Abstract
Autonomously replicating sequence binding factor 1 (ABF1) and repressor/activator protein 1 (RAP1) from budding yeast are multifunctional, site-specific DNA-binding proteins, with roles in gene activation and repression, replication, and telomere structure and function. Previously we have shown that RAP1 can prevent nucleosome positioning in the vicinity of its binding site and have provided evidence that this ability to create a local region of “open” chromatin contributes to RAP1 function at the HIS4 promoter by facilitating binding and activation by GCN4. Here we examine and directly compare to that of RAP1 the ability of ABF1 to create a region of open chromatin near its binding site and to contribute to activated transcription at the HIS4, ADE5,7, and HIS7 promoters. ABF1 behaves similarly to RAP1 in these assays, but it shows some subtle differences from RAP1 in the character of the open chromatin region near its binding site. Furthermore, although the two factors can similarly enhance activated transcription at the promoters tested, RAP1 binding is continuously required for this enhancement, but ABF1 binding is not. These results indicate that ABF1 and RAP1 achieve functional similarity in part via mechanistically distinct pathways.
Autonomously replicating sequence (ARS) binding factor 1 (ABF1) and repressor/activator protein 1 (RAP1) are multifunctional proteins expressed in the budding yeast Saccharomyces cerevisiae that have both been categorized as general regulatory factors (GRFs) (3). Both proteins play important roles in transcriptional activation and repression, gene silencing, recombination, and telomere structure, and both are abundant and essential for cell growth (12, 39, 45). Binding sites for both proteins are present in the promoter regions of numerous yeast genes, and ABF1 and RAP1 have been shown to contribute to transcriptional activation of genes involved in carbon source regulation, sporulation, amino acid biosynthesis, and ribosomal functions (21, 32, 33, 45). ABF1 and RAP1 act in concert to prevent gene expression at the silenced mating-type loci (7). ABF1 has also been implicated in gene silencing within subtelomeric regions (35) and nucleotide excision repair of silenced chromosomal regions (36), whereas RAP1 binds directly to telomere repeats to initiate formation of heterochromatin-like telomere structures (11).
Given their overlapping functions, it is not surprising that in several specific instances ABF1 and RAP1 have been found to be directly interchangeable. Both RAP1 and ABF1 can synergize with T-rich elements present in the rpS33 and rpL45 promoters to activate transcription (8), and they have also been reported to be interchangeable at the TRP3 promoter (23). Both RAP1 and ABF1 binding sites in ribosomal protein gene promoters are associated with recruitment of Esa1p and concomitant histone acetylation, as well as with recruitment of TFIID (24, 37). Furthermore, binding sites for RAP1 and ABF1 support ARS1 replication origin function equally well in a plasmid stability assay (22), and both proteins can function as insulator elements to establish boundaries between regions of silent (heterochromatic) and permissive (euchromatic) chromosomal regions (6, 60). Finally, both ABF1 and RAP1 possess functionally important C-terminal regions (10, 13, 26). Although these domains have only limited sequence homology (5), they are functionally interchangeable in cell viability assays and partly interchangeable in supporting transcriptional activation with specific promoters (9).
The precise mechanisms by which RAP1 and ABF1 contribute to the multiple processes in which they function are still being elucidated. Correspondingly, the basis for their functional similarity is not yet fully understood. One means by which ABF1 and RAP1 might contribute to disparate processes is by opening chromatin to allow access by other proteins (3, 4, 16, 19, 23, 29, 40, 58). This mode of action would also be consistent with findings that ABF1 and RAP1 are not able to stimulate robust transcription by themselves but synergize strongly with other transcription factors (4, 8, 23, 40, 42). Furthermore, direct observations show that both ABF1 and RAP1 can remodel chromatin near their binding sites, and RAP1 can outcompete histones for occupancy of its binding site (2, 16, 26, 54, 58, 59).
Although ABF1 and RAP1 are functionally similar, they differ in some regards. The two proteins share only a limited region of homology in their essential C-terminal regions. Both proteins have distinct DNA-binding domains and C-terminal regions that are functionally important. However, the C-terminal region of RAP1 contains domains that interact with various proteins involved in telomere and mating-type silencing, whereas interactions involving the ABF1 C-terminal region have not yet been identified, although regions important to transcription, chromatin remodeling, and viability have been mapped (26). The overall lack of homology in the functionally important C-terminal regions suggests that ABF1 and RAP1 may differ in some of the mechanistic aspects by which they perform their disparate functions. Furthermore, although ABF1 and RAP1 are equally effective at providing replication origin function at ARS1, replacement of the naturally occurring ABF1 binding site at ARS121 by a RAP1 binding site results in reduced replication efficiency, as assessed by mitotic stability of an ARS121-containing plasmid (22, 56).
Another indication that ABF1 and RAP1 may differ in the mechanism by which they function comes from experiments examining whether ABF1 or RAP1 binding is continually required to support activated transcription in which they participate. Using an abf1-1 ts mutant, Schroeder and Weil showed that, at a number of promoters requiring ABF1 sites to support normal transcriptional levels, little or no decrease in mRNA levels was seen at the restrictive temperature, although ABF1 binding was unambiguously lost (44). In contrast, RAP1 binding is continuously required at the HIS4 promoter to support normal levels of GCN4-mediated transcription (59).
Here, we directly compare ABF1 and RAP1 function in perturbing chromatin via nucleosomal binding sites, in synergizing with other transcription factors in transcriptional activation, and in whether their continuous binding is required to support activated transcription. Our results support the idea that these GRFs both perform their multiple tasks in part by a strong ability to perturb chromatin structure, and they possess very similar capabilities to synergize with other transcription factors in activating transcription. However, we also uncover differences between these two proteins that point to their using distinct mechanisms to support activated transcription.
MATERIALS AND METHODS
Plasmids.
The yeast plasmids TA/GCN1Δ80 and TAR/GCN1Δ80 are derived from the TRP1ARS1 plasmid and have been described previously (58). To create TAA/GCN1Δ80, the consensus ABF1 binding site was first inserted into pRS104-GCN1Δ80 (58) adjacent to the GCN4 binding site by two-step PCR (14) with primers A and B (Table 1) to create pRS104-A/GCN1Δ80, which was verified by DNA sequencing. Yeast DNA sequence was excised from this plasmid by SacI and HindIII, ligated with the complementary SacI-HindIII fragment of pRS110 (30), and then transformed into yeast (15) to create TAA/GCN1Δ80. Transformants were verified by Southern analysis. Similarly, TAAmut/GCN1Δ80, created with primers C and D (Table 1), contains a mutated ABF1 binding site (40) adjacent to the GCN4 binding site.
TABLE 1.
Primers used in this study
| Purpose | Primer | Restriction site created |
|---|---|---|
| Introduce wild-type ABF1 binding site in TA/GCN1Δ80 | (A) 5′-CGATCCGTCGGTAGTGACTTTTATGCTTGGTTTTCC | SphI |
| (B) 5′-GTCACTACCGACGGATCGATGACTATAAAAC | ||
| Introduce mutated ABF1 binding site in TA/GCN1Δ80 | ||
| (C) 5′-CGATCCGGTGGTAGTGACTTTTATGCTTGGTTTTC | SphI | |
| (D) 5′-GTCACTACCACCGGATCGATGACTCATAAAAC | ||
| Introduce wild-type ABF1 binding site into HIS4 promoter | (E) 5′-ATTGCCGTCGGTAGTGACGGCATGCACAGTGACTCACG | SphI |
| (F) 5′-GTCACTACCGACGGCAATTAATTAACTAATTTACCGGAGTC | ||
| Introduce mutated ABF1 binding site into HIS4 promoter | (G) 5′-ATTGCGGTCGGTAGTGACGGCATGCACAGTGACTCACG | SphI |
| (H) 5′-GTCACTACCACCGGCAATTAATTAACTAATTTACCGGAGTC | ||
| Introduce SacI site into HIS4 promoter | (I) 5′-CAATTGG AGCTCGAACGCAGAC | SacI |
| Introduce BamHI site into HIS4 promoter | (J) 5′-TTTGGATCCTATTGTATTACTATTACACAGCGCAG | BamHI |
| Introduce SacI site into HIS7 promoter | (K) 5′-CGCGATGAGCTCTGATTGACTACTCTCACGGTAACTCC | SacI |
| Introduce BamHI site into HIS7 promoter | (L) 5′-GCCATGGATCCTCTCTTTTTCTTTACTTGTAAATAATTAAAAACC | BamHI |
| Introduce RAP1 binding site into HIS7 promoter | (M) 5′-CATGATCACCTAATTTGTGCATGGGTTTAGCAAAAATAATCCCAAAGC | |
| (N) 5′-GCTTTGGGATTATTTTTGCTAAACCCATGCACAAATTAGGTGATCATG | ||
| Introduce mutated RAP1 binding site into HIS7 promoter | (O) 5′-CATGATCACCTAATTTGTGCATGGGTTTAGCAAAAATAATCCCAAAGC | |
| (P) 5′-GCTTTGGGATTATTTTTGCTAAACCCATGCACAAATTAGGTGATCATG | ||
| Introduce mutated ABF1 binding site into HIS7 promoter | (Q) 5′-CATGATCACCTAATTGTCACTACCACCGAAAAATAATCCCAAAGC | |
| (R) 5′-GCTTTGGGATTATTTTTCGGTGGTAGTGACAATTAGGTGATCATG | ||
| Introduce SacI site into ADE5,7 promoter | (S) 5′-GATCTAGAGCTCTGTTACACGCAGCATCGTTCTTTGG | SacI |
| Introduce BamHI site into ADE5,7 promoter | (T) 5′-GCTATGGATCCTGTGAGGGGAGGGAGAATGGTTCTC | BamHI |
| Introduce RAP1 binding site into ADE5,7 promoter | (U) 5′-GTCAGTCGGCACTTTGTGCATGGGTTTAGCGGGCGAGTCAACTG | |
| (V) 5′-CAGTTGACTCGCCCGCTAAACCCATGCACAAAGTGCCGACTGAC | ||
| Introduce mutated RAP1 binding site into ADE5,7 promoter | (W) 5′-GTCAGTCGGCACTTTGTGCATGGGTTTAGCGGGCGAGTCAACTG | |
| (X) 5′-CAGTTGACTCGCCCGCTAAACCCATGCACAAAGTGCCGACTGAC | ||
| Introduce mutated ABF1 binding site into ADE5,7 promoter | (Y) 5′-GTCAGTCGGCACTTGTCACTACCACCGGGGCGAGTCAACTG | |
| (Z) 5′-CAGTTGACTCGCCCCGGTGGTAGTGACAAGTGCCGACTGAC | ||
| ACT1 | (1) 5′-CCATGCCTAGACAAATCAAGGAAAGTATGTC | |
| (2) 5′-GGAGAGAGAGAGGCGAGTTTGGTTTCAAAACG | ||
| SPT15 | (3) 5′-CCCCTCTGATAGCTGAGATGTCGGGATTCC | |
| (4) 5′-CAGTAACTACTGTAATTTTCACGTCCCTTG | ||
| HIS4 | (5) 5′-GGATATGACTATGAACAGTAGTATACTGTG | |
| (6) 5′-TCCCAACCCATCTGTGGAGTGAG |
To construct HIS4-MEL1 reporter plasmids having a wild-type or mutant RAP1 binding site, the HIS4 promoter was amplified from genomic DNA from yeast strain LYY596 or LYY599, respectively (58), by use of primers shown in Table 1. These PCR products were cloned as BamHI-SacI fragments into 416-MEL1 (41), a plasmid that has a URA3 marker and the MEL1 coding sequence, to create 416-HIS4-MEL1. The HIS4 promoter sequences were confirmed by sequencing. Plasmids that contain modified HIS4 promoters with a wild-type or mutant ABF1 binding site in place of the RAP1 binding site were constructed by PCR amplification by use of primers shown in Table 1.
To construct reporter plasmids containing HIS7 and ADE5,7 promoters containing wild-type or mutant RAP1 or ABF1 binding sites fused to the MEL1 coding sequence, we first amplified promoter sequences from genomic DNA by using primers listed in Table 1. The resulting PCR products were cloned into pRS416-MEL1 (41) with the use of SacI and BamHI restriction sites. Construction of reporter plasmid variants in which the ABF1 binding site in HIS7 or ADE5,7 is replaced by a mutant ABF1 site, or a wild-type or mutant RAP1 binding site, was done by PCR with the use of primers shown in Table 1. Promoter sequences of these reporter plasmids were confirmed by DNA sequencing.
Strains and media.
The S. cerevisiae strains used in this study are listed in Table 2. Strains harboring ABF1 deletion mutations or the abf1-1 ts mutation were made by introducing the appropriate ABF1 gene in the pRS315 plasmid into TMY86, as described previously (26). Strains AY(1-731) and AY(1-592) were generated from strains 1-731 and 1-592 by replacement of the GCN4 open reading frame with the KanMX selectable marker. Yeast cells were grown at 30°C, unless stated otherwise, in complete synthetic medium (Bio 101) containing 2% glucose. Cell transformations were performed using a standard lithium acetate method (15).
TABLE 2.
Yeast strains used in this study
| Strain | Genotype or description | Reference or source |
|---|---|---|
| FY24 | MATα ura3-52 trp1Δ63 leu2Δ1 | 57 |
| LYY50 | Same as FY24 but gcn4Δ | 58 |
| YDS2 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | 18 |
| YDS408 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 rap1-2ts | 18 |
| TMY86 | mataade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 abf1Δ::HIS3MX6 [pRS416-ABF1] | 26 |
| 1-731 | TMY86-1/pRS415-ABF1-1-731 | 26 |
| 1-662 | TMY86-1/pTM211; pRS415-ABF1-1-662 | 26 |
| 1-633 | TMY86-1/pTM487; pRS415-ABF1-1-633 | 26 |
| 1-592 | TMY86-1/pTM629; pRS415-ABF1-1-592 | 26 |
| abf1 ts strain | TMY86-1/pTM629; pRS415-abf1-1 | 26 |
| YS18 | MATα his3-11 his3-15 leu2-3,112 ura3Δ5 canR | 1 |
| YS19 | MATα his3-11 his3-15 leu2-3,112 ura3Δ5 canR bas2− | 1 |
| MSY202 | MATα ura3-52 lys2Δ201 leu2-3,112 trp1-289 HHT1-HHF1 Δ(HHT2-HHF2) | 43 |
| AY(1-731) | Same as 1-731 but gcn4Δ | Present study |
| AY(1-592) | Same as 1-592 but gcn4Δ | Present study |
Analysis of chromatin structure.
Yeast cells were grown at 30°C to a density at 600 nm of between 0.6 and 1.3. Yeast spheroplast lysates were prepared and digested with micrococcal nuclease (MNase) and were analyzed by the indirect end-label technique as described elsewhere (17, 49). At least two independent transformants of each strain were tested.
α-Galactosidase assay.
α-Galactosidase activity was measured as described previously (41). Three independent clones were assayed for each reported value, and each experiment was repeated at least two times. Error bars shown in the figures represent standard deviations.
Northern analysis.
For Northern analysis involving temperature shift (see Fig. 6), cells were grown at 25°C overnight. The cultures were mixed with equal amounts of fresh medium preequilibrated to 25°C for analysis of cells grown at 25°C or to 50°C for analysis of cells shifted to 37°C. RNA was extracted from yeast cells, and approximately 8 to 10 μg was electrophoresed on agarose gels containing formaldehyde. Gels were blotted onto nylon membranes in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), UV cross-linked, and hybridized with probes labeled by random priming. Blots were stripped by boiling membranes in 0.015 M NaCl-0.1× SSC-1% sodium dodecyl sulfate before hybridization with another probe. Northern blots were quantitated using scanned images on a Molecular Dynamics PhosphorImager. The KpnI fragment from HIS7-MEL1 containing most of the MEL1 sequence was used as a MEL1 DNA probe. The BglII fragment of pGEM-PYK1 was used as a PYK1 DNA probe.
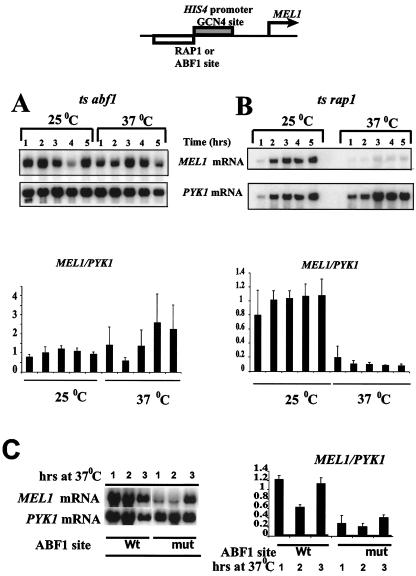
FIG. 6.
Continuous ABF1 binding to DNA is not required for efficient GCN4-mediated HIS4 activation. RNA was harvested from abf1 ts (A) and rap1-2 ts (B) cells harboring the HIS4-MEL1 reporter (schematized at the top) with either an ABF1 binding site (A) or a RAP1 binding site (B) after growth for the indicated times, in hours, at either 25 or 37°C. The RNA blots were hybridized with probes specific for MEL1 and PYK1 mRNA, and the signals were quantitated by PhosphorImager analysis. The bar graphs at the bottom show values derived from four independent experiments. Values for each experiment were normalized to the average MEL1/PYK1 value obtained for all time points at 25°C, and standard deviations are indicated. (C) Comparison of MEL1 mRNA levels (normalized to those of PYK1) from the modified HIS4 promoter having a wild-type or mutant ABF1 site, as indicated, after 1 to 3 h of growth at 37°C. All six lanes (for each mRNA) derive from a single exposure of the same blot. Quantitations in the bar graph on the right are from three independent experiments and were scaled by setting the average MEL1-PYK1 value for all three time points for the promoter having the wild-type ABF1 site to 1.
ChIP.
For chromatin immunoprecipitation (ChIP) involving temperature shift, cells were grown at 25°C overnight. The cultures were mixed with equal amounts of fresh medium preequilibrated to 25°C for analysis of cells grown at 25°C or to 50°C for analyzing cells shifted to 37°C for 1 h. ChIP was performed (see Fig. 7) essentially as described elsewhere (50), with the use of 20 μl of ABF1 antibody (SC6679; Santa Cruz Biotechnology). Cross-linked samples were sonicated using three repetitions of six pulses at 90% duty cycle, 20% output with a sonifier (model 250; Branson Ultrasonics, Danbury, Conn.), resulting in fragments ranging from 0.2 to 1.0 kb. Immunoprecipitated (IP) samples were eluted using standard protocols. For analysis, typically 1 to 2 μl of IP DNA or 1 μl of a 1:100 dilution of input DNA was amplified by using primers for HIS4, SPT15, and ACT1 (as negative control) (Table 1). PCR amplification was performed for 22 and 26 cycles, and aliquots were electrophoresed, Southern blotted, and hybridized to verify that amplification was in the linear range. We found that multiplexing primers used for analysis of SPT15, PYK1, and the modified HIS4 promoter resulted in spurious products that made quantitation difficult. We therefore performed amplification for each fragment in separate reactions and mixed aliquots from the separate reaction mixtures prior to gel electrophoresis. To guard against pipetting errors, each reaction was performed and analyzed at least three times, for two independent ChIP experiments. Quantitation was performed by using a Molecular Dynamics PhosphorImager.
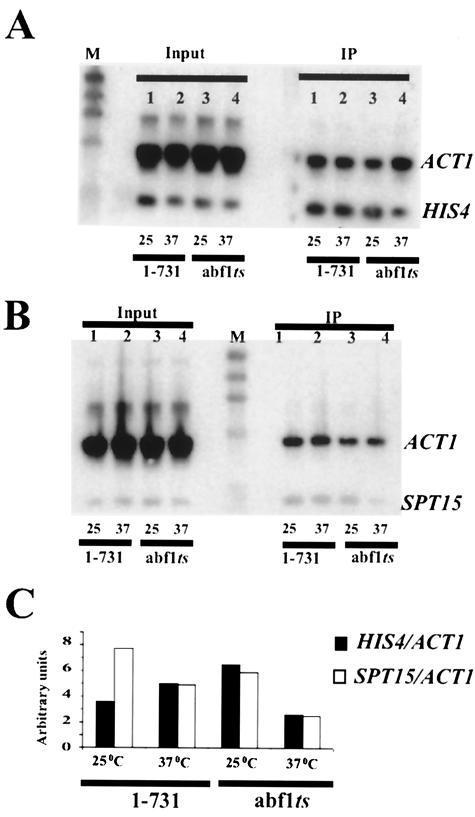
FIG. 7.
ChIP analysis of ABF1 binding to the modified HIS4-MEL1 promoter. ChIP was performed with ABF1 antibody in yeast carrying ABF1(1-731) or the abf1-1 ts mutant at 25°C or after 1 h at 37°C, as indicated. Input and IP samples were amplified in separate reactions with the use of primers for the HIS4-MEL1, SPT15, and ACT1 promoters. Samples were combined prior to electrophoresis, Southern blotting, and hybridization with appropriate probes. Although ACT1 was used as a negative control, its amplification was more efficient than that of HIS4-MEL1 or SPT15 promoters, for unknown reasons. Note, however, that ACT1 shows reduction in the IP lanes relative to the input lanes, and HIS4-MEL1 and SPT15 do not. Ratios of HIS4-MEL1 IP samples to input samples and SPT15 IP samples to input samples, relative to the ratio of ACT1 IP samples to input samples, are shown at the bottom. Similar results were obtained in this and one other independent experiment.
RESULTS
ABF1 perturbs nucleosome positioning via a nucleosomal binding site similarly to RAP1.
Previous work has indicated that one role of RAP1 is to assist activator binding by opening chromatin (4, 42, 58). Consistent with this idea, we have shown that RAP1 can perturb chromatin structure via a nucleosomal binding site in yeast (58). ABF1 is another general regulatory factor that shares many functions with RAP1 (see introduction). ABF1 has been shown to alter nucleosome positioning in the vicinity of a nonnucleosomal binding site in yeast (2, 16, 19, 54); however, a direct comparison of the ability of RAP1 and ABF1 to remodel chromatin has not been made, nor has the ability of ABF1 to perturb chromatin via a nucleosomal binding site been tested. To compare directly the abilities of RAP1 and ABF1 to perturb chromatin, we constructed four yeast episomes designed to place mutant or wild-type RAP1 or ABF1 binding sites into a positioned nucleosome (Fig. 1A). These plasmids are based on the yeast TRP1ARS1 plasmid, which is packaged into stably positioned nucleosomes in yeast (52). The chromatin structures of TA/GCN1Δ80, which has a GCN4 binding site in nucleosome I, and TAR/GCN1Δ80, which has a RAP1 binding site adjacent to the GCN4 binding site, have been documented in previous studies (58). TAA/GCN1Δ80 is identical to TAR/GCN1Δ80, except that an ABF1 binding site replaces the RAP1 binding site in nucleosome I. Since ABF1 is an essential gene (39), we introduced a mutated ABF1 site (40) into nucleosome I as a control. These four episomes were introduced into yeast, and nucleosome positioning was examined by indirect end labeling. This was done in GCN4+ yeast cells, as we have found that the chromatin structure of TA/GCN1Δ80, TAR/GCN1Δ80, and TARmut/GCN1Δ80 is not affected significantly by the presence or absence of GCN4 (data not shown). In the indirect end-labeling assay, MNase cleavage sites are compared in naked DNA and chromatin, and regions of 140 to 160 bp that are protected in chromatin, but not in naked DNA, are diagnostic of positioned nucleosomes (46, 51).
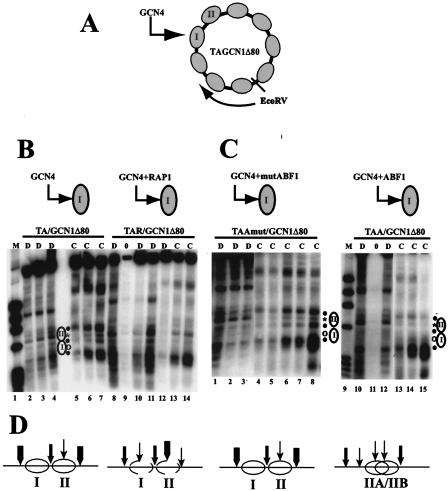
FIG. 1.
ABF1 perturbs nucleosome positioning via a nucleosomal binding site similarly to RAP1. (A) Schematic diagram of plasmid TA/GCN1Δ80. The gray ellipses represent positioned nucleosomes, and nucleosomes I and II are marked. Nucleosome I contains the GCN4 binding site, as shown, and chromatin structure was mapped clockwise from the EcoRV site, as indicated, by use of chromatin prepared from FY24 yeast cells containing the indicated plasmid episome. (B) Indirect end-label analysis of TA/GCN1Δ80 and TAR/GCN1Δ80 chromatin structure. TAR/GCN1Δ80 is similar to TA/GCN1Δ80 except for a RAP1 binding site introduced adjacent to the GCN4 binding site. MNase cleavage sites were mapped clockwise from the EcoRV site in naked DNA (D lanes) or in chromatin (C lanes) from cells grown in glucose medium under conditions that are noninducing for Gcn4p activation. Lane 1 contains φX/HaeIII marker DNA. Naked DNA in TA/GCN1Δ80 (lanes 2 to 4) and in TAR/GCN1Δ80 (lanes 8, 10, and 11) was digested using 2.5 (lanes 2 and 8), 5 (lanes 3 and 10), or 10 (lane 4 and 11) U of MNase per ml. Chromatin was digested using 0 (lane 9), 2.5 (lanes 5 and 12), 5 (lanes 6 and 13), or 10 (lanes 7 and 14) U/ml. (C) Indirect end-label analysis of TAA/GCN1Δ80 and TAAmut/GCN1Δ80 chromatin structure. TAA/GCN1Δ80 and TAAmut/GCN1Δ80 are similar to TA/GCN1Δ80, except for ABF1 or mutated ABF1 binding sites introduced adjacent to the GCN4 binding site. MNase cleavage sites were mapped clockwise from the EcoRV site in naked DNA (D lanes) or in chromatin (C lanes) from cells grown in glucose medium. Lane 9 contains φX/HaeIII marker DNA. Naked TAA/GCN1Δ80 and TAAmut/GCN1Δ80 DNA were digested using 2.5 (lanes 1 and 10), 5 (lanes 2 and 12), or 10 (lane 3) U/ml. Chromatin was digested using 0 (lane 11), 2.5 (lanes 4 and 13), 5 (lanes 5 and 14), 10 (lanes 6 and 15), 15 (lane 7), or 20 (lane 8) U of MNase per ml. All the chromatin samples were run on the same gel; the DNA lanes are derived from separate gels (Fig. 3 shows examples in which chromatin and naked DNA samples all derive from the same gel). The asterisks and open circles indicate cleavages in naked DNA that are protected by nucleosomes I and II in chromatin, and filled circles indicate the edges of nucleosomes I and II, which are present in TA/GCN1Δ80 and TAAmut/GCN1Δ80 but not in TAR/GCN1Δ80 and TAA/GCN1Δ80. The locations of positioned nucleosomes I and II are shown by ellipses. (D) Schematic diagram of nucleosome positioning in the region of nucleosomes I and II deduced from the MNase cutting patterns of TA/GCN1Δ80, TAR/GCN1Δ80, TAA/GCN1Δ80, and TAAmut/GCN1Δ80. The thickness of each vertical arrow indicates the relative strength of MNase cleavage.
Nucleosomes I and II were positioned in TAAmut/GCN1Δ80 as in TA/GCN1Δ80 (compare Fig. 1B, lanes 5 to 7, and 1C, lanes 4 to 8). The features of the MNase cleavage pattern diagnostic of this positioning are the two cleavage sites that are protected in chromatin relative to naked DNA (asterisk and open circle, Fig. 1B, lanes 2 to 7, and Fig. 1C, lanes 1 to 8) and the three cleavage sites enhanced in chromatin relative to naked DNA. The latter sites are separated by about 150 bp and mark the edges of positioned nucleosomes (closed circles) (28, 30, 52, 58). In contrast, the chromatin structure of TAR/GCN1Δ80 and TAA/GCN1Δ80 dramatically changed. As previously reported, the MNase cleavage pattern for TAR/GCN1Δ80 chromatin is essentially identical to that for the corresponding naked DNA in the regions of nucleosomes I and II, indicating that nucleosome positioning in this region is abolished (Fig. 1B, lanes 8 to 14) (58). The chromatin structure of TAA/GCN1Δ80 is similar to that of TAR/GCN1Δ80 but not identical (compare Fig. 1B, lanes 13 and 14, to Fig. 1C, lanes 13 to 15). Both TAA/GCN1Δ80 and TAR/GCN1Δ80 chromatin show MNase cleavage in the region of nucleosome I, at a site which is cleaved weakly in naked DNA but is completely protected in TA/GCN1Δ80 and in the mutABF1 plasmid chromatin (open circle in Fig. 1B and C). At the border between nucleosomes I and II, cleavage by MNase is enhanced in TA/GCN1Δ80 and in the mutABF1 plasmids relative to that in naked DNA (middle closed circle, Fig. 1B, lanes 2 to 8, and Fig. 1C, lanes 1 to 8) but is not enhanced relative to the naked DNA pattern in TAR/GCN1Δ80 (Fig. 1B, lanes 8 to 14). In TAA/GCN1Δ80, this cleavage site appears slightly protected relative to naked DNA, possibly indicating some nucleosomal protection (Fig. 1C, lanes 10 to 15). The cleavage site that is largely protected by nucleosome II in TA/GCN1Δ80 and TAAmut/GCN1Δ80 chromatin (Fig. 1B, lanes 2 to 7, and Fig. 1C, lanes 1 to 8) is not at all protected in TAR/GCN1Δ80 (Fig. 1B, lanes 8 to 14) but still shows some protection in TAA/GCN1Δ80 (Fig. 1C, lanes 2 to 7). Similarly, the enhanced cleavage site at the distal border of nucleosome II in TA/GCN1Δ80 and TAAmut/GCN1Δ80 (uppermost closed circle, Fig. 1B and C) is not at all enhanced in TAR/GCN1Δ80 but still shows some enhanced cleavage in TAA/GCN1Δ80. Taken together, these results demonstrate that ABF1, like RAP1, is effective in creating a localized region of open chromatin. However, whereas RAP1 abolishes positioning of two nucleosomes, ABF1 appears to clear the nucleosome in the immediate vicinity of its binding site but still allows positioning, or partial positioning, nearby (Fig. 1D). These results indicate that ABF1 may be able to function similarly to RAP1 in opening chromatin to allow transactivator access and thus facilitate transcription, although it may differ subtly in the mechanism by which it does so.
Contribution of C-terminal domains to chromatin perturbation by ABF1.
Having established that ABF1 shares with RAP1 an ability to perturb nucleosome positioning via a nucleosomal binding site in vivo (Fig. 1), we next wanted to test whether domains of ABF1 outside the DNA-binding domain were important for this property. Both RAP1 and ABF1 possess central DNA-binding domains and putative transactivation domains in their C-terminal regions (Fig. 2) (13, 20, 26). Furthermore, RAP1 and ABF1 share a short region of homology in their putative transactivation domains (5, 26). Somewhat surprisingly, we have found that the putative transactivation domain, and indeed most of the C-terminal region of RAP1, is dispensable for chromatin perturbation via a nucleosomal binding site as well as for activation of HIS4 (59). In contrast, the C-terminal region of ABF1 has been shown to contain two domains, CS1 and CS2, that contribute to transcriptional activation as well as to chromatin remodeling (but from binding sites located nearby and not within the remodeled nucleosomes) (26). To test the involvement of the C-terminal region of ABF1 in chromatin perturbation via a nucleosomal binding site, we tested the abilities of truncated C-terminal derivatives of ABF1 to perturb the chromatin structure of TAA/GCN1Δ80 relative to TAAmut/GCN1Δ80 in yeast.
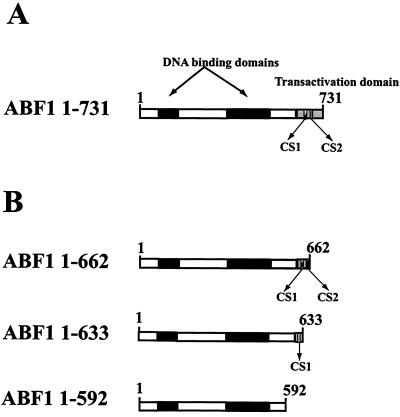
FIG. 2.
Schematic diagram of ABF1. (A) Full-length ABF1 is shown, with DNA-binding domains, transactivation domain, and the C-terminal sequences (CS1 and CS2) indicated. (B) The truncated versions of ABF1 used in this work are shown.
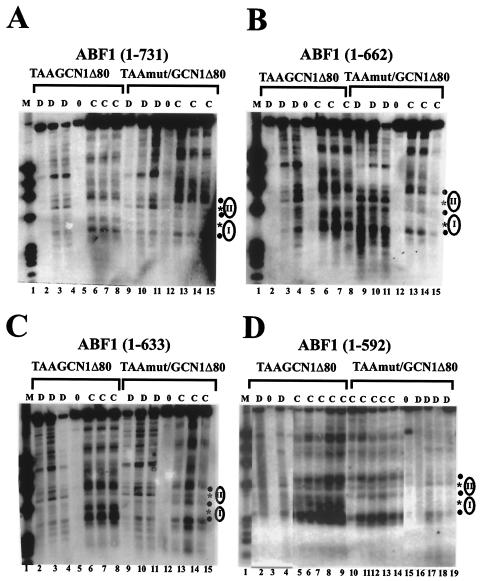
Figure 3A shows the results of indirect end-label analysis following MNase digestion of the episomes TAA/GCN1Δ80 and TAAmut/GCN1Δ80, which contain a wild-type or mutant ABF1 site near the center of positioned nucleosome I of TA/GCN1Δ80 (Fig. 1), in yeast cells in which the chromosomal ABF1 gene has been deleted and which express full-length ABF1(1-731) from a CEN plasmid. The MNase digestion pattern of TAA/GCN1 chromatin is essentially identical to that seen in Fig. 1, in which ABF1 was expressed from its normal chromosomal location. Cleavage is seen in TAA/GCN1Δ80 in the region of nucleosome I that is protected in TAAmut/GCN1Δ80 (compare lanes 6 to 8 and lanes 13 to 15 in Fig. 3A; band marked by asterisk), and the cleavage site between nucleosomes I and II in TAAmut/GCN1Δ80 is mostly protected in TAA/GCN1Δ80 (central closed circle in Fig. 3A). Thus, as seen in Fig. 1, full-length ABF1 evidently has a strong ability to perturb nucleosome positioning in vivo via a nucleosomal ABF1 binding site. The same MNase cleavage pattern was obtained using yeast expressing ABF1(1-662) (Fig. 3B). This was not surprising, as both CS1 and CS2 are still present in this construct, and no growth defect was observed in this yeast strain (26). Further deletion of ABF1 to remove the CS2 domain [ABF1(1-633)] leads to growth defects at 30 and 35°C and a weak ts phenotype at 37°C and also abolishes the ability of ABF1 to remodel a nucleosome from a nearby site in vivo (26). The ability of ABF1(1-633) to perturb TAA/GCN1Δ80 chromatin was, however, undiminished compared to that of wild-type ABF1 (Fig. 3C). Further deletion to remove both CS1 and CS2 domains [ABF1(1-592)] still did not impair the ability of ABF1 to perturb TAA/GCN1Δ80 nucleosome positioning, with cleavage in the region of nucleosome I and partial protection of the cleavage site between nucleosomes I and II being readily apparent (Fig. 3D). We conclude that, although CS1 and CS2 domains of ABF1 are needed to remodel chromatin from sites near a positioned nucleosome, they are not needed to outcompete histones for binding to the same sequence with concomitant perturbation of nucleosome positioning. This result also demonstrates that chromatin can be remodeled by more than one mechanism in vivo.
FIG. 3.
Contribution of C-terminal domains of ABF1 to chromatin perturbation via a nucleosomal binding site. (A) MNase cleavage sites in plasmids TAA/GCN1Δ80 and TAAmut/GCN1Δ80 were mapped in GCN4+ yeast harboring ABF1(1-731). Cleavage sites were mapped in naked DNA (D lanes) or in chromatin (C lanes) from the EcoRV site, as in Fig. 1. Lane 1 contains φX/HaeIII marker DNA. Locations of positioned nucleosomes I and II are indicated by ellipses. The filled circles to the right of the panel indicate cleavages enhanced in chromatin relative to DNA, and the stars indicate strong cleavages in naked DNA that are protected by nucleosomes I and II in chromatin of TAAmut/GCN1Δ80 but not TAA/GCN1Δ80. Each lane, beginning with lanes 2 to 4, differs only in the concentration of MNase used. Lanes 5 and 12 show controls not treated with MNase. (B) MNase cleavage sites in plasmids TAA/GCN1Δ80 and TAAmut/GCN1Δ80 were mapped in yeast harboring ABF1(1-662), as in panel A. (C) MNase cleavage sites in plasmids TAA/GCN1Δ80 and TAAmut/GCN1Δ80 were mapped in yeast harboring ABF1(1-633) as in panel A. (D) MNase cleavage sites in plasmids TAA/GCN1Δ80 and TAAmut/GCN1Δ80 were mapped in yeast harboring ABF1(1-592) as in panel A. All samples were run on the same gel, but some lanes are omitted from the figure, as seen from the visible “splice” marks.
ABF1 can contribute to activation of HIS4 similarly to RAP1.
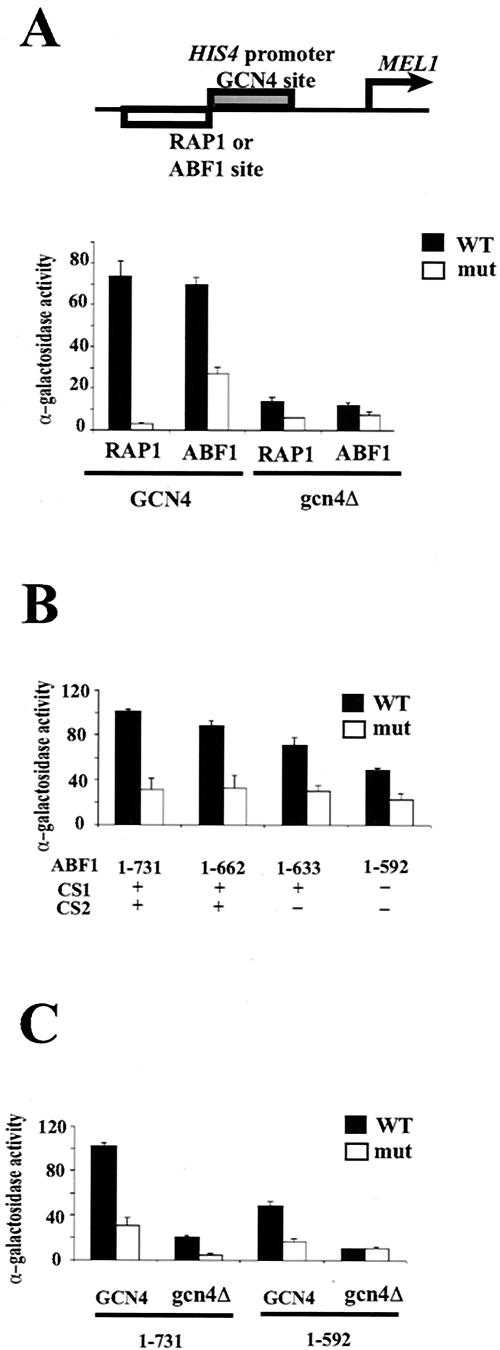
Previous work has provided strong evidence that RAP1 functions at the HIS4 promoter at least in part by opening chromatin to allow binding by the primary activators, GCN4 and Bas1p/Bas2p (4, 58). Correspondingly, RAP1 is very effective in outcompeting histones for binding to a nucleosome positioning sequence in yeast (58). Since ABF1 shares this latter property with RAP1 (Fig. 1 and 3), we wanted to test whether ABF1 could substitute for RAP1 to help GCN4 activate the HIS4 promoter. A MEL1 reporter gene, encoding α-galactosidase, was fused to the wild-type HIS4 promoter, or to the HIS4 promoter in which the RAP1 site has been mutated, and also to a modified HIS4 promoter having an ABF1 binding site or mutated ABF1 binding site adjacent to the GCN4 binding site. α-Galactosidase activity was monitored and compared between GCN4+ wild-type cells and gcn4Δ cells for all four promoters. The results shown in Fig. 4A demonstrate that, for the native HIS4 promoter, mutation of the RAP1 binding site or absence of GCN4 results in substantial loss of transcription, as expected (4). This synergism between RAP1 and GCN4 is consistent with RAP1 opening chromatin to facilitate access by GCN4, the principal activator. Replacement of the RAP1 binding site by a binding site for ABF1, or a mutated binding site, resulted in similar behavior (Fig. 4A). We do not know why the HIS4 promoter possessing a mutated ABF1 site has higher activity than that of the promoter with the mutated RAP1 site, as previous work indicated that ABF1 showed essentially no activity from this mutated site (40). In any event, these results are consistent with the notion that ABF1 shares with RAP1 an ability to facilitate GCN4 binding to the HIS4 promoter by overcoming the repressive effect of chromatin.
FIG. 4.
ABF1 can contribute to activation of the HIS4 promoter similarly to RAP1. (A) The MEL1 reporter gene fused to modified HIS4 promoter having an ABF1 or RAP1 binding site or mutant ABF1 or RAP1 binding sites adjacent to the GCN4 binding site, schematized at the top. MEL1 activity was monitored in GCN4+ cells or gcn4Δ cells and expressed as α-galactosidase activity. (B) Contribution of C-terminal domains to GCN4-mediated activation of the HIS4 promoter. The modified HIS4 promoter having an ABF1 binding site or mutant ABF1 binding site adjacent to the GCN4 binding site was introduced into yeast expressing ABF1 deletion mutants as indicated, and MEL1 activity was monitored. (C) MEL1 activity was monitored from the modified HIS4 promoter having an ABF1 binding site or mutant ABF1 binding site adjacent to the GCN4 binding site in GCN4+ or gcn4Δ yeast harboring full-length ABF1 (“1-731”) or truncated ABF1 lacking the CS1 and CS2 domains (“1-592”), as indicated. Standard deviations are indicated. WT, wild type.
The C-terminal portion of ABF1, and in particular the short CS2 domain, is evidently not required for perturbation of chromatin via a nucleosomal ABF1 binding site (Fig. 3). However, this domain is required for chromatin remodeling from sites near positioned nucleosomes and is also important for transcriptional activation (26). Thus, depending on the mechanism by which ABF1 perturbs chromatin at the modified HIS4 promoter, the C-terminal region and CS2 domain may or may not be required to facilitate HIS4 transcriptional activation. To examine this question, we introduced the modified HIS4-MEL1 reporter into cells expressing full-length or truncated ABF1 proteins, as in Fig. 3, and measured α-galactosidase activity. The results, shown in Fig. 4B, indicate that loss of amino acids 663 to 731 from ABF1 results in only a slight decrease in HIS4 activity. Loss of CS2 (amino acids 639 to 662) results in diminished transcriptional activity (P < 0.003 by Student t test), and loss of CS1 lowers activity even further (P < 0.001). However, ABF1 increases HIS4 activation by GCN4 even when both CS1 and CS2 are absent [ABF1(1-592)], although it can no longer stimulate transcription of HIS4 in the absence of GCN4 (Fig. 4C). This strongly suggests that ABF1 stimulates transcription from the modified HIS4 promoter in part through its CS1 and CS2 domains but also in part by remodeling chromatin in the immediate vicinity of its binding site via a mechanism independent of CS1 and CS2.
Comparison of ABF1 and RAP1 in facilitating transcriptional activation from the HIS7 and ADE5,7 promoters.
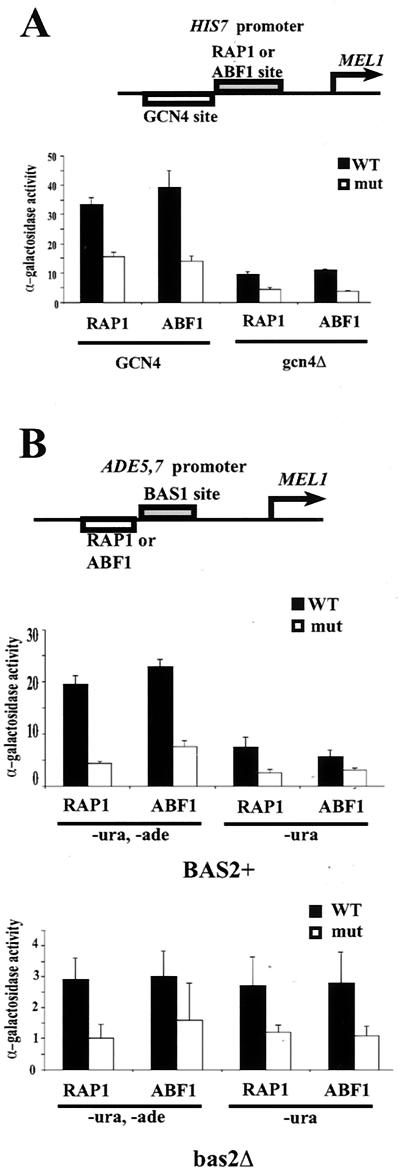
We next examined two additional promoters containing ABF1 binding sites, HIS7 and ADE5,7. The HIS7 promoter contains an ABF1 binding site approximately midway between two GCN4 binding sites at −231/−225 and −144/−139 bp relative to the translational start codon, and the region surrounding the ABF1 binding site and including both GCN4 binding sites has recently been shown to be nucleosome free (47, 53). In addition, Bas1p/Bas2p can activate HIS7 via the −144/−139 site (48). The ABF1 binding site was found to be essential for basal transcription of a HIS7-lacZ fusion gene in gcn4− yeast (47). We examined GCN4-dependent activation of a HIS7-MEL1 reporter gene and found modest stimulation by either ABF1 or GCN4 alone (Fig. 5A). Activation with the wild-type HIS7 promoter in GCN4+ yeast was about twofold stronger than the summed individual effects of GCN4 and ABF1 (Fig. 5A). Substitution of a RAP1 or mutated RAP1 binding site for the ABF1 binding site in the HIS7 promoter yielded similar results, although the synergism between RAP1 and GCN4 was somewhat less than that between ABF1 and GCN4 (Fig. 5A). We conclude that ABF1 and RAP1 can function interchangeably at the HIS7 promoter, with each showing modest (1.5- to 2-fold) synergism with GCN4. This suggests that ABF1 can facilitate GCN4-mediated activation, possibly by opening chromatin, in the context of a natural promoter and that RAP1 can provide this same function at a suitably modified promoter.
FIG. 5.
Comparison of ABF1 and RAP1 in facilitating transcriptional activation from the HIS7 and ADE5,7 promoters. (A) The MEL1 reporter gene fused to a modified HIS7 promoter having an ABF1 binding site or RAP1 binding site or mutant ABF1 or RAP1 site adjacent to the GCN4 binding site, schematized at the top. MEL1 activity was monitored and compared in GCN4+ wild-type cells and in gcn4Δ cells. Standard deviations are shown. (B) The MEL1 reporter gene fused to a modified ADE5,7 promoter having an ABF1 binding site or RAP1 binding site or mutant ABF1 or RAP1 binding site adjacent to the BAS1 binding site, schematized at the top. MEL1activity was monitored and compared in BAS1 and BAS2+ wild-type cells and in bas2Δ cells (note different scales) in the presence and absence of adenine. Standard deviations are shown. WT, wild type.
The HIS4 promoter can be activated by either GCN4 or, via an alternative pathway, BAS1/BAS2. In either case, the RAP1 binding site, which is located between binding sites for BAS1 and GCN4, is needed to facilitate activation (4). The ADE5,7 promoter is activated by BAS1/BAS2 in response to purine starvation, and an ABF1 binding site about 20 bp upstream of the proximal and more important of the two BAS1 binding sites is essential for this activation (40). If RAP1 and ABF1 both facilitate activation in part by opening chromatin, we would expect the two proteins to be able to assist different activators (as RAP1 can do at the HIS4 promoter) (4, 58). We therefore tested whether RAP1 could substitute for ABF1 to facilitate BAS1/BAS2-mediated activation of the ADE5,7 promoter. A MEL1 reporter gene was fused to the wild-type and modified versions of the ADE5,7 promoter having wild-type or mutant ABF1 or RAP1 binding sites (Fig. 5B). Both BAS1 and BAS2 are required for activation in response to adenine depletion (40); we found that, in the absence of BAS2, activation by RAP1 or ABF1 is very modest and independent of adenine depletion (Fig. 5B). In BAS2+ yeast, activation in response to adenine depletion depends strongly on the presence of a binding site for ABF1 or RAP1, with the two sites providing similar levels of stimulation (Fig. 5B). Thus, at three distinct yeast promoters, ABF1 and RAP1 can each synergize with GCN4, with BAS1/BAS2, or with both to activate transcription, most likely by opening chromatin to facilitate binding by the principal activator.
Continuous ABF1 binding to DNA is not required to maintain GCN4-mediated activation facilitated by ABF1.
Previous results have shown that continuous RAP1 binding is required for ongoing HIS4 transcription mediated by GCN4, even in nonreplicating cells (59). In contrast, several “ABF1-driven” genes (SPT15, RPL2A, RPL2B, TCM, and QCR8) apparently do not require continuous ABF1 occupancy of the ABF1 cis element for continuous high-level transcription (44). This could reflect differences in the mechanisms by which ABF1 and RAP1 contribute to transcriptional activation, or it could reflect differences between the HIS4 promoter and the ABF1-dependent promoters examined previously. To directly compare the requirements for continuous binding of RAP1 and ABF1 to stimulate transcriptional activation, we therefore examined whether continuous ABF1 binding is required for efficient GCN4-mediated activation of the modified HIS4 promoter in which an ABF1 site replaces the RAP1 site.
In this experiment, we used an abf1 ts mutant that is temperature sensitive for binding to DNA in vivo and in vitro (38, 44). The abf1-1 mutant encodes a mutation in the putative zinc finger motif in the DNA-binding domain of ABF1. Dimethyl sulfate footprinting has shown that this mutant ABF1 protein vacates its binding site in vivo within 1 h, and at some promoters within 3 to 4 min, following a shift to the nonpermissive temperature of 37°C (44). We examined MEL1 mRNA levels expressed from the normal and modified HIS4 promoters before and after shifting the cells from permissive temperature to the nonpermissive temperature. Cells were grown at 25°C overnight to log phase and were then inoculated into fresh medium preequilibrated to the appropriate temperature and grown at either 25 or 37°C. We then harvested mRNA from cells at various time points. We chose our first time point for mRNA sampling at 1 h after the temperature shift to allow ample time for the decay of preexisting MEL1 mRNA. As shown in Fig. 6A, the expression of MEL1 mRNA from the modified HIS4 promoter containing an ABF1 binding site in abf1-1ts cells was unchanged at 25 and 37°C. In contrast, levels of MEL1 mRNA expressed from the normal HIS4 promoter containing a RAP1 binding site declined substantially relative to that of a PYK1 control at 37°C in the rap1-2ts strain, consistent with previous results (59) (Fig. 6B). (We also showed previously that HIS3 mRNA, which has a shorter half-life than does the PYK1 message [23 min compared to about 60 min (55)], was similarly unaffected by the temperature shift in this strain [59].) Similar results were observed in four independent experiments. Furthermore, the results in Fig. 6C show that the level of MEL1 mRNA produced from the template having a mutant ABF1 site is reduced about fourfold relative to that of the template having a wild-type ABF1 site in yeast grown at 37°C, and a similar decrease was observed at 25°C (data not shown). Thus, if loss of ABF1 binding at 37°C in the ts mutant in the experiment whose results are shown in Fig. 6A led to a decrease in MEL1 mRNA levels corresponding to those produced from the template having a mutant ABF1 binding site (Fig. 6C), this decrease would be readily observable.
To test the possibility that ABF1 remains bound to the modified HIS4 promoter after a shift to the nonpermissive temperature, we performed ChIP using antibodies to ABF1. To visualize only the modified, plasmid-borne HIS4 promoter, we used one primer from the HIS4 promoter and another from the MEL1 coding sequence in our analysis. The results in Fig. 7A show that immunoprecipitation with the use of antibodies to ABF1 substantially enriched the modified HIS4 promoter compared to an ACT1 control, as expected. (The ACT1 product was amplified considerably more efficiently than was the modified HIS4 promoter or the SPT15 promoter [Fig. 7B], for reasons that we do not understand.) In cells expressing full-length, wild-type ABF1, no change is seen upon shifting the temperature to 37°C (Fig. 7A, lanes 1 and 2, and 7C). In contrast, shifting abf1-1 ts cells to 37°C resulted in about a twofold loss of IP HIS4 fragment compared to that for 25°C. The level of IP fragment relative to input seen after the temperature shift was comparable to that seen with a template having the mutant ABF1 binding site (data not shown), indicating that it represents nonspecific background. Quantitatively similar results were seen with the SPT15 promoter (Fig. 7B and C), indicating that loss of ABF1 at the restrictive temperature in the abf1-1 ts mutant is similar at the two promoters. Since loss of ABF1 from the SPT15 promoter was shown to be complete under these conditions by dimethyl sulfate footprinting (44), we conclude that complete or nearly complete loss of ABF1 also likely occurred from the modified HIS4 promoter at 37°C in the ts mutant. Thus, the requirement for continuous binding of RAP1, but not ABF1, to facilitate activated transcription reflects a fundamental mechanistic difference between these two otherwise functionally similar GRFs, rather than a difference between individual promoters either requiring or not requiring continuous GRF occupancy.
DISCUSSION
ABF1 and RAP1 are multifunctional proteins that are grouped together with GRF2/REB1 as GRFs (6). The functional similarities of ABF1 and RAP1 have been appreciated for some time, and the two factors have been compared with respect to their abilities to activate transcription from ribosomal protein gene promoters, to stimulate replication origin activity, to support cell viability, and to function as barriers between euchromatin and heterochromatin (6, 8, 9, 22, 24, 37, 60). However, other relevant properties of ABF1 and RAP1 have not been directly compared in previous work. First, although both ABF1 and RAP1 can perturb or remodel chromatin structure (2, 16, 54, 58), this function has not previously been compared for the same chromatin template. Second, the ability to open chromatin likely contributes to both ABF1 and RAP1 function at some promoters (3, 4, 16, 19, 23, 29, 40, 58), but the interchangeability of ABF1 and RAP1 binding sites at such promoters has not been previously examined. Third, although both ABF1 and RAP1 have carboxy-terminal regions with multiple important functions, the contributions of these regions to particular processes have not been directly compared. Fourth, ABF1 has been shown not to be continuously required at several ABF1-driven promoters for continued transcription, whereas RAP1 is needed for ongoing GCN4-mediated HIS4 activation (44, 59), but a comparison of this property at a single promoter was lacking.
In this paper, we have compared ABF1 and RAP1 in the functional assays delineated above. Previous work had shown that ABF1 could create a region of open chromatin in its immediate vicinity (although a direct test of its ability to outcompete histones for its binding site had not been performed) (16, 19, 26). We were therefore not surprised to find that ABF1, like RAP1, can perturb chromatin structure when its binding site is incorporated into the region of a positioned nucleosome (Fig. 1). However, although the C-terminal region is required for ABF1 to remodel nucleosomes from a nearby, accessible binding site (26), it is not required for chromatin perturbation via a nucleosomal binding site (Fig. 3). Similarly, RAP1 can perturb chromatin via a nucleosomal binding site independently of its C-terminal region (59). Previous work has indicated that DNA-binding proteins can compete with histones for the same binding site by mass action in vitro and in vivo (25, 34, 58), thus providing a mechanism by which abundant, high-affinity binding proteins such as ABF1 and RAP1 can outcompete histones for their binding sites without assistance by accessory proteins and therefore, by implication, without an activation domain (27).
Although both ABF1 and RAP1 could perturb chromatin structure via nucleosomal binding sites, there were some subtle differences in the structural changes elicited by the two proteins, as seen in the slightly different MNase cleavage patterns (Fig. 1). We do not at present understand the basis for this difference, although it does not appear to involve the C-terminal regions nor proteins interacting with these regions, since the MNase cleavage patterns induced by ABF1 and RAP1 do not depend on these regions (Fig. 3) (59). It also seems likely that these subtle differences will not, at least in most cases, lead to substantial differences between ABF1 and RAP1 in facilitating transcriptional activation, as the two function essentially interchangeably at the HIS4, HIS7, and ADE5,7 promoters. At these promoters, both ABF1 and RAP1 were able to synergize with either GCN4 (HIS4, HIS7) or BAS1/BAS2 (ADE5,7), consistent with a general function of opening chromatin to facilitate activator access. This notion is also supported by recent work showing that the region of the HIS7 promoter surrounding the ABF1 binding site is free of nucleosomes (53) and with the finding that even heterologous DNA-binding proteins expressed ectopically in yeast can facilitate GCN4 binding to nearby sites (25). Furthermore, ABF1 was able to facilitate GCN4-mediated activation at HIS4 even when lacking its transactivation domain, consistent with previous observations for RAP1 (59) (Fig. 4B). Taken together, these experiments and previous work point to ABF1 and RAP1 being prototypes of a category of DNA-binding proteins that mark chromatin by outcompeting histones for their binding sites and creating an open region of chromatin which other either less-abundant or lower-affinity DNA-binding proteins may then access. It seems likely that other proteins play this same role both in yeast (e.g., REB1, another GRF) and in higher eukaryotes (31). Such proteins, if identified in higher eukaryotes, could potentially be useful in maintaining open chromatin in transgenic applications.
We observed a notable and surprising difference between ABF1 and RAP1 when we tested whether continuous occupancy by ABF1 at the modified HIS4 promoter was required for ongoing transcription. As mentioned above, transcription of several ABF1-driven promoters (i.e., promoters containing ABF1 binding sites that had been shown to contribute to activation) had been reported to be unchanged upon loss of ABF1 binding in an abf1-1 ts mutant (44). In contrast, we had found that GCN4-mediated HIS4 activation greatly decreased upon loss of RAP1 binding (59). This contrasting behavior could have been due to differences in promoter structure rather than differences in ABF1 and RAP1 function. However, we found in a direct comparison at the HIS4 promoter that ABF1 could contribute to activation even without continuous binding, whereas continuous binding by RAP1 was required for it to facilitate activation (Fig. 4A and 6). ChIP demonstrated that ABF1 was lost to a quantitatively similar degree at HIS4 and SPT15 in the abf1-1 ts mutant at 37°C (Fig. 7). Since previous work had shown quantitative loss of ABF1 from the SPT15 promoter under these conditions, this suggests that ABF1 similarly is mostly or completely lost from the modified HIS4 promoter. We suggest that, in contrast to RAP1, ABF1 changes the promoter in a way that is stable to loss of ABF1 and that facilitates transcriptional activation. The most obvious possibility is that ABF1 modifies chromatin, either directly or via recruitment of chromatin-modifying enzyme(s), in a way different from that of RAP1. Future work will be directed at testing this possibility.
Acknowledgments
We thank Liuning Yu for providing the HIS4-MEL1 reporter gene, Wolfram Horz for providing yeast strains, and the Wadsworth Center Molecular Genetics Core for DNA sequencing and oligonucleotide synthesis.
This work was supported by grants from the National Institutes of Health (GM51993 to R.H.M. and GM57893 to R.L.).
REFERENCES
- 1.Barbaric, S., M. Munsterkotter, C. Goding, and W. Horz. 1998. Cooperative Pho2-Pho4 interactions at the PHO5 promoter are critical for binding of Pho4 to UASp1 and for efficient transactivation by Pho4 at UASp2. Mol. Cell. Biol. 18:2629-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bodmer-Glavas, M., K. Edler, and A. Barberis. 2001. RNA polymerase II and III transcription factors can stimulate DNA replication by modifying origin chromatin structures. Nucleic Acids Res. 29:4570-4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chasman, D. I., N. F. Lue, A. R. Buchman, J. W. LaPointe, Y. Lorch, and R. D. Kornberg. 1990. A yeast protein that influences the chromatin structure of UASG and functions as a powerful auxiliary gene activator. Genes Dev. 4:503-514. [DOI] [PubMed] [Google Scholar]
- 4.Devlin, C., K. Tice-Baldwin, D. Shore, and K. T. Arndt. 1991. RAP1 is required for BAS1/BAS2- and GCN4-dependent transcription of the yeast HIS4 gene. Mol. Cell. Biol. 11:3642-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diffley, J. F., and B. Stillman. 1989. Similarity between the transcriptional silencer binding proteins ABF1 and RAP1. Science 246:1034-1038. [DOI] [PubMed] [Google Scholar]
- 6.Fourel, G., T. Miyake, P. A. Defossez, R. Li, and E. Gilson. 2002. General regulatory factors (GRFs) as genome partitioners. J. Biol. Chem. 277:41736-41743. [DOI] [PubMed] [Google Scholar]
- 7.Gartenberg, M. R. 2000. The Sir proteins of Saccharomyces cerevisiae: mediators of transcriptional silencing and much more. Curr. Opin. Microbiol. 3:132-137. [DOI] [PubMed] [Google Scholar]
- 8.Goncalves, P. M., G. Griffioen, R. Minnee, M. Bosma, L. S. Kraakman, W. H. Mager, and R. J. Planta. 1995. Transcription activation of yeast ribosomal protein genes requires additional elements apart from binding sites for Abf1p or Rap1p. Nucleic Acids Res. 23:1475-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goncalves, P. M., K. Maurer, G. van Nieuw Amerongen, K. Bergkamp-Steffens, W. H. Mager, and R. J. Planta. 1996. C-terminal domains of general regulatory factors Abf1p and Rap1p in Saccharomyces cerevisiae display functional similarity. Mol. Microbiol. 19:535-543. [DOI] [PubMed] [Google Scholar]
- 10.Graham, I. R., R. A. Haw, K. G. Spink, K. A. Halden, and A. Chambers. 1999. In vivo analysis of functional regions within yeast Rap1p. Mol. Cell. Biol. 19:7481-7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grunstein, M. 1997. Molecular model for telomeric heterochromatin in yeast. Curr. Opin. Cell Biol. 9:383-387. [DOI] [PubMed] [Google Scholar]
- 12.Halfter, H., B. Kavety, J. Vandekerckhove, F. Kiefer, and D. Gallwitz. 1989. Sequence, expression and mutational analysis of BAF1, a transcriptional activator and ARS1-binding protein of the yeast Saccharomyces cerevisiae. EMBO J. 8:4265-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardy, C. F. J., L. Sussel, and D. Shore. 1992. Dissection of a carboxyl-terminal region of the yeast regulatory protein RAP1 with effects on both transcriptional activation and silencing. Mol. Cell. Biol. 12:1209-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill, J., K. A. Donald, D. E. Griffiths, and G. Donald. 1991. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 19:5791. (Erratum, 19:6688.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, Y. F., Z. L. Hao, and R. Li. 1999. Chromatin remodeling and activation of chromosomal DNA replication by an acidic transcriptional activation domain from BRCA1. Genes Dev. 13:637-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kent, N. A., L. E. Bird, and J. Mellor. 1993. Chromatin analysis in yeast using NP-40 permeabilised sphaeroplasts. Nucleic Acids Res. 21:4653-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurtz, S., and D. Shore. 1991. RAP1 protein activates and silences transcription of mating-type genes in yeast. Genes Dev. 5:616-628. [DOI] [PubMed] [Google Scholar]
- 19.Lascaris, R. F., E. Groot, P. B. Hoen, W. H. Mager, and R. J. Planta. 2000. Different roles for Abf1p and a T-rich promoter element in nucleosome organization of the yeast RPS28A gene. Nucleic Acids Res. 28:1390-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, R., D. S. Yu, M. Tanaka, L. Zheng, S. L. Berger, and B. Stillman. 1998. Activation of chromosomal DNA replication in Saccharomyces cerevisiae by acidic transcriptional activation domains. Mol. Cell. Biol. 18:1296-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lieb, J. D., X. Liu, D. Botstein, and P. O. Brown. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28:327-334. [DOI] [PubMed] [Google Scholar]
- 22.Marahrens, Y., and B. Stillman. 1992. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255:817-823. [DOI] [PubMed] [Google Scholar]
- 23.Martens, J. A., and C. J. Brandl. 1994. GCN4p activation of the yeast TRP3 gene is enhanced by ABF1p and uses a suboptimal TATA element. J. Biol. Chem. 269:15661-15667. [PubMed] [Google Scholar]
- 24.Mencia, M., Z. Moqtaderi, J. V. Geisberg, L. Kuras, and K. Struhl. 2002. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell 9:823-833. [DOI] [PubMed] [Google Scholar]
- 25.Miller, J. A., and J. Widom. 2003. Collaborative competition mechanism for gene activation in vivo. Mol. Cell. Biol. 23:1623-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyake, T., C. M. Loch, and R. Li. 2002. Identification of a multifunctional domain in autonomously replicating sequence-binding factor 1 required for transcriptional activation, DNA replication, and gene silencing. Mol. Cell. Biol. 22:505-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morse, R. H. 2003. Getting into chromatin: how do transcription factors get past the histones? Biochem. Cell Biol. 81:101-112. [DOI] [PubMed] [Google Scholar]
- 28.Morse, R. H. 1993. Nucleosome disruption by transcription factor binding in yeast. Science 262:1563-1566. [DOI] [PubMed] [Google Scholar]
- 29.Morse, R. H. 2000. RAP, RAP, open up! New wrinkles for RAP1 in yeast. Trends Genet. 16:51-53. [DOI] [PubMed] [Google Scholar]
- 30.Morse, R. H., S. Y. Roth, and R. T. Simpson. 1992. A transcriptionally active tRNA gene interferes with nucleosome positioning in vivo. Mol. Cell. Biol. 12:4015-4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pal, S., A. B. Cantor, K. D. Johnson, T. B. Moran, M. E. Boyer, S. H. Orkin, and E. H. Bresnick. 2004. Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc. Natl. Acad. Sci. USA 101:980-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pilpel, Y., P. Sudarsanam, and G. M. Church. 2001. Identifying regulatory networks by combinatorial analysis of promoter elements. Nat. Genet. 29:153-159. [DOI] [PubMed] [Google Scholar]
- 33.Pina, B., J. Fernandez-Larrea, N. Garcia-Reyero, and F. Z. Idrissi. 2003. The different (sur)faces of Rap1p. Mol. Genet. Genomics 268:791-798. [DOI] [PubMed] [Google Scholar]
- 34.Polach, K. J., and J. Widom. 1995. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 254:130-149. [DOI] [PubMed] [Google Scholar]
- 35.Pryde, F. E., and E. J. Louis. 1999. Limitations of silencing at native yeast telomeres. EMBO J. 18:2538-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reed, S. H., M. Akiyama, B. Stillman, and E. C. Friedberg. 1999. Yeast autonomously replicating sequence binding factor is involved in nucleotide excision repair. Genes Dev. 13:3052-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reid, J. L., V. R. Iyer, P. O. Brown, and K. Struhl. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6:1297-1307. [DOI] [PubMed] [Google Scholar]
- 38.Rhode, P. R., S. Elsasser, and J. L. Campbell. 1992. Role of multifunctional autonomously replicating sequence binding factor 1 in the initiation of DNA replication and transcriptional control in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:1064-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhode, P. R., K. S. Sweder, K. F. Oegema, and J. L. Campbell. 1989. The gene encoding ARS-binding factor I is essential for the viability of yeast. Genes Dev. 3:1926-1939. [DOI] [PubMed] [Google Scholar]
- 40.Rolfes, R. J., F. Zhang, and A. G. Hinnebusch. 1997. The transcriptional activators BAS1, BAS2, and ABF1 bind positive regulatory sites as the critical elements for adenine regulation of ADE5,7. J. Biol. Chem. 272:13343-13354. [DOI] [PubMed] [Google Scholar]
- 41.Ryan, M. P., R. Jones, and R. H. Morse. 1998. SWI-SNF complex participation in transcriptional activation at a step subsequent to activator binding. Mol. Cell. Biol. 18:1774-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryan, M. P., G. A. Stafford, L. Yu, and R. H. Morse. 2000. Artificially recruited TATA-binding protein fails to remodel chromatin and does not activate three promoters that require chromatin remodeling. Mol. Cell. Biol. 20:5847-5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sabet, N., F. Tong, J. P. Madigan, S. Volo, M. M. Smith, and R. H. Morse. 2003. Global and specific transcriptional repression by the histone H3 amino terminus in yeast. Proc. Natl. Acad. Sci. USA 100:4084-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schroeder, S. C., and P. A. Weil. 1998. Genetic tests of the role of Abf1p in driving transcription of the yeast TATA box binding protein-encoding gene, SPT15. J. Biol. Chem. 273:19884-19891. [DOI] [PubMed] [Google Scholar]
- 45.Shore, D. 1994. RAP1: a protean regulator in yeast. Trends Genet. 10:408-412. [DOI] [PubMed] [Google Scholar]
- 46.Simpson, R. T. 1991. Nucleosome positioning: occurrence, mechanisms, and functional consequences. Prog. Nucleic Acid Res. Mol. Biol. 40:143-184. [DOI] [PubMed] [Google Scholar]
- 47.Springer, C., S. Krappmann, M. Kunzler, C. Zmasek, and G. H. Braus. 1997. Regulation of the yeast HIS7 gene by the global transcription factor Abf1p. Mol. Gen. Genet. 256:136-146. [DOI] [PubMed] [Google Scholar]
- 48.Springer, C., M. Kunzler, T. Balmelli, and G. H. Braus. 1996. Amino acid and adenine cross-pathway regulation act through the same 5′-TGACTC-3′ motif in the yeast HIS7 promoter. J. Biol. Chem. 271:29637-29643. [DOI] [PubMed] [Google Scholar]
- 49.Stafford, G. A., and R. H. Morse. 1997. Chromatin remodeling by transcriptional activation domains in a yeast episome. J. Biol. Chem. 272:11526-11534. [DOI] [PubMed] [Google Scholar]
- 50.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11:83-93. [DOI] [PubMed] [Google Scholar]
- 51.Thoma, F. 1992. Nucleosome positioning. Biochim. Biophys. Acta 1130:1-19. [DOI] [PubMed] [Google Scholar]
- 52.Thoma, F., L. W. Bergman, and R. T. Simpson. 1984. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J. Mol. Biol. 177:715-733. [DOI] [PubMed] [Google Scholar]
- 53.Valerius, O., C. Brendel, C. Wagner, S. Krappmann, F. Thoma, and G. H. Braus. 2003. Nucleosome position-dependent and -independent activation of HIS7 expression in Saccharomyces cerevisiae by different transcriptional activators. Eukaryot. Cell 2:876-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Venditti, P., G. Costanzo, R. Negri, and G. Camilloni. 1994. ABFI contributes to the chromatin organization of Saccharomyces cerevisiae ARS1 B-domain. Biochim. Biophys. Acta 1219:677-689. [DOI] [PubMed] [Google Scholar]
- 55.Wang, Y., C. L. Liu, J. D. Storey, R. J. Tibshirani, D. Herschlag, and P. O. Brown. 2002. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. USA 99:5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiltshire, S., S. Raychaudhuri, and S. Eisenberg. 1997. An Abf1p C-terminal region lacking transcriptional activation potential stimulates a yeast origin of replication. Nucleic Acids Res. 25:4250-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winston, F., C. Dollard, and S. L. Ricupero-Hovasse. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53-55. [DOI] [PubMed] [Google Scholar]
- 58.Yu, L., and R. H. Morse. 1999. Chromatin opening and transactivator potentiation by RAP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:5279-5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu, L., N. Sabet, A. Chambers, and R. H. Morse. 2001. The N-terminal and C-terminal domains of RAP1 are dispensable for chromatin opening and GCN4-mediated HIS4 activation in budding yeast. J. Biol. Chem. 276:33257-33264. [DOI] [PubMed] [Google Scholar]
- 60.Yu, Q., R. Qiu, T. B. Foland, D. Griesen, C. S. Galloway, Y. H. Chiu, J. Sandmeier, J. R. Broach, and X. Bi. 2003. Rap1p and other transcriptional regulators can function in defining distinct domains of gene expression. Nucleic Acids Res. 31:1224-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]