2, 5′-AGGCCAACTAGGGCTACATG-3′), producing a 794-bp fragment; the targeted allele with H7.5-23/galneo-1 (galneo-1, 5′-TCATCAAGCTTATCGATACCG-3′) produced a 474-bp product. The PCR conditions were 95°C for 30 s, 60°C for 60 s, and 70°C for 60 s for 30 cycles.
β-Gal staining.
We used a protocol described by Mombaerts et al. (31) for β-Gal staining. Briefly, fresh tissues or tissues from animals perfused with 4% paraformaldehyde were incubated in 0.1 M phosphate buffer, pH 7.4, for 15 to 30 min at 4°C on ice, rinsed in buffer A (0.1 M phosphate buffer [pH 7.4], 2 mM MgSO4, 5 mM EGTA) at room temperature for 5 min, incubated in a fresh aliquot of the same buffer for 25 min, and soaked in buffer B (0.1 M phosphate buffer [pH 7.4], 2 mM MgSO4, 0.01% sodium deoxycholate, 0.02% NP-40) at room temperature twice for 5 min. The prepared tissue was stained in buffer C [0.1 M phosphate buffer (pH 7.4), 2 mM MgSO4, 0.01% sodium deoxycholate, 0.02% NP-40, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 1 mg of X-Gal/ml] at 37°C for 2 h to overnight. After staining, the tissue was incubated in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 1 h and washed with 1× PBS to complete the fixation.
Tissue sections affixed to glass slides were incubated in 4% paraformaldehyde for 10 min at room temperature, washed three times with 1× PBS plus (PBS plus 2 mM MgCl2), and stained in buffer C at 37°C for 2 h to overnight. The sections were then postfixed with 4% paraformaldehyde for 10 min before mounting coverslips. Where indicated, sections were counterstained with hematoxylin-eosin or eosin alone.
ERG recordings.
Mouse electroretinograms (ERGs) were obtained with an EPIC-2000 system (LKC Technologies Inc., Gaithersburg, Md.) as described previously (43).
Single-cell electrophysiology of rod photoreceptors.
Single-cell electrophysiology was as described by Yang et al. (47). Briefly, the mice were dark adapted overnight and killed by CO2 asphyxiation under dim red light. All subsequent procedures were performed under infrared light. The retina was isolated from the enucleated eye in chilled, oxygenated Leibovitz's L-15 medium (Invitrogen, Grand Island, N.Y.) and placed photoreceptor-side up on a glass capillary array (10-μm-diameter capillaries; Galileo Electro-Optics, Sturbridge, Mass.) on which the retina was held by suction, allowing the vitreous humor to be removed by slicing with a razor blade between the retina and the array. The retinal pieces were stored in L-15 medium on ice until use. When needed, a piece of retina was chopped under L-15 medium containing 8 μg of DNase (Sigma Chemical, St. Louis, Mo.)/ml with a razor blade mounted on a lever arm, and a suspension of small retinal fragments was transferred into the recording chamber. The chamber temperature was held at 36 to 38°C by continuously perfusing it with heated solution buffered with bicarbonate and bubbled with 95% O2-5% CO2, pH 7.4. The outer segment of an isolated rod or a rod projecting from a small fragment of retina was drawn into a suction electrode connected to a current-to-voltage converter. The recorded membrane current was filtered with a low-pass, eight-pole Bessel filter at 30 Hz and digitized.
The suction electrode was filled with a solution containing (in mM): 134.5 Na+, 3.6 K+, 2.4 Mg2+, 1.2 Ca2+, 136.3 Cl−, 3 succinate, 3 l-glutamate, 10 glucose, 10 HEPES, and 0.02 EDTA plus basal medium Eagle amino acid supplement and vitamin supplement (Invitrogen). The perfusion medium was the same except that 20 mM NaHCO3 replaced an equimolar amount of NaCl.
Auditory brain stem responses (ABR).
Using anesthetized mice, we placed electrodes at the vertex (active), bilaterally in the neighborhood of the postauricular bullae, and in the forehead (ground) as described by Xiang et al. (45). The acoustic stimulus consisted of a click of approximately 0.1 ms in duration at a rate of 11.4 per s. Responses were averaged over 1,000 stimuli. Auditory thresholds were determined by visual inspection of response traces obtained at stimulus intervals of 5 dB. Absolute stimulus intensities were calibrated to obtain the sound pressure level.
EOG recordings.
We followed the protocol described by Zhao et al. (48, 49) for electro-olfactogram (EOG) recordings. Briefly, stock odorant solution in 0.5 M dimethyl sulfoxide was diluted to the indicated concentration in water. The diluted solution (2 ml) was placed in a 10-ml glass test tube about 2 h before the experiments to allow the odorant to equilibrate in the vapor phase. The vapor was delivered as a 0.1-s pulse into a continuous stream of humidified air flowing over the tissue surface. The EOGs were initially recorded using amyl acetate as odorant at 10−6, 10−5, 10−4, and 10−3 M. Five additional odorants (carvone, cineole, citral, hexanal, and heptanol) were tested at 10−4 M. The magnitudes of the EOG responses were normalized to the response to 10−4 M amyl acetate measured at the beginning, midpoint, and completion of the experiment. This response declined by less than 10% over the course of the experiment.
Tyrosine hydroxylase detection.
As described by Baker et al. (4), we perfused anesthetized mice with 1× PBS for 5 min followed by 4% paraformaldehyde for 20 min and then removed and fixed the brain with 4% paraformaldehyde for another 2 h. After washing with 1× PBS, the brain was transferred to 30% sucrose in PBS overnight and then embedded in OCT (Tissue-Tek). We made coronal sections at 15 to 20 μm. The tyrosine hydroxylase was detected by rabbit anti-tyrosine hydroxylase polyclonal antibody (1:500; Chemicon, Temecula, Calif.) as primary antibody and biotinylated goat anti-rabbit immunoglobulin G as secondary antibody (1:200) in the Vectastain and DAB system (Vector, Burlingame, Calif.) according to the manufacturer's instructions.
RESULTS
The Phr1 targeting construct.
We utilized a 7-kb fragment of the Phr1 structural gene to assemble a targeting construct with a β-Gal/neor cassette inserted into exon 3 following the L22 codon (Fig. 1). The initiation methionine (M1) for photoreceptor-specific PHR1 isoforms is encoded in exon 2. A second methionine, M20, serves as the initiator for isoforms 3 and 4. Thus, insertion of the β-Gal/neor cassette following the L22 codon disrupts all four PHR1 isoforms and would be anticipated to bring the introduced β-Gal gene under the control of the native retina- and brain-specific Phr1 promoters. The next in-frame Phr1 methionine codon is M121. Initiation at this position would delete the N-terminal half of PHR1, including the PH domain. The β-Gal cassette has its own translation initiation site, 266 bp downstream of its 5′ end.
FIG. 1.
Targeted disruption of Phr1. (A) The targeting construct (top) and targeted region of Phr1 (bottom) are shown. We inserted the β-Gal/neor cassette into exon 3 of Phr1 following codon L22. The neor cassette includes a 5′ PGK promoter and is transcribed with the same orientation as Phr1. The black ovals indicate the location of the 5′ photoreceptor-specific (Pr) and the internal, general (Pi) promoter of Phr1. The positions of the 5′ and 3′ probes used for Southern blot screening of potential recombinant ES cell clones are shown below the Phr1 structural gene as solid black lines. (B) Southern blot, hybridized with both the 5′ and 3′ probes, of BclI-digested genomic DNA from parents heterozygous for the targeted Phr1 allele and their offspring. The 5′ probe recognizes an 8.5-kb fragment in the targeted allele and a 9.5-kb fragment in the wild-type allele. The 3′ probe detects a 3.5-kb fragment in the targeted allele and a 9.5-kb fragment in the wild-type allele. The filled symbol indicates a homozygote for the targeted allele, a symbol with a central black dot indicates a heterozygote for the targeted allele, and an open symbol indicates a homozygote for the wild-type allele.
Phr1 is highly expressed in primary sensory neurons.
We found intense β-Gal staining in the photoreceptors of Phr1β-Gal/+and Phr1β-Gal/β-Gal animals and in some cells in the retinal ganglion cell layer (Fig. 2). There was no staining in pigment epithelium (data not shown). The agreement of these results with those of our previous in situ hybridization and immunohistochemical studies indicates that β-Gal expression from the targeted Phr1 allele recapitulates the normal Phr1 expression pattern (46).
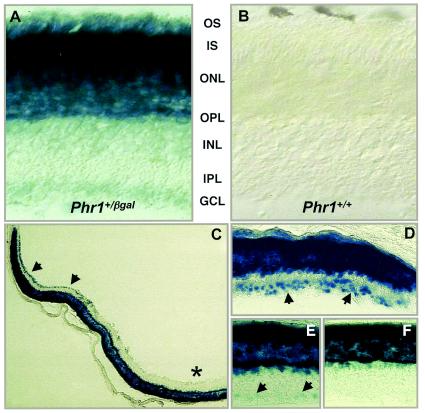
FIG. 2.
Expression of Phr1 as detected by β-Gal staining in adult mouse retina. Sections from comparable regions of retina from Phr1+/β-Gal (A) and Phr1+/+ (B) animals are shown following β-Gal staining. The retinal layers are indicated as follows: OS, photoreceptor outer segments; IS, photoreceptor inner segments; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. A lower power view of a retinal cross section (C) shows staining of cells in the peripheral (solid arrowheads) but not in the central (*) ganglion cell layer of the retina. (D to F) Higher-power views of the peripheral (D), medial (E), and central (F) retina. The arrowheads point to β-Gal-positive cells in the ganglion cell layer.
Interestingly, the pattern of β-Gal staining in the ganglion cell layer confirmed our impression from previous studies that not all cells in this layer express Phr1. To examine this in more detail, we stained retinal whole mounts and serial sections and found that the Phr1-expressing cells in the ganglion cell layer were greatly enriched in the peripheral retina and virtually absent in the central retina (Fig. 2C to F).
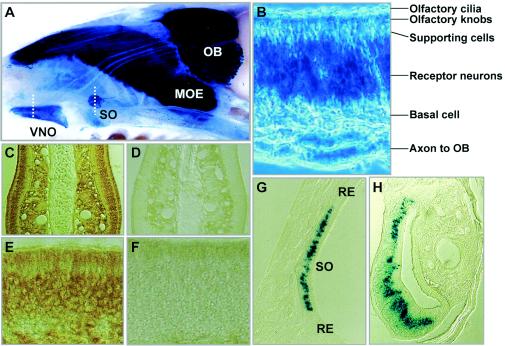
Unexpectedly, we also found that the olfactory epithelium (OE) and the olfactory bulb displayed intense β-Gal staining in Phr1β-Gal/+ and Phr1β-Gal/β-Gal animals (Fig. 3). All three patches of sensory epithelium in the nasal cavity, the major olfactory epithelium, the vomeronasal organ, and the septal organ of Masera, stained for β-Gal, as did most of the glomeruli and nerve fibers in both the main and accessory olfactory bulbs. The OE did not stain in control Phr1+/+ animals (data not shown), nor was there staining of respiratory epithelium in the Phr1β-Gal/+ animals (Fig. 3G). Thus, Phr1 is expressed in the ciliated primary sensory epithelium but not in the ciliated respiratory epithelium of the naso-pharynx.
FIG. 3.
Expression of Phr1 as detected by β-Gal staining and immunohistochemistry of olfactory structures in Phr1+/β-Gal mice. (A) Mid-sagittal view of a whole mount of the nasal septum. The dashed lines indicate the location of the sections shown in panels G and H. (B) Higher-power view of a section through the OE showing β-Gal staining of the receptor neurons and their axons. Immunohistochemical localization of PHR1 is shown in coronal sections of the nasal septum stained with anti-PHR1 antiserum (C) or preimmune serum (D) and in higher-power cross-sectional views of OE stained with anti-PHR1 antiserum (E) or preimmune serum (F). (G) Localization of β-Gal staining in a cross-section of the septal organ. Note the prominent β-Gal staining of receptor neurons but not of the surrounding respiratory epithelium. (H) Cross-section of the vomeronasal organ with β-Gal staining of the receptor neurons. Abbreviations: MOE, major olfactory epithelium; OB, olfactory bulb; RE, respiratory epithelium; SO, septal organ of Masera; VNO, vomeronasal organ.
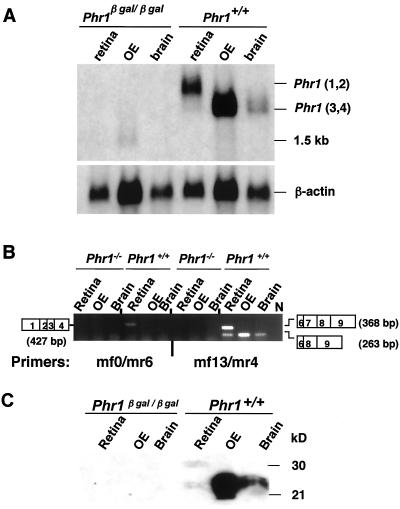
To confirm expression of Phr1 in the OE, we stained sections of the nasal cavity with anti-PHR1 antibody (Fig. 3C to F) and isolated RNA and protein from these tissues for RNA and immunoblot analysis (Fig. 4). The immunohistochemistry showed that PHR1 is present in the cell bodies and knobs of the mature olfactory receptor neurons (Fig. 3E and F). Northern blot analysis of RNA isolated from the OE showed that Phr1 mRNA was present in amounts that exceeded those in retina and that by size were transcript 3 or 4 (Fig. 4). Reverse transcription-PCR (RT-PCR) with Phr1 splice form-specific primers showed that transcript 4, driven by the internal promoter and lacking exon 7, is the major Phr1 transcript in OE (Fig. 4B). This result was consistent with immunoblotting studies showing that the short isoform of PHR1 was predominant in the OE (Fig. 4C).
FIG. 4.
The Phr1β-Gal allele is a null. (A) Northern blot of total cellular RNA (5 μg/lane) isolated from retina, OE, and brain of mice of the indicated genotypes and probed with a full-length Phr1 cDNA. Note that Phr1 transcripts were undetectable in the Phr1β-Gal/β-Gal mice and that Phr1 was highly expressed in OE of Phr1+/+ mice with a transcript size (Phr1 transcripts 3 and 4) similar to that in the brain and smaller than those (Phr1 transcripts 1 and 2) in retina. A faintly hybridizing 1.5-kb transcript was detected in OE RNA isolated from mice of both genotypes. The bottom panel shows the same filter hybridized with a β-actin probe to control for RNA quality and quantity. (B) RT-PCR analysis using RNA isolated from the indicated tissues of Phr1β-Gal/β-Gal (here designated as Phr1−/−) and Phr1+/+ mice. In Phr1+/+ mice, a primer pair (mf0 and mr6) that amplifies only Phr1 transcripts 1 and 2 detected transcript 1 in retina but not in RNA from other tissues, while a primer pair (mf13 and mr4) that detects all four Phr1 splice forms detected transcripts in RNA from all three tissues. N designates a lane in which no RT was added to an RT-PCR with retinal RNA. The open rectangles on each side show the Phr1 exons contributing to the segment of the transcript amplified. In brain and OE, transcript 4 lacking exon 7 predominates. In retina, transcript 1 including exon 7 predominates. No Phr1 transcripts were detected by either primer pair in RNA isolated from Phr1β-Gal/β-Gal mice. (C) Immunoblot analysis of extracts (50 μg of protein) of retina, OE, and brain. Abundant PHR1 was present in OE extracts, with the same size as PHR1 in brain. The loading for the retinal lane was only 5 μg. No PHR1 was detected in the tissue extracts from Phr1β-Gal/β-Gal mice.
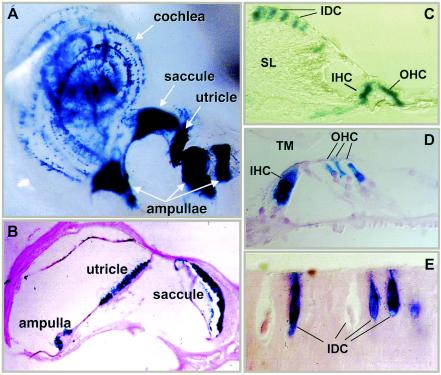
These results in photoreceptors and OE suggested that Phr1 might also be expressed in other sensory neurons. Using whole-mount staining of the inner ear of Phr1β-Gal/+ and Phr1β-Gal/β-Gal mice, we found strong β-Gal staining of the hair cells of the sensory epithelium of the cochlea and the vestibular organs, including the utricle, saccule, and ampullae (Fig. 5). RT-PCR of RNA isolated from Phr1+/+ inner ears confirmed Phr1 expression and showed that, as in the OE, Phr1 transcript 4 was predominant (data not shown). In cochlea, we also observed β-Gal staining of interdental cells at the surface of the spiral limbus facing the endolymph in the cochlear duct (Fig. 5C). Here, intensely staining interdental cells were intermixed with a similar number of nonstaining interdental cells (Fig. 5E).
FIG. 5.
Expression of Phr1 as detected by β-Gal staining in the inner ear of Phr1+/β-Gal mice. (A) β-Gal staining in a whole mount of the membranous labyrinth of a 4-week-old mouse. (B) Section through the vestibular organ of a 6-week-old mouse counterstained with eosin. (C) β-Gal staining in a section through the spiral limbus and organ of Corti of a 4-week-old mouse. (D) Higher-power view of the organ of Corti from a 6-week-old mouse, counterstained with eosin. (E) High-power view of a section through the spiral limbus of a 6-week-old mouse, showing the interdental cells, some of which are positive for β-Gal staining while others are not. Abbreviations: IDC, interdental cell; IHC, inner hair cell; OHC, outer hair cell; SL, spiral limbus; TM, tectorial membrane.
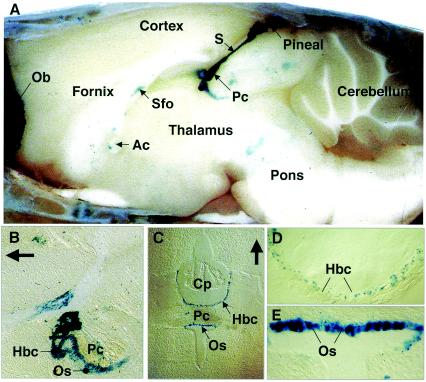
In brain, we found Phr1 expression in the pineal gland and in most, if not all, of the circumventricular organs (CVO), including the subfornical organ, subcommissural organ (SCO), and the organum vasculosum lamina terminalis (Fig. 6). Neurons in the CVO are thought to detect circulating peptide hormones, neuropeptides, and chemokines, as well as sensing changes in the osmotic pressure of the extracellular fluid (7, 16, 37). Thus, expression of Phr1 in these cells is consistent with its expression in primary sensory neurons. We also noted β-Gal staining in a few unidentified cells in the thalamus, hypothalamus, and other regions of the brain (data not shown). In contrast to the observations of Krappa et al. (22), we found no prominent staining in regions rich in oligodendrocytes.
FIG. 6.
Expression of Phr1 as detected by β-Gal staining in brain from a PHR1β-Gal/+ mouse. (A) Mid-sagittal view of a whole mount of the brain, showing dense staining of the pineal body and stalk (S). (B) Sagittal section through the region of the posterior commissure (Pc) and organus subcommisurale (Os). The arrow points rostrally. (C to E) Coronal sections through the CVO. Panels D and E are higher-power views of the structures shown in panel C with the same orientation. The arrow points dorsally. Abbreviations: Ac, anterior commissure; Cp, choroidal plexus; F, fornix; Hbc, habenular commissure; Ob, olfactory bulb; Os, organus subcommisurale; Pc, posterior commissure; Sfo, subfornical organ.
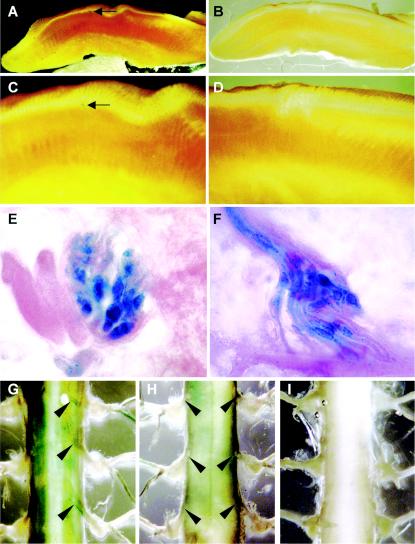
We also searched for β-Gal staining in taste buds and in the dorsal root ganglia. In tongue, we observed an occasional strongly stained taste bud, but most had no detectable staining (see the appendix, Fig. A1A to D). In contrast, the stretch receptors on the neuromuscular spindles of tongue muscle fibers were uniformly stained (see Fig. A1E and F). These structures have two or more myelinated afferent fibers entering each spindle and detect alterations in tension on the neuromuscular spindle (34). We also observed a few positive cells in the epithelia of the epiglottis, pharynx, and larynx that may represent sensory neurons in these areas (data not shown). In the dorsal roots, there was low but detectable staining of the axons but not the cell bodies, while there was no detectable staining in the ventral roots (see Fig. A1G to I). This modest β-Gal staining in the dorsal but not the ventral roots suggested low-level Phr1 expression in the sensory neurons.
FIG. A1.
Expression of Phr1 as detected by β-Gal staining of the tongue and spinal cord. Low-power (A and B) and high-power (C and D) views of sagittal sections of the tongue of Phr1β-Gal/β-Gal (A and C) and Phr+/+ (B and D) mice. The arrow in panel C points to a stained vallate papilla. (E and F) High-power views of a taste bud (E) and of the stretch receptor in the tongue (F) of a Phr1β-Gal/β-Gal animal. (G to I) Segments of thoracic spinal cord from a Phr1β-Gal/β-Gal animal (dorsal view) (G), a Phr1β-Gal/β-Gal animal (ventral view) (H), and a Phr1+/+ animal (dorsal view) (I). The black arrows indicate the stained dorsal roots in panel G and the nonstaining ventral roots in panel H.
The Phr1β-gal allele is a null.
Using RNA blotting or RT-PCR on total cellular RNA and immunoblot analyses of protein extracts from retina, OE, inner ear, and brain, we found no expression of Phr1 transcript or protein in Phr1β-Gal/β-Gal mice (Fig. 4). Thus, the targeted Phr1 allele is a null at both the mRNA and protein levels. We did detect a faintly hybridizing, smaller (∼1.5-kb) transcript in total cellular RNA isolated from the OE of both Phr1β-Gal/β-Gal and Phr1+/+ mice (Fig. 4A). Because this was present in animals of both genotypes and because we did not detect this transcript in RT-PCR experiments with primers complementary to the Phr1 open reading frame, we conclude that it represents a cross-hybridizing transcript that does not originate from the Phr1 locus.
Phenotypic assessment of Phr1β-Gal/β-Gal mice.
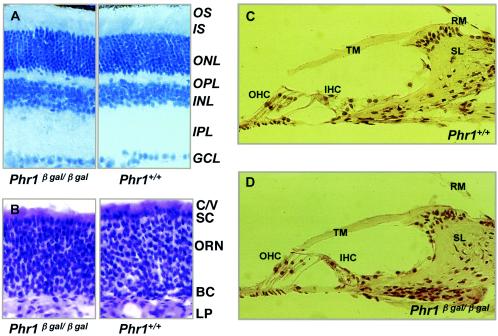
The Phr1β-Gal/β-Gal mice appeared normal at birth without obvious malformations or behavioral or growth abnormalities. The genotype of offspring of heterozygous matings fit Mendelian expectations. At ages 6 months and 1 year, we performed phenotypic studies focusing on the sensory systems with high Phr1 expression and using Phr1+/+ littermates as controls. Retinal histology and ERGs were normal, as were recordings of single rod photoreceptor recordings of cells isolated from a 2-month-old animal (Fig. 7; see also Fig. A2). Similarly, OE histology and tyrosine hydroxylase staining of the olfactory bulb, a neurotransmitter biosynthetic marker in the olfactory system (4), and EOG recordings were normal (Fig. 7; see also Fig. A3A to H). Also, rotorod tests for balance (data not shown), inner ear histology, and ABR were normal (Fig. 7; see also Fig. A4). Moreover, as a test of functions mediated by receptors in the CVO, we subjected 6-month-old Phr1β-Gal/β-Gal animals to a water deprivation test (30, 40). Their response as measured by changes in weight and water consumption at the end of 48 h without water was indistinguishable from that of controls (data not shown). Serum and urine electrolyte concentrations in animals eating and drinking ad libitum were also indistinguishable from those of controls (see Fig. A5).
FIG. 7.
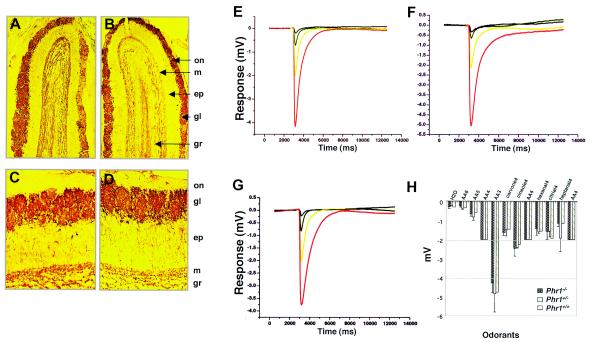
Histology of selected sensory organs. (A) Hematoxylin-eosin (HE)-stained sections of retina from 6-month-old mice of the indicated genotypes. Abbreviations: GCL, ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layer; IS, inner segment; ONL, outer nuclear layer; OPL, outer plexiform layer; OS, outer segment. (B) HE-stained sections of OE from 4-week-old mice of the indicated genotypes. In review of multiple sections from multiple animals, we saw no consistent difference between wild-type and Phr1β-Gal/-β-Gal animals. Abbreviations: C/V, olfactory cilia and microvilli of supporting cells; SC, supporting cells; ORN, olfactory receptor neurons; BC, basal cells; LP, lamina propria. (C and D) HE-stained sections of the organ of Corti from 6-month-old mice of the indicated genotypes. The sections show the basal turn of the cochlea. Similar results were observed in sections of the middle turn (data not shown). Abbreviations: IHC, inner hair cells; OHC, outer hair cells; RM, Reissner's membrane; SL, spiral limbus; TM, tectorial membrane.
FIG. A2.
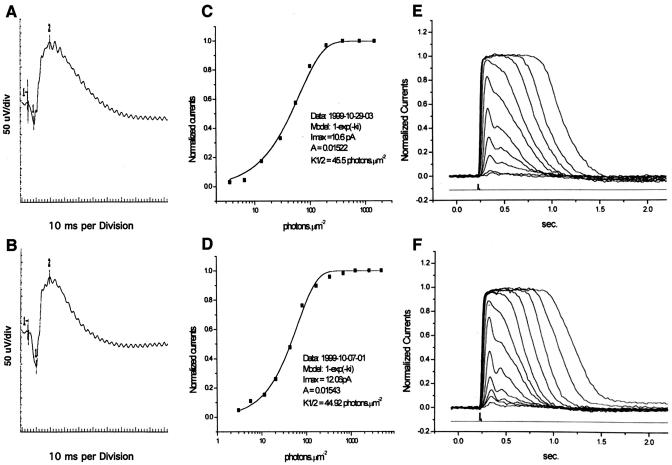
Retinal electrophysiology in Phr1β-Gal/β-Gal mice (A, C, and E) and Phr+/+ littermates (B, D, and F). (A and B) ERGs measured in 6-month-old animals. (C to F) Suction pipette recordings from single rod photoreceptors isolated from 2-month-old mice. Panels C and D show normalized responses of a rod (Phr1β-Gal/β-Gal [C] and Phr1+/+ [D]) to 500-nm flashes of increasing strength. Each trace is the averaged response from multiple flash trials (see Materials and Methods). Panels E and F show the relationship between the peak amplitude of flash response and the flash intensity for the rods shown in panels C and D.
FIG. A3.
Evaluation of olfaction in Phr1β-Gal/β-Gal mice. (A to D) Immunohistochemical localization of tyrosine hydroxylase in sections of OE from 6-week-old Phr1β-Gal/β-Gal mice (A and C) and normal littermates (B and D). Original magnification, ×50 (A and B) or ×100 (C and D). Abbreviations: ep, external plexiform layer; gl, glomerular layer; gr, granule cell layer; m, mitral cell layer; on, olfactory nerve layer. (E to G) EOG responses to 10−5(red), 10−4 (yellow), and 10−3 M (green) amyl acetate in Phr1β-Gal/β-Gal (E), Phr1β-Gal/+ (F), and Phr1+/+ (G) mice. The black curve shows the response to water as a negative control. (H) Peak responses to all odorants. AA6, AA5, AA4, and AA3 correspond to 10−6, 10−5, 10−4, and 10−3 M amyl acetate, respectively; carvone4, 10−4 M carvone; cineole4, 10−4 M cineole; hexanal4, 10−4 M hexanal; citrial4, 10−4 M citrial; heptanol4, 10−4 M heptanol.
FIG. A4.
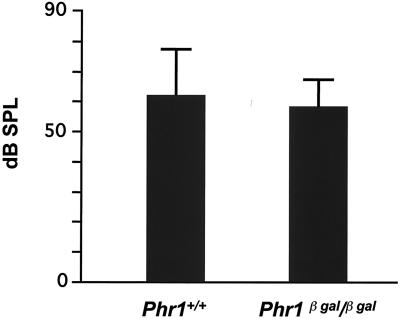
Auditory function as measured by ABR recordings for 6-month-old Phr1β-Gal/β-Gal mice and Phr+/+ littermate controls. The bars indicate the mean (± 1 standard deviation) sound pressure level (decibels) detected in three animals of the indicated genotypes.
FIG. A5.
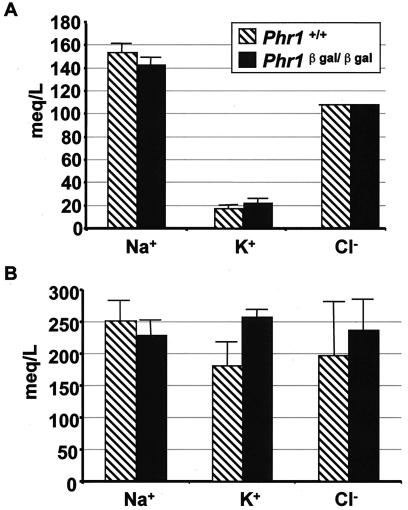
Electrolyte levels in serum (A) and urine (B) obtained from 7-month-old animals of the indicated genotypes. The heights of the bars indicate the means, and the thin lines indicate the ranges of determinations in three animals. The significance of the 43% increase in the mean urinary K+ concentration in the Phr1β-Gal/β-Gal animals is uncertain, particularly in light of the corresponding increase in serum K+.
Expression of Phr2 in Phr1β-Gal/β-Gal mice.
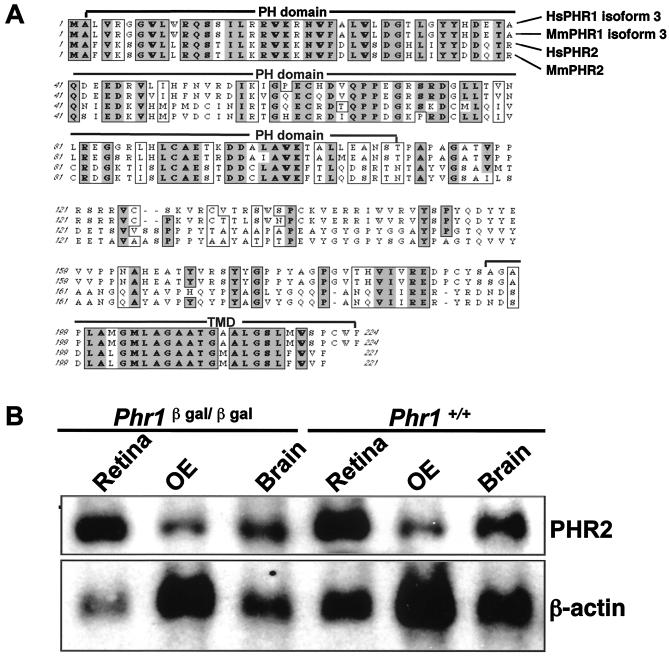
One possible explanation for the apparent lack of a phenotype in animals homozygous for targeted disruption of a specific gene is compensatory expression of another gene encoding a protein whose function is similar to that of the deficient protein. Phr2 is an obvious candidate suppressor of the phenotype in the Phr1β-Gal/β-Gal animals. Northern blotting of RNA from retina, OE, and brain, however, showed at most a modest increase in expression of Phr2 (Fig. 8). Using densitometry to quantitate the level of Phr2 expression by Northern blot analysis, we found the ratio of PHR2 to actin in brain RNA was 0.13 ± 0.02 (mean ± standard deviation) in wild-type animals (n = 4) and 0.16 ± 0.02 in Phr1β-Gal/β-Gal animals (n = 4) (+23%), while in retina RNA it was 0.23 ± 0.06 in wild-type animals (n = 2) and 0.29 ± 0.06 in Phr1β-Gal/β-Gal animals (n = 4) (+26%).
FIG. 8.
(A) Alignment of the amino acid sequences of human and murine PHR1 (isoform 3) and PHR2. Overall, murine PHR2 (isoform 3) has 38% amino acid identity with murine PHR1 isoform 3. The similarity is much higher in certain domains. For example, the N-terminal PH domains of PHR1 and PHR2 are 50% identical and 73% similar. Their C-terminal transmembrane domains (TMD) are 61% identical and 68% similar. Boxed bold or shaded letters are identical residues in three or four sequences, respectively. Boxed-only letters are similar residues. (B) Northern blot analysis of total cellular RNA (5 μg/lane) harvested from the indicated tissues from Phr1β-Gal/β-Gal and Phr1+/+ mice. A 3.2-kb Phr2 transcript is present in RNA from retina, OE, and brain, with levels in the following order: retina > brain > OE.
DISCUSSION
Evolution has produced many types of sensory neurons, each with unique structural and biochemical properties specifically suited for their function. Using a β-Gal reporter gene, we found that Phr1 is highly expressed in several of these highly specialized cells. In agreement with our previously published in situ hybridization and immunohistochemistry results (46), we observed strong β-Gal activity in the rod and cone photoreceptors, pineal body, and in some cells in the retinal ganglion cell layer of Phr1β-Gal mice. This result indicates that the expression patterns of Phr1 are preserved in our Phr1β-Gal mouse. In addition to these previously identified sites of Phr1 expression, we also found strong Phr1 expression in the receptor neurons of the OE and in the hair cells of the sensory epithelia of the inner ear. We confirmed the expression of Phr1 in these tissues by immunohistochemistry, RNA blotting, and RT-PCR studies. Our results in the inner ear were consistent with the recent report listing PHR1 among 52 genes preferentially expressed in the inner ear compared to a pool of mRNAs isolated from 29 other tissues (not including retina or OE) (1).
The major Phr1 transcript in the OE and inner ear is transcript 4, driven by the internal promoter and lacking exon 7, while in photoreceptors, transcripts 1 and 2 predominate. The latter is consistent with our group's previous conclusion that transcripts 1 and 2 are photoreceptor specific and expressed under the control of the 5′ or photoreceptor-specific Phr1 promoter. This promoter contains multiple copies of the TAATCC/A sequence recognized by CRX, a homeobox transcription factor whose expression is limited to photoreceptors and whose activity is necessary for photoreceptor development and function (13, 14, 46). Recent serial analysis of gene expression studies in the retina of mice with targeted disruption of Crx have shown that Phr1 expression is dependent on this transcription factor (6).
We also found β-Gal staining evidence for low-level Phr1 expression in dorsal root neurons, the stretch receptors of the tongue musculature, and in the neurons of at least a few taste receptors. These results indicate that Phr1 is expressed in many if not all of the classical sensory neurons, including those that utilize G-protein-coupled signal transduction systems (photoreceptors, OE, and taste receptors) and those that utilize a G-protein-independent transduction system (hair cells of the inner ear). These observations suggest that, despite our earlier in vitro observation of PHR1 binding to the βγ subunit of transducin, the function of PHR1 is something other than a modulator of G-protein-coupled signal transduction processes. Moreover, because the Phr1β-Gal/β-Gal animals did not manifest an overt phenotype at up to 1 year in age, PHR1 function clearly is not vital for sensory neuron survival.
Expression of Phr1 in cells other than classical sensory neurons is also of interest. In the retina some, but not all, cells in the ganglion cell layer express Phr1. There is a gradient of Phr1-expressing cells in the ganglion cell layer, increasing from nearly none in the central retina to nearly all in the peripheral retina (Fig. 2). In mouse retina, 40% of the cells in the ganglion cell layer are ganglion cells; most of the remainder are displaced amacrine cells (20). The density of ganglion cells decreases from central to peripheral retina (20), in a distribution inverse to that of the Phr1-expressing cells. This suggests that Phr1 expression marks a distinct cell type in the ganglion cell layer. In mammals, cells preferentially located in the peripheral ganglion cell layer include special melanopsin-containing photosensitive ganglion cells, whose function is necessary for entrainment of circadian rhythms (5, 9, 10, 18, 27). Given their overlapping distribution in the ganglion cell layer and the association of Phr1 expression with sensory neurons, it is tempting to speculate that the Phr1-expressing cells in the peripheral ganglion cell layer include the photosensitive ganglion cells; however, the density of these cells appears to be lower than those expressing Phr1 (18). Additional studies comparing the location of cells expressing melanopsin and PHR1 and the circadian behavior of Phr1β-Gal/β-Gal mice will be necessary to clarify this issue.
In the organ of Corti, we found Phr1 expression not only in the inner and outer hair cells, but also in a subset of interdental cells (Fig. 5C and E). These cells are long, spindle-shaped cells, oriented with their vertical axis perpendicular to the surface of the spiral limbus. Their functions are poorly understood, but recent studies suggest the presence of nerve fibers in the spaces between the cells (41). This observation, together with our recognition of preferential expression of Phr1 in primary sensory neurons, suggests the possibility that certain of the interdental cells may have a previously unrecognized sensory function. Moreover, Zheng and Gao (50) showed that overexpression of Math1, a mouse homolog of the Drosophila melanogaster gene atonal (2), in postnatal rat cochlear explants induces formation of extra hair cells in a region outside the sensory epithelium in the greater epithelial ridge. This is the same region that gives rise to the interdental cells in the spiral limbus. In additional studies of the organ of Corti of postnatal day 1 and 5 mice, we found that β-Gal-positive cells appeared in the greater epithelial ridge region as well as in the cochlear hair cells at this stage (Fig. 5C; S. Xu and D. Valle, unpublished observations). This result suggests the possibility that Phr1 expression marks a subset of interdental cells with the capacity to differentiate into primary sensory neurons in the organ of Corti.
We also found specific Phr1 expression in cells comprising the CVO of the mouse brain, including the pineal body, subfornical organ, organum vasculosum lamina terminalis, and SCO (Fig. 6). These cells are thought to provide an important link between the brain and the peripheral metabolic and endocrine systems (15). The CVOs are specialized structures near the surface of the third and fourth cerebral ventricle that are highly vascularized with special fenestrated capillaries (34). With the exception of the SCO, all CVOs lack a blood-brain barrier (34) and all have extensive afferent and efferent neural connections and a distinctive cytoarchitecture (33). Moreover, several peptide hormones, including angiotensin II, somatostatin, relaxin, leutinizing hormone releasing hormone, oxytocin, and vasopressin and their receptors have been detected in CVOs. For example, Liedtke et al. recently identified the vanilloid receptor-related osmotically activated channels expressed in the CVO neurons (26). These observations have lead to the hypothesis that the CVO contains special sensory neurons involved in osmoresponsive pathways, the baroreflexes, and in the control of food intake and salt and water homeostasis (15, 37). Expression of Phr1 is consistent with the hypothesized sensory function of these cells. Although we did not do extensive testing of phenotypes regulated by the CVO, we did find that Phr1β-Gal/β-Gal mice tolerated water deprivation and did not manifest abnormalities of body mass. Thus, the functional role of PHR1 in these cells is uncertain.
Finally, our Phr1 expression studies are also of interest for the cells in which we found no expression. Others have suggested, on the basis of in situ hybridization studies, that Phr1 is expressed in retinal pigment epithelium and oligodendrocytes and Schwann cells (22). We failed to find Phr1 expression in these cells. It may be that the in situ hybridization observed by Krappa et al. was to transcripts related to, but distinct from, Phr1.
Despite the specific and abundant expression of Phr1 in the primary sensory neurons, we did not detect an abnormal phenotype in the Phr1β-Gal/β-Gal mice. Expression of a related protein whose function overlaps with that of PHR1 could explain this apparent lack of phenotype. PHR2, a PHR1 homolog identified by Krappa et al. and designated EVT2 by them, is an obvious candidate for a functionally redundant suppressor of phenotype for a Phr1 null allele (22). Overall, PHR2 has 38.4% amino acid identity and 57.6% amino acid similarity to PHR1 isoform 3 (Fig. 8). In the PH domain and the C-terminal transmembrane domain, the resemblance was much higher, with 50% identity and 73% similarity in the PH domain and 61% identity and 68% similarity in the C-terminal transmembrane domain. Phr2 is expressed in many tissues, including brain, retina, heart, kidney, lung, muscle, and peripheral nerve, with the highest levels in brain and retina (22). We found that Phr2 is also expressed in OE (Fig. 8). Although these studies show that Phr2 is expressed in many of the same tissues as Phr1, they do not confirm that the two proteins are expressed in precisely the same cells. Moreover, in our Phr1β-Gal/β-Gal mice, the amounts of Phr2 mRNA were only modestly increased (∼25%) over controls (Fig. 8), suggesting that PHR2 does not compensate for the lack of PHR1. An unequivocal understanding of the role of PHR2 in mice lacking PHR1 will require creation and characterization of Phr2 mutant and Phr1/Phr2 double mutant mice.
Expression of Phr1 in the tongue and spinal cord as shown by β-Gal staining in mice homozygous to the targeted allele is shown in Fig. A1. Note the staining of an occasional taste bud and staining of neuromuscular spindles in muscle fibers of the tongue.
The remaining figures show various aspects of the phenotypes of mice homozygous for the targeted disruption of Phr1. ERG in intact 6-month-old mice and in single rod photoreceptors isolated from 2-month-old animals is shown in Fig. A2. Figure A3 shows studies to evaluate olfactory function in Phr1β-Gal/β-Gal animals, including immunohistochemical localization in OE and EOGs in response to a variety of odorants. Figure A4 shows auditory function as measured by ABR recordings in 6-month-old mice. Finally, Fig. A5 shows serum and urine electrolytes in 7-month-old animals who had ad libitum access to water and chow.
Acknowledgments
This work was supported in part by a grant from The Foundation Fighting Blindness (D.V.) and by National Institute on Deafness and Other Communication Disorders grants DC00232 (D.K.R.) and DC00276 for support for H.H. and E.G. David Valle is an Investigator in the Howard Hughes Medical Institute.
We thank Jeremy Nathans and Paul Fuchs for their comments on the manuscript and Sandy Muscelli for help with its preparation.
REFERENCES
- 1.Abe, S., T. Katagiri, A. Saito-Hisaminato, S.-I. Usami, Y. Inoue, T. Tsunoda, and Y. Nakamura. 2003. Identification of CRYM as a candidate responsible for nonsyndromic deafness, through cDNA microarray analysis of human cochlear and vestibular tissues. Am. J. Hum. Genet. 72:73-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akazawa, C., M. Ishibashi, C. Shimizu, S. Nakanshi, and R. Kageyama. 1995. A mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J. Biol. Chem. 270:8730-8738. [DOI] [PubMed] [Google Scholar]
- 3.Andrews, K., P. Potdar, P. Nettesheim, and L. E. Ostrowski. 2000. KPL1, which encodes a novel PH domain-containing protein, is induced during ciliated cell differentiation of rat tracheal epithelial cells. Exp. Lung Res. 26:257-271. [DOI] [PubMed] [Google Scholar]
- 4.Baker, H., D. M. Cummings, S. D. Munger, J. W. Margolis, L. Franzen, R. R. Reed, and F. L. Margolis. 1999. Targeted deletion of a cyclic nucleotide-gated channel subunit (OCNC1): biochemical and morphological consequences in adult mice. J. Neurosci. 19:9313-9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berson, D. M., F. A. Dunn, and M. Takao. 2002. Phototransduction by retinal ganglion cells that set the circadian clock. Science 295:1070-1074. [DOI] [PubMed] [Google Scholar]
- 6.Blackshaw, S., R. E. Fraioli, T. Furukawa, and C. L. Cepko. 2001. Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell 107:579-589. [DOI] [PubMed] [Google Scholar]
- 7.Buller, K. M. 2001. Role of circumventricular organs in pro-inflammatory cytokine-induced activation of the hypothalamic-pituitary-adrenal axis. Clin. Exp. Pharmacol. Physiol. 28:581-589. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson, K. M., J. M. Kavran, V. G. Sankaran, E. Fournier, S. J. Isakoff, E. Y. Skolnik, and M. A. Lemmon. 2000. Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol. Cell 6:373-384. [DOI] [PubMed] [Google Scholar]
- 9.Foster, R. G. 1998. Shedding light on the biological clock. Neuron 20:829-832. [DOI] [PubMed] [Google Scholar]
- 10.Freedman, M. S., R. J. Lucas, B. Soni, M. von Schantz, M. Munoz, Z. David-Gray, and R. Foster. 1999. Regulation of mammalian circadian behavior by non-rod, non-cone, ocular photoreceptors. Science 284:502-504. [DOI] [PubMed] [Google Scholar]
- 11.Friedrich, G., and P. Soriano. 1993. Insertional mutagenesis by retroviruses and promoter traps in embryonic stem cells. Methods Enzymol. 225:681-701. [DOI] [PubMed] [Google Scholar]
- 12.Friedrich, G., and P. Soriano. 1991. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 5:1513-1523. [DOI] [PubMed] [Google Scholar]
- 13.Furukawa, A., C. Koike, P. Lippincott, C. L. Cepko, and T. Furukawa. 2002. The mouse Crx 5′-upstream transgene sequence directs cell-specific and developmentally regulated expression in retinal photoreceptor cells. J. Neurosci. 22:1640-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furukawa, T., E. M. Morrow, and C. L. Cepko. 1997. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell 91:531-541. [DOI] [PubMed] [Google Scholar]
- 15.Ganong, W. F. 2000. Circumventricular organs: definition and role in the regulation of endocrine and autonomic function. Clin. Exp. Pharmacol. Physiol. 27:422-427. [DOI] [PubMed] [Google Scholar]
- 16.Harré, E.-M., J. Roth, U. Pehl, M. Kueth, R. Gerstberger, and T. Hübschle. 2002. Role of IL-6 in LPS-induced nuclear STAT3 translocation in sensory circumventricular organs during fever in rats. J. Appl. Physiol. 92:2657-2666. [DOI] [PubMed] [Google Scholar]
- 17.Haslam, R. J., H. B. Koide, and B. A. Hemmings. 1993. Why microtubules grow and shrink. Nature 363:309-310. [DOI] [PubMed] [Google Scholar]
- 18.Hattar, S., H.-W. Liao, M. Takao, D. M. Berson, and K.-W. Yau. 2002. Melanopsin-containing retinal ganglion cells: architecture, projections and intrinsic photosensitivity. Science 295:1065-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 20.Jeon, C.-J., E. Strettoi, and R. H. Masland. 1998. The major cell populations of the mouse retina. J. Neurosci. 18:8936-8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin, T., N. Zhang, Y. Long, C. A. Parent, and P. N. Devreotes. 2000. Localization of the G protein bg complex in living cells during chemotaxis. Science 287:982-983. [DOI] [PubMed] [Google Scholar]
- 22.Krappa, R., A. Nguyen, P. Burrola, D. Deretic, and G. Lemke. 1999. Evectins: vesicular proteins that carry a pleckstrin homology domain and localize to post-Golgi membranes. Proc. Natl. Acad. Sci. USA 96:4633-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemmon, M. A. 2003. Phosphoinositide recognition domains. Traffic 4:201-213. [DOI] [PubMed] [Google Scholar]
- 24.Lemmon, M. A., and K. M. Ferguson. 2000. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem. J. 350:1-18. [PMC free article] [PubMed] [Google Scholar]
- 25.Lemmon, M. A., K. M. Ferguson, and C. S. Abrams. 2002. Pleckstrin homolog domains and the cytoskeleton. FEBS Lett. 513:71-76. [DOI] [PubMed] [Google Scholar]
- 26.Liedtke, W., Y. Choe, M. A. Marti-Renom, A. M. Bell, S. C. Denis, A. Sali, A. J. Hudspeth, J. M. Friedman, and S. Heller. 2000. Vanilloid receptor-related osmotically activated channel (VR-OAC), a candidate vertebrate osmoreceptor. Cell 103:525-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lucas, R. J., M. Freedman, M. Muñoz, J.-M. Garcia-Fernández, and R. G. Foster. 1999. Regulation of the mammalian pineal by non-rod, non-cone ocular photoreceptors. Science 284:505-507. [DOI] [PubMed] [Google Scholar]
- 28.Maffucci, T., and M. Falasca. 2001. Specificity in pleckstrin homology (PH) domain membrane targeting: a role for a phosphoinositide-protein co-operative mechanism. FEBS Lett. 506:173-179. [DOI] [PubMed] [Google Scholar]
- 29.Mayer, B. J., R. Ren, K. L. Clark, and D. Baltimore. 1993. A putative modular domain present in diverse signaling proteins. Cell 73:629-630. [DOI] [PubMed] [Google Scholar]
- 30.McKinley, M. J., M. L. Mathai, G. Pennington, M. Rundgren, and L. Vivas. 1999. Effect of individual or combined ablation of the nuclear groups of the lamina terminalis on water drinking in sheep. Am. J. Physiol. 276:R673-R683. [DOI] [PubMed] [Google Scholar]
- 31.Mombaerts, P., F. Wang, C. Dulac, S. K. Chao, A. Nemes, M. Mendelsohn, J. Edmondson, and R. Axel. 1996. Visualizing an olfactory sensory map. Cell 87:675-686. [DOI] [PubMed] [Google Scholar]
- 32.Musacchio, A., T. Gibson, P. Rice, J. Thompson, and M. Saraste. 1993. The PH domain: a common piece in the structural patchwork of signalling proteins. Trends Biochem. Sci. 18:343-348. [DOI] [PubMed] [Google Scholar]
- 33.Oldfield, B. J., D. K. Hards, and M. J. McKinley. 1992. Neurons in the median preoptic nucleus of the rat with collateral branches to the subfornical organ and supraoptic nucleus. Brain Res. 586:86-90. [DOI] [PubMed] [Google Scholar]
- 34.Parent, A. 1996. Carpenter's human neuroanatomy, 9th ed. Williams & Wilkins, Philadelphia, Pa.
- 35.Parent, C. A., and P. N. Devreotes. 1999. A cell's sense of direction. Science 284:765-770. [DOI] [PubMed] [Google Scholar]
- 36.Shaw, G. 1996. The pleckstrin homology domain: an intriguing multifunctional protein module. Bioessays 18:35-46. [DOI] [PubMed] [Google Scholar]
- 37.Summy-Long, J. Y., and M. Kadekaro. 2001. Role of circumventricular organs (CVO) in neuroendocrine responses: interactions of CVO and the magnocellular neuroendocrine system in different reproductive states. Clin. Exp. Pharmacol. Physiol. 28:590-601. [DOI] [PubMed] [Google Scholar]
- 38.Swanson, D. A., C. L. Freund, J. M. Steel, S. Xu, L. Ploder, R. R. McInnes, and D. Valle. 1997. A differential hybridization scheme to identify photoreceptor-specific genes. Genome Res. 7:513-521. [DOI] [PubMed] [Google Scholar]
- 39.Swanson, D. A., J. Steel, and D. Valle. 1998. Identification and characterization of the human ortholog of at STXBP1, a protein implicated in vesicle trafficking and neurotransmitter release. Genomics 48:373-376. [DOI] [PubMed] [Google Scholar]
- 40.Thrasher, T. N., L. C. Keil, and D. J. Ramsay. 1982. Lesions of the organum vasculosum of the lamina terminalis (OVLT) attenuate osmotically-induced drinking and vasopressin secretion in the dog. Endocrinology 110:1837-1839. [DOI] [PubMed] [Google Scholar]
- 41.Ulfendahl, M., E. Scarfone, Å. Flock, S. LeCalvez, and P. Conradi. 2000. Perilymphatic fluid compartments and intercellular spaces of the inner ear and the organ of Corti. Neuro Image 12:307-313. [DOI] [PubMed] [Google Scholar]
- 42.Wang, T., A. M. Lawler, G. Steel, I. Sipila, A. H. Milam, and D. Valle. 1995. Mice lacking ornithine-d-aminotransferase have paradoxical neonatal hypoornithinaemia and retinal degeneration. Nat. Genet. 11:185-190. [DOI] [PubMed] [Google Scholar]
- 43.Wang, T., A. H. Milam, G. Steel, and D. Valle. 1996. A mouse model of gyrate atrophy of the choroid and retina: early retinal pigment epithelium damage and progressive retinal degeneration. J. Clin. Investig. 97:2753-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, Y., J. P. Macke, S. L. Merbs, D. J. Zack, B. Klaunberg, J. Bennett, J. Gearhart, and J. Nathans. 1992. A locus control region adjacent to the human red and green visual pigment genes. Neuron 9:429-440. [DOI] [PubMed] [Google Scholar]
- 45.Xiang, M., L. Gan, D. Li, Z.-Y. Chen, L. Zhou, B. W. O'Malley, W. Klein, and J. Nathans. 1997. Essential role of POU-domain factor Brn-3c in auditory and vestibular hair cell development. Proc. Natl. Acad. Sci. USA 94:9445-9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu, S., R. Ladak, D. A. Swanson, A. Soltyk, H. Sun, L. Ploder, D. Vidgen, A. M. V. Duncan, D. Valle, and R. R. McInnes. 1999. PHR1 encodes an abundant PH-domain containing integral membrane protein in the photoreceptor outer segment. J. Biol. Chem. 27:35676-35685. [DOI] [PubMed] [Google Scholar]
- 47.Yang, R.-B., S. W. Robinson, W.-H. Xiong, K.-W. Yau, D. G. Birch, and D. L. Garbers. 1999. Disruption of a retinal guanylyl cyclase gene leads to cone-specific dystrophy and paradoxical rod behavior. J. Neurosci. 19:5889-5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao, H., L. Ivic, J. M. Otaki, M. Hashimoto, K. Mikoshiba, and S. Firestein. 1998. Functional expression of a mammalian odorant receptor. Science 279:237-242. [DOI] [PubMed] [Google Scholar]
- 49.Zhao, H., and R. R. Reed. 2001. X inactivation of the OCNC1 channel gene reveals a role for activity-dependent competition in the olfactory system. Cell 104:651-660. [DOI] [PubMed] [Google Scholar]
- 50.Zheng, J. L., and W.-Q. Gao. 2000. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat. Neurosci. 3:580-586. [DOI] [PubMed] [Google Scholar]