Abstract
The histone amino termini have emerged as key targets for a variety of modifying enzymes that function as transcriptional coactivators and corepressors. However, an important question that has remained largely unexplored is the extent to which specific histone amino termini are required for the activating and repressive functions of these enzymes, Here we address this issue by focusing on the prototypical histone deacetylase, Rpd3p, in the budding yeast Saccharomyces cerevisiae. We show that targeting Rpd3p to a reporter gene in this yeast can partially repress transcription when either the histone H3 or the histone H4 amino terminus is deleted, indicating that the “tails” are individually dispensable for repression by Rpd3p. In contrast, we find that the effect of rpd3 gene disruption on global gene expression is considerably reduced in either a histone H3Δ1-28 (H3 lacking the amino-terminal 28 amino acids) or a histone H4(K5,8,12,16Q) (H4 with lysine residues 5, 8, 12, and 16 changed to glutamine residues) background compared to the wild-type background, indicating a requirement for one or both of these histone tails in Rpd3p-mediated regulation for many genes. These results suggest that acetylation of either the H3 or the H4 amino terminus could suffice to allow the activation of such genes. We also examine the relationship between H3 tails and H4 tails in global gene expression and find substantial overlap among the gene sets regulated by these histone tails. We also show that the effects on genome-wide expression of deleting the H3 or H4 amino terminus are similar but not identical to the effects of mutating the lysine residues in these same regions. These results indicate that the gene regulatory potential of the H3 and H4 amino termini is substantially but not entirely contained in these modifiable lysine residues.
DNA in eukaryotes is packaged into chromatin by the histone proteins. The four core histones, H2A, H2B, H3, and H4, have amino-terminal “tails” that extend from the structured interior and that are subject to numerous posttranslational modifications (7, 54). Acetylation and deacetylation of the histone H3 and H4 amino termini are particularly important for gene regulation, but basic questions about the mechanism by which they affect transcription remain unanswered. For instance, a variety of enzymes can acetylate or deacetylate the H3 and/or H4 tails with different specificities, but it is not known whether modifications of specific lysine residues vary in importance at particular promoters or whether acetylation activates transcription by removing a repressive influence (unacetylated histone tails) or by providing a required positive stimulus (e.g., providing a binding surface for general transcription factors) (1). Furthermore, histone-modifying enzymes may also posttranslationally target other, nonhistone targets, complicating analysis of their functions (6, 22, 43).
In an effort to understand how histone modifications affect gene regulation, we have focused in this study on histone deacetylase Rpd3p from the budding yeast Saccharomyces cerevisiae. This protein was first identified as a histone deacetylase in a screen for rat proteins binding to trapoxin, a histone deacetylase inhibitor; sequence analysis then showed it to be a homolog of the known yeast corepressor Rpd3p (46). Subsequent work demonstrated that the histone deacetylase activity of Rpd3p is needed for it to function as a corepressor and that promoter-bound histones show decreased acetylation upon recruitment of Rpd3p to nearby sites (18, 20, 35). The loss of Rpd3p results in a global increase in the acetylation of lysine residues 5, 8, 12, and 16 of histone H4 and lysine residues 9 or 18 and 14 of histone H3 (32, 34, 50), whereas promoter-bound nucleosomes near sites of Rpd3p recruitment have been reported in various studies to show the deacetylation of principally K5 of histone H4 (35); K5 and K12 of H4 and undetermined sites of histone H3 (20); and K5, K8, and K12 of H4, K9, K14, K18, K23, and K27 of H3, K27 of histone H2A and, to somewhat lesser extent, K11 and K16 of histone H2B (45).
Rpd3p affects the regulation of a substantial number of genes in S. cerevisiae (3). It can be recruited to gene promoters by the repressor Ume6p but evidently by other mechanisms as well (19, 23, 32), and it is also responsible for a lower level of untargeted histone deacetylation throughout the genome in S. cerevisiae (50). Chromatin immunoprecipitation experiments have shown that repression by Rpd3p is associated with reduced levels of promoter-bound TATA binding protein, SWI/SNF, and SAGA components, and it has been suggested that reduced acetylation of histone amino termini could weaken the interactions of components of SWI/SNF and/or SAGA with promoter-bound nucleosomes, resulting in decreased levels of transcription (9, 41). However, other histone-modifying enzymes have been shown to target nonhistone proteins with functional consequences (22, 43), and it is possible that Rpd3p does so as well.
A prediction that has not yet been tested is that if the histone amino termini are the principal functionally important targets of Rpd3p, then the deletion of Rpd3p should have a reduced (or no) effect in their absence. A similar approach was used previously to test the importance of the histone H3 and H4 amino termini for regulation by Gcn5p in S. cerevisiae (53). Here we report experiments testing this prediction at the gene-specific and genome-wide levels. We also compare, by microarray analysis, the roles of the H3 and H4 amino termini in gene regulation at the genome-wide level and examine the extent to which the loss of the H3 and H4 tails and the mutation of lysines to glutamines results in overlapping phenotypes with respect to transcriptional regulation. Our results provide a global view of the roles of the H3 and H4 amino termini in gene regulation in S. cerevisiae and their relationship to regulation by Rpd3p.
MATERIALS AND METHODS
Plasmids.
Plasmids CHA1-LexA-MEL1, GALl0-LexA-MEL1, and pRS414GAL4-ER-VP16 were described previously (37, 42). Plasmids pRS425-LexARPD3 and pRS425-LexARPD3(H188A) were created by ligating the SacI-PstI fragment from YEplac112-Rpd3-LexA and the H188A mutant (18), generously provided by David Kadosh and Kevin Struhl, into pRS425 (8). Plasmids pNS329, pNS338, and pNS358, harboring HHF1-HHT1, hhf1-8(H4Δ2-26)-HHT1, and HHF1-hht1-2(H3Δ1-28), respectively, on pRS414, were described previously (38). To construct pNS491 {H4 with lysine residues 5, 8, 12, and 16 changed to glutamine residues [H4(K5,8,12,16Q)]}, the EcoRI-SalI fragment from pMS329 carrying HHF1-HHT1 (30) was subcloned into pRS414 (8) from which the BamHI site had been removed by filling in with the Klenow fragment (pRS414ΔBamHI); the BamHI fragment harboring HHF1 then was replaced with hhf1-10 from pMS391, generously provided by M. M. Smith. Plasmid pCL460 was constructed by first replacing the EcoRI-SmaI fragment from pMS358 containing hht1-2 (30) with hht1-3 from pMS391 and then cloning the EcoRI-SalI fragment from the resulting plasmid into pRS414ΔBamHI. Mutant histone genes were verified by sequencing.
Yeast strains.
The strains used in this study are listed in Table 1. Disruption of the LEU2 gene in strains MHY308 and PKY918 (generously provided by Michael Grunstein) was done by two-step disruption with a URA3-marked leu2Δ vector. Disruption of RPD3 was accomplished by one-step disruption with a LEU2 marker and verified by PCR and Southern blotting. Strains carrying histone mutations were constructed by transforming MX1-4C {MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hhf1-hht1)(hhf2-hht2) pMS329 [CEN ARS URA3 HHT1-HHF1]} (30), in which wild-type HHF1-HHT1 is carried on a URA3-marked plasmid, with TRP1-marked plasmids harboring the appropriate mutant histone genes and selecting for 5-fluoroorotic acid (5-FOA)-resistant colonies. Histone replacement was verified by PCR amplification and sequencing.
TABLE 1.
Yeast strains used in this study
| Strain | Description | Reference or source |
|---|---|---|
| NSY429 | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hhf1-hht1)(hhf2-hht2) pNS329 [CEN TRP1 HHF1-HHT1] | 38 |
| NSY430 | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hhf1-hht1)(hhf2-hht2) pNS329 [CEN TRP1 HHF1-HHT1] rpd3::LEU2 | This work |
| NSY438 | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hhf1-hht1)(hhf2-hht2) pNS338 [CEN TRP1 hhf1-8 (H4Δ2-26) HHT1] | 38 |
| NSY458 | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hhf1-hht1)(hhf2-hht2) pNS358 [CEN TRP1 HHF1 hht1-2 (H3Δ1-28)] | 38 |
| NSY459 | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hhf1-hht1)(hhf2-hht2) pNS358 [CEN TRP1 HHF1 hht1-2 (H3Δ1-28)] rpd3::LEU2 | This work |
| CLY460 | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hhf1-hht1)(hhf2-hht2) pCL460 [CEN TRP1 HHF1 hht1-3 H3(K4,9,14,18,23,27Q)] | This work |
| RMY491 | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hhf1-hht1)(hhf2-hht2) pNS491 [CEN TRP1 hhf1-10 H4(K5,8,12,16Q) HHT1] | This work |
| RMY492 | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hhf1-hht1)(hhf2-hht2) pNS491 [CEN TRP1 hhf1-10 H4(K5,8,12,16Q) HHT1] rpd3::LEU2 | This work |
| MHY308 | MATaade2-101 arg4-1 his3-Δ200 leu2-3,112 lys2-801 trp1-Δ901 ura3-52 thr tyr hhf1::HIS3 hhf2::LEU2 pUK499 (HHF2) | 16 |
| RMY308 | As for MHY308 but leu2 | This work |
| PKY918 | MATaade2-101 arg4-1 his3-Δ200 leu2-3,112 lys2-801 trp1-901 ura3-52 thr tyr hhf1::HIS3 hhf2::LEU2 pPK618 (hhf2Δ4-19) | 21 |
| RMY919 | As for PKY918 but leu2 | This work |
Alpha-galactosidase assays.
Yeast cells were grown at 30°C in complete synthetic dropout medium (CSM; Bio 101) containing 2% dextrose or 1.5% raffinose plus 2% galactose (for GAL10 induction) (33). The CHA1 promoter was induced by supplementing cultures with 1 mg of serine/ml for >4 h. GAL4-ER-VP16 was induced by the addition of 0.1 μM β-estradiol (42). Alpha-galactosidase activity (encoded by MEL1) was measured as previously described (36).
Microarrays.
Yeast cultures were grown in yeast extract-peptone-dextrose medium (33) to mid-logarithmic phase, and RNA was prepared by hot phenol extraction (40) and purified with RNeasy kits (Qiagen). Processing and hybridization to Affymetrix (Santa Clara, Calif.) S98 microarrays were performed according to the manufacturer's protocol as described previously (38). At least two independent RNA preparations were used for each mutant, and three were used for the wild-type yeast strain. Scatter plots include only genes which were identified as being present by Affymetrix software (meaning that they were expressed above background levels) and depict the expression of a single mRNA preparation (the ordinate) against averaged values from two [H3Δ1-28 (H3 lacking the amino-terminal 28 amino acids) or H4(K5,8,12,16Q)] or three [wild type or H3(K4,9,14,18,23,27Q)] independent mRNA preparations (the abscissa). Changes in gene expression used for the construction of tables and for cluster analysis were derived by averaging log2 changes in expression (compared to the averaged wild-type expression values) for each mutant.
Statistical significance was assessed by performing an unpaired t test for each mutant and the corresponding wild type; P values were adjusted by using the Benjamini-Hochberg statistic, which is closely coupled to the false discovery rate (2). Analysis was performed by using Microsoft Excel, Genespring (Affymetrix), GeneTraffic (Iobion Informatics), and Cluster and Treeview (14). Munich Information Center for Protein Sequences (MIPS) categorization was done by using the web-based tool FunSpec (http://funspec.med.utoronto.ca/) (31).
Nucleotide sequence accession number.
Microarray gene expression data have been submitted to Gene Expression Omnibus under accession number GSE1639 and are also accessible at www.wadsworth.org/resnres/bios/morse.htm.
RESULTS
Effect of targeted recruitment of Rpd3p on activated transcription.
We first examined the importance of the histone H3 and H4 amino termini as functional targets for Rpd3p-mediated repression of activated transcription. For this purpose, we used two reporter genes in which promoters containing activator binding sites and LexA binding sites were fused to the MEL1 coding sequence (Fig. 1). The MEL1 gene encodes alpha-galactosidase, which is easily quantifiable (36). The two promoters, from the GAL10 and CHA1 genes, each contain positioned nucleosomes that incorporate their TATA elements under uninduced conditions; these nucleosomes are disrupted upon activation (24, 29, 37). Ryan et al. previously showed that the chromatin structures of these two promoters are retained in MEL1 fusions (36, 37). The LexA binding sites were placed close to these nucleosomal sequences so that the recruitment of Rpd3p as a LexA-Rpd3p fusion could target these promoter-proximal nucleosomes, as previous work indicated that histone deacetylation accompanying Rpd3p-mediated repression is localized to the proximal-promoter region of affected genes (20, 35). Furthermore, by comparing the effects of recruitment of Rpd3p to that of a mutant version of Rpd3p (H188A) that lacks deacetylase activity, we could specifically monitor the contribution of deacetylation to transcriptional repression (18). Our rationale was that if we found conditions under which the recruitment of enzymatically active Rpd3p could repress activator-mediated transcription, then we could examine whether this repression was compromised in yeast cells lacking the histone H3 or H4 amino terminus.
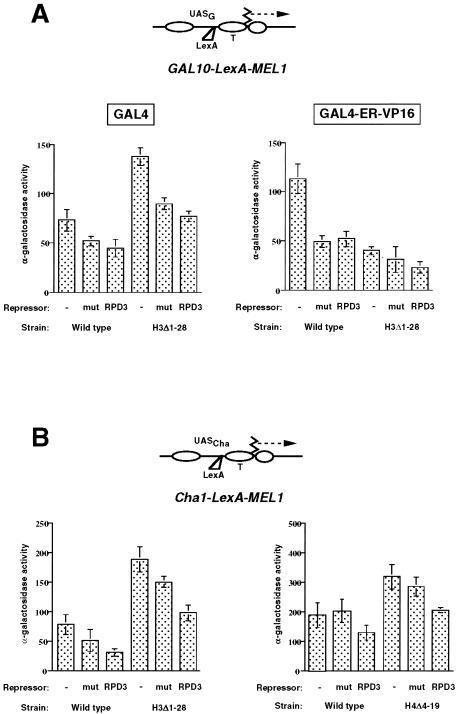
FIG. 1.
Effect of Rpd3p recruitment on activated transcription of two reporter genes. (A) The expression of a GAL10-MEL1 reporter gene containing a LexA binding site (top) was measured in galactose-containing medium (GAL4 activation) (left) or in the presence of GAL4-ER-VP16 and β-estradiol (right). Activity was measured in wild-type (NSY429) or H3Δ1-28 (NSY458) cells and in the presence of an empty vector (pRS415) (−), LexA-Rpd3p (RPD3), or the H188A mutant of LexA-Rpd3p (mut). With glucose, activity was less than 5 U in both wild-type and H3Δ1-28 yeast cells in the presence of an empty vector (data not shown). (B) The expression of a CHA1-MEL1 reporter gene containing a LexA binding site (top) was measured in the presence of serine (induced conditions). Activity was measured in wild-type (NSY438) or H3Δ1-28 (NSY458) yeast cells (left) or in matched wild-type (RMY308) or H4Δ4-19 (RMY919) yeast cells (right) and in the presence of an empty vector (−), LexA-Rpd3p (RPD3), or the H188A mutant of LexA-Rpd3p (mut). In the absence of serine, activities were less than 20% induced levels, except for H3Δ1-28 yeast cells (see the text). All measurements were performed with at least three independent colonies, and standard deviations are indicated. UAS, upstream activation sequence; T, TATA element.
Figure 1A shows that the deacetylase activity of Rpd3p did not significantly inhibit transcriptional activation of the GAL10 promoter by either Gal4p or GAL4-ER-VP16; similar results were obtained for a GAL4-HAP4 fusion protein (data not shown). Although significantly reduced transcription was seen in the presence of LexA-Rpd3p, especially for GAL4-ER-VP16, inhibition was the same for the catalytically inactive H188A mutant version of Rpd3p (fused to LexA) as for the native Rpd3p. These results indicate that the inhibition observed was likely due to steric interference caused by LexA-Rpd3p binding between the binding sites for Gal4p and the TATA elements (4) and either that histone deacetylation is not inhibitory at the GAL10 promoter or that the activators are able to overcome the activity of Rpd3p by recruiting histone acetyltransferase(s).
In contrast, the recruitment of catalytically active Rpd3p to the CHA1 promoter was significantly more repressive than the recruitment of the H188A mutant version of Rdp3p (P value determined by the Student t test, <0.001) (Fig. 1B). This result indicates that the deacetylase activity of Rpd3p, when targeted to the CHA1 promoter, is capable of inhibiting Cha4p-mediated activation (38). Consistent with this inhibition, chromatin immunoprecipitation experiments revealed a modest (about twofold) decrease in the acetylation of histone H4 at the CHA1-LexA-MEL1 promoter in the presence of LexA-Rpd3p relative to the empty vector control, whereas no such decrease was measured in the presence of the inactive H188A mutant version or at the endogenous CHA1 promoter (which lacks a LexA binding site) (data not shown).
To examine whether the amino termini of histones H3 and H4 were critical targets for the Rpd3p-mediated repression of CHA1 activation, we repeated this experiment with a congenic yeast strain lacking the histone H3 amino terminus (H3Δ1-28) and with another matched set of strains expressing either wild-type histone H4 or histone H4 lacking the amino terminus (H4Δ4-19). In both cases, significantly more repression was observed upon recruitment of catalytically active Rpd3p than upon recruitment of the inactive H188A mutant version of Rpd3p (P < 0.001) (Fig. 1B). A similar effect was seen under uninduced conditions with H3Δ1-28, in which the CHA1 promoter is strongly derepressed (38) (data not shown). Activation by GAL4 and GAL4-ER-VP16 was reduced similarly by LexA-Rpd3p and the H188A mutant version of Rpd3p in H3Δ1-28, as was seen with the wild type (Fig. 1A). We do not understand the reason for the increased activation by GAL4 seen in H3Δ1-28, although it is consistent with an earlier report of increased GAL gene activation under partially derepressing conditions in yeast cells lacking the H3 amino terminus (25). We conclude that the H3 and H4 amino termini are individually dispensable for the mild repression of the CHA1 promoter caused by the targeted recruitment of Rpd3p.
Global analysis of interactions between the H3 and H4 amino termini and Rpd3p.
To assess the functional importance of the histone H3 and H4 amino termini as targets for Rpd3p-mediated repression on a genome-wide basis, we performed microarray experiments with Affymetrix glass slides. We first examined the effect of RPD3 deletion on global gene expression in wild-type and H3Δ1-28 yeast cells (Fig. 2B and C). Deletion of RPD3 in wild-type yeast cells affects the regulation of about 5% of the total yeast genome by twofold or more when cells are grown in rich medium (yeast extract-peptone-dextrose medium), with a 2.5-fold bias toward increased expression in rpd3Δ yeast cells (Fig. 2B and E). These results are consistent with the view that Rpd3p is generally repressive, although they contrast somewhat with previously reported microarray results (3). The number of genes affected by RPD3 deletion is greatly reduced in H3Δ1-28 yeast cells (Fig. 2B, C, and E). The smaller effect of RPD3 deletion in H3Δ1-28 yeast cells than in wild-type yeast cells was seen whether the histone H3 genes were genomic or carried on a CEN-containing plasmid (data not shown). These results indicate that on a genome-wide basis, the number of genes regulated by Rpd3p in yeast cells grown in rich medium is considerably reduced by removal of the histone H3 amino terminus. These results are consistent with the H3 tail being a critical regulatory target of Rpd3p.
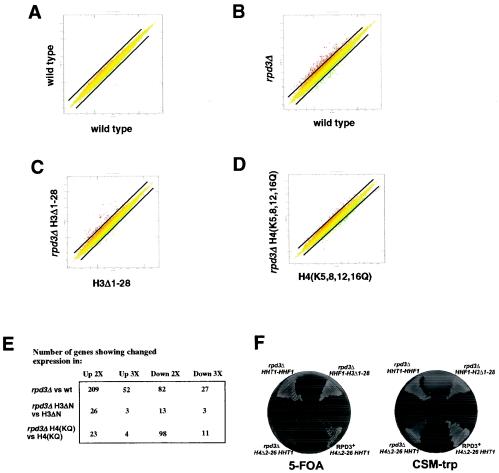
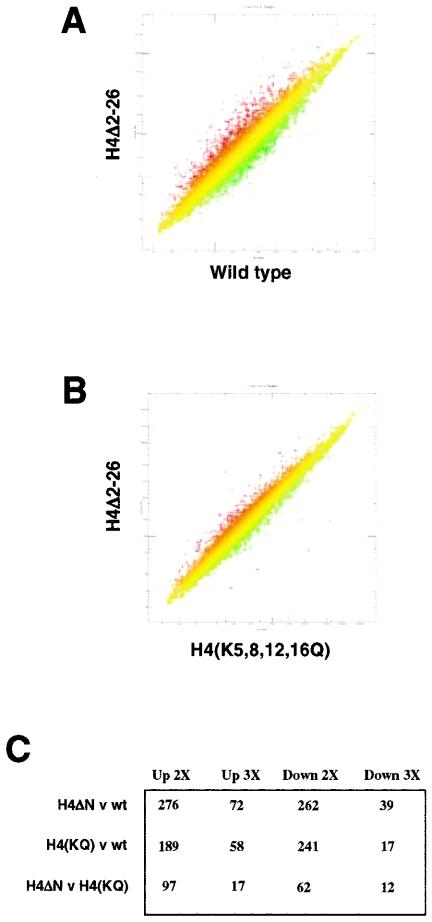
FIG. 2.
The effect of rpd3 deletion on genome-wide gene expression is reduced when the H3 tail is deleted or the H4 tail lysines are mutated. (A) Scatter plot of gene expression in one mRNA preparation from wild-type yeast cells (NSY429) compared to averaged values from three independent mRNA preparations from wild-type yeast cells (including the one being compared). Each point represents the expression of an individual gene. The diagonal lines represent twofold changes in expression. For this and all subsequent scatter plots, only genes indicated by Affymetrix software as being present (i.e., expressed above background levels) are shown. (B) Scatter plot of gene expression in rpd3Δ (NSY430) and wild-type (NSY429) yeast cells. (C) Scatter plot of gene expression in rpd3Δ H3Δ1-28 (NSY459) and H3Δ1-28 (NSY458) yeast cells. (D) Scatter plot of gene expression in rpd3Δ H4(K5,8,12,16Q) (RMY492) and H4(K5,8,12,16Q) (RMY491) yeast cells. (E) Table of gene expression changes for the indicated comparisons. Entries are derived from averaged values from at least two independent mRNA preparations and have Benjamini-Hochberg-derived P values (2) of <0.05. wt, wild type. (F) Synthetic growth defect of rpd3Δ and H4Δ2-26 yeast cells. Wild-type or rpd3Δ yeast cells harboring HHT1-HHF1 on a TRP1-marked plasmid and the indicated combination of hht1 and hhf1 alleles on a URA3-marked plasmid were streaked on 5-FOA and CSM-trp plates and allowed to grow for 2 days at 30°C.
We wished to similarly examine the effect of RPD3 deletion in yeast cells lacking the H4 amino terminus but were unable either to create an rpd3 deletion in H4Δ2-26 yeast cells or to replace the wild-type HHF1 gene with hhf mutant genes encoding either H4Δ4-19 or H4Δ2-26 by plasmid shuffling in rpd3Δ yeast cells (Fig. 2F and data not shown). In those few cases in which we obtained 5-FOA-resistant colonies, sequencing showed that the TRP1-marked plasmid that originally carried the mutant hhf gene now harbored HHF1, which had presumably been “repaired” by recombination. Similarly, deletion of GCN5 has been reported to be synthetically lethal with the loss of either the H3 or the H4 amino terminus (53). Somewhat surprisingly, however, we were able to construct an rpd3Δ yeast strain expressing hhf1-10; in this strain, the codons for Lys-5, Lys-8, Lys-12, and Lys-16 were mutated to code for Gln, thereby mimicking the uncharged, acetylated state at these residues (27). As with the deletion of the H3 amino terminus, mutation of Lys-5, Lys-8, Lys-12, and Lys-16 at the H4 amino terminus resulted in substantially fewer genes being affected by the deletion of RPD3 than in cells expressing wild-type histone H4 (Fig. 2B, D, and E). This effect was particularly pronounced for genes showing increased expression upon loss of Rpd3p. We conclude that for many genes regulated by Rpd3p, loss of the modifiable lysine residues in either the H3 amino terminus (through its deletion) or the H4 amino terminus (by mutation) results in loss of this regulation.
The above results suggest that for many genes, Rpd3p-mediated regulation, particularly repression, requires the presence of the deacetylated amino terminus of either H3 or H4 or both. To study further the relationship of the genes repressed by Rpd3p and by the H3 and H4 tails, we examined the overlap between the genes showing increased expression upon deletion of the repressor Rpd3p and those showing increased expression (with a P value of <0.05 as a cutoff) upon removal of the H3 or H4 tail (Fig. 3A). Substantial overlap was observed among these data sets, with 159 genes showing increased expression in all three sets. Although indirect effects cannot be ruled out and doubtless pertain in some cases (see Discussion), this high degree of overlap strongly suggests that Rpd3p acts directly on many of these genes by targeting the histone tails. Even for cases in which indirect effects occur, the overlap suggests that many of these effects are likely to occur via the same or similar upstream events caused by the loss of Rpd3p or the H3 or H4 tail.
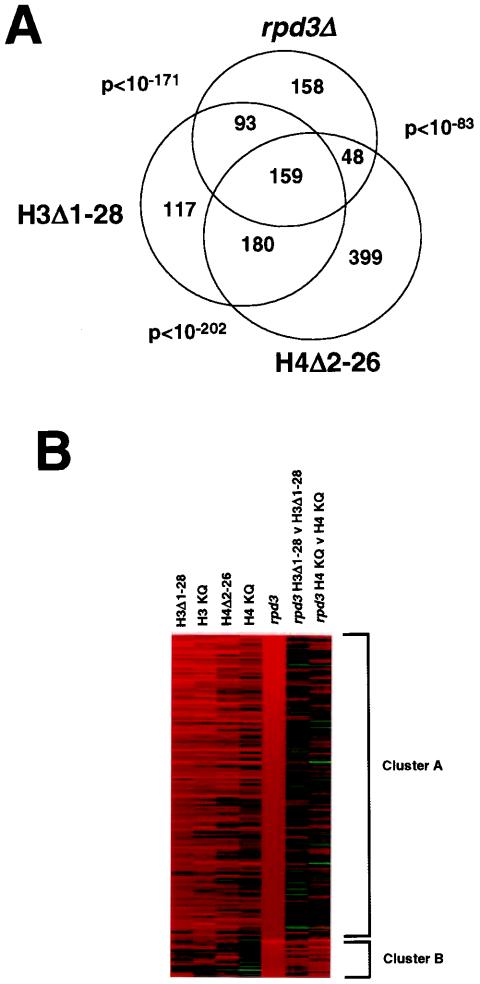
FIG. 3.
(A) Overlap among genes up-regulated twofold or more in rpd3Δ, H3Δ1-28, and H4Δ2-26 yeast cells. Only genes having altered expression values in rpd3Δ yeast cells with Benjamini-Hochberg-derived P values of <0.05 were considered. P values for the significance of overlap between pairwise-considered data sets were based on a hypergeometric distribution. (B) K-means cluster analysis (14) of the 209 genes showing at least a twofold increase in expression (P < 0.05) in rpd3Δ versus (v) wild-type yeast cells. Red represents increased gene expression and green represents decreased gene expression, with intensity being proportional to the magnitude of the change and the range of expression being from fourfold up to fourfold down (i.e., changes of greater than fourfold up or fourfold down are shown at maximal intensity).
A detailed comparison of the data with published work indicates some variability in the requirement for the H3 and H4 tails among genes likely to be direct targets of Rpd3p-mediated repression. Bernstein et al. (3) reported finding Ume6p binding sites upstream of 31 genes that were found to be up-regulated by the loss of Sin3p in a microarray analysis. We found that 18 of these genes showed an increase in expression of at least 1.6-fold (P < 0.05) in rpd3Δ yeast cells, but only 5 of these also showed an increase in expression (1.6-fold) (P < 0.05) in both H3Δ1-28 and H4Δ2-26 yeast cells, like the known Rpd3p-regulated genes INO and SPO11. One possible explanation for these results is that some of these genes may be regulated by Sin3p even in the absence of Rpd3p and therefore may be less dependent on the H3 or H4 tail for repression. We also found 96 genes that showed an increase (2-fold) in expression in rpd3Δ yeast cells but not (<1.3-fold) in rpd3Δ H3Δ1-28 (relative to H3Δ1-28) or rpd3Δ H4(K5,8,12,16Q) [relative to H4(K5,8,12,16)] yeast cells (that is, requiring both histone amino termini for Rpd3p-mediated repression). However, for only seven genes (PGM1, FOX2, ECI1, YGL250W, YJL163C, YLR446W, and YNL033W) were the individual intact histone amino termini dispensable for Rpd3p-mediated repression, as seen earlier for Rpd3p recruited to the CHA1 promoter. Future examination of these genes and other genes with anomalous behavior should provide new insights into gene regulation by the histone tails.
Cluster analysis provides additional support for the close relationship between genes regulated by Rpd3p and the histone H3 and H4 amino termini. Figure 3B shows the results of clustering of the 209 genes showing at least a twofold increase in expression in rpd3Δ versus wild-type yeast cells (P < 0.05). K-means cluster analysis (14) showed that these genes were divided into two principal groups when clustered according to the effect of removing or mutating the H3 or H4 tail. The majority (Fig. 3B, cluster A) showed a high degree of correlation with increased gene expression in yeast cells in which the H3 or H4 amino terminus was absent or in which lysines were mutated to glutamines. In contrast, little change was seen in the expression of these genes when RPD3 was deleted from cells lacking the H3 amino terminus or containing H4(K5,8,12,16Q). These results are not surprising in light of the already increased expression, relative to that in wild-type yeast cells, of this gene set in H3Δ1-28 or H4(K5,8,12,16Q) yeast cells. Nevertheless, they reinforce the genetic link, on a genome-wide scale, between Rpd3p-mediated repression and regulation by the histone H3 and H4 amino termini. Furthermore, these results strongly suggest that for many genes that are repressed by Rpd3p, acetylation of either the H3 or the H4 amino terminus could suffice to overcome this repression. As indicated above, a smaller set of genes showed increased expression when RPD3 was deleted from H3Δ1-28 or H4(K5,8,12,16Q) yeast cells and less change upon removal or mutation of the H3 or H4 tail (Fig. 3B, cluster B). This set may include genes for which a deacetylated H3 or H4 amino terminus suffices for repression; it is possible that the acetylation of both tails is normally required for the activation of such genes. The existence of such genes is consistent with the results of our analysis with CHA1-LexA-MEL1 (Fig. 1B), with which we found that Rpd3p-mediated repression could occur in the absence of either the H3 or the H4 amino terminus.
In summary, the microarray analysis presented here supports the idea that the histone H3 and H4 amino termini are important functional targets of Rpd3p. Many genes require intact H3 and H4 amino termini for Rpd3p-mediated repression, although some require only one or the other, and for a few genes, neither individual amino terminus is required. These findings suggest that the precise molecular mechanisms by which Rpd3p represses transcription differ for particular gene targets.
Comparison of histone amino-terminal deletions with mutations of amino-terminal lysine residues.
The histone H3 and H4 amino termini can be modified by acetylation or methylation of lysine residues, phosphorylation of serine residues, and methylation of arginine residues (7, 54). To assess the extent to which the effect of the loss of the H3 or H4 amino terminus on gene expression was due to the loss of modifiable lysine residues, we compared the effect of histone tail deletion to the effect of mutation of lysines to glutamines.
Sabet et al. reported on microarray studies of genome-wide changes in gene expression in H3Δ1-28 and H4Δ2-26 yeast cells relative to wild-type cells (38). In that work, slightly different strain backgrounds were used for the H3Δ1-28 and H4Δ2-26 (and corresponding wild-type) strains. Here, all strains investigated, including those containing wild-type histones, had one copy each of HHT1 and HHF1 on a CEN-containing plasmid with a TRP1 marker.
A comparison of gene expression in H3(K4,9,14,18,23,27Q) yeast cells against averaged values from three independent mRNA preparations from H3(K4,9,14,18,23,27Q) yeast cells showed little variation in gene expression, with most genes differing by less than twofold (Fig. 4A). Considerably more variability was seen in a comparison between H3Δ1-28 or H3(K4,9,14,18,23,27Q) and wild-type yeast cells (Fig. 4B and D and data not shown). Similar to the effect of the loss of the H3 amino terminus on gene expression (38), considerably more genes showed increased rather than decreased expression in the H3(K4,9,14,18,23,27Q) mutant strain (Fig. 4D). Similar results have been obtained for mutation of these lysine residues to glycine (26). A comparison of gene expression in H3Δ1-28 and H3(K4,9,14,18,23,27Q) yeast cells showed considerably fewer changes in gene expression (Fig. 4C and D). These results indicate that for many genes, a loss of charge in the H3 amino terminus has the same effect as a loss of the H3 tail. In a similar vein, linker histone H1 has been found to affect gene expession in Tetrahymena via a negatively charged patch created by phosphorylation or mutation (11).
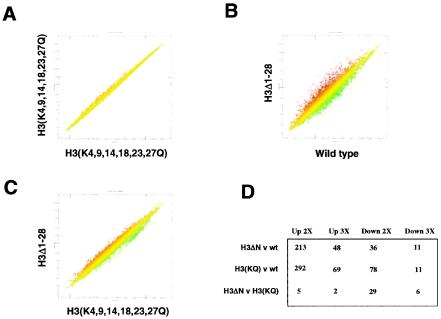
FIG. 4.
Comparison of genome-wide gene expression with H3 tail deletion and with H3 lysine-to-glutamine substitution mutation. (A) Scatter plot of gene expression in one mRNA preparation from H3(K4,9,14,18,23,27Q) (CLY601) yeast cells compared to averaged values from three independent mRNA preparations from H3(K4,9,14,18,23,27Q) yeast cells (including the one being compared). Each point represents the expression of an individual gene. (B) Scatter plot of gene expression in H3Δ1-28 (NSY458) and wild-type (NSY429) yeast cells. (C) Scatter plot of gene expression in H3Δ1-28 (NSY458) and H3(K4,9,14,18,23,27Q) yeast cells. (D) Table of gene expression changes for the indicated comparisons. Entries are derived from averaged values from each yeast strain, with Benjamini-Hochberg-derived P values for altered expression of <0.05. v, versus; wt, wild type.
Similar to our results obtained from an analysis of the H3 amino terminus, a comparison of H4Δ2-26 and H4(K5,8,12,16Q) yeast cells showed fewer changes in genome-wide expression than were seen when either individual mutant was compared with the wild type (Fig. 5). However, a substantial number of genes showed different effects of deletion and mutation of the H4 tail (Fig. 5C), suggesting that in these cases, the regulation of gene expression by the H4 tail depends on more than a simple charge effect. Also, in contrast to our results obtained from an analysis of the H3 amino terminus, deletion and mutation of the H4 amino terminus result in approximately equal numbers of genes showing increased expression and decreased expression, consistent with earlier results obtained from an analysis of H4 tail deletion (38).
FIG. 5.
Comparison of genome-wide gene expression with H4 tail deletion and with H4 lysine-to-glutamine substitution mutation. (A) Scatter plot of gene expression in H4Δ2-26 (NSY438) and wild-type (NSY429) yeast cells. (B) Scatter plot of gene expression in H4Δ2-26 (NSY438) and H4(K5,8,12,16Q) (RMY491) yeast cells. (C) Table of gene expression changes for the indicated comparisons. Entries are derived from averaged values from each yeast strain, with Benjamini-Hochberg-derived P values for altered expression of <0.05. v, versus; wt, wild type.
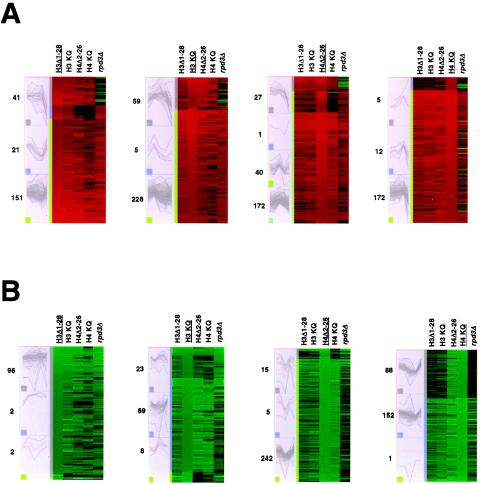
A high degree of correlation was also observed for the effects of the H3 and H4 tails on gene expression, as noted previously (Fig. 3A). To test these findings further, we performed K-means cluster analysis of the sets of genes most affected (up- or down-regulated) in the various histone tail mutants relative to the wild type (14). The results revealed strong correlations for the effect of deletion and the effect of mutation of the H3 and H4 amino termini on gene expression (Fig. 6). Correlations between tail deletions and mutations were stronger for the histone tail mutant strains than for the rpd3Δ strain. Notably, there was a substantial correlation among genes down-regulated in H4Δ2-26 and H4(K5,8,12,16Q) yeast cells, but there was considerably less correlation between either of these mutant strains and either strains with H3 tail mutations or the rpd3Δ yeast strain. The activation of specific genes has been shown to depend on an intact H4 amino terminus (12); perhaps some genes that are active during growth in rich medium exhibit a similar specific dependence on the H4 tail.
FIG. 6.
K-means cluster analyses. Genes showing expression increased (A) or decreased (B) by at least twofold, with Benjamini-Hochberg-derived P values of <0.05, were used for each group, except for those showing decreased expression in H3Δ1-28 or H3(K4,9,14,18,23,27Q) (H3 KQ) yeast cells, for which the 100 genes showing expression decreased by at least 1.6- or 1.9-fold, respectively, were used. H4 KQ, H4(K5,8,12,16Q). The underlined histone designation indicates the data set on which clustering was based. The panels on the left show the patterns of expression for the genes in each cluster, with the number of genes in each cluster indicated to the left; the red line in each panel indicates the average expression pattern within that cluster. Increasing K above the number shown did not significantly affect the resulting clustering. In the panels on the right, red represents increased gene expression and green represents decreased gene expression, with intensity being proportional to the magnitude of the change and the range of expression being from fourfold up to fourfold down (i.e., changes of greater than fourfold up or fourfold down are shown at maximal intensity).
DISCUSSION
In this study we examined the relationship between the histone H3 and H4 amino termini and Rpd3p in gene regulation in budding yeast. We found that the ability to repress gene activation through Rpd3p recruitment to a proximal promoter region is variable (Fig. 1). Repression of activation by Gal4p or GAL4-ER-VP16 by Rpd3p recruitment was independent of its deacetylase activity, indicating primarily steric effects (4) or repression by the Rpd3p-Sin3p complex independent of deacetylase activity (18). These data show that under some circumstances, Rpd3p recruitment is not sufficient for repression, and are consistent with chromatin immunoprecipitation experiments showing an association of Rpd3p with highly transcribed genes (23). In contrast, repression of activation by Cha4p (using the CHA1 promoter) depended strongly on Rpd3p deacetylase activity and was observed in the absence of either the H3 or the H4 amino terminus, indicating that these targets of Rpd3p deacetylase activity are individually dispensable for Rpd3p repressor function in at least some cases. Deckert and Struhl (9) also reported a variable ability of Rpd3p to repress transcription, depending on the activator. Their study did not include an analysis of the effect of an enzymatically deficient Rpd3p mutant, but since Rpd3p recruitment in their experiments was to a site upstream of the activator, steric effects are unlikely to have come into play.
Microarray experiments showed a strong dependence on both the histone H3 and the histone H4 amino termini for genome-wide regulation by Rpd3p, supporting the idea that the H3 and H4 tails are functionally important targets of Rpd3p on a genome-wide scale (Fig. 2 and 3). One important question that arises in such experiments is whether genes showing altered expression are directly or indirectly affected by the relevant mutation. One way in which indirect effects could occur is if one or a few genes responsible for controlling a major cellular process, such as cell cycle, were affected by a given mutation (e.g., RPD3 deletion or mutation of a histone tail). Analysis of the genes up-regulated by the loss of the H3 or H4 amino terminus or by the loss of Rpd3p by use of the MIPS database showed enrichment of genes involved in carbon compound metabolism and energy but not in the cell cycle or transcription (data available on request). Enrichment of genes involved in the stress response was seen among the set of genes up-regulated by the loss of the H4 amino terminus, and it is possible that this effect is largely indirect, if the loss of the H4 amino terminus results in the cells sensing stress in some way. Genes down-regulated by the H3 or H4 amino-terminal deletions or substitutions included many pheromone-responsive genes, a result expected on the basis of the loss of mating-type silencing observed in these mutants (21, 47). Interestingly, loss or mutation of the H4 tail causes a disproportionate down-regulation of genes involved in transcription (data available on request), suggesting a unique role for this amino terminus in regulating this subset of genes. Understanding the basis for this role will require further investigation.
Although the enrichment of genes affected by loss or mutation of the H3 or H4 amino terminus or of Rpd3p was seen in some categories, generally the affected genes were spread over many functional categories, suggesting that many of the affected genes are directly regulated by the histone tails and/or Rpd3p rather than that there are a few “master control” genes whose altered expression affects the remainder. Furthermore, the high concordance among genes affected by the histone tail mutations and the loss of Rpd3p also suggests a set of commonly controlled genes. Finally, as mentioned in Results, genes known to be directly regulated by Rpd3p are also affected by loss or mutation of the H3 and H4 amino termini. Together, these findings strongly suggest a sizeable family of genes that require Rpd3p and the H3 and H4 amino termini for their proper regulation.
Several investigators have combined chromatin immunoprecipitation with microarrays to identify promoters that show higher acetylation of the H3 and/or H4 amino termini in rpd3Δ yeast cells than in wild-type cells and to identify promoters associated with Rpd3p (23, 32). We tested the overlap between genes showing increased acetylation in rpd3Δ yeast cells and those up-regulated upon the loss of Rpd3p or of the H3 or H4 amino terminus and found the degree of overlap to be only slightly higher than that expected by chance (data not shown). Similarly, genes whose promoters have been found to be physically associated with Rpd3p (more than twofold) also were not enriched in groups of genes that were up-regulated more than twofold upon the loss of Rpd3p or of the H3 or H4 amino terminus (data not shown). Although these previous studies did report modest correlations between genes physically associated with Rpd3p and those regulated by Rpd3p or Sin3p, as well as those showing increased H3 K18 acetylation upon the loss of Rpd3p, examination of the data in these studies for the simple overlap of genes associated with Rpd3p (twofold or higher enrichment) and those showing increased acetylation in rpd3Δ yeast cells (twofold or higher for both H4 K5 and H4 K12) reveals the degree of overlap to be no greater than that expected by chance (data not shown). These findings do not imply that the data in those studies or in our studies are flawed; indeed, an impressive correlation was found among genes showing increased acetylation in rpd3Δ yeast cells at H4 K5 and K12 and H3 K18 in the data of Kurdistani et al. (23), vouching for the robustness of these data (data not shown). Similarly, the substantial overlap seen among genes that were up-regulated upon the loss of the H3 or H4 amino terminus or of Rpd3p strongly suggests that these genes are regulated by a common mechanism, which may be direct or indirect for individual instances but likely stems from the action of Rpd3p on the target histone tails. The weak correlation between Rpd3p binding and its effects on acetylation and gene regulation may reflect its nontargeted activity as a histone deacetylase and/or the regulation of Rpd3p activity by mechanisms that are not yet understood (39, 50).
Although we constructed an rpd3 H4(K5,8,12,16Q) yeast strain, we were unable to generate rpd3Δ H4Δ4-19 or rpd3Δ H4Δ2-26 yeast strains in the S288C background, indicating that some essential function in yeast cells requires either Rpd3p or the H4 amino terminus. These results could reflect independent regulation of a gene or genes by Rpd3p and the H4 tail; however, they could also be due to the loss of some essential checkpoint function, as the H4 amino terminus has been implicated in cell cycle checkpoint regulation (28).
We also found a high degree of correlation between genes affected by the loss of the H3 and H4 amino termini, as well as those affected by mutation of H3 lysine residues 4, 9, 14, 18, 23, and 27 to glutamine residues and those affected by mutation of H4 lysine residues 5, 8, 12, and 16 to glutamine residues (Fig. 3 to 6). We draw two conclusions from these findings. First, for many genes, regulation by the histone H3 and H4 amino termini is exerted principally via mechanisms specific to the lysine residues. This scenario, of course, likely reflects the involvement of chromatin-modifying enzymes, whose involvement in transcriptional regulation is well known. Second, many genes evidently require both the histone H3 and the histone H4 amino termini for proper regulation, particularly repression (Fig. 6A and B). For such genes, the acetylation of either the H3 or the H4 amino terminus may suffice to allow activation. This scenario would permit activation by recruitment of histone acetyltransferases that target either the H3 or the H4 tail, for example, Esa1p or Gcn5p (5, 43), thus providing flexibility for gene activation. Genes requiring both amino termini may depend on charge-specific effects of the histone tails for their proper regulation, whereas those exhibiting regulation specific to the H3 or H4 amino terminus may require the recognition of modifications in one or the other tail by transcription factors or other transcriptional regulatory proteins, as postulated by the histone code hypothesis (44, 48, 49).
Various activators and repressors are known to act at least in part via the histone amino termini; these include Tup1p, Gcn5p, Esa1p, Hda1p, Hos1p, and Hos2p (13, 15, 43, 51, 52). Novel pathways of gene regulation via the histone amino termini may still remain to be identified (38). A comparison of genes regulated via the H3 and H4 amino termini, as described in this work, with known targets of regulators that function via the histone tails (3, 10, 17) may facilitate the discovery of such novel pathways.
Acknowledgments
We thank Michael Grunstein, M. M. Smith, David Stillman, David Kadosh, Kevin Struhl, and Steve Triezenberg for plasmids and yeast strains; Ping Ye for technical help; Mike Ryan and Robin Pietropaolo of the Wadsworth Center Microarray Facility for assistance; and members of the Wadsworth Center Molecular Genetics Core for assistance. We also thank Chip Lawrence and Mike Palumbo for helpful discussions of statistical issues and John Wyrick for communicating unpublished results.
We gratefully acknowledge financial support from the NSF (grant MCB-0133399) and the NIH (grant GM51993).
Footnotes
This work is dedicated to the memory of Robert Simpson.
REFERENCES
- 1.Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111:381-392. [DOI] [PubMed] [Google Scholar]
- 2.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57:289-300. [Google Scholar]
- 3.Bernstein, B. E., J. K. Tong, and S. L. Schreiber. 2000. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 97:13708-13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brent, R., and M. Ptashne. 1984. A bacterial repressor protein or a yeast transcriptional terminator can block upstream activation of a yeast gene. Nature 312:612-615. [DOI] [PubMed] [Google Scholar]
- 5.Brown, C. E., T. Lechner, L. Howe, and J. L. Workman. 2000. The many HATs of transcription coactivators. Trends Biochem. Sci. 25:15-19. [DOI] [PubMed] [Google Scholar]
- 6.Brunet, A., L. B. Sweeney, J. F. Sturgill, K. F. Chua, P. L. Greer, Y. Lin, H. Tran, S. E. Ross, R. Mostoslavsky, H. Y. Cohen, L. S. Hu, H. L. Cheng, M. P. Jedrychowski, S. P. Gygi, D. A. Sinclair, F. W. Alt, and M. E. Greenberg. 2004. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303:2011-2015. [DOI] [PubMed] [Google Scholar]
- 7.Cheung, P., C. D. Allis, and P. Sassone-Corsi. 2000. Signaling to chromatin through histone modifications. Cell 103:263-271. [DOI] [PubMed] [Google Scholar]
- 8.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 9.Deckert, J., and K. Struhl. 2002. Targeted recruitment of Rpd3 histone deacetylase represses transcription by inhibiting recruitment of Swi/Snf, SAGA, and TATA binding protein. Mol. Cell. Biol. 22:6458-6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 11.Dou, Y., and M. A. Gorovsky. 2000. Phosphorylation of linker histone H1 regulates gene expression in vivo by creating a charge patch. Mol. Cell 6:225-231. [DOI] [PubMed] [Google Scholar]
- 12.Durrin, L. K., R. K. Mann, P. S. Kayne, and M. Grunstein. 1991. Yeast histone H4 N-terminal sequence is required for promoter activation in vivo. Cell 65:1023-1031. [DOI] [PubMed] [Google Scholar]
- 13.Edmondson, D. G., M. M. Smith, and S. Y. Roth. 1996. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 10:1247-1259. [DOI] [PubMed] [Google Scholar]
- 14.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galarneau, L., A. Nourani, A. A. Boudreault, Y. Zhang, L. Heliot, S. Allard, J. Savard, W. S. Lane, D. J. Stillman, and J. Cote. 2000. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell 5:927-937. [DOI] [PubMed] [Google Scholar]
- 16.Han, M., and M. Grunstein. 1988. Nucleosome loss activates yeast downstream promoters in vivo. Cell 55:1137-1145. [DOI] [PubMed] [Google Scholar]
- 17.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 18.Kadosh, D., and K. Struhl. 1998. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 12:797-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadosh, D., and K. Struhl. 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89:365-371. [DOI] [PubMed] [Google Scholar]
- 20.Kadosh, D., and K. Struhl. 1998. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 18:5121-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kayne, P. S., U. J. Kim, M. Han, J. R. Mullen, F. Yoshizaki, and M. Grunstein. 1988. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell 55:27-39. [DOI] [PubMed] [Google Scholar]
- 22.Kouzarides, T. 2000. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurdistani, S. K., D. Robyr, S. Tavazoie, and M. Grunstein. 2002. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 31:248-254. [DOI] [PubMed] [Google Scholar]
- 24.Lohr, D., and J. Lopez. 1995. GAL4/GAL80-dependent nucleosome disruption/deposition on the upstream regions of the yeast GAL1-10 and GAL80 genes. J. Biol. Chem. 270:27671-27678. [DOI] [PubMed] [Google Scholar]
- 25.Mann, R. K., and M. Grunstein. 1992. Histone H3 N-terminal mutations allow hyperactivation of the yeast GAL1 gene in vivo. EMBO J. 11:3297-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin, A. M., D. J. Pouchnik, J. L. Walker, and J. J. Wyrick. 2004. Redundant roles for histone H3 N-terminal lysine residues in subtelomeric gene repression in Saccharomyces cerevisiae. Genetics 167:1123-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Megee, P. C., B. A. Morgan, B. A. Mittman, and M. M. Smith. 1990. Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science 247:841-845. [DOI] [PubMed] [Google Scholar]
- 28.Megee, P. C., B. A. Morgan, and M. M. Smith. 1995. Histone H4 and the maintenance of genome integrity. Genes Dev. 9:1716-1727. [DOI] [PubMed] [Google Scholar]
- 29.Moreira, J. M., and S. Holmberg. 1998. Nucleosome structure of the yeast CHA1 promoter: analysis of activation-dependent chromatin remodeling of an RNA-polymerase-II-transcribed gene in TBP and RNA pol II mutants defective in vivo in response to acidic activators. EMBO J. 17:6028-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan, B. A., B. A. Mittman, and M. M. Smith. 1991. The highly conserved N-terminal domains of histones H3 and H4 are required for normal cell cycle progression. Mol. Cell. Biol. 11:4111-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson, M. D., J. Grigull, N. Mohammad, and T. R. Hughes. 2002. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics 3:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robyr, D., Y. Suka, I. Xenarios, S. K. Kurdistani, A. Wang, N. Suka, and M. Grunstein. 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109:437-446. [DOI] [PubMed] [Google Scholar]
- 33.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Rundlett, S. E., A. A. Carmen, R. Kobayashi, S. Bavykin, B. M. Turner, and M. Grunstein. 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 93:14503-14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rundlett, S. E., A. A. Carmen, N. Suka, B. M. Turner, and M. Grunstein. 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392:831-835. [DOI] [PubMed] [Google Scholar]
- 36.Ryan, M. P., R. Jones, and R. H. Morse. 1998. SWI-SNF complex participation in transcriptional activation at a step subsequent to activator binding. Mol. Cell. Biol. 18:1774-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryan, M. P., G. A. Stafford, L. Yu, and R. H. Morse. 2000. Artificially recruited TATA-binding protein fails to remodel chromatin and does not activate three promoters that require chromatin remodeling. Mol. Cell. Biol. 20:5847-5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sabet, N., F. Tong, J. P. Madigan, S. Volo, M. M. Smith, and R. H. Morse. 2003. Global and specific transcriptional repression by the histone H3 amino terminus in yeast. Proc. Natl. Acad. Sci. USA 100:4084-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandmeier, J. J., S. French, Y. Osheim, W. L. Cheung, C. M. Gallo, A. L. Beyer, and J. S. Smith. 2002. RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J. 21:4959-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimizu, M., K. Takahashi, T. M. Lamb, H. Shindo, and A. P. Mitchell. 2003. Yeast Ume6p repressor permits activator binding but restricts TBP binding at the HOP1 promoter. Nucleic Acids Res. 31:3033-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stafford, G. A., and R. H. Morse. 1998. Mutations in the AF-2/hormone-binding domain of the chimeric activator GAL4.estrogen receptor.VP16 inhibit hormone-dependent transcriptional activation and chromatin remodeling in yeast. J. Biol. Chem. 273:34240-34246. [DOI] [PubMed] [Google Scholar]
- 43.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 45.Suka, N., Y. Suka, A. A. Carmen, J. Wu, and M. Grunstein. 2001. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8:473-479. [DOI] [PubMed] [Google Scholar]
- 46.Taunton, J., C. A. Hassig, and S. L. Schreiber. 1996. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272:408-411. [DOI] [PubMed] [Google Scholar]
- 47.Thompson, J. S., X. Ling, and M. Grunstein. 1994. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature 369:245-247. [DOI] [PubMed] [Google Scholar]
- 48.Turner, B. M. 1993. Decoding the nucleosome. Cell 75:5-8. [PubMed] [Google Scholar]
- 49.Turner, B. M. 2000. Histone acetylation and an epigenetic code. Bioessays 22:836-845. [DOI] [PubMed] [Google Scholar]
- 50.Vogelauer, M., J. Wu, N. Suka, and M. Grunstein. 2000. Global histone acetylation and deacetylation in yeast. Nature 408:495-498. [DOI] [PubMed] [Google Scholar]
- 51.Watson, A. D., D. G. Edmondson, J. R. Bone, Y. Mukai, Y. Yu, W. Du, D. J. Stillman, and S. Y. Roth. 2000. Ssn6-Tup1 interacts with class I histone deacetylases required for repression. Genes Dev. 14:2737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, J., N. Suka, M. Carlson, and M. Grunstein. 2001. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell 7:117-126. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, W., J. R. Bone, D. G. Edmondson, B. M. Turner, and S. Y. Roth. 1998. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 17:3155-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, Y., and D. Reinberg. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15:2343-2360. [DOI] [PubMed] [Google Scholar]