SUMMARY
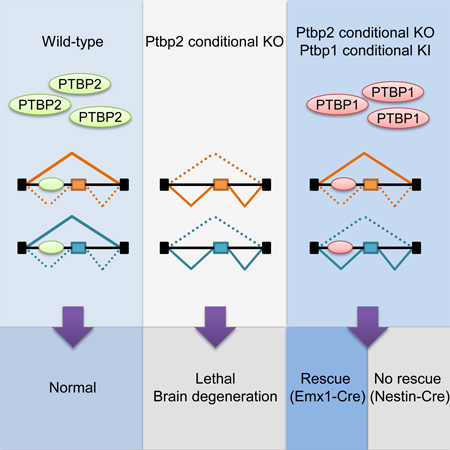
Families of alternative splicing regulators often contain multiple paralogs presumed to fulfill different functions. Polypyrimidine tract binding proteins PTBP1 and PTBP2 reprogram developmental pre-mRNA splicing in neurons, but how their regulatory networks differ is not understood. To compare their targeting, we generated a knock-in allele that conditionally expresses PTBP1. Bred to a Ptbp2 knockout, the transgene allowed us to compare the developmental and molecular phenotypes of mice expressing only PTBP1, only PTBP2, or neither protein in the brain. This knock-in Ptbp1 rescued a forebrain-specific but not a pan-neuronal Ptbp2 knockout, demonstrating both redundant and distinct roles for the proteins. Many developmentally-regulated exons exhibited different sensitivities to PTBP1 and PTBP2. Nevertheless, the two paralogs displayed similar RNA binding across the transcriptome indicating that their differential targeting does not derive from their RNA interactions but from possible different cofactor interactions.
Keywords: PTBP1, PTBP2, neuronal differentiation, brain development, RNA binding proteins, RNA-Seq, CLIP, alternative splicing, splicing reprogramming
Graphical Abstract
eTOC Blurb
Using engineered mice, Vuong et al. reveal partial phenotypic compensation and overlapping exon targeting by PTBP1 and PTBP2, two essential RNA binding proteins regulating neuronal splicing. Unexpectedly, many developmentally-controlled exons exhibit different sensitivity yet similar binding to the two proteins, shedding light on the mechanisms determining their target specificity.
INTRODUCTION
Brain development is accompanied by large-scale changes in spliced isoform expression (Pan et al., 2008; Wang et al., 2008). These neural specific splicing events are regulated by families of special pre-mRNA binding proteins including PTBP, RBFOX, NOVA, MBNL, SRRM4, KHDRBS and TDP43 (Raj and Blencowe, 2015; Vuong et al., 2016). Most of these regulator families include multiple paralogs whose functional redundancy and specificity of targeting are poorly understood. Differences in expression patterns and different phenotypes from mutations indicate that family members serve different roles. However, splicing regulators of the same family often show similar RNA binding and similar activity in regulating individual exons (Amir-Ahmady et al., 2005; Damianov et al., 2016; Lambert et al., 2014). It is difficult to define targets that are differentially affected by the paralogs and to understand how their differential activity is achieved. Assessing the specificity of targeting is also complicated by the crossregulation of splicing factors, with depletion of one protein inducing the expression of another family member. For example, depletion of the polypyrimidine tract binding protein PTBP1 (PTB or hnRNP I) induces PTBP2 (brPTB or nPTB) expression in most cells (Boutz et al., 2007; Makeyev et al., 2007; Spellman et al., 2007). Some targets require depletion of both proteins to alter splicing (Han et al., 2014; Linares et al., 2015). Other exons that change upon the depletion of PTBP1 alone could be responding to either PTBP1 loss or PTBP2 gain.
PTBP1 is widely expressed outside the nervous system and is a repressor of the neural-specific splicing program (Keppetipola et al., 2012; Makeyev et al., 2007). At the onset of neurogenesis, PTBP1 reduction allows PTBP2 expression and causes a large-scale shift in alternative splicing patterns (Linares et al., 2015). Ptbp1 depletion with the associated upregulation of PTBP2 can promote transdifferentiation of fibroblasts into neurons (Xue et al., 2013). These data demonstrate different activities of the two proteins and particularly that PTBP2 does not fulfill all the functions of PTBP1.
Although playing different roles in neuronal differentiation, PTBP1 and PTBP2 have many common targets. One example is exon 18 in the Psd-95 mRNA, encoding a key component of the postsynaptic density at glutamatergic synapses, that is repressed by both PTB proteins (Zheng et al., 2012). Exon 18 skipping causes a frameshift with premature translation termination leading to nonsense-mediated mRNA decay (NMD) of Psd-95 transcripts in early neurons and non-neuronal cells (Zheng, 2016). After its early expression in neurons, PTBP2 is reduced before synaptogenesis to allow exon 18 splicing and PSD-95 production. Many exons sensitive to both proteins exhibit a similar developmental profile to Psd-95 exon 18, maintaining their repression until late in neuronal maturation when PTBP2 levels drop (Licatalosi et al., 2012; Li et al., 2014). However, other exons appear to be more sensitive to PTBP1 than PTBP2 and exhibit a variety of regulatory patterns (Han et al., 2014; Linares et al., 2015; Zhang et al., 2016).
To define the PTBP1 and PTBP2 target networks and to assess their redundancy and specificity, we created mice that express each protein alone in brains of the same developmental age. Comparing the neurons of these mice to Ptbp2−/− neurons that do not express either PTBP1 or PTBP2 allows the direct comparison of PTBP1 and PTBP2 targets in the same physiological context.
RESULTS
Generation of Ptbp1 conditional knock-in mice
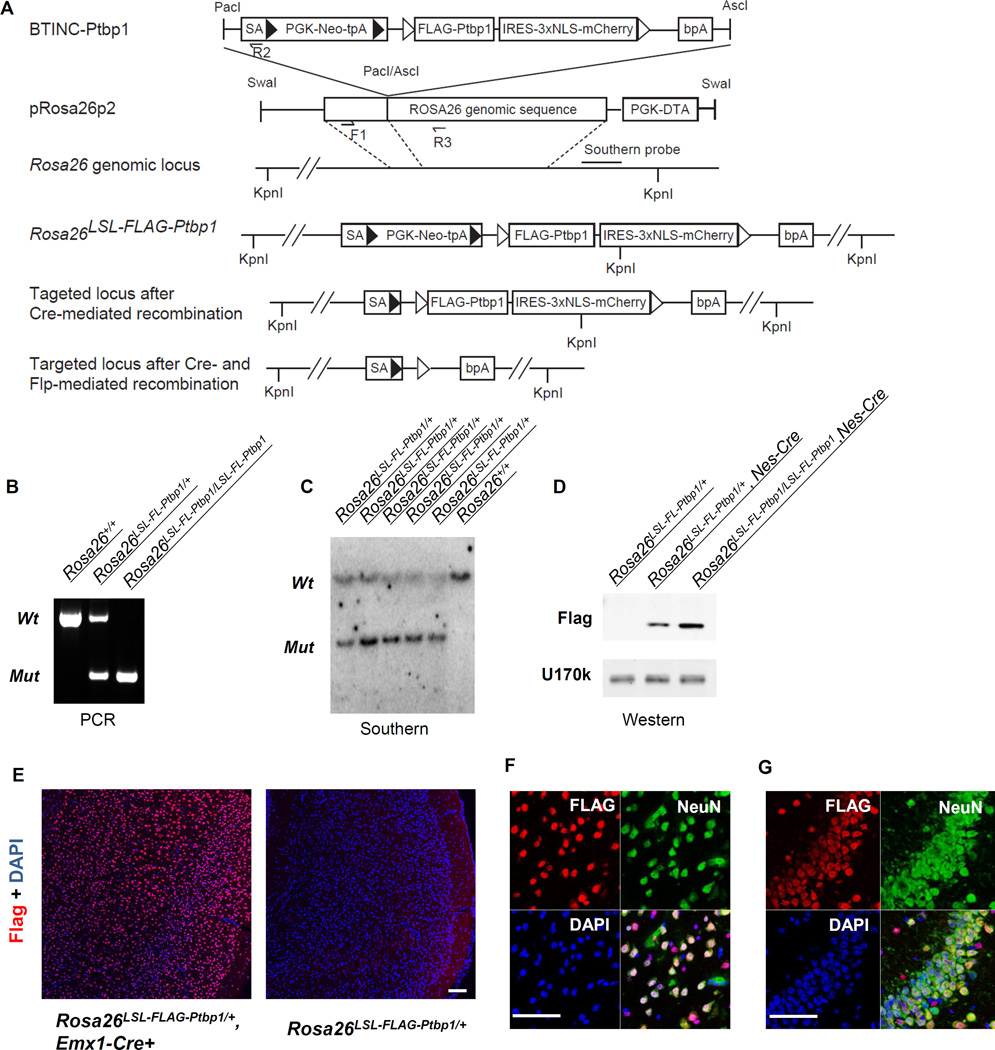
We created a targeted gain-of-function allele for Ptbp1 at the ubiquitously-expressed but dispensable Rosa26 locus (Zambrowicz et al., 1997). An expression cassette containing a FLAG-tagged mouse Ptbp1 (FL-Ptbp1) coding sequence was inserted via homologous recombination so that Rosa26 transcripts would splice from the first Rosa26 exon to a new splice acceptor site (Rosa26LSL-FL-Ptbp1, Figure 1A). Between this splice acceptor site and the FL-Ptbp1 cDNA, a loxp-STOP-loxp (LSL) cassette was inserted to prevent FLAG-PTBP1 from expression until the LSL cassette is removed by Cre recombinase. IRES-mCherry was added immediately after FL-Ptbp1 to fluorescently label FLAG-PTBP1 expressing cells. Finally, FRT sites flanking the FL-Ptbp1-IRES-mCherry cassette allow FLP-mediated excision and conditional silencing of transgene expression.
Figure 1. A Ptpb1 conditional knock-in mouse expresses inducible FLAG-PTBP1 from the Rosa26 locus.
(A) Schematic of the targeting strategy. SA: splicing acceptor. PGK-Neo-tpA: PGK promoter-driven neomycin with PGK polyA site. NLS: nuclear localization signal. bPA: bovine growth hormone polyA site. F1, R2 and R3: genotyping primers. ► Loxp sites. 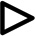 Frt sites. (B) Genotyping shows distinct amplicons for the wildtype, Ptbp1-KI heterozygous and homozygous animals. (C) Southern blot of Kpn1-treated genomic DNA from the wildtype and Ptbp1-KI heterozygous animals. The wildtype allele is 37 kb and the mutant allele is 7.3kb. (D) Western blot shows the expression of Flag-PTBP1 protein using an anti-Flag antibody. Anti-U1-70k is used as a control. (E) Immunofluorescence staining shows the expression of FLAG-PTBP1 in the cortex of Emx1-Ptbp1 KI mice. Co-staining of FLAG-PTBP1 and neuronal marker NeuN in the neocortex (F) and hippocampus (G) of Emx1-Ptbp1 KI mice. Scale bars: 100 µm.
Frt sites. (B) Genotyping shows distinct amplicons for the wildtype, Ptbp1-KI heterozygous and homozygous animals. (C) Southern blot of Kpn1-treated genomic DNA from the wildtype and Ptbp1-KI heterozygous animals. The wildtype allele is 37 kb and the mutant allele is 7.3kb. (D) Western blot shows the expression of Flag-PTBP1 protein using an anti-Flag antibody. Anti-U1-70k is used as a control. (E) Immunofluorescence staining shows the expression of FLAG-PTBP1 in the cortex of Emx1-Ptbp1 KI mice. Co-staining of FLAG-PTBP1 and neuronal marker NeuN in the neocortex (F) and hippocampus (G) of Emx1-Ptbp1 KI mice. Scale bars: 100 µm.
Correct targeting in ES cells and germ line transmission were confirmed by genotyping PCR and Southern blot of genomic DNA (Figure 1B–C). Conditional FLAG-PTBP1 protein expression was confirmed via crossing Rosa26LSL-FL-Ptbp1/LSL-FL-Ptbp1 mice to a Nestin-Cre transgenic line (Figure 1D). In the conditional homozygous neocortices, FLAG-PTBP1 was expressed at about the same level as the endogenous PTBP1 at E14.5 (Figure S1A), and mice homozygous for the conditional knock-in (cKI) allele expressed higher levels of protein than heterozygous mice. The dependency of FLAG-PTBP1 expression on Cre recombinase was further confirmed in an Emx1-Cre transgenic line expressing Cre in excitatory neurons of the forebrain particularly throughout the neocortex and hippocampus (Figure 1E). Normally, PTBP1 is only expressed in the progenitor cells at the ventricular and subventricular zone of the developing cortex, but not in the outer layers of mature neurons (Figure S1C). Widespread induction of FLAG-PTBP1 was observed in the cortices of Rosa26LSL-FL-Ptbp1/+; Emx1-Cre animals. In the neocortex, over 92% of FLAG-PTBP1 positive cells were immunoreactive for the neuron specific marker NeuN and over 97% of NeuN positive neurons expressed FLAG-PTBP1 (Figure 1F). The overlap was even higher in the hippocampus (Figure 1G). These data confirm the induction of PTBP1 upon Cre expression in neurons, where it is usually absent.
PTBP1 partially compensates for PTBP2 loss during development
To test whether FLAG-PTBP1 expression could compensate for the loss of PTBP2 in vivo, we bred the cKI allele into the Ptbp2 conditional knockout (KO) mice in either the Nestin-Cre or Emx1-Cre backgrounds in order to generate homozygous Ptbp2 conditional null animals carrying the FLAG-Ptbp1 and a Cre transgene (Rosa26LSL-FL-Ptbp1/+; Ptbp2loxp/loxp; Cre). In these mice, induction of FLAG-PTBP1 expression and depletion of endogenous PTBP2 occurs in the same cells and at the same time, allowing assessment of their phenotypic compensation.
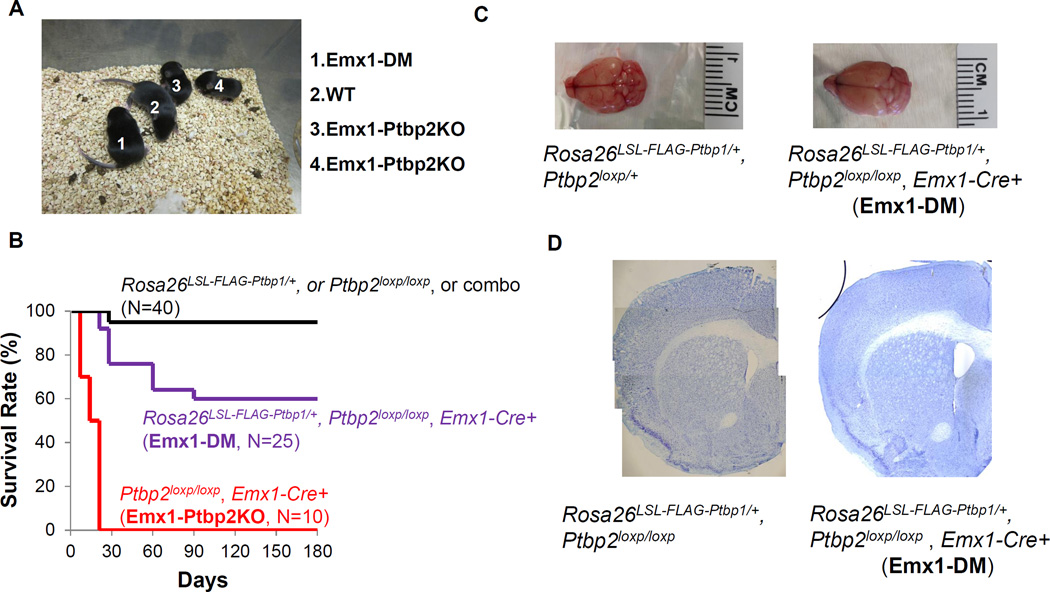
We found that FLAG-PTBP1 expression could rescue some phenotypic features of Ptbp2 deletion. As seen previously, Emx1-Ptbp2KO mice exhibited retarded growth, progressive degeneration of the cerebral cortex and died by the weaning age (Li et al., 2014). In contrast, most Rosa26LSL-FL-Ptbp1/+; Ptbp2loxp/loxp; Emx1-Cre (henceforth called Emx1-DM) mice survived past the first three postnatal weeks and appeared grossly normal (Figure 2A–B). Some Emx1-DM animals died between the third and eighth postnatal week from unknown causes. Those that survived the first three months usually lived a normal life span and were fertile (Figure 2B). Unlike the Emx1-Ptbp2KO brains whose cerebral cortices degenerated as early as postnatal day 5, the Emx1-DM brain, even at 18-months of age, displayed a normal size and structure (Figure 2C–D). Although not 100 percent penetrant, FLAG-PTBP1 was sufficient to rescue the Emx1-Ptbp2KO mutation with regard to neuronal survival, development, body growth and life span.
Figure 2. The FLAG-PTBP1 transgene partially rescues the slow growth and early mortality of Emx1-Ptbp2KO mice.
(A) Postnatal day 10 animals of (1) Rosa26LSL-FLAG-Ptbp1/+ Ptbp2loxp/loxp Emx1-Cre+ (Emx1-DM), (2) Ptbp2loxp/loxp (considered as WT), and (3–4) Ptbp2loxp/loxp Emx1-Cre+ (Emx1-Ptbp2KO). (B) Survival curves of wildtype-like (black), Emx1-DM (purple) and Emx1-Ptbp2KO (red) mice. The overall brain sizes (C) and structures shown by Nissl staining (D) were similar between Rosa26LSL-FLAG-Ptbp1/+ Ptbp2loxp/loxp Emx1-Cre+ (Emx1-DM) mouse and wildtype-like mice at 18 months of age.
We also found that this FLAG-PTBP1 transgene was not sufficient to rescue other conditions of Ptbp2 depletion. Pan-neuronal Ptbp2 knockout mice (Ptbp2loxp/loxp; Nes-Cre) die at birth (Li et al., 2014). Rosa26LSL-FL-Ptbp1/+; Ptbp2loxp/loxp; Nes-Cre mice (henceforth called Nes-DM) also exhibited complete perinatal lethality, with none (out of 57) surviving past the first postnatal day. Higher levels of FLAG-PTBP1 expression did not improve survival, as no double homozygous mutant (Rosa26LSL-FL-Ptbp1/LSL-FL-Ptbp1; Ptbp2loxp/loxp; Nes-Cre) animals (out of 104) survived beyond the first postnatal day. Thus, PTBP1 can compensate for the loss of PTBP2 in some developmental contexts but not others.
PTBP1 and PTBP2 regulate distinct but overlapping sets of alternative exons
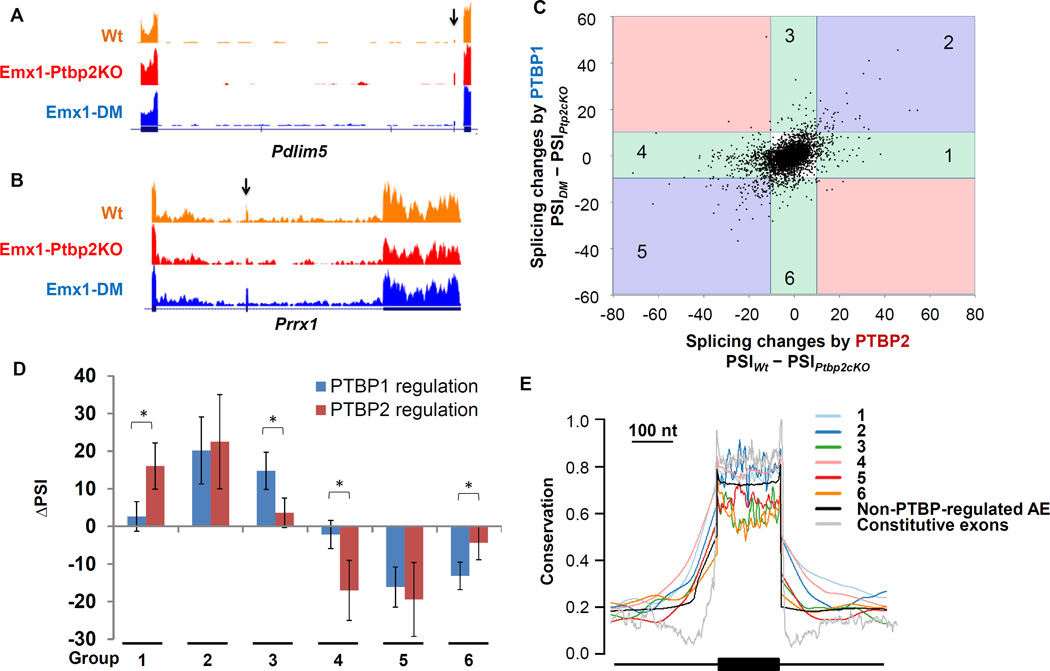
The Ptbp2−/− brains showed extensive changes in alternative splicing (Licatalosi et al., 2012; Li et al., 2014). To assess whether these changes were being reversed by FLAG-PTBP1 expression in the Emx1-DM mice, we performed deep RNA-Seq on cortical RNA from wildtype (Wt), Emx1-Ptbp2KO and Emx1-DM animals (Experimental Procedures). We used an analysis pipeline (GeneSplice) that quantitatively estimates cassette exon inclusion (PSI) for 13,097 Gencode annotated cassette exons (release m6, GRCm38.p4; see Experimental Procedures), and subsequently validated these measurements by RT-PCR (Figure S2 and Table S2). Setting a threshold of |ΔPSI| > 10, we found that 42% of the exons dysregulated in the Emx1-Ptbp2KO neocortices were rescued by FLAG-PTBP1 expression. For example, Pdlim5 exon 8 was included in 12% of the mRNA transcripts in wildtype, 36% in Emx1-Ptbp2KO, and reverted back to 14% inclusion with FLAG-PTBP1 expression (Figure 3A). Similarly, 61% of Prrx1 mRNA transcripts included exon 4 in the wildtype. Exon inclusion decreased to 28% in the Emx1-Ptbp2KO, but was restored to 69% in the Emx1-DM cortex (Figure 3B). Thus, FLAG-PTBP1 compensated for some splicing changes occurring with PTBP2 loss, while other PTBP2-dependent exons exhibited smaller or minimal responses to the FLAG-PTBP1 transgene.
Figure 3. PTBP1 and PTBP2 regulate overlapping but distinct sets of alternative exons.
RNA-Seq expression tracks show examples of exons that are (A) up-regulated or (B) repressed in the Emx1-Ptbp2KO (Ptbp2loxp/loxp; Emx1-Cre+) neocortices and reversed in the Emx1-DM (Rosa26LSL-FLAG-Ptbp1/+; Ptbp2loxp/loxp; Emx1-Cre+) neocortices. Arrows point to the regulated exons. (C) A scatter plot of cassette exons shows splicing changes caused by FLAG-PTBP1 (y axis) and PTBP2 (x axis). Using a threshold of |ΔPSI| larger than 10, six exon groups in the colored sectors were identified by their responses to PTBP1 and PTBP2. (D) The averaged splicing changes induced by PTBP1 or PTBP2 were plotted for each exon group. Error bars represent standard deviation; with * P value < 1×10−13, using a paired Student’s t-test. (E) Conservation of each exon group, as well as non-PTBP-regulated alternative exons (AE) and constitutive exons.
In addition to comparing each mutant to wildtype, we also directly compared the Emx1-DM to the Emx1-Ptbp2cKO. This comparison where PTBP2 is absent from both conditions allows attribution of splicing changes to the FLAG-PTBP1 alone. These splicing changes induced by PTBP1 (PSIDM - PSIPtbp2cKO) were plotted against changes caused by PTBP2 alone (PSIWt - PSIPtbp2cKO; Figure 3C). Cassette exons typically responded to PTBP1 and PTBP2 in the same direction but with different magnitudes. Very few exons gave opposite responses to the two proteins and these responses were small and not significant (pink sectors in Figure 3C). Using a cutoff of |ΔPSI| > 10 and P value < 0.05, we defined exons (Table S1) activated by PTBP2 alone (group 1, N=78), by PTBP1 alone (group 3, N=44) or by both proteins (group 2, N=24), as well as exons repressed by PTBP2 alone (group 4, N=357), PTBP1 alone (group 6, N=47) or both proteins (group 5, N=61). These exon groups defined by their sensitivity to the two proteins in vivo were used for further analyses. Exons in group 1 and 4 showed respectively increased and decreased inclusion in the presence of PTBP2 but were minimally affected by FLAG-PTBP1 (Figure 3D). Similarly, PTBP1-activated exons in group 3 and PTBP1-repressed exons in group 6 reacted weakly to PTBP2.
We examined the evolutionary conservation of exons and their flanking introns from the different groups (Figure 3E). Intronic sequence immediately flanking PTBP-regulated exons was in general more conserved than that adjacent to non-PTBP-regulated alternative exons or constitutive exons consistent with previous studies (Chen and Zheng, 2008). Introns flanking exons sensitive to PTBP2 alone (groups 1 and 4) exhibited the highest conservation and the longest stretches of conserved nucleotides (up to 150 nucleotides flanking the exons).
PTBP1 and PTBP2 exhibit very similar RNA binding profiles
Exons may respond differently to PTBP1 and PTBP2 due to differences in RNA binding by the two proteins, differences in protein-protein interactions or both. To examine RNA binding by PTBP1 and PTBP2 across the transcriptome, we performed iCLIP-Seq for the two paralogs using a previously published protocol (König et al., 2010; Stork and Zheng, 2016). Each sample containing multiple neocortices of the same genotype (WT, KI, or DM) was divided in half for immunoprecipitation with either FLAG or PTBP2 antibodies (Figure S3 and Experimental Procedures). FLAG iCLIP in the WT cortices served as a negative control. After IP, micrococcal nuclease digestion, and isolation of the crosslinked RNA, the bound RNA fragments were sequenced, aligned to the mouse genome and groups of fragments assessed for enrichment over background (König et al., 2010). We identified 351,602 and 265,953 PTBP1 crosslinking sites showing significant enrichment over the background in the KI and DM mice respectively. The PTBP2 iCLIP identified 155,447, and 74,416 crosslinking sites enriched over background in WT and KI respectively. Applying the Bedtools intersect tool to these libraries, we found significant overlap between the sites of FLAG-PTBP1 and PTBP2 binding. For example, 66.9% of the PTBP1 crosslinking sites in DM overlapped with those in KI, while 60% of those in KI overlapped with those in DM. Between the two paralogs, 42.5% of the PTBP1 crosslinking sites in KI mice overlapped with PTBP2 sites in WT mice, and 62.4% of PTBP2 crosslinking sites in the WT mice overlapped with PTBP1 sites in the KI. Since the iCLIP reads are not expected to saturate all the binding sites, some differences in PTBP1 and PTBP2 binding likely result from incomplete coverage. For example, some exons with significant PTBP1 or 2 crosslinking sites showed crosslinked fragments for the other paralog at the same locations, but which did not pass the threshold for statistical significance.
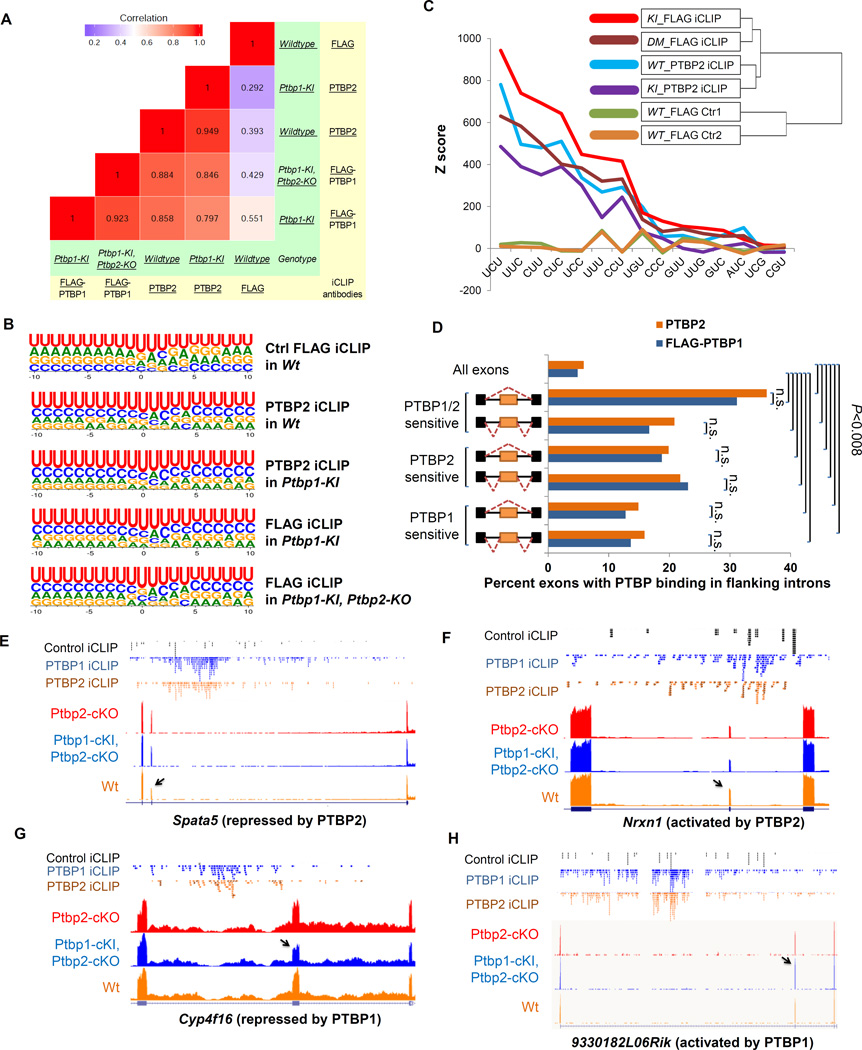
To further compare transcriptome-wide RNA-binding of FLAG-PTBP1 and PTBP2, we collapsed the crosslinked fragments into clusters (Experimental Procedures). We calculated the number of unique and significant iCLIP tags within each cluster and derived the coefficients of determination (R2) for these values for the clusters common to the two iCLIP-Seq libraries (Figure 4A). As expected, the control FLAG iCLIP in WT brain lacked most clusters identified in the PTBP iCLIP libraries (Figure S4), and these clusters correlated poorly with both PTBP2 and PTBP1 clusters. By contrast, PTBP1 and PTBP2 iCLIP clusters were highly correlated with R2 values ranging from 0.797 to 0.949 (Figure 4A). These data indicated extensive overlap in the RNA binding sites of PTBP1 and PTBP2.
Figure 4. PTBP1 and PTBP2 exhibit very similar RNA binding independent of regulation specificity.
(A) A heat map shows the similarity between iCLIP libraries generated with anti- FLAG or PTBP2 antibodies (in yellow) from mouse neocortices of different genotypes (in green). The number of unique and significant iCLIP tags for each cluster was compared between iCLIP-Seq experiments to derive the R2 values. (B) Logos plots of FLAG-PTBP1 or PTBP2 binding sites in each iCLIP experiment. Predicted crosslinked nucleotides are at the zero position. (C) Z scores of the triplets enriched within 30 nucleotides upstream or downstream of the crosslinking sites for each iCLIP experiment. A clustering dendrogram shows the similarity of the iCLIP libraries regarding Z scores of all possible motifs. (D) Bar graph showing within each group the percentage of exons containing FLAG-PTBP1 (blue) and PTBP2 (brown) significant binding sites in the flanking introns. n.s. indicates P value > 0.7. (E–H) Aligned genome browser tracks of RNA-Seq of Emx1-Ptbp2-cKO (red), Emx1-DM (Ptbp1-cKI, Ptbp2-cKO, dark blue) and Wt (orange), control iCLIP-Seq (black), PTBP1 iCLIP-seq (blue) and PTBP2 iCLIP-Seq (brown) for exons exhibiting different sensitivity to PTBP1 and PTBP2 but similar PTBP1 and PTBP2 binding. Arrows point to the regulated exons.
To examine features of the binding sites, we aligned the predicted crosslinked nucleotide and measured the nucleotide frequency at each adjacent position (Figure 4B). As expected, uridine was the most common nucleotide at the crosslink sites and was enriched at the flanking positions. Cytidine was the second most preferred nucleotide at almost all positions for both PTBP1 and PTBP2, but was least frequent in the control FLAG iCLIP. At most positions the third and fourth most frequent nucleotides were also identical for PTBP1 and PTBP2.
We examined nucleotide triplet frequencies in the sequences surrounding the crosslink sites (−30 nt to +30 nt). The top nine enriched triplets for FLAG-PTBP1 or PTBP2 iCLIP included all possible combinations of three pyrimidines plus UGU (Figure 4C). None of the pyrimidine triplets were enriched in the FLAG control iCLIP except UUU, which presumably reflects the intrinsic crosslinking bias for uridines. Only these triplets consistently exhibited significant enrichment in the PTBP iCLIP sites over the control FLAG iCLIP. Unsupervised hierarchical clustering showed that motif frequencies were similar between PTBP1 and PTBP2 crosslinking sites but very different from the control iCLIP sites (Figure 4C).
Differential RNA binding is not a major determinant of specific regulation by PTBP1 or PTBP2
To assess whether regulation specificity could be attributed to differences between PTBP1 and PTBP2 binding, we examined their binding in exon groups exhibiting different sensitivities to the two proteins. For each exon group defined in Figure 3, we calculated the percent of exons with PTBP1 and PTBP2 binding sites in flanking introns (Figure 4D). Approximately 5% of all exons had PTBP1 or PTBP2 binding in the flanking introns. PTBP-regulated exon groups all exhibited significantly higher proportions of PTBP1 or PTBP2 binding than the total exon set (hypergeometric test, P<0.008 for groups 2, 3 and 6, P<10−7 for all other groups). Among all groups, exons sensitive to repression by both proteins exhibited similar PTBP1 and PTBP2 binding and had the highest percentages of exons bound to either PTBP. Most notably, the exons that were preferentially targeted by one PTBP also exhibited binding by PTBP1 and PTBP2 that was not significantly different (Fisher’s exact test, P>0.7).
The similar binding of PTBP1 and PTBP2 to exons that were nevertheless differentially regulated by the two proteins as seen in the global analyses could also be observed on individual exons. For example, PTBP2-repressed Spata5 exon 16 was insensitive to PTBP1, but contained similar iCLIP tags (Figure 4E) and similar iCLIP clusters (Figure S3B) for the two proteins in its downstream introns. Nrxn1 exon 7 was activated by PTBP2 but not by PTBP1. The introns flanking this exon had very similar distributions of iCLIP reads (Figure 4F) and iCLIP clusters (Figure S3C) for the two proteins. Similar binding was also seen for exons more strongly regulated by PTBP1 than PTBP2 (Figure 4G–H, Figure S3D–E). There were observable differences in the peak heights and sometimes the peak positions for the PTBP1 and PTBP2 iCLIP clusters. However, these could be due to incomplete coverage of the iCLIP and were difficult to connect to consistent differences in regulation. Additional exons repressed or activated more strongly by one PTBP but bound similarly by the two proteins are shown in Figure S5C–F.
We also observed iCLIP clusters in CU-rich regions adjacent to exons unaffected by changes in PTBP expression, including Celf2, Nrg3, Nlgn1, Nrnx3, Rbfox3, Xist, and others (Figure S5G–H). The regulation of these exons is presumably dominated by additional factors, at least in the cortex. We also did not see a clear effect of the position of PTBP binding in activating or repressing splicing. Many PTBP-activated exons exhibited upstream binding by PTBP1 or PTBP2 (e.g., Figure 4H and Figure S5D, F). This is consistent with previous observations with PTBP1/2, and in contrast to results with splicing regulators such as Rbfox and Nova that exhibit position dependent regulation (Damianov et al., 2016; Han et al., 2014; Licatalosi et al., 2012, 2008; Li et al., 2014; Yeo et al., 2009).
PTBP-control of developmentally-regulated exons
Next, we asked how the PTBP dependent exons changed over brain development when PTBP1 and PTBP2 expression undergo substantial changes. PTBP target exons are presumably affected by multiple regulatory factors, and the extent to which PTBP targeting alone can predict developmental regulation has not been clarified. To examine these questions, we performed RNA-Seq analysis of polyA+ RNA from mouse neocortex at E14, E17, P2 and P10 (Experimental Procedures). Within each PTBP-regulated exon group, we performed unsupervised hierarchical clustering of PSI values from the four developmental ages. The PSI values of individual exon clusters were examined for significant changes between the four developmental ages.
Exon clusters exhibiting developmental regulation were consistent with the developmental downregulation of PTBP1 and PTBP2 (Figure 5). For example, among exons repressed by PTBP2 alone, four of six clusters exhibited developmental regulation, but the timing and magnitude of their changes differed (Figure 5A, D-G, Table S3–6). Clusters 2 and 3 displayed large increases in exon inclusion between E14 and E17, and between E17 and P2. Cluster 3 continued to increase in splicing between P2 and P10, but Cluster 2 plateaued at P2. The other two clusters (5 and 6) exhibited a shallower increase across the time course and like Cluster 2 did not change between P2 and P10. For all these clusters, the period exhibiting the most significant splicing change was between E17 and P2 coincident with PTBP2 downregulation (P values of 3.5e-11, 5.4e-14, 0.07 and 2.3e-05 for clusters #2, 3, 5 and 6; Wilcoxon signed rank test).
Figure 5. Developmental regulation of PTBP1 and PTBP2 target exons.
(A–C) Heat maps show the hierarchical clustering of cassette exon groups based on their splicing levels at embryonic days 14 and 17 (E14, E17) and postnatal days 2 and 10 (P2, P10). Numbers on the dendrogram branches indicate individual exon clusters. Exon clusters exhibiting developmental-control were selected and their PSI values at E14, E17, P2 and P10 are plotted (D–J). * P value < 0.02., ** P value < 10−4, and *** P value < 10−9, determined using a Wilcoxon signed rank test.
The exon group repressed by both PTBP proteins segregated into 5 clusters (Figure 5B). One of these (cluster 3) showed distinct developmental regulation with a remarkable increase in PSI values between E14 and P2 before declining at P10 (Figure 5H, Table S7). Between E14 and P2 the ΔPSI of Cluster 3 was 41, the largest among all clusters in all exon groups. These splicing changes coincided with the loss of PTBP1 around E15 and the decrease in PTBP2 after birth. The decrease in PSI at P10 could result from increased numbers of PTBP1-positive non-neuronal cells in the P10 transcriptome, or from new factors expressed late in development.
Developmentally-controlled clusters of the PTBP-activated exons generally showed decreased splicing over the time course (Figure 5C). Exons activated by PTBP2 had two developmentally-controlled clusters (Figure 5I–J, Table S8–9). Other groups had low exon numbers and did not produce robust segregation into developmentally-regulated exon clusters, although individual exons within the groups exhibited developmental changes consistent with PTBP expression.
The patterns of coregulation identified in the clustering points to the complex combinatorial regulatory processes occurring during neuronal development. Exons in other clusters than those described above are likely controlled by additional factors that maintain their relatively stable inclusion ratios between E14 and P10. It will be interesting to determine profiles of factor sensitivity for these exon clusters in additional mutant mice and knockdown experiments. This should identify factors both counteracting and enhancing PTBP action over development.
DISCUSSION
Partial genetic compensation for Ptbp2 null by a Ptbp1 transgene
We found both redundancy and specificity in the action of the PTBP1 and PTBP2. The Ptbp1 transgene expressed from the Rosa26 locus rescued the lethality of the Emx1-Ptbp2KO but not the Nes-Ptbp2KO, indicating that the commonality of function between the proteins varies with development and brain region. The Flag-Ptbp1 cKI mice exhibited a similar life span to wildtype, indicating that the transgene on its own did not affect survival and that the failure to rescue the loss of PTBP2 was likely not due to PTBP1 toxicity. Failure to rescue the Nes-Ptbp2KO was also unlikely to be caused by insufficient transgene expression, as the Flag-PTBP1 was expressed at fairly constant levels across brain regions including the forebrain (Figure S1B). Nevertheless, we can’t exclude the possibility that a small subset of neurons expresses inadequate FLAG-PTBP1.
Exon sensitivity to PTBP is likely to vary among cell types and brain regions and this may contribute to the differences in the ability of PTBP1 to rescue the loss of PTBP2. Nearly half of the mis-spliced cassette exons in Emx1-Ptbp2KO brains were reverted to near normal splicing by the Ptbp1 transgene (|PSIDM - PSIWt|<10). It is tempting to attribute the phenotypic rescue to these reverted exons, but connecting this developmental phenotype to particular transcripts will be a challenge. It will also be interesting to profile non-cortical structures in the Nestin-Cre mice and to identify differentially spliced exons between the wildtype, Ptbp2KO and DM.
Determinants of the specificity of PTBP regulation
A majority of the Ptbp1-rescued exons exhibited intermediate inclusion levels in the DM, between those found in the wildtype and the Ptbp2-cKO. This may result from lower expression from one copy of the Ptbp1 transgene than the endogenous Ptbp2 locus. However, PTBP2-regulated exons were not all affected by the Ptbp1 transgene to the same degree, and it appears that PTBP2 is more active than PTBP1 in the perinatal neocortex.
Since RNA binding does not correlate with PTBP1 and PTBP2 target specificity, regulatory co-factors likely determine an exon’s response to the two proteins. This is in agreement with a recent analysis of a target exon that exhibited greater repression by PTBP1 than PTBP2 (Keppetipola et al., 2016). Determinants of the higher repressive activity were found at multiple locations across the PTBP1 protein, including the flexible linker between RRM2 and RRM3. Notably, the known PTBP1 cofactor, Raver1, was found to bind more strongly to PTBP1 than to PTBP2 (Keppetipola et al., 2016). The developmental regulation of Raver1 is not known, but determinants conferring PTBP1 specific regulation could, like Raver1, promote PTBP1 activity or could antagonize PTBP2 function. One possibility for the latter is NSR100, which has been shown to antagonize PTBP1 on certain exons, but whose expression more closely aligns with PTBP2 during development (Raj et al., 2014). Similarly, we previously observed a class of exons in ES cells that are relatively insensitive to PTBP2 induction upon PTBP1 knockdown, possibly due to lack of positive acting PTBP2 co-factors (Linares et al., 2015). Modulators of chromatin states could be another class of co-factors that determine PTBP target specificity. PTBP1 was reported to interact with MRG15 in coupling H3K36 methylation and alternative splicing (Luco et al., 2010). We have not observed MRG15 interactions with PTBP1 or PTBP2 in brain. The extensive chromatin changes during neuronal differentiation could have many effects on the activity of PTBP1 and PTBP2 through changes in cofactor expression (Dixon et al., 2015).
As neuronal progenitor cells exit mitosis and begin to differentiate, some exons switch their splicing, in part due to greater sensitivity to PTBP1 than PTBP2 (Boutz et al., 2007; Linares et al., 2015; Makeyev et al., 2007; Zhang et al., 2016). Later in development as PTBP2 expression decreases, exons regulated by both proteins transition to adult patterns of splicing. Many other regulatory factors are changing over this time, and by identifying exons altered in the brains of DM mice and assessing their normal developmental profiles, we can discern more complex splicing behaviors. Overlapping these developmental profiles with the expression of other factors we can begin to define intricate interactions of splicing regulatory programs in vivo.
EXPERIMENTAL PROCEDURES
Ptbp1 conditional knock-in mice
A 3xNLS sequence was inserted into the CAGIG vector with the restriction sites MscI and BsrgI. The vector was then digested with BsrgI, blunt-ended with Klenow fragment followed by ClaI digestion. The linearized vector was treated with PNK and ligated with a ClaI-treated mCherry fragment, followed by insertion of a FRT sequence using the XmaI and SacI sites. The other FRT sequence was inserted into the BigT vector (Srinivas et al., 2001) using the NheI and SalI sites. The IRES-3XNLS-mCherry-FRT sequence was then subcloned into the FRT-containing BigT vector using the NotI and SacI sites. FLAG-Ptbp1 was subsequently inserted using the XhoI site. The resulting BigT-PTBP1 vector was digested with PacI and AscI and cloned into the pROSA26pm1 vector to generate the targeting vector pROSA26pm1-FRT-FLAG-Ptbp1-IRES-3XNLS-mCherry-FRT. The targeting vector was linearized with SwaI and electroporated into 129-derived ES cells. After ES cell selection, recombinant clones were identified for correct homologous recombination by PCR and Southern blot, and injected into blastocysts. Chimeric mice were propagated to obtain germline-transmitted Ptbp1 conditional knockin mice identified by PCR and Southern blot. Nissl staining was performed as previously described (Zheng et al., 2010). All animal procedures conformed to the regulatory standards and were approved by the Institutional Animal Care and Use Committee at UCR and UCLA. The Rosa26LSL-FL-Ptbp1 mice are available at The Jackson Laboratory as Stock No. 029838. F1: AAAGTCGCTCTGAGTTGTTAT. R2: GCGAAGAGTTTGTCCTCAACC (310bp). R3: GGCGGATCACAAGCAATAAT (413bp)
Statistical methods
Student’s t-test, Fisher’s exact test, hypergeometric test and Wilcoxon signed rank test, as stated in the Results, were performed using the corresponding test functions in R.
RNA-Seq and iCLIP-Seq
RNA-Seq and iCLIP-Seq were performed following Illumina’s Truseq protocol and the original iCLIP protocol with some modifications. Details of the experimental procedures and analysis are in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
PTBP1 partially rescues PTBP2 loss during brain development
PTBP1 and PTBP2 regulate overlapping but distinct sets of alternative exons in vivo.
Exons exhibit developmental regulation following their sensitivity to PTBP1/2.
Differential targeting is likely determined by cofactors rather than RNA binding.
Acknowledgments
We thank Andrey Damianov for sharing his unpublished iCLIP protocol. This work was supported by the NIH grants R01 GM097230 (to L.C.), R01 GM049662 (to D.L.B.) and R00 MH096807 (to S.Z.), and by earlier support to D.L.B. from the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization, S.Z. and D.L.B.; Methodology, S.Z.; Formal Analysis, C-H.L., C.L., S.Z. and J.V.; Investigation, S.Z., J.V., and M.Z.; Data Curation, C-H.L.; Writing – Original Draft, S.Z. and D.L.B.; Writing – Review and Editing, S.Z. and D.L.B.; Supervision, D.L.B. and S.Z.; Funding Acquisition, D.L.B. and S.Z.
REFERENCES
- Amir-Ahmady B, Boutz PL, Markovtsov V, Phillips ML, Black DL. Exon repression by polypyrimidine tract binding protein. RNA. 2005;11:699–716. doi: 10.1261/rna.2250405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin C-H, Chawla G, Ostrow K, Shiue L, Ares M, Jr, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zheng S. Identify alternative splicing events based on position-specific evolutionary conservation. PLoS One. 2008;3:e2806. doi: 10.1371/journal.pone.0002806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damianov A, Ying Y, Lin C-H, Lee J-A, Tran D, Vashisht AA, Bahrami-Samani E, Xing Y, Martin KC, Wohlschlegel JA, Black DL. Rbfox Proteins Regulate Splicing as Part of a Large Multiprotein Complex LASR. Cell. 2016;165:606–619. doi: 10.1016/j.cell.2016.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, Diao Y, Liang J, Zhao H, Lobanenkov VV, Ecker JR, Thomson JA, Ren B. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A, Stoilov P, Linares AJ, Zhou Y, Fu X-D, Black DL. De novo prediction of PTBP1 binding and splicing targets reveals unexpected features of its RNA recognition and function. PLoS Comput. Biol. 2014;10:e1003442. doi: 10.1371/journal.pcbi.1003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppetipola NM, Yeom K-H, Hernandez AL, Bui T, Sharma S, Black DL. Multiple determinants of splicing repression activity in the polypyrimidine tract binding proteins, PTBP1 and PTBP2. RNA. 2016 doi: 10.1261/rna.057505.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppetipola N, Sharma S, Li Q, Black DL. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit. Rev. Biochem. Mol. Biol. 2012;47:360–378. doi: 10.3109/10409238.2012.691456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert N, Robertson A, Jangi M, McGeary S, Sharp PA, Burge CB. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol. Cell. 2014;54:887–900. doi: 10.1016/j.molcel.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Yano M, Fak JJ, Mele A, Grabinski SE, Zhang C, Darnell RB. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev. 2012;26:1626–1642. doi: 10.1101/gad.191338.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares AJ, Lin C-H, Damianov A, Adams KL, Novitch BG, Black DL. The splicing regulator PTBP1 controls the activity of the transcription factor Pbx1 during neuronal differentiation. Elife. 2015;4:e09268. doi: 10.7554/eLife.09268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zheng S, Han A, Lin C-H, Stoilov P, Fu X-D, Black DL. The splicing regulator PTBP2 controls a program of embryonic splicing required for neuronal maturation. Elife. 2014;3:e01201. doi: 10.7554/eLife.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Raj B, Blencowe BJ. Alternative Splicing in the Mammalian Nervous System: Recent Insights into Mechanisms and Functional Roles. Neuron. 2015;87:14–27. doi: 10.1016/j.neuron.2015.05.004. [DOI] [PubMed] [Google Scholar]
- Raj B, Irimia M, Braunschweig U, Sterne-Weiler T, O’Hanlon D, Lin Z-Y, Chen GI, Easton LE, Ule J, Gingras A-C, Eyras E, Blencowe BJ. A global regulatory mechanism for activating an exon network required for neurogenesis. Mol. Cell. 2014;56:90–103. doi: 10.1016/j.molcel.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman R, Llorian M, Smith CWJ. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol. Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork C, Zheng S. Genome-Wide Profiling of RNA-Protein Interactions Using CLIP-Seq. Methods Mol. Biol. 2016;1421:137–151. doi: 10.1007/978-1-4939-3591-8_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong CK, Black DL, Zheng S. The neurogenetics of alternative splicing. Nat. Rev. Neurosci. 2016;17:265–281. doi: 10.1038/nrn.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y, Jiang L, Cai Z, Sun H, Zhang K, Zhang Y, Chen J, Fu X-D. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152:82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu X-D, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG, Soriano P. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl. Acad. Sci. U. S. A. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen MH, Wu X, Kodani A, Fan J, Doan R, Ozawa M, Ma J, Yoshida N, Reiter JF, Black DL, Kharchenko PV, Sharp PA, Walsh CA. Cell-Type-Specific Alternative Splicing Governs Cell Fate in the Developing Cerebral Cortex. Cell. 2016;166:1147.e15–1162.e15. doi: 10.1016/j.cell.2016.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S. Alternative splicing and nonsense-mediated mRNA decay enforce neural specific gene expression. Int. J. Dev. Neurosci. 2016 doi: 10.1016/j.ijdevneu.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Eacker SM, Hong SJ, Gronostajski RM, Dawson TM, Dawson VL. NMDA-induced neuronal survival is mediated through nuclear factor I-A in mice. J. Clin. Invest. 2010;120:2446–2456. doi: 10.1172/JCI33144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Gray EE, Chawla G, Porse BT, O’Dell TJ, Black DL. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat. Neurosci. 2012;15:381–388. doi: 10.1038/nn.3026. S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.