Abstract
Mitogen-activated protein kinase kinase kinase-Ste11 (MAPKKK-Ste11), MAPKK-Ste7, and MAPK-Kss1 mediate pheromone-induced mating differentiation and nutrient-responsive invasive growth in Saccharomyces cerevisiae. The mating pathway also requires the scaffold-Ste5 and the additional MAPK-Fus3. One contribution to specificity in this system is thought to come from stimulus-dependent recruitment of the MAPK cascade to upstream activators that are unique to one or the other pathway. To test this premise, we asked if stimulus-independent signaling by constitutive Ste7 would lead to a loss of biological specificity. Instead, we found that constitutive Ste7 promotes invasion without supporting mating responses. This specificity occurs because constitutive Ste7 activates Kss1, but not Fus3, in vivo and promotes filamentation gene expression while suppressing mating gene expression. Differences in the ability of constitutive Ste7 variants to bind the MAPKs and Ste5 account for the selective activation of Kss1. These findings support the model that Fus3 activation in vivo requires binding to both Ste7 and the scaffold-Ste5 but that Kss1 activation is independent of Ste5. This scaffold-independent activation of Kss1 by constitutive Ste7 and the existence of mechanisms for pathway-specific promoter discrimination impose a unique developmental fate independently of any distinguishing external stimuli.
In response to the appropriate environmental stimulus, haploid cells of Saccharomyces cerevisiae are capable of two developmental fates. One entails differentiation into a mating-competent form, and the other is a switch from vegetative to pseudohyphal growth. Mitogen-activated protein kinase (MAPK) pathways comprised of the same enzymes mediate both transitions. This situation offers an excellent opportunity for studies into regulatory mechanisms that impart biological specificity to signaling pathways.
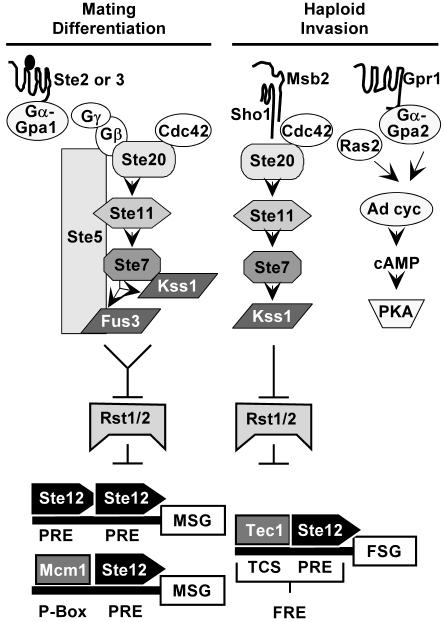
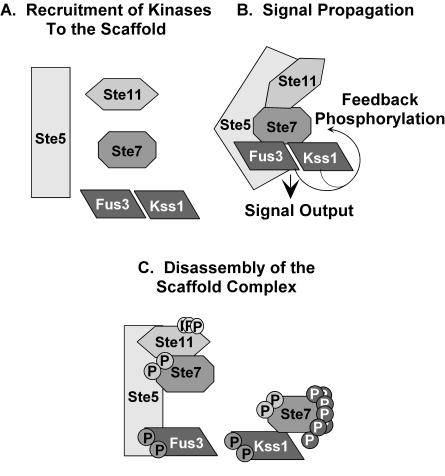
Mating is the fusion of two haploid cells of opposite mating types to produce a diploid cell. Differentiation to a mating-competent form is triggered when pheromone from the opposite mating type binds to a G-protein-coupled receptor. The activated G-protein stimulates a MAPK module comprised of the p21 (Cdc42)-associated kinase-Ste20, MAPKKK-Ste11, MAPKK-Ste7, and two MAPKs, Fus3 and Kss1 (Fig. 1) (see reference 12 for review). The scaffold-Ste5 organizes the core enzymes of this pathway by directly binding to Ste11, Ste7, and Fus3 (12). Kss1 does not bind directly to Ste5 but is part of this complex through binding to Ste7 (7). Although Fus3 and Kss1 kinase activities are induced to an equivalent extent in pheromone-treated cells, Fus3, but not Kss1, has specificity for critical substrates, such as Far1, that are dedicated to the mating differentiation response (7, 23, 44). Fus3 and Kss1 are equivalent with respect to promoting a mating-specific transcription program through their activation of the Ste12 transcription factor (7, 41, 57). MAPK activation of Ste12 involves phosphorylation-dependent inactivation of two inhibitors called Rst1 (Dig1) and Rst2 (Dig2) (51). Ste12 functions at mating-specific genes (MSG) either as a homo-multimer at promoters with reiterated pheromone response elements (PREs) or in conjunction with Mcm1 at promoters with P-box elements in proximity to PREs (14, 18) (Fig. 1).
FIG. 1.
Signal transduction pathways with common components mediate two life cycle transitions in haploid S. cerevisiae. See text for explanation.
The transition from vegetative to pseudohyphal growth is induced in diploid cells on solid medium containing a good carbon source but limiting in nitrogen (27). A related transition causing haploid cells to invade solid medium occurs on rich medium in response to glucose depletion or high alcohol (see reference 37 for review). In the pseudohyphal and invasive growth modes, chains of elongated cells remain attached at the mother-daughter neck to form the pseudohyphal filaments that emanate from the periphery of colonies (37). Regulation of the transition to filamentous growth involves a network of several signaling pathways (Fig. 1). Glucose-sensing G-protein-coupled receptor-Gpr1 stimulates one branch of the network through Gα-Gpa2, which together with Ras2 regulates cyclic AMP production and protein kinase A activity (37). The Sho1 and Msb2 transmembrane proteins are upstream regulators of the MAPK-mediated branch of the network (11, 36; P. Cullen and G. F. Sprague, Jr., personal communication). With the exception of Fus3, the same kinases used in the mating pathway comprise the filamentous growth MAPK activation module (Fig. 1) (37). Another notable difference is that there is no known scaffold for the invasive pathway. The Kss1 branch of the pseudohyphal signaling network regulates the filamentation-specific transcription program through activation of the Ste12 transcription factor by phosphorylation-dependent inactivation of the Rst1 (Dig1) and Rst2 (Dig2) inhibitors (5, 9, 51). Ste12 acts at filamentation-specific genes cooperatively with Tec1 at promoters that have a Tec1 consensus binding sequence in proximity to the PRE binding element for Ste12 (6, 31) (Fig. 1).
Thus, the transcriptional output for the mating and invasive response depends on the same transcription factor and inhibitor complex, which can be activated by either Kss1 or Fus3. How and whether transcriptional specificity occurs in this circumstance are still poorly understood. Recent evidence from genome-wide location analyses suggests that regulation of Ste12 binding at the different promoters rather than selective activation by the two MAPKs regulates the distinct gene expression programs associated with mating differentiation and filamentous growth (57).
While filamentous growth is persistent, mating differentiation is transient. The transient signal results because cells reenter the cell cycle to resume vegetative growth whether or not mating occurs. Accordingly, the responsiveness of cells to persistent pheromone induction diminishes with time. This phenomenon occurs through several desensitization or adaptation mechanisms that impinge on different components of the signaling pathway upstream of the MAPK activation module (see reference 12 for review). Rapid attenuation of MAPK activation is reinforced through pheromone-dependent mechanisms that directly affect the MAPK module enzymes themselves. Examples include dephosphorylation of the MAPKs by phosphatases, one of which is induced by pheromone, ubiquitin-dependent turnover of Ste11, which apparently depletes activated Ste11 from the Ste5-organized complex, and ubiquitination of Ste7 upon prolonged pheromone exposure (13, 21, 52, 54, 55, 58).
Pheromone induction also regulates the phosphorylation status of Ste7. Similar to other MAPKKs, Ste7 activation requires phosphorylation of two residues in the activation loop (20, 35, 59). In addition, both Fus3 and Kss1 mediate hyperphosphorylation of Ste7 in a feedback reaction that is contemporaneous with their activation (19, 20, 60, 61). Our group subsequently showed that Fus3 directly phosphorylates multiple residues in the Ste7 N-terminal regulatory domain and a single residue C-terminal to the catalytic domain (20; see also data in the supplemental material). Because Ste7 is also a direct substrate for Kss1, it is likely that both MAPKs target the same subset of residues (4).
Little is known about how feedback phosphorylation of Ste7 affects its activity and subsequent MAPK activation. Early observations indicated that the hyperphosphorylated Ste7 is less represented than the hypophosphorylated form in the Ste5-scaffold complex (8). Additionally, Ste7 in vitro kinase activity is significantly lower when the enzyme is isolated from cells expressing Fus3 and Kss1 than from cells lacking both MAPKs (19, 20). These observations suggest that feedback phosphorylation contributes to attenuation of signaling in the mating differentiation pathway. It has not been resolved whether the decrease in pheromone-stimulated Ste7 activity occurs because the hyperphosphorylated form has attenuated specific activity, is excluded from the interactions with components of the MAPK module, or is targeted for ubiquitination.
In this study we employed Ste7 activation loop and feedback phosphorylation site substitution variants to examine the above questions. We show that feedback phosphorylation of Ste7 does not attenuate its kinase activity nor influence its susceptibility to pheromone-induced ubiquitination. Unexpectedly, we found that constitutive Ste7 specifies an invasive response through selective activation of Kss1 and filamentation-specific gene expression. The selective transcriptional output occurs because mating-specific gene expression is suppressed through mechanisms that are dependent on the feedback phosphorylation status of Ste7.
MATERIALS AND METHODS
Yeast genetic procedures and strains.
Unless otherwise specified, yeast growth media and genetic manipulations were employed as described by Sherman et al. (46). Yeast transformations and gene replacements were done using standard procedures (25, 43). All gene replacements or integrations were confirmed by PCR analysis of genomic DNA. Table 1 lists the strains used in these studies and their complete genotypes. The details of how they were constructed are provided in the on-line supplemental material for this paper.
TABLE 1.
S. cerevisiae strains
| Strain | Genotype | Source or reference | |
|---|---|---|---|
| C699a | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | K. Nasmyth | |
| C699-5 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1Δ::HisG | 21 | |
| C699-15 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1Δ::HisG FUS1-LacZ::LEU2 | This work | |
| C699-31 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1Δ::HisG FUS1-LacZ::LEU2 ste5Δ::ADE2 | This work | |
| C699-32 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1Δ::HisG FUS1-LacZ::LEU2 ste7Δ::ADE2 | This work | |
| C699-33 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1Δ::HisG FUS1-LacZ::LEU2 fus3Δ::ADE2 | This work | |
| C699-47 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1Δ::HisG FUS1-LacZ::LEU2 ste11Δ::ADE2 | This work | |
| C699-80 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1Δ::HisG FUS1-LacZ::LEU2 ste7Δ3::ura3Δ58 fus3Δ::ADE2 | This work | |
| C699-81 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1Δ::HisG FUS1-LacZ::LEU2 ste11Δ::ADE2 ste7Δ3::ura3Δ58 | This work | |
| C699-82 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1Δ::HisG FUS1-LacZ::LEU2 ste5Δ::ADE2 ste7Δ3::ura3Δ58 | This work | |
| C699-87 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1Δ::HisG FUS1-LacZ::LEU2 ste5Δ::ura3Δ58 ste7Δ3::ura3Δ58 fus3Δ::ADE2 | This work | |
| C699-89 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1Δ::HisG ste7Δ4::HisG | This work | |
| C699-90 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1Δ::HisG FUS1-LacZ::LEU2 ste5Δ::ura3Δ58 ste7Δ3::ura3Δ58 fus3Δ:ADE2 kss1Δ::kanMX6 | This work | |
| C699-91 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1Δ::HIS3MX6 ura3-1 bar1Δ::HisG FUS1-LacZ::LEU2 ste5Δ::ura3Δ58 ste7Δ3::ura3Δ58 fus3Δ::ADE2 kss1Δ::kanMX6 | This work | |
| C699-92 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1Δ::HIS3MX6 ura3-1 bar1Δ::HisG FUS1-LacZ::LEU2 ste5Δ::ura3Δ58 ste11Δ6::ura3Δ58 ste7Δ3::ura3Δ58 fus3Δ::ADE2 kss1Δ::kanMX6 | This work | |
| C699-93 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1Δ::HisG FUS1-LacZ::LEU2 ste11Δ6::ura3Δ58 fus3Δ::ADE2 | This work | |
| C699-106 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1Δ::HIS3MX6 ura3-1 bar1Δ::HisG ste7Δ4::HisG | This work | |
| C699-108 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1Δ::HisG FUS1-LacZ::LEU2 ste11Δ6::ura3Δ58 ste7Δ3::ura3Δ58 fus3Δ::ADE2 | This work | |
| C699-112 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1Δ::HisG FUS1-LacZ::LEU2 ste7Δ4::HisG | This work | |
| C699-114 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1Δ::HisG FUS1-LacZ::LEU2 ste7Δ::ADE2 kss1Δ::kanMX6 | This work | |
| C699-115 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1Δ::HisG FUS1-LacZ::LEU2 ste11Δ::ADE2 ste7Δ3::ura3Δ58 kss1Δ::kanMX6 | This work | |
| C699-116 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1Δ::HisG FUS1-LacZ::LEU2 ste5Δ::ADE2 ste7Δ3::ura3Δ58 kss1Δ::kanMX6 | This work | |
| C699-127 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar 1Δ::HisG FUS1-LacZ::LEU2 ste5Δ::ADE2 ste7Δ::ura3Δ58 ste11Δ6::URA3 | This work | |
| C699-140 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1Δ::HisG FUS1-LacZ::LEU2 ste7Δ3::ura3Δ58 fus3Δ::ADE2 kss1Δ::kanMX6 | This work | |
| C699-145 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1Δ::HIS3MX6 ura3-1 bar1Δ::HisG ste7Δ4::HisG fus3Δ6::LEU2 | This work | |
| C699-146 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1Δ::HIS3MX6 ura3-1 bar1Δ::HisG ste7Δ4::HisG kss1Δ::kanMX6 | This work | |
| K2149b | MATaHLMaHMRacan1-100 leu2-3,112 trp1-1 ura3 ho::lacZ bar1::HISG ade2-1 met his3 | 24 | |
| K2313b | MATaHLMaHMRacan1-100 leu2-3,112 trp1-1 ura3 ho::lacZ bar1::HISG ade2-1 met his3 fus3Δ::LEU2 kss1::URA3 | 24 | |
| KZ8-1D | MATα cyc1-1 CYC7 his4-38 ura1-1 | K. Zaret and F. Sherman | |
| KZ8-5C | MATacyc1-1 CYC7 his4-38 ura1-1 | K. Zaret and F. Sherman | |
| MLY218ac | MATaleu2 ura3 ste7Δ::kanMX6 | M. Lorenz and J. Heitman | |
| MLY218αc | MATα leu2 ura3-52 ste7Δ::kanMX6 | M. Lorenz and J. Heitman | |
| SM-001c | MATaleu2 ura3 ste7Δ::kanMX6 fus3Δ-6::LEU2 | This work | |
| SM-002c | MATα leu2 ura3-52 ste7Δ::kanMX6 fus3Δ-6::LEU2 | This work | |
| YHD1001 | MATaura3-52 lys2-801am ade2-1oc trp1Δ63 his3-Δ200 leu2-Δ1 ste7::ADE2 ubp3::kanMX6 | 55 |
Isogenic to W303-1A.
Isogenic to K1107.
Derived from the Σ1278 background.
Recombinant DNA procedures and plasmids.
Bacterial transformations, bacterial DNA preparations, plasmid constructions, and DNA restriction enzyme digestions were performed by standard methods (45). A list of the plasmids used in these studies is provided in Table 2. The details of site-directed mutagenesis for generating alanine or glutamate substitutions at the phosphorylation sites of Ste7 and for different plasmid constructions are provided in the methods section of the supplemental material.
TABLE 2.
Plasmids
| Plasmid | Allele | Vector base (reference) | Source or reference |
|---|---|---|---|
| pNC318 | PCYC1-STE7M | pNC160 [TRP1 CEN3 ARS1] (39) | 61 |
| pNC318-R220 | PCYC1-STE7M-R220 | pNC160 (39) | 61 |
| pNC318-N349 | PCYC1-STE7M-N349 | pNC160 (39) | This work |
| pNC571 | PCYC1-STE7M-AA-S4T3 | pNC160 (39) | This work |
| pNC586 | PCYC1-STE7M-ST-A7 | pNC160 (39) | This work |
| pNC589 | PCYC1-STE7M-EE-S4T3 | pNC160 (39) | This work |
| pNC597 | PCYC1-STE7M-EE-A7 | pNC160 (39) | This work |
| pNC892 | PCYC1-STE7M-ST-E7 | pNC160 (39) | This work |
| pNC893 | PCYC1-STE7M-EE-E7 | pNC160 (39) | This work |
| pNC894 | PCYC1-STE7M-AA-A7 | pNC160 (39) | This work |
| pNC895 | PCYC1-STE7M-AA-E7 | pNC160 (39) | This work |
| pNC752 | STE7M | pNC160 (39) | This work |
| pNC766 | STE7M | pRS316 [URA3 CEN6 ARS4] (48) | This work |
| pNC767 | STE7M-AA-S4T3 | pRS316 (48) | This work |
| pNC768 | STE7M-EE-S4T3 | pRS316 (48) | This work |
| pNC769 | STE7M-ST-A7 | pRS316 (48) | This work |
| pNC770 | STE7M-EE-A7 | pRS316 (48) | This work |
| pNC771 | STE7M-N349 | pRS316 (48) | This work |
| pNC781 | STE7M-AA-A7 | pRS316 (48) | This work |
| pNC790 | STE7M-AA-E7 | pRS316 (48) | This work |
| pNC791 | STE7M-EE-E7 | pRS316 (48) | This work |
| pNC793 | STE7M-ST-E7 | pRS316 (48) | This work |
| pNC809 | PGAL-STE7M-EE-E7 | pNC160 [TRP1 CEN3 ARS1] (39) | This work |
| pNC896 | PGAL-STE7M-AA-A7 | pNC160 (39) | This work |
| pNC897 | PGAL-STE7M-EE-A7 | pNC160 (39) | This work |
| pNC898 | PGAL-STE7M-AA-E7 | pNC160 (39) | This work |
| pNC906 | PGAL-STE7M | pNC160 (39) | This work |
| pEG-KT | PGAL-GST | pEMBLyex2 [LEU2-d URA3 2μm] (2) | 33 |
| pNC901 | PGAL-GST-FUS3 | pEMBLyex2 (2) | This work |
| pNC903 | PGAL-GST-KSS1 | pEMBLyex2 (2) | This work |
| pRD-GST-STE111-717 | PGAL-GST-STE11 | pRS316 [URA3 CEN6 ARS4] (48) | 35 |
| pYBS293 | PGAL-GST-STE5 | YEpLac181 [LEU2 URA3 2μm] (26) | 8 |
| pNC977 | PTPI1-KSS1M-TADH1 | pRS313 [HIS3-CEN6 ARS4] (48) | This work |
| pNC979 | PTPI1-FUS3M-TADH1 | pRS316 [URA3 CEN6 ARS4] (48) | This work |
| pNC756 | FUSI-LacZ | Modified pLGΔ178 [LEU2 2μm] (28) | This work |
| pNC343 | Ty-LacZ | pLGΔ178 [URA3 2μm] (28) | 6 |
Mating response and pheromone induction assays.
Mating competence was scored by an assay for prototrophic diploid formation between MATa strain C699-89 (ste7Δ4::HisG) expressing STE7 alleles from plasmids as specified and MATα tester strain KZ8-1D (50). As a control to confirm mating dependence of prototroph formation, parallel mating mixtures were made using MATa tester strain KZ8-5C. Semiquantitative comparisons were made as follows. Mid-log-phase cultures of tester strains were used to make grids with 106 cells per spot on complete medium (yeast extract-peptone-dextrose). Strains to be tested were grown to mid-log phase in selective medium (-Ura). Ten-fold dilutions were made and used for applying 105, 104, 103, and 102 cells from each culture to a tester strain spot on a grid on the solid medium. Mixtures were incubated at 30°C and then replica plated onto medium for selection of prototrophic diploids synthetic dextrose. Growth was scored after overnight incubation at 30°C.
FUS1-lacZ expression is a standard reference for pheromone-induced and mating pathway-specific transcription (50). Reporter gene expression was measured by activity of the lacZ gene product (β-galactosidase) in whole-cell extracts as previously described (40). Pheromone-induced G1 arrest was monitored by the percentage of unbudded cells in the sample by using a microscope and a hemacytometer grid to count cells. Pheromone-induced mating differentiation was monitored by the percentage of cells with mating projections using the same procedure. At least 200 individual cells were scored for each sample.
Invasive growth and filamentation reporter gene assays.
Growth chambers for visualizing invasive growth were made from two 3- by 1-in. glass microscope slides separated by a 5-mm-wide and 1.5-mm-thick U-shaped spacer. The spacer and slides were held in place using a 3/4-in.-wide binder clip at each end of the assembly. The chamber assemblies were autoclaved without media in glass petri dishes. Immediately before use, sterilized yeast extract-peptone-dextrose medium with 1.5% agar was liquefied by microwave heating and delivered into each sterilized assembly using a syringe with an 18-gauge needle. Cultures of strains to be tested were grown to mid-log phase in selective medium. A 25-μl aliquot of a 104 cell/ml dilution was applied to the solid medium in the trough between the microscope slides. After inoculation, the chambers were wrapped in saran, placed vertically in plastic boxes containing wet 3-mm paper to maintain humidity, and incubated at 30°C. After ∼48 h, images of colonies that grew beneath the agar surface were visualized using an Axioplan research microscope with a 2.5× objective. Images were captured using an Axio Cam camera with Zeiss Axiovision software. Images were taken at ∼4-mm intervals, allowing a sampling of 8 to 10 separate fields from each growth chamber. The areas of individual colonies from images at each interval were measured using NIH ImageJ software.
Ty-lacZ expression is a standard reference for filamentation pathway-specific transcription (31, 32, 34). The pNC343 plasmid version of this reporter that we employed has the composite Tec1 Ste12 binding element from Ty1 replacing the upstream activating sequence of the CYC1 promoter in pLGΔ178 (6, 29). This context differs from the TDH3 context of the FG-TyA-LacZ (pIL30) reporter that has been more generally used (30, 34). In our hands the FG-TyA-LacZ reporter has high basal expression compared with the pNC343 reporter plasmid. Additionally, the pNC343 Ty-LacZ reporter shows a more stringent dependence on components of the MAPK cascade, including Ste7 and Kss1. Reporter gene expression was measured by activity of the lacZ gene product (β-galactosidase) in whole-cell extracts as previously described (40).
Immune precipitation kinase assays.
In vitro assays of Ste7 catalytic activity were performed directly on immune complexes of myc epitope-tagged Ste7 (Ste7M) with [γ-32P]ATP and catalytically inactive Fus3-R42 as substrate. The source of Ste7M or variant proteins was from strain K2149 (bar1Δ FUS3 KSS1) or K2313 (bar1Δ fus3Δ kss1Δ) transformed with plasmids expressing Ste7M (or variants) from the CYC1 promoter. Strains were grown in selective medium (-Trp) to a density of ∼1 × 107 cells/ml. Samples were removed before and after 30 min of induction with mating pheromone (α-factor; 50 nM). Native protein extracts were prepared from samples as previously described (19). Fus3-R42 substrate polypeptide was prepared as a glutathione S-transferase (GST) fusion protein produced in Escherichia coli. The fusion protein was isolated on glutathione-agarose columns, eluted, and treated with thrombin to release the GST polypeptide as described elsewhere (49). The resulting mixture was used as substrate without further purification.
Immune complexes were isolated using buffers and wash conditions as previously described (19). For each assay we used 400 μg of extract with 22 μg of anti-myc 9E10 antibodies (22) and 50 μl of protein A-Sepharose beads (Pharmacia-LKB). These specific conditions were empirically determined to be in the linear range for activity versus immune complex (beads). Immune complex beads were suspended in 6 μl of kinase assay buffer (25 mM HEPES [pH 7.2], 15 mM MgCl2, 5 mM EGTA, 1 mM dithiothreitol, 0.1 mM orthovanadate, 15 mM 4-nitrophenylphosphate, 1 mM phenylmethylsulfonyl fluoride [PMSF], 2 μg of leupeptin per ml, 40 μg of aprotinin per ml). Approximately 250 ng of thrombin-treated GST-Fus3R42 substrate in a volume of 2 to 5 μl was added to the above suspension. The reactions were initiated by addition of an ATP mix (1 μl of 250 mM HEPES [pH 7.2], 2 μl of 1 mM ATP, 0.2 μl of [γ-32P]ATP [10 μCi/μl; Dupont-NEN], 2.4 μl of 100 mM MgCl2) and water to give a final reaction volume of 20 μl. The mixtures were incubated at 30°C for 20 min, and reactions were terminated by addition of an equal volume of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer and boiling for 3 min. Proteins in the reaction were fractionated by SDS-PAGE (7.5%). The portion of the gels that included the immune-precipitated Ste7 (above the 45-kDa molecular size marker) was transferred to nitrocellulose membranes for detection of the amount of Ste7M in the immune-precipitated complex with anti-myc antibodies. The lower portion of the gels that included Fus3-R42 and immunoglobulin chains was dried to determine the amount of 32P incorporated into Fus3-R42 by PhosphorImager analysis (Molecular Dynamics).
GST fusion protein copurification assays.
Either strain C699-92 (ste5Δ ste11Δ ste7Δ fus3Δ kss1Δ) or C699-93 (ste11Δ fus3Δ) was cotransformed with a plasmid for GAL1,10 promoter expression of one of the Ste7M phosphorylation site variant proteins (pNC809, pNC896, pNC897, pNC898, or pNC906) and GST (pEG-KT) or GST fusion proteins (pNC901, pNC903, pRD-GST-STE11 or pYBS293). Cultures of each cotransformed strain were grown in selective medium (-Trp -Ura) with raffinose (2%) as the carbon source. When cultures reached a density of ∼1 × 107 cells/ml, galactose (2%) was added to the medium to induce expression of the Ste7M variant and the GST fusion protein. After 6 h of induction, cultures were harvested and cell pellets were suspended in an equal volume of modified H-buffer (25 mM Tris [pH 7.4], 15 mM ethylene glycol tetraacetic acid [EGTA], 15 mM MgCl2, 1 mM dithiothreitol, 1 mM sodium azide, 0.1% Triton X-100) made with 10% glycerol and containing phosphatase and protease inhibitors (0.25 mM orthovanadate, 1 mM PMSF, and 5 μg each of pepstatin A, chymostatin, antipain, and aprotinin/ml). Cells in the suspensions were broken by vigorous shaking with an equal volume of glass beads on an IKA platform mixer at 4°C for a total of 12 min. Glass beads and cellular debris were removed by centrifugation at 4°C, and the supernatant was transferred to a new tube. The protein concentration of the resulting whole-cell protein extract was determined by the Bradford method using the Bio-Rad protein assay reagent.
GST fusion protein purifications were carried out using batch purification essentially as described by Choi et al. (8). In this study, 3 mg of protein extract was brought to a 300-μl final volume using modified H-buffer. A 20-μl sample was removed for immune blot analysis to determine the amount of GST fusion protein and Ste7 variant protein in the input fraction for each pull-down assay. The remaining sample was adjusted to 150 mM NaCl and 1% ovalbumin (or bovine serum albumin) and added to an Eppendorf tube containing 75 μl (packed volume) of glutathione agarose beads. Binding mixtures were incubated for 1 h at 4°C on a roller wheel. Beads were washed three times by addition of 1 ml of modified H-buffer made with 10% glycerol. After the final wash, beads were suspended in 40 μl of 2× SDS-PAGE buffer and boiled for 5 min. The protein supernatant was subjected to immune blot analysis to determine the amount of GST fusion protein and Ste7M variant protein in the eluant fraction for each pull-down assay.
Input and eluant samples were fractionated on duplicate gels by SDS-10% PAGE and transferred to nitrocellulose filters. One of the filters was probed for detection of the Ste7M variants by using the primary monoclonal anti-myc 9E10 antibodies (2.1 μg/ml). The duplicate filter was probed for detection of the GST fusion protein by using the primary monoclonal anti-GST antibodies (1:1,000; Promega). Secondary antibodies for both were goat anti-mouse immunoglobulin G (IgG)-alkaline phosphatase conjugate (1:7,000; Promega). Immunoreactive species on the blots were detected by colorimetric methods according to procedures detailed for the Promega Protoblot system. Signals from the scanned blots were quantified using NIH ImageJ software.
Immune blotting conditions for analysis of dual-phosphorylated Fus3M and Kss1M.
Strains C699-90 (ste5Δ ste7Δ fus3Δ kss1Δ) and C699-140 (ste7Δ fus3Δ kss1Δ) were transformed with plasmids for TPI1-promoted expression of myc-tagged Kss1 (Kss1M; pNC977) and/or myc-tagged Fus3 (Fus3M; pNC979). These two strains were also cotransformed with pRS313 and pRS316 to provide references for potential cross-reacting species. The resulting strains were then transformed with plasmids for CYC1-promoted expression of either wild-type Ste7 (pNC318), Ste7-EE-A7 (pNC597), or Ste7-EE-E7 (pNC893).
Cultures of each strain were grown in selective medium (-Trp -His and/or -Ura) with sucrose (1%) as the carbon source to a density of 5 × 106 cells/ml. A sample of the culture from strain C699-140 expressing wild-type Ste7 was used for the uninduced reference. The remaining portion of this wild-type Ste7 culture was pheromone induced by addition of α-factor (50 nM). Samples were removed after addition of pheromone for comparison of 10-, 15-, and 20-min induction times. Cells were harvested by centrifugation and suspended in lysis buffer [0.2 M Tris (pH 8.0), 0.4 M (NH4)2SO4, 1 mM EDTA (pH 8.0), 10% glycerol, 7 mM β-mercaptoethanol, 1 mM orthovanadate, 1 mM PMSF, and 5 μg each of pepstatin A, chymostatin, antipain, leupeptin, and leupeptin/ml]. Cell lysis, extract preparation, and trichloroacetic acid precipitation were done according to procedures described by Sabbaugh et al. (44) except for the composition of the lysis buffer. Pellets from trichloroacetic acid precipitation were suspended and adjusted to a protein concentration of 7 μg/μl in 3× SDS-PAGE loading buffer and boiled for 10 min.
Samples were fractionated by SDS-PAGE on duplicate 10% gels and transferred to nitrocellulose filters. One filter was used for detection of the total amount of Fus3M and Kss1M with anti-c-myc goat polyclonal antibodies (Santa Cruz) at 0.2 μg/ml with rabbit anti-goat alkaline phosphatase-conjugated IgG (Santa Cruz) at 0.04 μg/ml. The duplicate filter was used to detect the fraction of dual-phosphorylated Fus3M and Kss1M by using anti-phospho p44/p42 MAPK rabbit polyclonal antibodies (Cell Signaling Technology) at a 1:500 dilution with goat anti-rabbit alkaline phosphatase-conjugated IgG (Santa Cruz) at 0.04 μg/ml. The anti-phospho p44/p42 MAPK rabbit polyclonal antibodies specifically recognize the dually phosphorylated forms of Fus3 and Kss1 (5, 44). Immunoreactive species were detected by colorimetric methods according to procedures detailed by the Promega Protoblot system. Signals from the scanned blots were quantified using NIH ImageJ software. The fold induction for phospho-Kss1 and phospho-Fus3 in each sample was determined by normalizing the ratio of the anti-p44/p42 signal to the anti-myc signal for each MAPK to the corresponding ratio for the uninduced reference sample.
Immune blotting conditions for detection of Ste7 phosphorylation and ubiquitination status.
Pheromone induction results in the accumulation of hyperphosphorylated Ste7, which has a 10-kDa-slower SDS-PAGE mobility than unphosphorylated Ste7 (61). To assess SDS-PAGE mobility, Ste7M and different substitution variants as specified were expressed from the CYC1 promoter in strain C699-89 (ste7Δ trp1-1) or C699-106 (ste7Δ trp1Δ::HIS3MX6). Eighty micrograms of each whole-cell extract was fractionated by SDS-10% PAGE and transferred to nitrocellulose. Ste7M and substitution variant proteins were detected by using goat anti-myc antibodies (Santa Cruz) at a 1:1,000 dilution with rabbit anti-goat alkaline phosphatase-conjugated IgG (Santa Cruz) at a 1:10,000 dilution. Immunoreactive species were detected by colorimetric methods according to procedures detailed for the Promega Protoblot system.
Ubiquitination of Ste7 results in the formation of a high-molecular-weight species (Ubi-Ste7) (54, 55). For detection of this species, wild-type Ste7 and Ste7 variant proteins were expressed from the STE7 promoter in strain YHD1001 (ste7Δ ubp3Δ) or from the CYC1 promoter in strain C699-32 (ste7Δ) expressing a dominant-negative form of Ubp3 (pYES-Ubp3C469S). Cells were grown to mid-log phase in selective medium (-Ura or -Trp), treated with 3 μM (for YHD1001) or 50 nM (for C699-32) α-factor for 1 h, lysed directly in SDS-PAGE buffer for fractionation by SDS-7.5% PAGE, and transferred to nitrocellulose. Membranes were developed by using primary anti-Ste7 antibodies (yN-18; Santa Cruz) at 1:200 dilution and secondary anti-goat IgG conjugated to horseradish peroxidase (Santa Cruz) at 1:6,000 dilution. Immunoreactive species were visualized using the enhanced chemiluminescence detection system (Pierce) as directed by the vendor.
RESULTS
Ste7 phosphorylation site variant proteins.
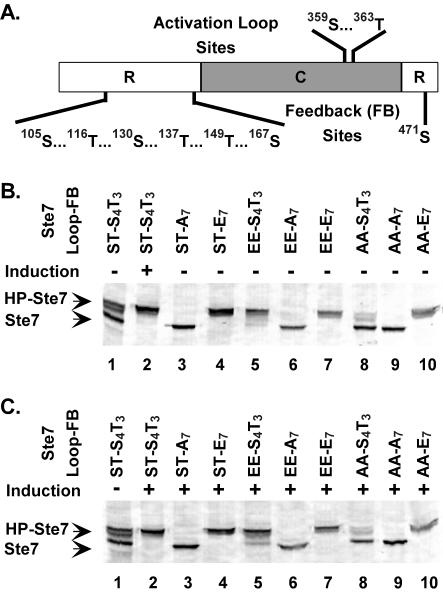
Pheromone induction leads to activation of Ste7 through phosphorylation of two residues at the activation loop of the catalytic domain (Fig. 2A) (20, 35, 59). Once activated, Ste7 phosphorylates and activates the downstream MAPKs, either of which mediate feedback hyperphosphorylation of Ste7 (61). Using an in vitro reconstitution system with E. coli-purified components, our group previously showed that Fus3 directly phosphorylates both the N- and C-terminal domains of Ste7 (20). The evidence reported in Fig. S1 to S3 in the supplemental material provides documentation that S105, T116, S130, T137, T149, S167, and S471 account for the major sites of Ste7 that are feedback phosphorylated by Fus3 in vitro and for the mobility shift of Ste7 that occurs during pheromone induction (Fig. 2).
FIG. 2.
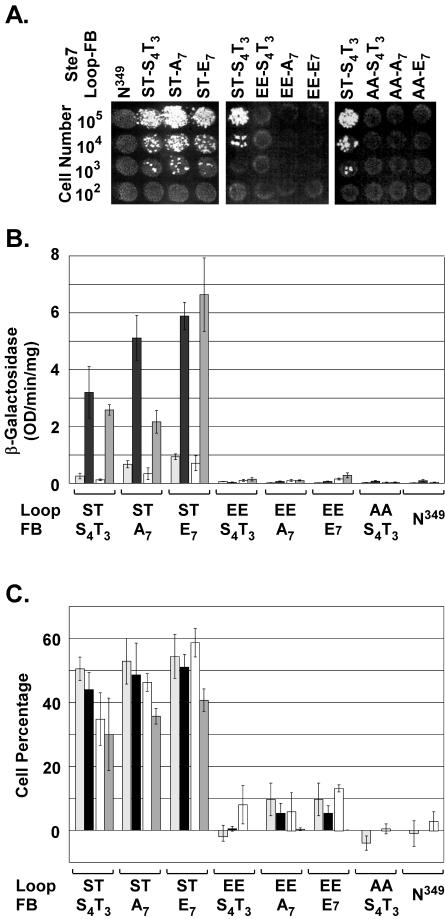
Location of phosphorylation sites in Ste7 and SDS-PAGE mobility of variant proteins with phosphorylation site substitution mutations. (A) Residues phosphorylated in the activation loop of the catalytic domain (C) and feedback sites phosphorylated by Fus3/Kss1 in the regulatory domains (R). (B and C) Western blots show mobility of Ste7 species that are characteristic of hyperphosphorylated (HP-Ste7) or hypophosphorylated (Ste7) forms after SDS-PAGE fractionation of protein extracts from uninduced (−) or induced (+) cultures as indicated. Residues at the activation loop (Loop) and feedback (FB) sites denote the different Ste7 variants. Ste7M variants were expressed from the CYC1 promoter on plasmids in strain C699-89 (ste7Δ) or C699-106 (ste7Δ) using the following plasmids: pNC318 (Ste7M), pNC586 (Ste7M-A7), pNC892 (Ste7M-E7), pNC589 (Ste7M-EE), pNC597 (Ste7M-EE-A7), pNC893 (Ste7M-EE-E7), pNC571 (Ste7M-AA), pNC894 (Ste7M-AA-A7), and pNC895 (Ste7M-AA-E7).
To explore systematically the function and regulation of the different Ste7 phosphoforms, we constructed a panel of Ste7 variants with substitutions at residues that become phosphorylated in the activation loop, feedback sites, or both. Glutamate substitutions have a negative charge and should mimic phosphorylation, whereas alanine substitutions preclude phosphorylation and mimic the unphosphorylated state. Consistent with these assumptions, previous evidence established that glutamate substitutions at the activation loop residues make Ste7 a constitutively active kinase and that alanine substitutions at these residues render Ste7 inactive (20, 59).
MAPK-dependent hyperphosphorylation of Ste7 causes slow mobility of this species on SDS-PAGE (61). To assess whether glutamate and alanine substitutions at the feedback residues similarly confer conformations corresponding to hyper- and hypophosphorylated Ste7, respectively, we compared the SDS-PAGE mobilities of the variants to that of Ste7 from uninduced and pheromone-induced cultures (Fig. 2B and C). A single species with mobility identical to that of hyperphosphorylated Ste7 was observed for any variant with glutamate substitutions at the feedback residues, whether or not cultures were treated with pheromone (Fig. 2B and C, Ste7-E7, Ste7-AA-E7, and Ste7-EE-E7). Also, regardless of pheromone induction, a single species with mobility identical to that of hypophosphorylated Ste7 was observed for variants with alanine substitutions at the feedback residues (Fig. 2B and C, Ste7-A7, Ste7-AA-A7, and Ste7-EE-A7). The constitutively active and inactive variants with serine and threonine residues at the feedback sites also exhibited mobility on SDS-PAGE that was independent of pheromone induction. Ste7-EE induced Fus3/Kss1 activity and became hyperphosphorylated even in the absence of pheromone induction. Ste7-AA failed to stimulate Fus3/Kss1 activity above background and remained largely hypophosphorylated, even in the presence of pheromone induction (Fig. 2B and C, Ste7-EE and Ste7-AA). From these correlations, we infer that glutamate and alanine substitutions at the feedback residues correctly confer conformations corresponding to hyper- and hypophosphorylated Ste7, respectively.
MAPK-mediated feedback regulation of Ste7 activity.
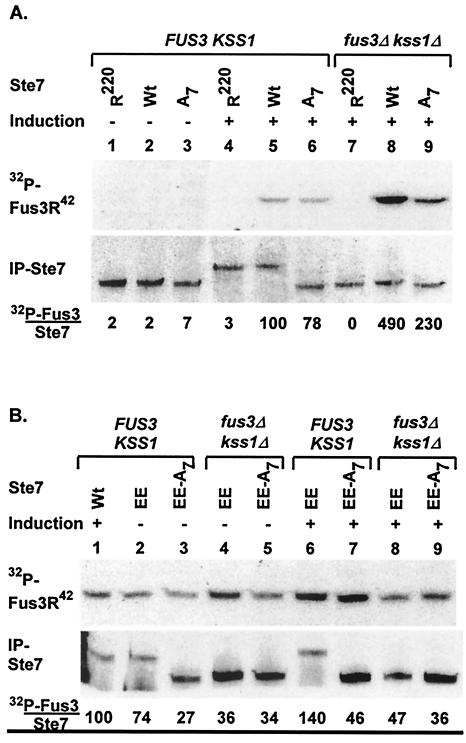
Our group previously observed attenuation of Ste7 activity in cells expressing Fus3 and Kss1 compared with cells deficient in both MAPKs (19, 20). One explanation for the observation is that the specific activity of Ste7 is lower after feedback phosphorylation than before modification. To test this hypothesis, we compared in vitro kinase activity of the variant that cannot be phosphorylated at the feedback sites (Ste7-A7) to that of wild-type Ste7 and catalytically inactive Ste7-R220. For this comparison, myc epitope-tagged Ste7, Ste7-A7, and Ste7-R220 proteins were expressed from the CYC1 promoter and immune precipitated from extracts of uninduced or pheromone-induced cells either expressing Fus3 and Kss1 (FUS3 KSS1) or deficient in both MAPKs (fus3Δ kss1Δ). Assays were done directly in immune complexes using [γ-32P]ATP with catalytically inactive Fus3 (Fus3-R42) as the protein substrate. Contrary to our prediction, Ste7-A7 had lower activity than the wild-type reference (Ste7) (Fig. 3A, lanes 5 and 6). Activity of Ste7-A7 was higher when isolated from fus3Δ kss1Δ cells than from FUS3 KSS1 cells (Fig. 3A, lanes 6 and 9). This activity was still lower than that of the wild-type reference in this background (Fig. 3A, lanes 8 and 9). These findings show that MAPK-dependent suppression of Ste7 activity occurs through a mechanism that is distinct from MAPK-catalyzed Ste7 feedback phosphorylation.
FIG. 3.
In vitro kinase activities of Ste7 phosphorylation site substitution derivatives. Upper panels show the PhosphorImage of 32P incorporation into Fus3R42, and lower panels show immune blots detecting immune-precipitated Ste7 after SDS-PAGE fractionation of proteins from immune complex kinase assay reactions. Cultures for these analyses were either untreated (−) or treated with pheromone (+), as indicated. Activities for each sample are calculated as ([32P]Fus3 signal)/(IP-Ste7 signal). The numbers below the immune blot report the activity for each reaction as a percentage of that obtained for the sample with pheromone-induced Ste7 (WT). (A) Samples in lanes 1 to 6 are from strain K2149 (FUS3KSS1), and those in lanes 7 to 9 are from strain K2313 (fus3Δ kss1Δ) expressing catalytically inactive Ste7M-R220 (R220; pNC318-R220), wild-type Ste7M (Wt; pNC318), or Ste7M that cannot be feedback phosphorylated (A7; pNC586). (B) Samples in lanes 1 to 3, 6, and 7 are from strain K2149 (FUS3 KSS1), and those in lanes 4, 5, 8, and 9 are from strain K2313 (fus3Δ kss1Δ) expressing Ste7M (Wt; pNC318), constitutively active Ste7M (EE; pNC589), or constitutively active Ste7M that cannot be feedback phosphorylated (EE-A7; pNC597).
Fus3- and Kss1-mediated signal attenuation mechanisms that target components upstream of Ste7 could diminish accumulation of active Ste7. For example, the MAPKKK-Ste11 is targeted for MAPK- and ubiquitin-dependent degradation during pheromone induction (21). Consequently, a greater fraction of Ste7 can be activated in a fus3Δ kss1Δ background than with the FUS3 KSS1 background, because active Ste11 can accumulate in the former but not the latter (21). According to this model, the kinase activity of constitutive Ste7 variants should be independent of Fus3 and Kss1 genetic background, because they do not require modification by an upstream component for their activation. Therefore, we compared in vitro kinase activities of constitutive Ste7 derivatives that can and cannot be feedback phosphorylated using immune complex kinase assays as before.
As expected for a constitutive enzyme, Ste7-EE isolated from cultures without pheromone induction readily catalyzes phosphorylation of catalytically inactive Fus3 (Fus3-R42) (Fig. 3B, lane 2). The effect of pheromone induction on activity of the constitutive enzymes is minimal when isolated from either FUS3 KSS1 or fus3Δ kss1Δ cells (∼2-fold or less) (Fig. 3B, compare lanes 2 and 3 with 6 and 7 and lanes 4 and 5 with 8 and 9). Constitutive Ste7 that is hyperphosphorylated (Ste7-EE) has higher activity than non-feedback-phosphorylated constitutive Ste7 (Ste7-EE-A7) (Fig. 3B, compare lanes 2 and 6 to lanes 3 to 5, 8, and 9). As seen with wild-type Ste7, feedback phosphorylation had a positive effect on the ability of constitutive Ste7 to phosphorylate Fus3-R42 in vitro. The comparisons made with Ste7-EE-A7, which eliminates the effect of feedback phosphorylation, showed that constitutive Ste7 has essentially the same activity regardless of pheromone induction and of FUS3 KSS1 genetic background (Fig. 3B, compare lanes 3, 5, 7, and 9). This outcome is consistent with the assessment that MAPK-dependent suppression of wild-type Ste7 activity occurs through attenuation mechanisms that target upstream components of the mating pathway.
Based on the above model, we infer that much of the Ste7 isolated from pheromone-induced FUS3 KSS1 cultures is still in the inactive state. Therefore, the amount of Fus3-R42 phosphorylation catalyzed by enzymes isolated from pheromone-induced fus3Δ kss1Δ cultures should be used to assess their relative specific activities. Using this criterion, the specific activity of Ste7 with glutamates at the activation loop was only 10 to 15% of that for Ste7 activated by phosphorylation (Fig. 3A and B, lanes 8 and 9). Yet, the amount of Fus3-R42 phosphorylation catalyzed in these in vitro reactions by constitutive Ste7 was similar to that for wild-type Ste7 isolated from pheromone-induced FUS3 KSS1 cultures (Fig. 3B, lanes 1, 2, and 6). Thus, despite low specific activity, expression of constitutive Ste7 from the CYC1 promoter yields a total activity comparable to what is achieved by the fraction of wild-type Ste7 that becomes activated during pheromone induction.
MAPK-mediated feedback regulation of Ste7 ubiquitination.
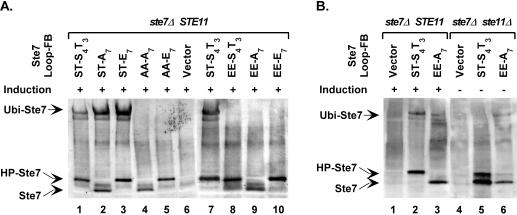
Recent studies revealed that Ste7 undergoes SCF (Skp1/Cullin/F-box)-dependent ubiquitination and proteosome-mediated degradation upon prolonged stimulation by pheromone (54, 55). To examine whether feedback phosphorylation or constitutive activity preferentially targets Ste7 for ubiquitination, we expressed wild-type Ste7 and different phosphorylation site variants from their own promoter in a mutant strain background known to accumulate ubiquitinated Ste7 (ubp3Δ) (54, 55). The ubiquitin-modified protein accumulates for wild-type Ste7 and for derivatives with serine and threonine at the activation loop, regardless of what residues are present at the feedback sites (Fig. 4A, lanes 1 to 3). The ubiquitin-modified species fails to accumulate for derivatives with either alanines or glutamates at the activation loop (Fig. 4A, lanes 4, 5, and 8 to 10). Therefore, the ubiquitin modification of Ste7 is dependent on phosphorylation at the activation loop but is independent of the feedback phosphorylation status of Ste7.
FIG. 4.
Ubiquitin modification of Ste7 phosphorylation site variants. Western blots show mobility of Ste7 species that are characteristic of the ubiquitinated (Ubi-Ste7), hyperphosphorylated (HP-Ste7), and hypophosphorylated (Ste7) forms. (A) Ste7 proteins were expressed from the STE7 promoter in strain YHD1001 (ste7Δ ubp3Δ) by using plasmids pNC766 (Ste7M), pNC768 (Ste7M-EE), pNC770 (Ste7M-EE-A7), pNC791 Ste7M-EE-E7), pNC781 (Ste7M-AA-A7), or pNC790 (Ste7M-AA-E7). The same strain with vector (pRS316) was used as a negative reference. (B) Ste7 proteins were expressed from the CYC1 promoter in strain C699-32 (ste7Δ) or strain C699-81 (ste7Δ ste11Δ) carrying pYES-UBP3C469S by using plasmids pNC318 (Ste7M) or pNC597 (Ste7M-EE-A7). The same strain with vector (pNC160) was used as a negative reference.
The failure to accumulate ubiquitinated Ste7 when glutamates occupy the activation loop sites could be explained if glutamates were an inadequate mimic for phosphorylated residues with regard to recognition by the ubiquitination machinery. Alternatively, signal output from the mating pathway may be critical to the mechanism for Ste7 ubiquitination. We learned that the glutamate substitution variants expressed from the endogenous STE7 promoter are functional for invasive growth but not for mating. However, overexpression of Ste7-EE-A7 from the CYC1 promoter restored some level of mating pathway output (see below). To test the idea that signal output is critical for ubiquitination, we compared wild-type Ste7 and Ste7-EE-A7 when expressed from the CYC1 promoter for ubiquitination in ste7Δ and ste7Δ ste11Δ strains. Both strains also expressed a dominant-negative form of Ubp3 (Ubp3C469S) to allow accumulation and visualization of ubiquitinated species as before. Ubiquitinated Ste7 and ubiquitinated Ste7-EE-A7 were detected in the ste7Δ strain (Fig. 4B, lanes 2 and 3). However, the ratio of the ubiquitinated species to the unmodified species for Ste7-EE-A7 (0.08) was markedly less than that for the pheromone-induced wild-type Ste7 reference (0.31). The comparison in the ste7Δ ste11Δ strain was striking, because Ste7-EE-A7 became ubiquitinated in the absence of pheromone treatment and under conditions where wild-type Ste7 remained unmodified (Fig. 4B, lanes 5 and 6). The fraction of ubiquitinated Ste-EE-A7 (0.21) under these conditions was comparable to that for the pheromone-induced wild-type reference (0.31) (Fig. 4B, lanes 2 and 6). These results establish that Ste7 variants with glutamates at the activation loop are competent substrates for ubiquitination and that signal output from the pathway is required for the modification.
Mating response and invasive growth phenotypes of Ste7 phosphorylation site variants.
Ste7 mediates mating differentiation and invasive growth in haploid S. cerevisiae. To determine if expression of constitutive Ste7 and feedback phosphorylation influence the establishment of one or the other of these fates, we quantified mating and invasive phenotypes of strains expressing the different variants. For evaluation of mating pathway function, strains were compared for mating efficiency and pheromone-induced transcriptional, G1 arrest, and morphological responses (Fig. 5). A strain expressing a Ste7 variant (Ste7-N349) that is inactive because of an Asn substitution for a catalytically essential Asp residue was included as a negative control for these comparisons. As expected for catalytically inactive enzymes, the variants with Ala substitutions at the activation loop (Ste7-AA, Ste7-AA-E7, and Ste7-AA-A7) failed to mate and to support pheromone-induced responses (Fig. 5). The variants with Ser and Thr at the activation loop supported mating independently of the charge status of the feedback phosphorylation residues (Fig. 5A, Ste7, Ste7-E7, and Ste7-A7). These variants also functioned similarly with respect to the pheromone-induced reporter gene expression, G1 arrest, and mating projection formation (Fig. 5B and C).
FIG. 5.
Mating and pheromone response assays comparing the in vivo function of Ste7 phosphorylation site substitution derivatives. (A) Semiquantitative mating assays. Photographs show diploid colonies selected for growth on minimal medium after an overnight incubation of mating mixtures at 30°C. Tester strain KZ8-1D (MATα; 106 cells) was mixed with the indicated number of cells from strain C699-89 (MATa bar1Δ ste7Δ) with a plasmid expressing the specified Ste7 variant from the STE7 promoter. (B) Pheromone-induced reporter gene expression. The bar graph shows the average β-galactosidase activity for expression of the mating-specific FUS1-LacZ reporter gene (pNC756) in two different strains carrying the specified Ste7 M variant expressed from the STE7 promoter. Averages are from four independent cultures of each variant in strain C699-89 (bar1Δ ste7Δ) before (░⃞) and after (▪) 2-h induction with 50 nM α-factor and from four independent cultures of each variant in strain MLY218a (MATa ste7Δ) before (□) and after (▩) 2.5-h induction with 3 μM α-factor. Error bars show the average deviations in measurements. (C) Pheromone-induced G1 arrest and mating differentiation. The bar graph shows the average G1 arrest and mating differentiation indices for samples from the same cultures used to measure reporter gene expression in panel B. The G1 arrest index (C699-89 ░⃞; MLY218a [□]) is the percentage of unbudded cells in each culture after pheromone induction minus the percentage of unbudded cells in the starting vegetative culture. The differentiation index (C699-89 [▪]; MLY218a [▩]) is the percentage of cells that formed mating projections. Error bars show the average deviations in measurements. Ste7 variants were expressed from the STE7 promoter by using the following plasmids: pNC766 (Ste7M), pNC769 (Ste7M-A7), pNC793 (Ste7M-E7), pNC768 (Ste7M-EE), pNC770 (Ste7M-EE-A7), pNC791 (Ste7M-EE-E7), pNC767 (Ste7M-AA), and pNC771 (Ste7M-N349).
Despite constitutive in vitro kinase activity, the variants with glutamate substitutions at the activation loop failed to support mating, transcriptional, G1 arrest, and morphological responses to pheromone (Fig. 5, Ste7-EE, Ste7-EE-E7, and Ste7-EE-A7). The defective responses did not depend on which residues occupied the feedback sites. Therefore, it appears that the constitutive negative charge at the activation loop underlies the failure of these variants to support a mating response. We further infer that the problem must arise at the level of maintaining signal output, because Ste7-EE caused sufficient activation of Fus3 and/or Kss1 in vivo to promote phosphorylation of the feedback residues (Fig. 2B, lane 5).
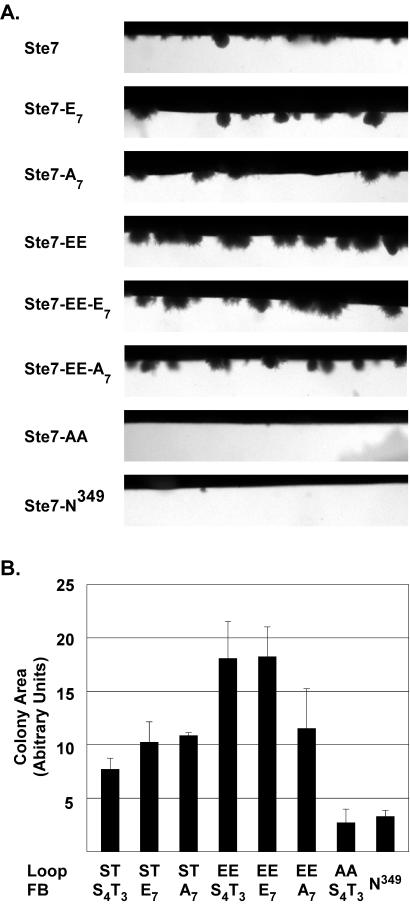
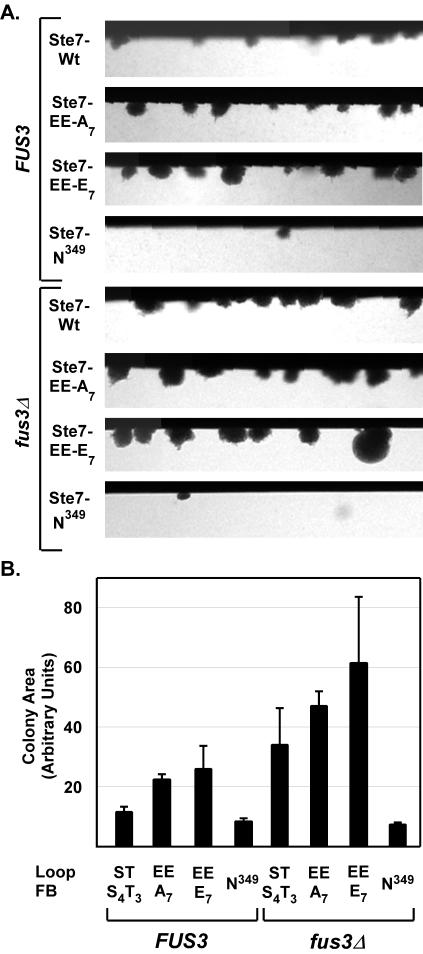
To examine the function of Ste7 phosphorylation site variants in the filamentous growth pathway, each variant was expressed in a ste7Δ strain derived from a background (Σ1278) that is competent for pseudohyphal and invasive growth. Cells were cultured using complex medium in chambers that allow direct microscopic visualization of colonies as they invade and grow beneath the agar surface (Fig. 6A). To quantify this response, we calculated the mean area of individual colonies below the agar surface. The average of the mean colony area from three independent experiments is shown (Fig. 6B).
FIG. 6.
Invasion assays comparing the in vivo functions of Ste7 phosphorylation site substitution derivatives. (A) Micrographs show invasive colonies from a section of growth chambers inoculated with strain MLY218a (ste7Δ) carrying plasmids for expression of the Ste7 variants as specified in Fig. 4. (B) Bar graph shows the average and average deviation of the invasive colony mean area from three independent experiments. Measurements were made on 25 or more individual colonies in a given chamber after 46 to 48 h of growth at 30°C. Because few invasive colonies formed with the strains expressing the catalytically inactive Ste7M-N349 and Ste7M-AA variants, the values are an average from measurement of only 9 and 11 total colonies, respectively.
Strains expressing the Ste7 variants with inducible activity invaded the agar substrate and grew into colonies with pseudohyphal filaments at the periphery (Fig. 6A, Ste7, Ste7-A7, and Ste7-E7). As expected, strains expressing the catalytically inactive Ste7 variants invaded the agar poorly (Fig. 6A, Ste7-N349 and Ste7-AA). There were few invasive colonies in these cultures, and those that formed below the agar surface were in general smaller than those of the wild-type reference (Fig. 6B). Also, the invasive colonies of the strains with inactive Ste7 did not elongate or form filaments as did the colonies from strains with active Ste7 (see Fig. S4 in the supplemental material). In contrast to their null mating phenotype, the constitutive Ste7 variants supported invasive growth (Fig. 6A, Ste7-EE, Ste7-EE-A7, and Ste7-EE-E7). Indeed, the invasive colonies of strains expressing the constitutive variants had a larger mean area than strains expressing inducible Ste7 (Fig. 6B). They also had a hyperfilamented phenotype relative to the wild-type reference (Fig. S4 in the supplemental material).
The above comparison shows that the constitutive Ste7 variants are functional under in vivo conditions, and it eliminates trivial explanations for their defective mating response phenotypes. Also, the failure to function in the mating pathway cannot be solely a function of strain background, because the constitutive variants were defective for mating responses in the same Σ1278 strain that was used for these invasion assays (Fig. 5B and C). Therefore, these observations establish that constitutive Ste7 uniquely specifies a filamentous growth response.
Constitutive Ste7 phosphorylates Kss1 but not Fus3 in vivo.
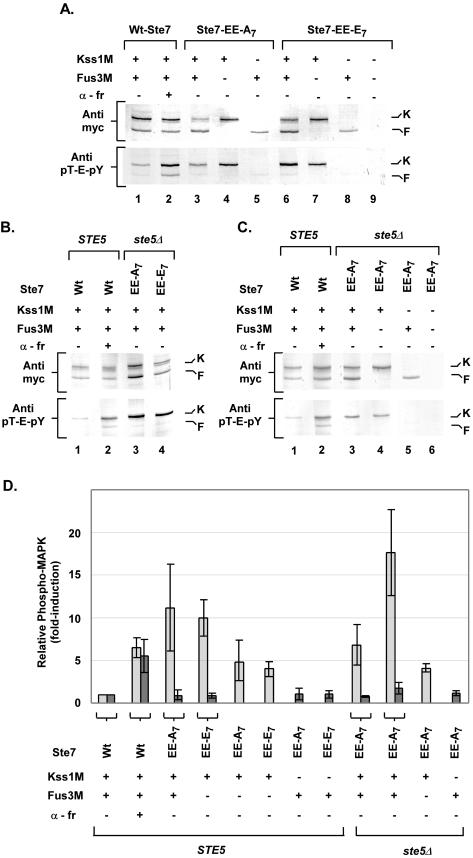
Although constitutive Ste7 variants are functional kinases in vitro (Fig. 3), it is possible that their activity is insufficient to support high levels of active MAPK. In this case the observed signal specificity could simply be that a lower threshold of MAPK activation is needed for the invasion response than for the mating response. To examine this possibility, we used immune blot detection methods to compare the fraction of phosphorylated Fus3 and Kss1 that was sustained in vivo by constitutive Ste7 (Ste7-EE-A7 or Ste7-EE-E7) to that achieved by wild-type Ste7 during pheromone induction (5, 44). Fus3M and Kss1M were expressed from the constitutive TPI1 promoter in the strains used for this comparison. Because endogenous Fus3 (but not Kss1) expression is induced by pheromone, their expression from the TPI1 promoter ensured equivalent transcription of the two MAPKs independently of a mating pathway-induced response (16). Extracts of the different cultures were fractionated on duplicate gels by SDS-PAGE, which resolves Kss1 and Fus3, and transferred to nitrocellulose for immune blot analysis. Anti-myc antibodies were used on one filter to determine the total amount of Fus3M and Kss1M in each extract. The duplicate filter was used to detect the amount of dual-phosphorylated (pT-E-pY) Fus3M and Kss1M by using anti-phospho Erk antibodies (5, 44).
Normal pheromone induction leads to an increase in the relative amount of dually phosphorylated Fus3 and Kss1 (7, 44). Extracts of cultures expressing either Ste7-EE-A7 or Ste7-EE-E7 without pheromone induction were compared directly to those from the uninduced (t = 0) and pheromone-induced (t = 15 min) wild-type Ste7 reference (Fig. 7A ). Constitutive Ste7 promoted in vivo phosphorylation of Kss1 to levels that were as high or higher than those seen for the pheromone-induced reference (Fig. 7A and D). By contrast, there was no increase in Fus3 phosphorylation above the basal levels seen for the uninduced reference (Fig. 7A and D). We also examined Kss1M phosphorylation when coexpressed with Ste7-EE-A7 or Ste7-EE-E7 in the absence of Fus3. We found that constitutive Ste7 promoted levels of phospho-Kss1 that were lower in the absence than in the presence of Fus3. But the amount of phospho-Kss1 was nevertheless comparable to the pheromone-induced levels (Fig. 7A, lanes 4, 5, 7, and 8, and D). The amount of phospho-Fus3 remained at the basal levels seen for the uninduced reference whether or not Kss1 was also present (Fig. 7A and D). Similar results were seen with extracts from the corresponding ste5Δ strains (Fig. 7B, C, and D). These results showed that the ability of constitutive Ste7 to phosphorylate Kss1 is independent of the scaffold-Ste5. They also suggest that Fus3 has some positive role in allowing hyperaccumulation of phosphorylated Kss1. The finding that constitutive Ste7 promotes high levels of phospho-Kss1 rules out the possibility that the substitution variants of Ste7 are ineffective kinases in vivo. Further, the activation of Kss1 is consistent with the observed stimulation of an invasive response by constitutive Ste7.
FIG. 7.
Comparison of Fus3 and Kss1 phosphorylation induced by pheromone or constitutive Ste7. (A to C) The upper panel shows the total amount of Fus3M and Kss1M by detection with anti-myc antibodies. The lower panel shows the fraction of dually phosphorylated Fus3M and Kss1M by detection with anti-Erk (pT-E-pY) antibodies. The signals corresponding to Kss1M (K) and Fus3M (F) are indicated to the right of the blots. (A) Protein extracts (33 μg) were from strain C699-140 (STE5 ste7Δ fus3Δ kss1Δ) expressing wild-type (Wt) Ste7 (pNC318), Ste7-EE-A7 (pNC597), or Ste7-EE-E7 (pNC893) with Kss1M (pNC977), Fus3M (pNC979), or both, as indicated. Lanes 1 and 2 show extracts from the Wt-Ste7 strain before (−) and after (+) 15-min pheromone (α-fr; 50 nM) induction. (This time was empirically shown to result in optimal induction of Fus3 and Kss1 phosphorylation.) Lane 9 shows an extract from the Ste7-EE-E7 strain with vectors (pRS313 and pRS316) as a negative reference. (B and C) Lanes 1 and 2 show extracts (66 μg) from strain C699-140 (STE5 ste7Δ fus3Δ kss1Δ) expressing Wt-Ste7 with Kss1M and Fus3M before (−) and after (+) induction with pheromone as specified in panel A. Lanes 3 and 4 (B) or 3 to 5 (C) show extracts (33 μg) from strain C699-90 (ste5Δ ste7Δ fus3Δ kss1Δ) expressing Ste7-EE-A7 or Ste7-EE-E7 with Kss1M, Fus3M, or both as specified. Lane 6 (C) shows an extract from strain C699-90 expressing Ste7-EE-A7 with vectors (pRS313 and pRS316) as a negative reference. (D) Bar graph comparing the relative fold induction of phospho-Kss1 (░⃞) and phospho-Fus3 (▩) in strains as defined for panels A to C. Values are the average fold induction determined from three experiments. Induction is the fraction of phospho-MAPK [(anti-myc signal)/(anti-pT-E-pY signal)] for each sample relative to that from the uninduced reference. The lines with bars show the average deviations in measurements.
We did not anticipate the finding that constitutive Ste7 would phosphorylate Kss1 but not Fus3 in vivo. This outcome brings up two questions. What mechanisms contribute to the selective activation of Kss1, and what suppresses the mating response expected from the activation of Kss1? We addressed each of these issues in the experiments described below.
Phosphorylation status of Ste7 determines its selective binding to components of the MAPK activation module.
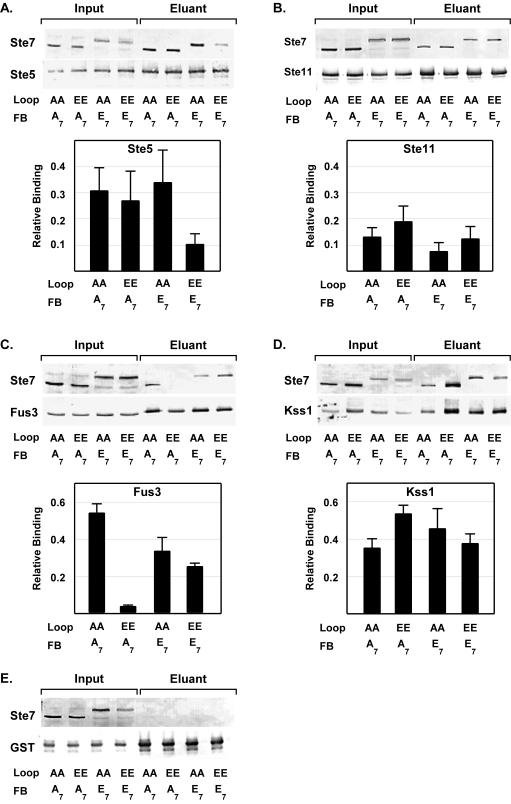
To delineate how constitutively active Ste7 selectively phosphorylates Kss1 in vivo, we compared Ste7 variants for relative binding to each of the relevant components of this pathway by using a standard GST fusion protein copurification assay. We limited the comparison to those variants that represent the four possible combinations of loop and feedback site phosphorylation states of Ste7 (Ste7-AA-A7, Ste7-EE-A7, Ste7-AA-E7, and Ste7-EE-E7). This selection was made to ensure that the population of Ste7 species in different binding reactions would be homogenous and unaffected by the spurious actions of phosphatases or kinases. Additionally, the substitution variants allow assessment of interactions with species of Ste7 that would otherwise be transient and thus of too low abundance to be detected in comparisons of wild-type Ste7 from uninduced and pheromone-induced extracts.
A host strain that is deficient in all MAPK module components (ste5Δ ste11Δ ste7Δ fus3Δ kss1Δ) was used so that only pairwise interactions would be assessed. (The exception was for comparisons of Ste7 binding to GST-Ste5 [see below].) The GST fusion proteins and the myc-tagged Ste7 variant proteins in the input and eluant fractions of these assays were detected by using anti-GST antibodies and anti-myc antibodies, respectively (Fig. 8). Relative binding efficiencies are reported as the ratio of the anti-myc signal to the anti-GST signal in the eluant fraction. Because there was no binding of Ste7 variants to GST alone, the presence of Ste7 in an eluant fraction was due to its binding to the specified MAPK pathway component (Fig. 8E). For reference, the magnitude of the relative binding of Ste7-AA-A7 to each of the GST fusion proteins reported below was the same or slightly greater than what we observed for wild-type Ste7 in the corresponding strains (data not shown.)
FIG. 8.
Copurification assays comparing the in vivo binding of Ste7 phosphorylation site substitution derivatives to other components of the MAPK activation module. Representative Western blots show the amount of indicated Ste7 variant and GST fusion protein in the starting extracts (Input) and in the glutathione-agarose-immobilized fraction (Eluant). Proteins were fractionated by SDS-PAGE. Monoclonal anti-myc 9E10 antibodies were used for detection of myc epitope-tagged Ste7 (Ste7M) derivatives, and monoclonal anti-GST antibodies were used for detection of GST or the indicated GST fusion protein. Bar graphs show the average relative binding and average deviation for the specified Ste7M variant and GST fusion protein. Relative binding is the ratio of the anti-myc signal (Ste7) to the anti-GST signal (fusion protein) in the eluant fraction. (A) Strain C699-93 (ste11Δ fus3Δ) was used to coexpress each Ste7M variant with GST-STE5 (pYBS298). Strain C699-92 (ste7Δ ste5Δ ste11Δ fus3Δ kss1Δ) was used to coexpress each Ste7M variant with the following: GST-STE11 (pRD-GST-Ste111-717) (B); GST-FUS3 (pNC901) (C); GST-KSS1 (pNC903) (D); or GST (pEG-KT) (E). Indicated Ste7M variant proteins were expressed using the plasmids pNC896 (Ste7M-AA-A7), pNC897 (Ste7M-EE-A7), pNC898 (Ste7M-AA-E7), and pNC809 (Ste7M-EE-E7).
We failed to detect a binding interaction between GST-Ste5 and Ste7 (or any substitution variant) when the proteins were coexpressed in the strain deficient in the scaffold and all the core enzymes of the MAPK activation module. The failure to detect a complex is consistent with published two-hybrid studies that found that the Ste7-Ste5 interaction was substantially lower in a strain lacking endogenous Ste5 (8). Additionally, the published copurification assay for wild-type Ste7 and GST-Ste5 used a strain deficient only in Ste11 and Fus3. We suspect that endogenous Ste5 oligomerizes with GST-Ste5 and provides some essential binding function that is missing for the GST fusion protein. Therefore, we used the analogous strain (ste11Δ fus3Δ) to coexpress GST-Ste5 with the different Ste7 variant proteins. Under these conditions we found that Ste7-AA-A7, Ste7-EE-A7, and Ste7-AA-E7 bound equally well to Ste5 but Ste7-EE-E7 bound less effectively (Fig. 8A). The latter observation is consistent with the report that hyperphosphorylated wild-type Ste7 binds poorly to Ste5 (8). However, the inactive but hyperphosphorylated Ste7 variant (Ste7-AA-E7) binds to Ste5 as well as the variants without charge at the feedback phosphorylation sites. To reconcile this observation with that of Choi et al. (8), we assume that inactive but hyperphosphorylated Ste7 is a minor species in the wild-type population. The relatively poor binding of Ste7-EE-E7 to Ste5 could contribute to inefficient colocalization with Fus3. This deficiency could explain the failure of Ste7-EE-E7 to phosphorylate Fus3 in vivo. However, we note that Ste7-EE-A7 binds well to Ste5 even though this variant also fails to phosphorylate Fus3 in vivo (Fig. 7). These results suggest that simply tethering Ste7 on the scaffold is not sufficient to ensure effective phosphorylation of Fus3 (see below).
All four Ste7 phosphorylation site substitution variants bound to the MAPKKK-Ste11 (Fig. 8B). In principle, the binding of Ste11 to constitutive forms of Ste7 could negatively affect signaling by competing with the MAPKs for access to Ste7. However, it is unclear whether this potential competition would be significant in the scaffold-organized complex.
The most surprising finding is that the phosphorylation state dependence of Ste7 binding to the MAPK-Fus3 is different from that for binding to the MAPK-Kss1. All four Ste7 phosphorylation site substitution variants bound similarly to Kss1 (Fig. 8D). By contrast, the strongest binding to Fus3 was seen with Ste7-AA-A7, which represents inactive Ste7 without feedback phosphorylation (i.e., naïve Ste7) (Fig. 8C). Ste7-EE-A7 essentially showed no binding to Fus3 (Fig. 8C). This variant can be thought of as the transition state intermediate on the pathway from active Ste11 to active Fus3 and Kss1. The failure of this intermediate to bind to Fus3 reveals that the scaffold-Ste5 probably provides the major route for colocalizing active Ste7 with Fus3. The binding characteristics of the different Ste7 variants are also consistent with Ste5 being dispensable for the activation of Kss1 by Ste7.
Constitutive Ste7 variants promote high levels of filamentation-specific reporter gene expression.
Invasive growth and Kss1 activation promoted by constitutive Ste7 predict that these variants should stimulate filamentation-specific gene expression above the basal levels seen in a wild-type Ste7 background. To confirm this prediction, we measured expression of a filamentation-specific reporter gene in ste7Δ, ste7Δ fus3Δ, and ste7Δ kss1Δ strains that expressed Ste7 and variant proteins from the CYC1 promoter. The reporter gene (Ty-LacZ) for this comparison uses a Tec1-Ste12 composite binding site derived from the Ty1 sequence to drive LacZ expression (Fig. 1) (6).
Consistent with this prediction, constitutive Ste7 variants promoted 9- to 20-fold-higher β-galactosidase activities relative to the amount promoted by the wild-type Ste7-reference (Table 3, strain C699-106). In a strain deficient in the MAPK-Kss1, reporter gene activity remained at background amounts whether or not the strain expressed wild-type or constitutive Ste7 variants (Table 3, strain C699-146). This outcome is consistent with the effects of constitutive Ste7 being mediated by phosphorylation and activation of Kss1. Deleting the mating pathway-specific MAPK-Fus3 significantly enhances Ty-LacZ expression promoted by the wild-type Ste7 but not by constitutive Ste7-variants (Table 3, strain C699-145). The negative effect that Fus3 had on Ty-LacZ expression in the wild-type Ste7 strain is consistent with previously reported findings (5, 32, 44, 57). The finding that Fus3 has no dramatic effect on output from constitutive Ste7 variants could be either because Fus3 does not accumulate in the activated state in these strains or because hyperactivation of Kss1 is sufficient to overcome any negative effects that active Fus3 mediates.
TABLE 3.
Ste7-promoted reporter gene expression in strains with deletions of MAPK activation module components
| Relevant genotype and strain | β-Galactosidase activity (mOD min−1 mg−1)a for plasmid, variant (induction) |
||||||
|---|---|---|---|---|---|---|---|
| pNC318, Ste7 (+) | pNC318, Ste7 (−) | pNC597, Ste7-EE-A7 (−) | pNC589, Ste7-EE (−) | pNC893, Ste7-EE-E7 (−) | pNC160, none (−) | ||
| Ty-LacZ | |||||||
| C699-106 | ste7Δ | 400 ± 20 | 8,100 ± 70 | 3,500 ± 100 | 5,400 ± 500 | 20 ± 2 | |
| C699-145 | ste7Δ fus3Δ | 3,000 ± 300 | 2,400 ± 70 | 4,700 ± 500 | 7,100 ± 300 | 20 ± 10 | |
| C699-146 | ste7Δ kss1Δ | 50 ± 10 | 20 ± 10 | 20 ± 10 | 10 ± 10 | 10 ± 10 | |
| FUS1-LacZ | |||||||
| C699-32 | ste7Δ | 2,900 ± 400 | 40 ± 4 | 80 ± 10 | 3 ± 3 | 6 ± 3 | 1 ± 1 |
| C699-82 | ste7Δ ste5Δ | 10 ± 3 | 50 ± 5 | 8 ± 6 | 6 ± 3 | 1 ± 1 | |
| C699-81 | ste7Δ ste11Δ | 1 ± 1 | 400 ± 40 | 1 ± 2 | 4 ± 1 | 1 ± 1 | |
| C699-80 | ste7Δ fus3Δ | 1,000 ± 100 | 50 ± 7 | 200 ± 20 | 20 ± 4 | 30 ± 3 | 9 ± 3 |
| C699-127 | ste7Δ ste11Δ ste5Δ | 5 ± 3 | 350 ± 10 | 8 ± 3 | 9 ± 3 | 2 ± 2 | |
| C699-87 | ste7Δ ste5Δ fus3Δ | 10 ± 2 | 4,700 ± 300 | 10 ± 4 | 20 ± 2 | 6 ± 3 | |
| C699-108 | ste7Δ ste11Δ fus3Δ | 10 ± 2 | 400 ± 100 | 9 ± 3 | 6 ± 1 | 7 ± 2 | |
| C699-114 | ste7Δ kss1Δ | 1,600 ± 400 | 50 ± 5 | 6 ± 1 | 4 ± 1 | 8 ± 2 | 6 ± 1 |
| C699-116 | ste7Δ ste5Δ kss1Δ | 4 ± 1 | 6 ± 2 | 8 ± 3 | 8 ± 2 | 9 ± 3 | |
| C699-115 | ste7Δ ste11Δ kss1Δ | 4 ± 2 | 8 ± 1 | 7 ± 1 | 3 ± 1 | 6 ± 1 | |
Values are averages ± average deviations. OD, optical density.
The negative effect that Fus3 has on the transcriptional output for wild-type Ste7 is also apparent for the invasion phenotype. Using the same assay as before, we found the average invasion colony area for wild-type Ste7 was ∼3-fold greater in the fus3Δ strain compared with the FUS3 strain (Fig. 9). The constitutive Ste7 variants also showed a more robust invasion phenotype in fus3Δ strains compared with FUS3 strains (Fig. 9). This result shows that Fus3 has a more negative effect on invasion compared with the Ty-LacZ reporter gene expression. We infer that other branches of the pseudohyphal network (and/or downstream events required for invasion) are more sensitive than the transcriptional output to inhibitory effects of Fus3.
FIG. 9.
Invasion assays comparing wild-type and constitutive Ste7 variants in FUS3 and fus3Δ genetic backgrounds. (A) Micrographs show invasive colonies from a section of the invasion chambers inoculated with strain MLY218α (FUS3 ste7Δ) or SM002 (fus3Δ ste7Δ) carrying plasmids for expression of wild-type (Wt) Ste7 (pNC766), Ste7-EE-A7 (pNC770), Ste7-EE-E7 (pNC791), or catalytically inactive Ste7-N349 (pNC771) as indicated. (B) Bar graph shows the average and average deviation of the invasive colony mean area from three independent experiments. Measurements were made after 46 to 48 h of growth at 30°C. The number of colonies measured in each experiment depended on the Ste7 variant and FUS3 background. Chambers of the FUS3 strain with functional Ste7 variants contained 29 to 127 invasive colonies; those of the corresponding fus3Δ strain contained 53 to 230 invasive colonies. Chambers of the negative reference (Ste7-N349) strains contained 7 to 27 invasive colonies.
Overexpression of Ste7-EE-A7 in strains deficient in Fus3 and Ste5 promotes high levels of mating-specific gene expression.
Although Fus3 and Kss1 have nonoverlapping roles in the mating response, they are largely equivalent with respect to activating the pheromone-induced mating transcription program (7, 57). Therefore, constitutive Ste7, which potently activates Kss1, is expected to promote mating specific gene expression. The failure to do so suggests the existence of inhibitory mechanisms that selectively block activation of mating-specific genes under conditions of persistent Kss1 activation. There have been various studies revealing different circumstances where Fus3 and Kss1 have inhibitory effects on gene expression (5, 10, 23, 32, 51, 57). These reports prompted us to test whether deletions of MAPK activation module components might rescue the ability of constitutive Ste7 to promote mating-specific gene expression.
To make this test, wild-type and constitutive Ste7 variants were expressed from the CYC1 promoter in ste7Δ strains that also had deletions of other MAPK activation module components. β-Galactosidase activity was measured to assess pheromone-independent activation of a mating pathway reporter gene (FUS1-LacZ). This reporter uses the previously described upstream activating sequence from the FUS1 promoter that encompasses three Ste12 binding sites to drive LacZ expression (Fig. 1) (18). Pheromone-treated ste7Δ FUS3 KSS1, ste7Δ fus3Δ KSS1, and ste7Δ FUS3 kss1Δ cultures expressing wild-type Ste7 were included in the comparison to establish reference values for induced reporter gene activation in the three different MAPK genetic backgrounds.
Pheromone treatment of a wild-type strain caused a 3,000-fold increase in reporter gene expression compared with the reference strain lacking Ste7 (Table 3, strain C699-32, pNC318 Ste7 induced, compared with strain C699-32, pNC160). Only the constitutive Ste7 variant that cannot be feedback phosphorylated (Ste7-EE-A7) promoted FUS1-lacZ expression above the background amount (Table 3, strain C699-32, pNC597, compared with strain C699-32, pNC160). Deletion of certain components of the MAPK activation cascade further enhanced this activity. Eliminating the MAPK-Fus3 or the MAPKKK-Ste11 allowed significant enhancement of Ste7-EE-A7-promoted reporter gene expression above that in the otherwise wild-type background (Table 3, pNC597 data for strains C699-81 and C699-80 compared with data for strain C699-32). These findings showed that Ste11 and Fus3 are negative regulators of constitutive mating pathway transcription. Ste7-EE-A7 failed to promote reporter gene expression above background amounts in the corresponding strains expressing Fus3 but lacking Kss1 (Table 3, pNC597, strains C699-114, C699-116, and C699-115). This outcome corroborates the finding that Ste7-EE-A7 fails to phosphorylate and activate Fus3 in vivo (Fig. 7). This outcome further shows that Kss1 is the sole mediator of any transcriptional response promoted by Ste7-EE-A7.
The most striking finding was that Ste5 and Fus3 synergistically mediate inhibition of mating-specific gene expression. Eliminating both Ste5 and Fus3 allowed Ste7-EE-A7 to promote reporter gene expression that was ∼5-fold higher than that seen for pheromone-induced Ste7 in the fus3Δ reference strain (Table 3, pNC597, strain C699-87 versus pNC318 induced, strain C699-80). This synergism was not a simple consequence of deleting two components with negative effects, because eliminating Ste11 and Fus3 or Ste5 and Ste11 had greater no greater effect than eliminating Ste11 alone (Table 3, pNC597, strains C699-81, C699-127, and C699-108). These findings suggest that the scaffold-Ste5 and the MAPK-Fus3 collaborate in a mechanism that restricts mating pathway output under conditions of persistent Kss1 activation. This collaborative mechanism is notable because these are the two components that distinguish the mating pathway from the invasive growth pathway.
We observed that constitutive Ste7 variants with and without charge at the feedback sites promote comparable levels of phospho-Kss1 (Fig. 7). Thus, it is perplexing that the constitutive variants with negative charge at the feedback sites (Ste7-EE or Ste7-EE-E7) remained defective for promoting FUS1-lacZ expression even in the deletion strains that expose activity promoted by Ste7-EE-A7 (Table 3, pNC597, pNC589, and pNC893 versus pNC160). This outcome points to an inhibitory mechanism imposed by Ste7-EE-E7 that is independent of Fus3 and Ste5. Thus, two distinct mechanisms act to selectively block transcription from mating-specific genes. Whether the Ste5- and Fus3-mediated mechanism or Fus3- and Ste5-independent mechanism is predominant depends on whether constitutive Ste7 is without or with charge at the feedback sites. The potential relevance for the existence of two different inhibitory mechanisms is discussed below.
DISCUSSION
Ste7 is the MAPKK in pathways mediating two distinct developmental programs in haploid S. cerevisiae: mating differentiation and invasive (or filamentous) growth. By using substitution variants of Ste7 with constitutive activity we learned that pathway discrimination occurs independently of distinguishing external stimuli. Constitutive Ste7 completely fails to support mating but promotes an invasive response. This biological outcome is consistent with the finding that constitutive Ste7 promotes phosphorylation of the filamentous growth pathway MAPK-Kss1, but not the mating pathway-specific MAPK-Fus3 in vivo. Based on patterns of reporter gene expression, it appears that persistent activation of Kss1 stimulated by constitutive Ste7 selectively specifies a transcriptional program for invasive growth.
Constitutive Ste7 selectively activates Kss1.
We found a direct correlation between phospho-MAPK levels in vivo and the ability of different constitutive Ste7 variants to bind to Fus3, Kss1, and Ste5 (Table 4). The correlations suggested that activation of Fus3 in vivo requires binding to both Ste7 and the scaffold-Ste5 but that activation of Kss1 is independent of Ste5. Constitutive Ste7 with or without negative charge at the feedback sites (Ste7-EE-E7 and Ste7-EE-A7) promoted accumulation of phospho-Kss1 in vivo independently of Ste5. Also, both Ste7-EE-E7 and Ste7-EE-A7 bound well to Kss1. Neither Ste7-EE-A7 nor Ste7-EE-E7 promoted accumulation of active phospho-Fus3 in vivo. Ste7-EE-A7 bound to Ste5 but did not bind to Fus3. Ste7-EE-E7 bound to Fus3 but interacted poorly with Ste5. Taking mass action into account, we expect the equilibrium distribution of Ste7-EE-E7 expressed from either the CYC1 or endogenous promoter would favor a pool that is localized away from the Ste5 complex. Thus, both forms of constitutive Ste7 miss one or the other of the two binding interactions that we postulate is crucial for efficient Fus3 phosphorylation and activation.
TABLE 4.
Summary of activities promoted by constitutive Ste7 variants
| Activity | Ste7-EE-A7 | Ste7-EE-E7 |
|---|---|---|
| Phospho-Kss1 accumulation | + | + |
| Phopsho-Fus3 accumulation | − | − |
| Ste5 binding | + | − |
| Kss1 binding | + | + |
| Fus3 binding | − | + |
| Ty-LacZ reporter expression (filamentation specific) | + | + |
| FUS1-LacZ reporter expression (mating specific), STE5 FUS3 background | − | − |
| FUS1-LacZ reporter expression (mating specific), ste5Δ fus3Δ background | + | − |
Observations in the literature support this proposed difference in the way Ste7 activates Kss1 compared with Fus3. Using constitutively active Ste11, Andersson et al. (1) showed that Ste5 is essential for Fus3 activation but dispensable for Kss1 activation. Bardwell et al. (3) showed that direct binding of Ste7 to Fus3 cooperates with the scaffold protein to enhance signal transmission. In addition to these binding interactions, a specific stereochemical orientation of Fus3 and Ste7 on the scaffold appears to be important for optimal activation of Fus3 and mating responses. Park et al. (38) showed that Fus3 activation is 5- to 10-fold lower than normal and mating efficiencies are 2 to 3 logs lower than normal when Fus3 or Ste7 are tethered at an artificial site through a PDZ domain binding at the C terminus of Ste5.
The differential amounts of active Kss1 and Fus3 reflect not only differences in their phosphorylation by constitutive Ste7 but also potential differences in dephosphorylation by phosphatases. Msg5 is a dual-specificity MAPK phosphatase that regulates Fus3 and Kss1 through dephosphorylation of the phospho-tyrosine and phospho-threonine residues at their activation loop (11, 13). Andersson et al. (1) recently reported that Msg5 selectively inhibits Fus3 under basal conditions where signal intensity is low. Therefore, it is likely that Msg5 contributes to maintaining low basal levels of phopsho-Fus3 under conditions of inefficient activation by the constitutive Ste7 variants.
Implications for Ste7 function on the Ste5-scaffold during pheromone-induced signaling.
Constitutive Ste7 variants with or without negative charge at the feedback sites (Ste7-EE-E7 and Ste7-EE-A7) are defective for activating Fus3 and sustaining mating responses. By contrast, their counterparts that have serine and threonine residues at the activation loop (Ste7-E7 and Ste7-A7) function indistinguishably from wild-type Ste7 and, therefore, must efficiently activate Fus3 in vivo. We conclude from this contrasting behavior that the glutamate substitutions at the activation loop and not the phosphorylation status of Ste7 feedback sites are responsible for the failure of the constitutive variants to activate Fus3.
A higher specific activity of phospho-Ste7 compared with the glutamate-mimic could explain different efficiencies of Fus3 activation in vivo. Yet, differences in specific activity of the inducible and glutamate derivatives have little effect on levels of Kss1 activation in vivo. Therefore, we believe a more important consideration is whether a Ste7 variant can function properly on the scaffold to phosphorylate Fus3. In contrast to the active Ste7 mimics, Ste7-AA-A7 (which mimics naïve Ste7) and Ste7-AA-E7 (which mimics feedback-phosphorylated but inactive Ste7) efficiently bind to both Fus3 and Ste5. Thus, the requirements for function on the scaffold could be satisfied for Ste7-A7 and Ste7-E7 in their naïve state or after dephosphorylation at the activation loop. The failure of glutamate substitution counterparts to activate Fus3 could be viewed as a consequence of their being impervious to phosphatases and thus unable to achieve the correct stereochemical orientation for activation of Fus3 on the scaffold.
This interpretation for function of phospho-Ste7 compared with the glutamate-mimic suggests the following model for activation of Fus3 during normal pheromone induction (Fig. 10). We propose that pheromone stimulates the recruitment of inactive Ste7 and other core enzymes of the mating pathway cascade to the scaffold complex for activation by upstream components (Fig. 10A). This recruitment allows Ste7 to make the contacts with Ste5 and Fus3 that are necessary for the correct stereochemical orientation of the scaffold assembly (Fig. 10B). This configuration is compatible with Ste11 activation of Ste7 and subsequent efficient activation of Fus3. Upon MAPK activation, feedback phosphorylation of active Ste7 promotes its dissociation from the scaffold (Fig. 10C). An implication of this model is that continuous recruitment of naïve Ste7 to the scaffold or action of a Ste7 phosphatase would be critical for sustaining the ability of Ste7 to continue activating Fus3.
FIG. 10.
Model for regulation of the mating pathway MAPK cascade. See text for explanation.
The precedent for positive regulation of MAPK activation by a phosphatase comes from genetic studies of photoreceptor development in Drosophila melanogaster and vulval development in Caenorhabditis elegans (47, 56). The first indication that PP2A is a positive regulator in the C. elegans pathway came from the discovery that mutation of a PP2A B-targeting subunit (sur-6) reduced vulval induction (47). We similarly observed that pheromone-induced MAPK activation was impeded by mutation of the Tap42 targeting subunit for PP2A (A. Scott and B. Errede, unpublished data). Except for the subset of targets involved in stress regulation, Tap42 is a positive regulator of PP2A-mediated signaling (15, 53). These observations suggest that a PP2A family enzyme may be a positive regulator of the mating pathway MAPK cascade in yeast and provide some support for the proposal that dephosphorylation is one route to sustaining Fus3 activation in the scaffold organized complex.
Constitutive Ste7 promotes invasion and a filamentation-specific transcriptional output.
Genome-wide expression profiles of pheromone-induced strains deficient in either Fus3 or Kss1 provide evidence that either MAPK is sufficient to promote the mating transcriptional program through activation of Ste12 (7, 41). However, the two MAPKs have different effects on Ste12-mediated induction of the filamentation transcription program (57). The kinase activity of Fus3, but not Kss1, prevents Ste12 from binding to filamentation-specific genes in pheromone-stimulated cells (32, 44, 57). The inhibitory effects of Fus3 at these promoters are dependent on Tec1 (32, 44, 57). Our analyses suggest the existence of at least two reciprocal mechanisms that prevent Ste12 from acting at mating-specific genes under conditions that otherwise promote an invasive growth response.
The finding that overproduction of the constitutive Ste7 without feedback phosphorylation (Ste7-EE-A7) restored expression of a mating pathway reporter gene opened avenues for us to identify components that suppressed Kss1-mediated activation at these promoters. The comparison revealed that Ste7-EE-A7 in a ste5Δ fus3Δ double mutant background promoted levels of FUS1-lacZ that were even greater than what is typically seen for a pheromone-induced wild-type reference strain (Tables 3 and 4). While essential for mating responses, Ste5 and Fus3 are not required for the invasive response (42). Thus, it appears these mating pathway-designated components can limit a mating response under conditions where Kss1 accumulates in the active form but Fus3 activity remains at basal levels. The pool of inactive Fus3 could interfere with active Kss1 by binding to common downstream targets, such as the Ste12-Rst1/2 (Dig1/2) transcription complex (5, 10, 17, 32, 51). Alternatively, the basal activity of Fus3 could be sufficient to modify components of the mating-specific promoter complex in a way that prevents activation at these promoters. Although there is no evidence for the scaffold-Ste5 participating in either process, such a role would explain the synergistic effect of eliminating both Fus3 and Ste5.
Ste7-EE-A7 is essentially equivalent to Ste7-EE-E7 with respect to promoting Kss1 activation and filamentation-specific gene expression, but the mechanism responsible for suppression of mating-specific gene expression is different for the two constitutive Ste7 variants. In contrast to the outcome for Ste7-EE-A7, there was no activation of FUS1-LacZ expression promoted by Ste7-EE-E7 in the ste5Δ fus3Δ background (Tables 3 and 4). This result rules out a Fus3- and Ste5-mediated mechanism as the sole means for transcriptional suppression at mating-specific promoters in strains expressing Ste7-EE-E7. We speculate that a separate Ste7-EE-E7-dependent effect predominates in this circumstance and that it may involve a binding and/or substrate specificity that pertains only to active Ste7 with charge at the feedback sites. For example, because of this postulated specificity, Ste7-EE-E7 might modify the localization or activity of yet-to-be identified targets that mediate the inhibition at mating-specific genes.
The Ste5- and Fus3-dependent and -independent mechanisms for suppressing transcription at mating-specific promoters may be important under different physiological conditions where one or the other feedback form of Ste7 is predominant. For example, Ste5 and Fus3 may contribute to maintaining a low transcriptional output from the mating pathway under basal conditions where feedback-phosphorylated Ste7 does not accumulate. By contrast, suppression mediated by the Ste5- and Fus3-independent mechanism may be more important for inhibiting mating output when the invasive pathway is stimulated and there is likely to be a persistent and high level of active and feedback-phosphorylated Ste7.
Supplementary Material
Acknowledgments
We thank Richard A. Firtel for valuable discussions and comments on the manuscript.
This work was supported by NIH grants GM39852 (B.E.) and GM 5059167 (H.G.D.) and by American Heart Association fellowship 02253390U (Y.W.).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Andersson, J., D. M. Simpson, M. Qi, Y. Wang, and E. A. Elion. 2004. Differential input by Ste5 scaffold and Msg5 phosphatase route a MAPK cascade to multiple outcomes. EMBO J. 23:2564-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldari, C., J. A. Murray, P. Ghiara, G. Cesareni, and C. L. Galeotti. 1987. A novel leader peptide which allows efficient secretion of a fragment of human interleukin 1 beta in Saccharomyces cerevisiae. EMBO J. 6:229-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell, A. J., L. J. Flatauer, K. Matsukuma, J. Thorner, and L. Bardwell. 2001. A conserved docking site in MEKs mediates high-affinity binding to MAP kinases and cooperates with a scaffold protein to enhance signal transmission. J. Biol. Chem. 276:10374-10386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bardwell, L., J. G. Cook, E. C. Chang, B. R. Cairns, and J. Thorner. 1996. Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol. Cell. Biol. 16:3637-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bardwell, L., J. G. Cook, V. Depak, D. M. Baggott, A. R. Martinez, and J. Thorner. 1998. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev. 12:2887-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baur, M., R. K. Esch, and B. Errede. 1997. Cooperative binding interactions required for function of the Ty1 sterile responsive element. Mol. Cell. Biol. 17:4330-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breitkreutz, A., L. Boucher, and M. Tyers. 2001. MAPK specificity in the yeast pheromone response independent of transcriptional activation. Curr. Biol. 11:1266-1271. [DOI] [PubMed] [Google Scholar]
- 8.Choi, K. Y., B. Satterberg, D. M. Lyons, and E. A. Elion. 1994. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell 78:499-512. [DOI] [PubMed] [Google Scholar]
- 9.Cook, J. G., L. Bardwell, S. J. Kron, and J. Thorner. 1996. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 10:2831-2848. [DOI] [PubMed] [Google Scholar]
- 10.Cook, J. G., L. Bardwell, and J. Thorner. 1997. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature 390:85-88. [DOI] [PubMed] [Google Scholar]
- 11.Davenport, K. D., K. E. Williams, B. D. Ullmann, and M. C. Gustin. 1999. Activation of the Saccharomyces cerevisiae filamentation/invasion pathway by osmotic stress in high-osmolarity glycogen pathway mutants. Genetics 153:1091-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dohlman, H. G., and J. W. Thorner. 2001. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70:703-754. [DOI] [PubMed] [Google Scholar]
- 13.Doi, K., A. Gartner, G. Ammerer, B. Errede, H. Shinkawa, K. Sugimoto, and K. Matsumoto. 1994. MSG5, a novel protein phosphatase promotes adaptation to pheromone response in S. cerevisiae. EMBO J. 13:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolan, J. W., C. Kirkman, and S. Fields. 1989. The yeast STE12 protein binds to the DNA sequence mediating pheromone induction. Proc. Natl. Acad. Sci. USA 86:5703-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duvel, K., A. Santhanam, S. Garrett, L. Schneper, and J. R. Broach. 2003. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol. Cell 11:1467-1478. [DOI] [PubMed] [Google Scholar]
- 16.Elion, E. A., P. L. Grisafi, and G. R. Fink. 1990. FUS3 encodes a cdc2+/CDC28-related kinase required for the transition from mitosis into conjugation. Cell 60:649-664. [DOI] [PubMed] [Google Scholar]
- 17.Elion, E. A., B. Satterberg, and J. E. Kranz. 1993. FUS3 phosphorylates multiple components of the mating signal transduction cascade: evidence for STE12 and FAR1. Mol. Biol. Cell 4:495-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Errede, B., and G. Ammerer. 1989. STE12, a protein involved in cell-type-specific transcription and signal transduction in yeast, is part of protein-DNA complexes. Genes Dev. 3:1349-1361. [DOI] [PubMed] [Google Scholar]
- 19.Errede, B., A. Gartner, Z. Zhou, K. Nasmyth, and G. Ammerer. 1993. MAP kinase-related FUS3 from S. cerevisiae is activated by STE7 in vitro. Nature 362:261-264. [DOI] [PubMed] [Google Scholar]
- 20.Errede, B., and Q. Y. Ge. 1996. Feedback regulation of MAP kinase signal pathways. Philos. Trans. R. Soc. London B 351:143-148. [DOI] [PubMed] [Google Scholar]
- 21.Esch, R. K., and B. Errede. 2002. Pheromone induction promotes Ste11 degradation through a MAPK feedback and ubiquitin-dependent mechanism. Proc. Natl. Acad. Sci. USA 99:9160-9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evan, G. I., G. K. Lewis, G. Ramsay, and J. M. Bishop. 1985. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 5:3610-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farley, F. W., B. Satterberg, E. J. Goldsmith, and E. A. Elion. 1999. Relative dependence of different outputs of the Saccharomyces cerevisiae pheromone response pathway on the MAP kinase Fus3p. Genetics 151:1425-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gartner, A., K. Nasmyth, and G. Ammerer. 1992. Signal transduction in Saccharomyces cerevisiae requires tyrosine and threonine phosphorylation of FUS3 and KSS1. Genes Dev. 6:1280-1292. [DOI] [PubMed] [Google Scholar]
- 25.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS- DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 26.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 27.Gimeno, C. J., P. O. Ljungdahl, C. A. Styles, and G. R. Fink. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077-1090. [DOI] [PubMed] [Google Scholar]
- 28.Guarente, L., B. Lalonde, P. Gifford, and E. Alani. 1984. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell 36:503-511. [DOI] [PubMed] [Google Scholar]
- 29.Guarente, L., and M. Ptashne. 1981. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 78:2199-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laloux, I., E. Jacobs, and E. Dubois. 1994. Involvement of SRE element of Ty1 transposon in TEC1-dependent transcriptional activation. Nucleic Acids Res. 22:999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madhani, H. D., and G. R. Fink. 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275:1314-1317. [DOI] [PubMed] [Google Scholar]
- 32.Madhani, H. D., C. A. Styles, and G. R. Fink. 1997. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91:673-684. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell, D. A., T. K. Marshall, and R. J. Deschenes. 1993. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast 9:715-722. [DOI] [PubMed] [Google Scholar]
- 34.Mosch, H. U., R. L. Roberts, and G. R. Fink. 1996. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5352-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neiman, A. M., and I. Herskowitz. 1994. Reconstitution of a yeast protein kinase cascade in vitro: activation of the yeast MEK homologue STE7 by STE11. Proc. Natl. Acad. Sci. USA 91:3398-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Rourke, S. M., and I. Herskowitz. 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12:2874-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palecek, S. P., A. S. Parikh, and S. J. Kron. 2002. Sensing, signalling and integrating physical processes during Saccharomyces cerevisiae invasive and filamentous growth. Microbiology 148:893-907. [DOI] [PubMed] [Google Scholar]
- 38.Park, S. H., A. Zarrinpar, and W. A. Lim. 2003. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science 299:1061-1064. [DOI] [PubMed] [Google Scholar]
- 39.Rhodes, N., M. Company, and B. Errede. 1990. A yeast-Escherichia coli shuttle vector containing the M13 origin of replication. Plasmid 23:159-162. [DOI] [PubMed] [Google Scholar]
- 40.Rhodes, N., L. Connell, and B. Errede. 1990. STE11 is a protein kinase required for cell-type-specific transcription and signal transduction in yeast. Genes Dev. 4:1862-1874. [DOI] [PubMed] [Google Scholar]
- 41.Roberts, C. J., B. Nelson, M. J. Marton, R. Stoughton, M. R. Meyer, H. A. Bennett, Y. D. He, H. Dai, W. L. Walker, T. R. Hughes, M. Tyers, C. Boone, and S. H. Friend. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287:873-880. [DOI] [PubMed] [Google Scholar]
- 42.Roberts, R. L., and G. R. Fink. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8:2974-2985. [DOI] [PubMed] [Google Scholar]
- 43.Rothstein, R. J. 1983. One-step gene disruption in yeast. Methods Enzymol. 101:202-211. [DOI] [PubMed] [Google Scholar]
- 44.Sabbagh, W., Jr., L. J. Flatauer, A. J. Bardwell, and L. Bardwell. 2001. Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation. Mol. Cell 8:683-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 46.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Sieburth, D. S., M. Sundaram, R. M. Howard, and M. Han. 1999. A PP2A regulatory subunit positively regulates Ras-mediated signaling durin Caenorhabditis elegans vulval induction. Genes Dev. 13:2562-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 50.Sprague, G. F., Jr. 1991. Assay of yeast mating reaction. Methods Enzymol. 194:77-93. [DOI] [PubMed] [Google Scholar]
- 51.Tedford, K., S. Kim, D. Sa, K. Stevens, and M. Tyers. 1997. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr. Biol. 7:228-238. [DOI] [PubMed] [Google Scholar]
- 52.van Drogen, F., S. M. O'Rourke, V. M. Stucke, M. Jaquenoud, A. M. Neiman, and M. Peter. 2000. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 10:630-639. [DOI] [PubMed] [Google Scholar]
- 53.Wang, H., X. Wang, and Y. Jiang. 2003. Interaction with Tap42 is required for the essential function of Sit4 and type 2A phosphatases. Mol. Biol. Cell 14:4342-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, Y., and H. G. Dohlman. 2002. Pheromone-dependent ubiquitination of the mitogen-activated protein kinase kinase Ste7. J. Biol. Chem. 277:15766-15772. [DOI] [PubMed] [Google Scholar]
- 55.Wang, Y., Q. Ge, D. Houston, J. Thorner, B. Errede, and H. G. Dohlman. 2003. Regulation of Ste7 ubiquitination by Ste11 phosphorylation and the SCF (Skp1/Cullin/F-box) complex. J. Biol. Chem. 278:22284-22289. [DOI] [PubMed] [Google Scholar]
- 56.Wassarman, D. A., N. M. Solomon, H. C. Chang, F. D. Karim, M. Therrien, and G. M. Rubin. 1996. Protein phosphatase 2A positively and negatively regulates Ras1-mediated photoreceptor development in Drosophila. Genes Dev. 10:272-278. [DOI] [PubMed] [Google Scholar]
- 57.Zeitlinger, J., I. Simon, C. T. Harbison, N. M. Hannett, T. L. Volkert, G. R. Fink, and R. A. Young. 2003. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113:395-404. [DOI] [PubMed] [Google Scholar]
- 58.Zhan, X. L., R. J. Deschenes, and K. L. Guan. 1997. Differential regulation of FUS3 MAP kinase by tyrosine-specific phosphatases PTP2/PTP3 and dual-specificity phosphatase MSG5 in Saccharomyces cerevisiae. Genes Dev. 11:1690-1702. [DOI] [PubMed] [Google Scholar]
- 59.Zheng, C. F., and K. L. Guan. 1994. Activation of MEK family kinases requires phosphorylation of two conserved Ser/Thr residues. EMBO J. 13:1123-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou, Z. 1993. Mating pheromone induced signal transduction in Saccharomyces cerevisiae: characterization of the Ste7 kinase. Ph.D. dissertation. University of North Carolina at Chapel Hill, Chapel Hill.
- 61.Zhou, Z., A. Gartner, R. Cade, G. Ammerer, and B. Errede. 1993. Pheromone-induced signal transduction in Saccharomyces cerevisiae requires the sequential function of three protein kinases. Mol. Cell. Biol. 13:2069-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.