Abstract
Crop species exhibit an astounding capacity for environmental adaptation, but genetic bottlenecks resulting from intense selection for adaptation and productivity can lead to a genetically vulnerable crop. Improving the genetic resiliency of temperate maize depends upon the use of tropical germplasm, which harbors a rich source of natural allelic diversity. Here, the adaptation process was studied in a tropical maize population subjected to 10 recurrent generations of directional selection for early flowering in a single temperate environment in Iowa, USA. We evaluated the response to this selection across a geographical range spanning from 43.05° (WI) to 18.00° (PR) latitude. The capacity for an all-tropical maize population to become adapted to a temperate environment was revealed in a marked fashion: on average, families from generation 10 flowered 20 days earlier than families in generation 0, with a nine-day separation between the latest generation 10 family and the earliest generation 0 family. Results suggest that adaptation was primarily due to selection on genetic main effects tailored to temperature-dependent plasticity in flowering time. Genotype-by-environment interactions represented a relatively small component of the phenotypic variation in flowering time, but were sufficient to produce a signature of localized adaptation that radiated latitudinally, in partial association with daylength and temperature, from the original location of selection. Furthermore, the original population exhibited a maladaptive syndrome including excessive ear and plant heights along with later flowering; this was reduced in frequency by selection for flowering time.
Introduction
Genetic diversity is essential for the breeding of resilient and productive crop varieties. The cultivation of a crop such as maize across a tremendous geographical and ecological range (Kuleshov, 1933; Ruiz Corral et al., 2008) reflects the broad adaptability of some crop species. However, concomitant with the domestication of crops and the subsequent breeding to create elite modern cultivars is a reduction in genetic diversity (Tanksley, 1997; Wright et al., 2005). For instance, the founding and prevailing source of germplasm used for US Corn Belt maize traces to only a few key populations (Troyer, 1999) from only two races (Anderson and Brown, 1952); yet, over 300 races, not to mention their many populations, have been documented for maize (for a phenotypic perspective: Committee on The Preservation of Indigenous Strains of Maize monographs on Races of Maize, 1952; for a molecular genetic perspective: Vigouroux et al., 2008). Concerns about genetic vulnerability prompted various efforts beginning in the 1940s to broaden the genetic base of US maize, through the introduction of ‘exotic' germplasm (for example, Melhus, 1948; Wellhausen, 1965 (Rockefeller Foundation Agricultural Program); Pollak, 1997 (Latin American Maize Project) and Pollak, 2003 (Genetic Enhancement of Maize project)). Given worldwide reliance on maize as a food and feed crop coupled with increasing concerns about climate change and the emergence of new pests and diseases, this objective seems even more pertinent today.
Broadening the genetic base of maize in the USA and other parts of the world requires the use of tropical germplasm, which harbors unique (Liu et al., 2003) and favorable (Holland et al., 1996) genetic diversity. However, assessing the intrinsic genetic merit of tropical germplasm through phenotypic evaluation in subtropical and, particularly, temperate environments is virtually meaningless due to a maladaptive syndrome that includes weak and variable seedling vigor, excessively late flowering, high ear and plant heights (PLHs) and poor standability, which mask the value of potentially favorable alleles (Goodman, 1999; Betŕan et al., 2003; Tarter and Holland, 2006). The adaptation of exotic germplasm to a new production environment is one approach to capitalizing on unique diversity; and knowing about the adaptive potential of a population and the adaptive process is of fundamental interest.
Flowering is a key characteristic of adaptation, the timing of which is influenced by both genetics and the environment (Aitken, 1977; Taiz and Zeiger, 2006; Jung and Müller, 2009; Manel et al., 2012). Among traits in the maladaptive syndrome, flowering is the most important because it impacts the timing of seed production in relation to climate. The phenomena referred to as photoperiodism (first mentioned in Garner and Allard, 1923) describes plant responses, including flowering time, to the length of the period of daily light or photoperiod. A plant's transition from vegetative to reproductive phase can be sensitive to photoperiod, and the level of sensitivity measured in terms of flowering time can vary substantially among different genotypes within a species (for example, Quinby, 1967; Yano et al., 2001; Gouesnard et al., 2002), resulting at one extreme in delayed flowering or failure to flower. Tropical maize germplasm often exhibits photoperiod sensitivity at daylengths exceeding 10–13.5 h, depending on the genotype (Kiniry et al., 1983; Warrington and Kanemasu, 1983), leading to an important, although tractable (via breeding improvement) barrier to temperate environment adaptation (Holley and Goodman, 1988). Photoperiod-insensitive tropical germplasm has been described (Gouesnard et al., 2002), but such germplasm may still be poorly adapted in terms of flowering time. Other mechanisms such as response to temperature, which varies among maize germplasm sources (Ellis et al., 1992), are also thought to be important. Thus, photoperiod insensitivity is required but not sufficient to alleviate the spectrum of symptoms associated with the poor adaptation of tropical germplasm to temperate environments.
The genetics of standing variation in flowering time has been studied in a number of species. In maize, a polygenic architecture of mostly small-effect alleles has been described to underlie flowering time per se (Chardon et al., 2004; Buckler et al., 2009), whereas a smaller number of loci appear to underlie photoperiodism (Hung et al., 2012b). Understanding how the genetic variation associated with flowering time interacts with the environment and responds to selection for adaptation is of basic interest and can help breeders address challenges of food security (Godfray et al., 2010; Tester and Langridge, 2010). Combining the study of selection response in climate-characterized multi-environment trials (METs) can be used to improve this understanding. Analysis of selected breeding populations is attractive because of the opportunity to interpret results in terms of a well-established theoretical foundation on response to selection (Falconer and Mackay, 1996; Orr, 2005; Hallauer et al., 2010). Simultaneously, developments in statistical modeling are making it feasible to interpret some components of genotype-by-environment and gene-by-environment interactions in terms of specific environmental factors (Crossa et al., 1999; Epinat-Le Signor et al., 2001; Reymond et al., 2003; Malosetti et al., 2013).
Here, the response to selection was studied by testing the environmental range performance of Hallauer's Tusón, a maize population of tropical origin phenologically adapted to one location in central Iowa through a decade of phenotypic selection for early flowering. This study is part of a broader effort to develop a framework for genetically dissecting the response to selection (Wisser et al., 2011); here, the focus is on modeling and inference on the basis of tropical-to-temperate adaptation using MET data on families from generations 0–10. To assess the importance of photoperiod sensitivity across generations of temperate adaptation, nine environments encompassing a daylength cline were chosen for our MET design. We examined the response to selection across these environments, modeled variation in flowering time associated with environments and genotype-by-environment interactions in terms of specific environmental variables, tested the hypothesis that selection against photoperiodism was the basis for population improvement in flowering time and studied the response of other traits composing the maladaptive syndrome.
Materials and methods
Population development
During the 1990s at Iowa State University, Dr Arnel Hallauer adapted a lowland tropical maize landrace, Tusón, to central Iowa. The base population used for selection was produced by one generation of intermating (in an open-pollinated isolation block in Iowa, without intentional selection) of five accessions: PI 449556 and PI 583912 (collected from Brazil); NSL 283507 (collected from Cuba); PI 487940 (tentatively) (collected from Ecuador); and PI 498583 (collected from Guatemala). Subsequently, 10 generations of phenotypic mass selection were performed with a major emphasis on early female flowering or silking and a secondary emphasis on reduced stalk and root lodging or standability. Each generation of selection was allowed to open-pollinate in an isolation block with ~10 000 individuals sampled from an equal bulk of seed from 300 to 500 selected individuals from the previous generation. Following the design of Wisser et al. (2011), a random sample of plants from remnant seed of the even-numbered generations were self-pollinated in DE and FL to produce S0:1 families and S0:2 families (random sib mating of S0:1 families) used in the MET: g0 (n=18), g2 (n=56), g4 (n=56), g6 (n=56), g8 (n=56) and g10 (n=55). A low germination rate in g0 limited its sample size for this study (seed quality was expected to be low due to a cold seed storage problem, and this had specifically affected g0 grown under field conditions).
Experimental design and data collection
An MET was performed during 2009 and 2010 across nine US locations ranging in latitude from 43.05° (Madison, Wisconsin) to 18.00° (Ponce, Puerto Rico) (Table 1). A set of 301 entries were evaluated, including the 297 S0:1/S0:2 families and two temperate (relatively photoperiod-insensitive B73 and B97) and two tropical (photoperiod-sensitive Ki14 and CML254) inbred line checks. The experimental design was a 17 × 17 α-lattice with two replications at each location. With more entries (n=301) than experimental units per complete block (n=289), replicates were imbalanced; 277 of the entries were assigned to both replications and 24 of the entries were assigned to only one of the two replications (12 from g0, 2 from g2, 2 from g4, 3 from g6, 3 from g8 and 2 from g10). This was done to maximize the number of families evaluated from g0, because not enough seed was available for them to be tested in both replications across all environments; only six g0 families could be tested in both replications across all environments. Entries were grown in single-row plots with roughly equal plant spacing (~0.35 m) across locations, which were managed under the standard agronomic practices for each location.
Table 1. Description of the environments used for the MET.
| Descriptor |
Environments (Tusón DTA-referenced) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| WI | IA | DE | MO | NC | nTX | cTX | FL | PR | |
| Planting date | 5/20/09 | 5/12/09 | 5/14/10 | 6/2/09 | 4/21/09 | 4/10/09 | 3/18/10 | 10/15/09 | 10/27/09 |
| Planting day of year | 140 | 132 | 134 | 153 | 111 | 100 | 77 | 288 | 300 |
| DTAa | 87 | 82 | 77 | 71 | 72 | 83 | 77 | 65 | 54 |
| Soil type | Silt loam | Silt clay loam | Sandy loam | Silt loam | Sandy clay | Clay loam | Clay | Loam | Clay loam |
| Latitude (decimal degree) | 43.05° | 42.03° | 39.67° | 38.89° | 35.67° | 33.60° | 30.55° | 25.50° | 18.00° |
| Longitude (decimal degree) | −89.53° | −93.77° | −75.75° | −92.20° | −78.49° | −101.91° | −96.43° | −80.50° | −66.51° |
| Altitude (m) | 331 | 332 | 33 | 271 | 107 | 990 | 118 | 2 | 10 |
| DLseb,c | 0.020 | 0.028 | 0.026 | 0.011 | 0.032 | 0.032 | 0.031 | −0.023 | −0.015 |
| DLstb,c | −0.010 | 0.001 | 0.001 | −0.012 | 0.019 | 0.020 | 0.026 | −0.019 | −0.011 |
| DLhtb,c | 15.3 | 15.2 | 15.0 | 14.8 | 14.3 | 14.1 | 13.4 | 10.9 | 11.2 |
| Daylength classificationd | LD | LD | LD | LD | LD | LD | NC | SD | SD |
| SRtb,c | 5983 | 6587 | 6246 | 6912 | 6403 | 6716 | 5851 | 4123 | 5305 |
| SATnb,c | 13.4 | 14.6 | 16.6 | 17.7 | 17.7 | 17.0 | 15.7 | 18.7 | 20.4 |
| SATmb,c | 17.9 | 19.4 | 20.4 | 23.6 | 22.3 | 23.0 | 20.7 | 23.0 | 26.1 |
| SATxb,c | 22.0 | 24.0 | 24.3 | 29.7 | 27.4 | 29.3 | 26.1 | 27.3 | 31.8 |
| SATrb,c | 4.4 | 4.9 | 4.3 | 6.5 | 5.4 | 6.5 | 5.7 | 4.4 | 5.8 |
| RHmb,c | 80.4 | 75.7 | 79.1 | 68.9 | 74.7 | 42.9 | 70.7 | 80.9 | 80.7 |
| ETtb,c | 0.24 | 0.23 | 0.25 | 0.27 | 0.23 | 0.33 | 0.33 | 0.14 | 0.15 |
| SADu06 (days under 6 °C)b | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| SADo30 (days over 30 °C)b | 3 | 10 | 1 | 35 | 27 | 49 | 27 | 11 | 47 |
| SAGDDb | 1014 | 1073 | 1115 | 1139 | 1129 | 1213 | 1042 | 1097 | 966 |
| SAGDDu06b | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| SAGDDo30b | 4 | 20 | 0 | 121 | 62 | 211 | 109 | 7 | 120 |
Abbreviations: cTX, central TX; DTA: days to anthesis; DLht: hour of daylength at transition; DLse, slope of daylength at emergence; DLst: slope of daylength at transition; ETt, average of daily total evapotransipiration; LD, long day; MET, multi-environment trial; NC, not classified; nTX, north TX; RHm: average of daily relative humidity; SATm: average of daily mean soil-air temperature; SATn: average of daily minimum soil-air temperature; SRt, average of daily total solar radiation; SATr, average of daily range (max–min) in soil-air temperature; SATx, average of daily maximum soil-air temperature; SD, short day.
Mean DTA of the Tusón population.
Environment variable summarized as described in Materials and methods; here, based on the period from planting to the mean DTA date of the Tusón population.
Variable used for elastic net regession (see Materials and Methods: DLse; DLst; DLht; SRt; SATn; SATm; SATx; SATr; RHm; ETt).
Classification used for LD vs SD analysis; cTX was excluded due to uncertainty about LD/SD classification.
Six primary traits were measured in each single-row plot, excluding the end plants from measurement: number of days from planting to when 50% of the plants in a plot were silking (days to silking (DTS); measured at all locations except MO) and extruding anthers (days to anthesis (DTA); measured at all locations); mean distance from the ground to the node of the uppermost ear (ear height (ERH); measured at all locations) and to the last leaf node (PLH); measured at all locations except that in Lubbock, TX height was measured to the tip of the tassel); mean dry weight of the whole tassel cut from immediately above the node of the peduncle (tassel weight (TSW); measured at DE, NC, FL and PR); and mean length of the central spike of the dried tassel (spike length (SPL); measured at DE, NC, FL and PR). Derived traits included: anthesis-silking interval (ASI: DTS−DTA); PLH above the ear (PEH: PLH−ERH); and DTS and DTA in terms of growing degree days (GDD) to silking and anthesis (GDDTS and GDDTA) based on the SM(B,30−) equation described by Bonhomme et al. (1994). Specifically, GDD=([Tmin+Tmax]/2)−Tb; where Tb is the base temperature of 6 °C, Tmin is the daily minimum temperature set to Tb when Tmin<Tb, Tmax is the maximum daily temperate up to Topt=30 °C, and for Tmax>30 °C Topt is [Topt−(Tmax−Topt)].
Data analysis
Model fitting
Mixed linear model analysis was performed using ASReml v.3 (Gilmour et al., 2009). Data from plots with fewer than four plants were excluded from analysis (~3% of the data). Modeling of the MET data began by fitting single-environment models. For each of two different MET style models (generation model [1] and family model [2] described below), single-environment models were fitted with the corresponding effect structure as defined for each (excluding across environment effects). This pre-modeling procedure was used to examine outliers in the data, test the significance of the experimental design effects (replications and incomplete block nested in replications) based on the likelihood-ratio test (α=0.10), and to obtain estimates of variance components to use as starting values for fitting the MET model. Outliers representing a clear data entry error or improbable inconsistencies between replications were removed (for a given trait this never exceeded 0.18% of the MET data). In the final step, only the environment-specific experimental design effects that were significant were included in the respective MET model.
Two different linear mixed models were fitted to the data: (1) a generation model in which inference was made at the level of generations; (2) a family model in which inference was made at the level of the whole population (generation structure ignored). The data were coded and models were specified to partition the variability due to entries into a fixed difference between the mean of all inbred checks and the mean of all Tusón families, fixed effects of the check lines, fixed effects of the generations for the generation model (1), and random effects of the families for the family model (2) (Oakey et al., 2006; Piepho et al., 2006).
Generation model:
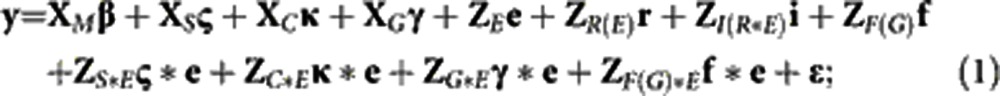 |
Family model:
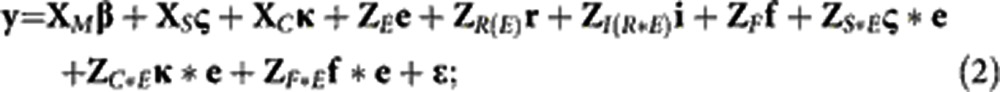 |
where y is the vector of observations; fixed effects are: β, the overall mean; ς, the vector of ‘germplasm set' effects (mean of check entries (set 1) and mean of Tusón entries (set 2)); κ, the vector of check effects; and γ, the vector of generation effects; random effects are: e, the vector of environment effects; r, the vector of replications nested in environment effects; i, the vector of incomplete blocks nested in replications-by-environment effects; f, the vector of families nested in generation (1) or non-nested family (2) effects; ς*e, the vector of set-by-environment effects; κ*e, the vector of check-by-environment effects; γ*e, the vector of generation-by-environment (gEI) effects; f*e, the vector of families nested in generation-by-environment effects (1) or non-nested family-by-environment (fEI) effects (2); and ɛ, the vector of residual effects. The design matrices, MM, XS, XC, XG, ZE, ZR(E), ZI(R*E), ZF, ZS*E, ZC*E, ZG*E, ZF(G)*E and ZF*E, relate the vector of observations to the corresponding vectors of effects.
The generation model was defined in two different ways. A generation-means model was fitted with the levels of generation treated as a categorical variable. Alternatively, a generation-slopes model was used to estimate the response to selection using a second-order polynomial function, where the levels of generation were treated as a continuous variable of non-orthogonal (to describe the response profile) or orthogonal (to test associations between the estimates and other variables) input values. For the generation-slopes model, γ and γ*e were vectors of estimated regression coefficients instead of vectors of mean effects.
The initial models (that is, single-environment models) were reduced models that assumed that the random effects were distributed 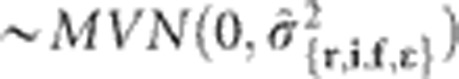 . The MET model was initiated under the assumption that the random-effect terms were distributed
. The MET model was initiated under the assumption that the random-effect terms were distributed 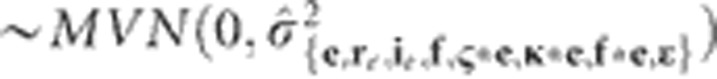 , where re and ie correspond to environment-specific variances for replications and incomplete blocks, respectively. However, modeling environment-specific residual variances
, where re and ie correspond to environment-specific variances for replications and incomplete blocks, respectively. However, modeling environment-specific residual variances
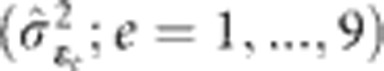 |
always led to better fitting models (based on the Bayesian Information Criterion (Wolfinger, 1993), so all of the MET models were fitted with environment-specific residual variances.
Alternative covariance or correlation structures were also fitted to the DTS and DTA data. For the generation-means model (1), the data were fitted treating generations as a random effect and assuming heterogeneous variances for γ, providing estimates of variance among families within each generation. For the family model, the fit of the following matrices modeled on the f*e term was examined: IDV (default), CORUV, CORUH, CORBU, CORBH, CORGU, CORGH, AR1U, AR1H, EXPU, EXPH, IEXPU, IEXPH, AIEXPU, AIEXPH, MAT2V, MAT2H and FA1 (matrix structures described in Gilmour et al., 2009, pp. 132–145). For each model in which a non-IDV structure for f*e was assumed, the family main effect was removed from the model, as required to define a non-singular model. Latitude and longitude coordinates were used in formulating the exponents for the exponential (those with EXP in their name) and Matérn (those with MAT in their name) class matrices. The Bayesian Information Criterion was used to select the best-fitting model.
The generation-means model (1) was further extended as a bivariate model for the analysis of pairs of traits, with the primary aim of estimating genetic correlations between traits. This model included the same effects structure as the univariate model, but for the bivariate model, following notation by Apiolaza et al. (2000), y=[y1′ y2′]′, corresponding to the vector of phenotypes for each pair of traits, y1 (DTS or DTA) and y2 (one of the other traits). Notating the independent variables also extends as, for example, MMβ; where 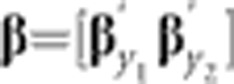 , the vector mean effects for each trait, and MM is the design matrix relating β to y. Each random term was assumed to be distributed ~BVN(0, Σi), where:
, the vector mean effects for each trait, and MM is the design matrix relating β to y. Each random term was assumed to be distributed ~BVN(0, Σi), where:
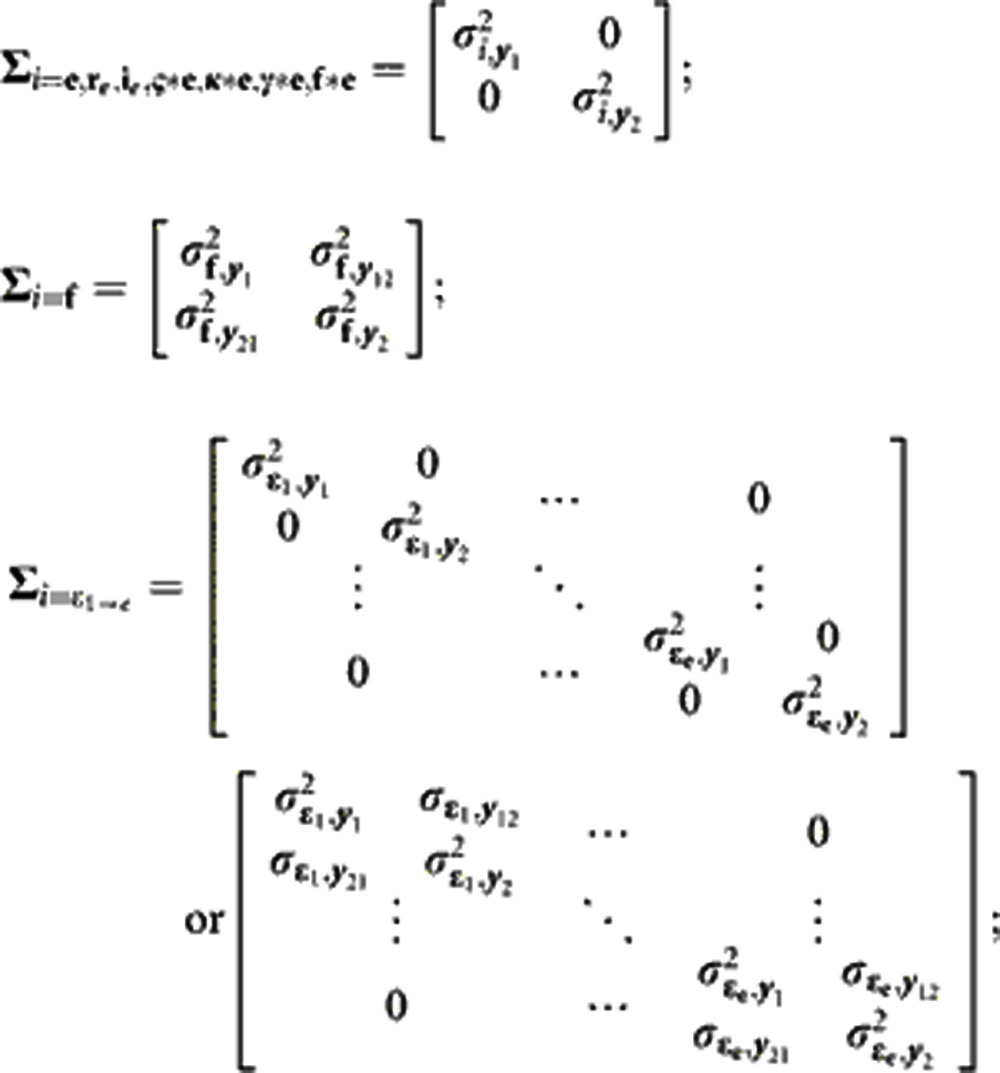 |
with subscripts for variables the same as described for the generation model (1). 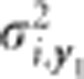 is the genotypic variance of trait y1 for variable i,
is the genotypic variance of trait y1 for variable i, 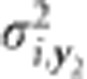 is the genotypic variance of trait y2 for variable i, and σi,y.. is the genotypic covariance between trait y1 and y2 for variable i. The residual,
is the genotypic variance of trait y2 for variable i, and σi,y.. is the genotypic covariance between trait y1 and y2 for variable i. The residual,  , was modeled on a trait- and environment-specific basis, where e is the number of environments in which both traits were measured (e=4 for DTA/DTS versus TSW and SPL; e=8 for DTS versus ASI, PLH, ERH and PEH; e=9 for DTA versus ASI, PLH, ERH and PEH). On the basis of the Bayesian Information Criterion, the within-environment residual effects were assumed to have no correlation (DTA and DTS versus TSW, SPL, PLH, ERH and PEH) or some correlation (DTA and DTS versus ASI) between traits.
, was modeled on a trait- and environment-specific basis, where e is the number of environments in which both traits were measured (e=4 for DTA/DTS versus TSW and SPL; e=8 for DTS versus ASI, PLH, ERH and PEH; e=9 for DTA versus ASI, PLH, ERH and PEH). On the basis of the Bayesian Information Criterion, the within-environment residual effects were assumed to have no correlation (DTA and DTS versus TSW, SPL, PLH, ERH and PEH) or some correlation (DTA and DTS versus ASI) between traits.
Analysis of estimated variance components
Variances estimated by the generation-means model (1) fitted with environment-specific variance (IDH structure) for ɛ and uniform variance and zero correlation (IDV structure) for f*e were used to calculate broad-sense heritability on a single-plot basis within a single generation 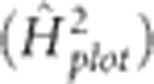 . This was estimated to reflect the heritability associated with mass selection; here
. This was estimated to reflect the heritability associated with mass selection; here 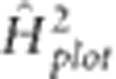 is an inflated estimate as it represents that for the mean of a single-row plot of plants and not an individual plant on which selection was actually applied. Heritability was also calculated on an MET family mean basis
is an inflated estimate as it represents that for the mean of a single-row plot of plants and not an individual plant on which selection was actually applied. Heritability was also calculated on an MET family mean basis 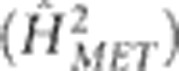 , using variances estimated by the family model (2). This was estimated to reflect the maximum amount of heritable variation that could be ascribed to loci if the data were used for genetic mapping of average effects.
, using variances estimated by the family model (2). This was estimated to reflect the maximum amount of heritable variation that could be ascribed to loci if the data were used for genetic mapping of average effects.
 |
 |
where the variance components are as described above, eh is the harmonic mean of the number of environments each family was evaluated in, and ph is the harmonic mean of the total number of plots each family was evaluated in (Holland et al., 2003).
Analysis of estimated slopes and means
Bias-corrected distance correlation (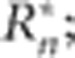 Székely and Rizzo, 2013) was used as a general test of the null hypothesis of independence between a pair of variables, using the R package energy v. 1.6.0 (http://cran.r-project.org/web/packages/energy/index.html)(function:dcor.ttest).The statistic was used to test for a geography-dependent pattern in the response profiles across environments, by testing whether the environment-specific first- and second-degree coefficients estimated by orthogonal polynomial regression were independent of latitude, longitude and altitude.
Székely and Rizzo, 2013) was used as a general test of the null hypothesis of independence between a pair of variables, using the R package energy v. 1.6.0 (http://cran.r-project.org/web/packages/energy/index.html)(function:dcor.ttest).The statistic was used to test for a geography-dependent pattern in the response profiles across environments, by testing whether the environment-specific first- and second-degree coefficients estimated by orthogonal polynomial regression were independent of latitude, longitude and altitude.
Elastic net (Zou and Hastie, 2005; R package glmnet v. 1.9–5) was used to regress environment main effects and gEI effects per se (estimated by the generation-means model (1)) on environmental variables. Prior to fitting the models, the e and γ*e effects for DTS and DTA (response variables) and the environmental variables (predictor variables) were Z-transformed across environments per generation for each trait, providing estimated coefficients for the predictors that could be compared within and across generations and traits. The elastic net models were fitted with α=αp, a p-specific α-value for each model; where αp was determined based on minimizing the cross-validation prediction error based on a grid search of α from 0–1 in increments of 0.05. For the model fitted to environment main effects αp=0.05 for DTS and αp=0.15 for DTA. For the 12 (generation x trait combination) models fitted to gEI effects αp was 0 in all cases except one. For DTS g2 αp=0.9 and explained 99% of the variation in gEI effects; we considered this an overfitted model and therefore fit all gEI elastic net models using the more consistent αp=0 value (equivalent to ridge regression).
The environment variables used for elastic net, which are described in detail in the following paragraphs, included: (1) DLse=slope of the daylength at emergence; (2) DLst=slope of the daylength at reproductive transition; (3) DLht=hour of the daylength at reproductive transition; and planting to flowering period averages of: (4) SRt=total daily solar radiation; (5) SATn=minimum daily soil or air temperature; (6) SATm=mean daily soil or air temperature; (7) SATx=maximum daily soil or air temperature; (8) SATr=difference between maximum and minimum daily soil or air temperature (that is, temperature range); (9) RHm=mean daily relative humidity; (10) ETt=total daily potential evapotranspiration.
Daylength data (variables 1–3 above) were calculated for each environment according to the function described by Forsythe et al. (1995). The slope of the change in daylength across days (DLse and DLst) was calculated as the first derivative of the function at specified days after planting (more on this below). Climate data (variables 4–10 from above) were obtained from ZedX Inc. (Bellefonte, PA, USA). For all of the variables except DL variables, the environment data were summarized across the flowering periods estimated for each generation in each environment. To estimate the day of emergence used for several calculations, we relied upon a separate field experiment where it was found that 115 GDDs were required for germination (data not shown). Then, based on 2-inch soil temperature data for each environment, the specific day after planting when 115 GDD had been accumulated was referenced when summarizing the environment variables. Pre-emergence soil temperatures and post-emergence air temperatures were integrated when calculating SATn, SATm, SATx, SATr and GDD. The day of predicted emergence was used to calculate DLse and to define the growth period from emergence to flowering for summarizing the remaining variables (SRt, RHm and ETt). The day of reproductive transition assumed for summarizing DLst and DLht was calculated for each environment as half of the GDD accumulated by B73 from GDD-predicted emergence to flowering (an approximation of the date for tassel initiation or the start of photoperiod sensitivity (Kiniry et al., 1983)).
Temperature data used to back-predict flowering time in 2001–2011 were obtained from the nearest weather station of the trial locations (WI, IA, MO, NC, cTX and FL) or from the Weather Underground web service using the GPS coordinates of the trial locations (http://www.wunderground.com/history/; DE, nTX, PR).
Results
Response to selection
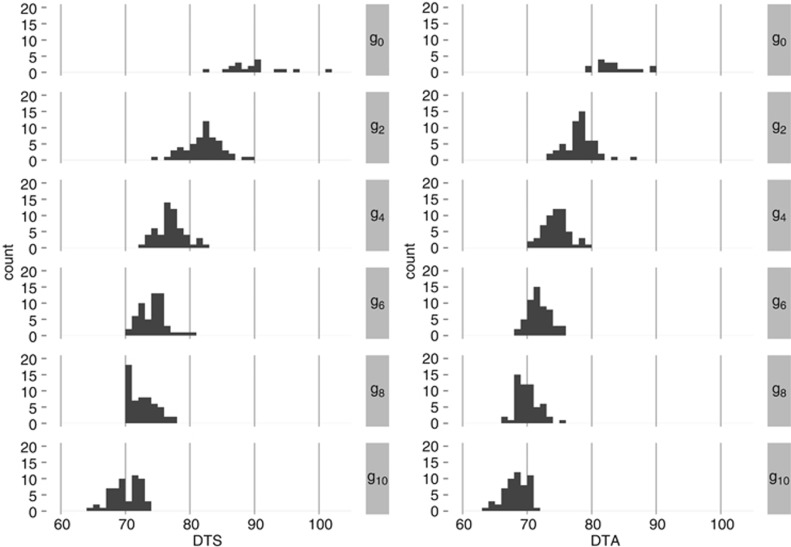
The multi-generational response to primary selection for early flowering was evaluated at Ames, IA, the location at which the Tusón population was originally adapted, and at eight other locations diverging −1.02° to 24.03° latitude and −27.26° to 8.14° longitude relative to Ames (Table 1). The overall response to selection was striking: the mean of g10 for DTS and DTA was 20 and 15 days earlier than g0, respectively (Figure 1). Furthermore, the generational distributions of family effects were distinct, with an 8–9-day separation in flowering time between the earliest family from g0 and the latest family from g10 (Figure 1).
Figure 1.
Distributions of flowering time variation among families within generations averaged across environments. Family effects were calculated as the linear combination of effects (β+ςs=2+γg+ff(g)) estimated by the generation-means model (1).
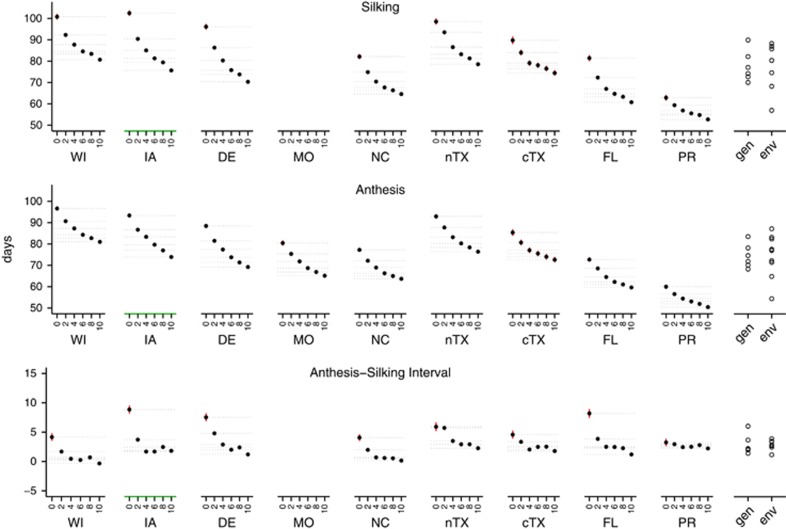
Figure 2 highlights several features about the response to selection in DTS and DTA: (i) the environment contributed substantially to the MET variation in flowering time; (ii) the magnitude of the response tended to be greater in the first few generations than in the latter generations; (iii) the direction of response to selection was consistent in each environment—generation effects were more pronounced than gEI effects; (iv) nevertheless, the response profiles exhibited variability across environments in terms of shape and magnitude; and (v) the magnitude of the response appeared to roughly follow a latitudinal pattern diverging from IA and DE where the greatest responses were observed.
Figure 2.
Generational changes in flowering time across environments. Dot plots show trait means (points) and s.e. (red lines) per generation (x-axes) for each environment (ordered by latitude; DTS and ASI data were not available for MO). The x-axis for IA is colored green to highlight the location where selection was originally performed. The horizontal dashed gray lines intersect each point to facilitate visualization of the non-linear nature of the response profiles. The right-most plots show the main effects of generations (gen) and environments (env).
Environment main effects were associated with ~70% of the variation in flowering time in the MET. These effects were roughly twice as large (1.6 × for DTS and 2.2 × for DTA) as generation main effects, which translated into an environmental impact that stretched the range in flowering time of the population by ~30 days (Figure 2). Performance of the population in PR contributed substantially to this effect, but even upon removing the data from PR the environment effect was still at least as large as the generation main effects. The heritability of single-row plots was 0.47±0.02 for DTS and 0.53±0.02 for DTA, whereas the family mean-basis heritabilities were extremely high for DTS 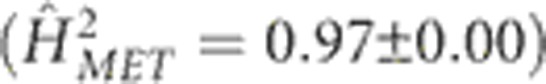 and DTA
and DTA 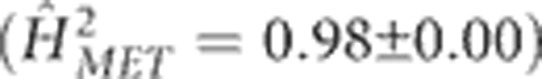 , reflecting the importance of genotypic main effects coupled with extensive replication across environments in the MET. Averaged across all environments, 39%, 35% and 51% of the overall response for DTS, DTA and ASI, respectively, occurred by g2 (compared with a change of 10% per generation expected if the response were equal across generations). This non-linear response to selection over generations was observed consistently across environments, as indicated by a significant second-order polynomial relationship (Wald test: P<0.001) between these flowering time traits and generations of selection within each environment. Comparing DTS and DTA, the response associated with DTS was somewhat larger in its overall magnitude (~20 days) and curvilinearity (−3.89[gen]+0.211[gen2]) than it was for DTA (~15 days; × 2.68[gen]+0.126[gen2]). This difference was reflected in the polynomial response profiles of ASI (a function of the two traits) in most environments, which also indicated that the larger, early-generational changes in DTS were the most important contribution to the reduction of ASI as a response to selection.
, reflecting the importance of genotypic main effects coupled with extensive replication across environments in the MET. Averaged across all environments, 39%, 35% and 51% of the overall response for DTS, DTA and ASI, respectively, occurred by g2 (compared with a change of 10% per generation expected if the response were equal across generations). This non-linear response to selection over generations was observed consistently across environments, as indicated by a significant second-order polynomial relationship (Wald test: P<0.001) between these flowering time traits and generations of selection within each environment. Comparing DTS and DTA, the response associated with DTS was somewhat larger in its overall magnitude (~20 days) and curvilinearity (−3.89[gen]+0.211[gen2]) than it was for DTA (~15 days; × 2.68[gen]+0.126[gen2]). This difference was reflected in the polynomial response profiles of ASI (a function of the two traits) in most environments, which also indicated that the larger, early-generational changes in DTS were the most important contribution to the reduction of ASI as a response to selection.
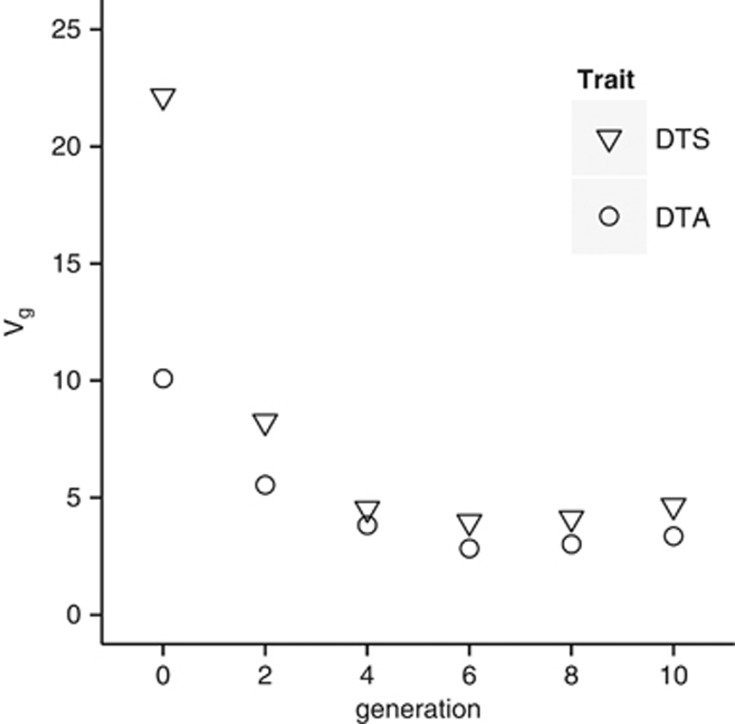
The more pronounced change in the mean of DTS and DTA across the initial generations of selection also coincided with a reduction in genotypic variance among families within those generations (Figure 3). At g4, the variance was reduced by 20–40% of that in g0 (Figure 3), but at g6 the variance inflected upward and showed a continuous although subtle increase across g6 to g10. In summary, under constant directional selection a sustained change in the population mean for DTS and DTA (Figure 1) was observed. This was associated with an initially strong decrease in genotypic variance followed by a plateau, if not an increase, in the variance.
Figure 3.
Generational changes in variance in genotypic main effects (Vg).
On the basis of the generation-slopes model (1), the slopes of the selection responses for DTS and DTA interacted significantly (P<0.001) with environments. The nine environment-specific regression equations for selection response were tested for association with latitude, longitude and altitude. Only the first-order coefficients for DTA exhibited a significant association with latitude 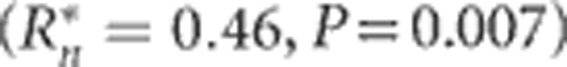 , whereby the selection response tended to be greater at higher rather than lower latitudes (Supplementary Figure S1). The association of DTS and latitude was much weaker
, whereby the selection response tended to be greater at higher rather than lower latitudes (Supplementary Figure S1). The association of DTS and latitude was much weaker 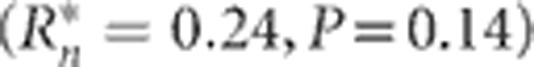 . Inspection of the relationships showed that the response to selection did not increase monotonically with latitude, and the overall response was greatest in the original selection location (IA) and decreased with latitude changes in either direction from IA (Figure 2). There was response data from only one location (WI) at higher latitude than IA, but the response there was lower than in IA.
. Inspection of the relationships showed that the response to selection did not increase monotonically with latitude, and the overall response was greatest in the original selection location (IA) and decreased with latitude changes in either direction from IA (Figure 2). There was response data from only one location (WI) at higher latitude than IA, but the response there was lower than in IA.
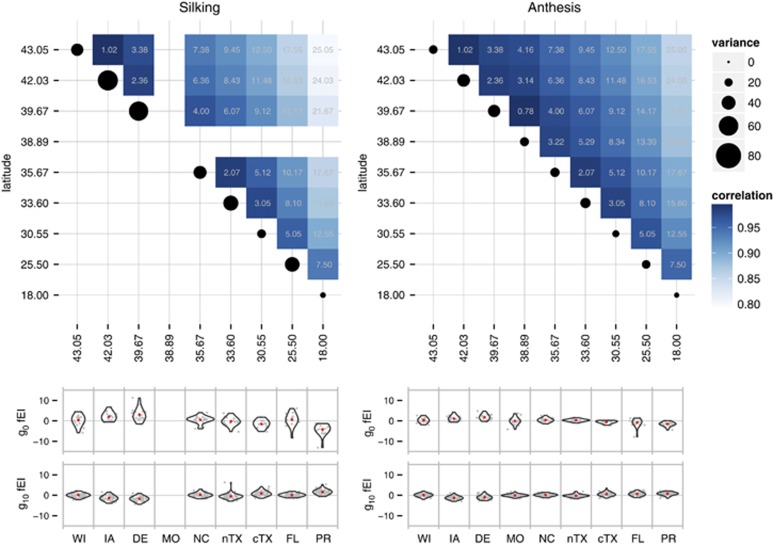
The family model (2) was used to determine whether pairwise correlations among families between environments followed any discernible pattern, including patterns dependent on geography. For both DTS and DTA, the overall fEI variance was significant and best modeled with a latitude-dependent exponential covariance matrix (Supplementary Table S1), implying that variation in DTS and DTA among families was more highly correlated between pairs of environments nearest to each other in terms of latitude (Figure 4a). Taken together, the results stemming from polynomial regression, tests of independence and analysis of variance of DTS and DTA suggested that the variation in genotype-by-environment effects partially followed a geographical trend, with the strength of the selection response radiating latitudinally from IA.
Figure 4.
Family-by-environment interaction variation in flowering time. (a) Tile plots are based on a heterogeneous exponential variance-covariance structure (EXPH VCOV) modeled on the fEI effects (see Materials and methods) with the base correlation estimated as 0.9912 for DTS and 0.9938 for DTA and the exponents (gray text within each tile) calculated as pairwise latitudinal distances. (b) Violin plots showing the density of fEI effects estimated from the family model with an IDV VCOV structure for fEI and subsequently partitioned by generation and environment. Gray points show the fEI effects jittered in the x-direction and red dots show the mean of each distribution. One estimated effect from a family in g0 (fEI estimate: 23.14) was beyond the y-axis scale and is not shown (see Supplementary Figure S3).
Examining the distributions of fEI effects across generations and environments could shed light on the adaptive process and the apparent specificity of adaptation to IA. For this, we fitted the family model (2) assuming the fEI variable as independent and identically distributed (IDV covariance) and then examined the fEI effects per se, partitioned by generations and environments. Under this assumption, there is no expectation for the model to produce trends in the estimates of fEI effects (unlike modeling the data under the exponential-latitude structure, which gleans information from between environments). Visualizing the distributions of these effects revealed that g0 contributed substantially more fEI variance than other generations, but that some variation in fEI effects was still present in g10 (Figure 4). The redistribution of fEI effects across generations and environments (Supplementary Figures S2 and S3) suggested a transition to temperate adaptation that included a footprint of IA-specific adaptation also evident in DE, the environment south of IA with the most similar latitude. In both IA and DE there was a more pronounced change in the fEI effects across generations than in other environments (Supplementary Figure S2). By g10, the median of fEI effects in IA and DE was significantly less than that in any other environment (Wilcoxon signed-rank test, 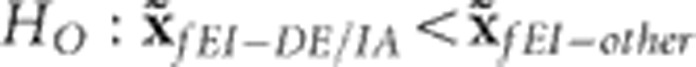 , P<0.001), which included environments broadly classified as having similar temperate climatic conditions (MO, NC and cTX; according to Kottek et al., 2006). The distributions of IA fEI effects were neither significantly less than nor greater than those of DE. In summary, fEI effects were neither uniformly nor randomly distributed across generations and environments: families belonging to g0 and g2 tended to flower later than expected in temperate environments and earlier than expected in tropical environments, whereas families belonging to g8 and g10 tended to exhibit an opposite trend. An excess of high and low family-by-IA and family-by-DE effects in the initial and latter generations, respectively, underlay the stronger response to selection observed in these environments (Supplementary Figure S2).
, P<0.001), which included environments broadly classified as having similar temperate climatic conditions (MO, NC and cTX; according to Kottek et al., 2006). The distributions of IA fEI effects were neither significantly less than nor greater than those of DE. In summary, fEI effects were neither uniformly nor randomly distributed across generations and environments: families belonging to g0 and g2 tended to flower later than expected in temperate environments and earlier than expected in tropical environments, whereas families belonging to g8 and g10 tended to exhibit an opposite trend. An excess of high and low family-by-IA and family-by-DE effects in the initial and latter generations, respectively, underlay the stronger response to selection observed in these environments (Supplementary Figure S2).
Environmental-variable associations with environment and genotype-by-environment interaction effects
We sought to gain insight into environmental variables (Table 1) underlying the variation in environment main effects and gEI effects, but these variables exhibited considerable multicollinearity. More than half of the 45 pairwise environmental variable correlation coefficients had absolute values greater than |r|=0.5. In particular, daylength and temperature variables had strong negative associations (rmin=−0.83). This, in addition to the fact that the number of variables exceeded the number of observations (nine environments) challenged our ability to make inference on specific variables; therefore, we used a modeling procedure that handles high levels of multicollinearity (Zou and Hastie, 2005).
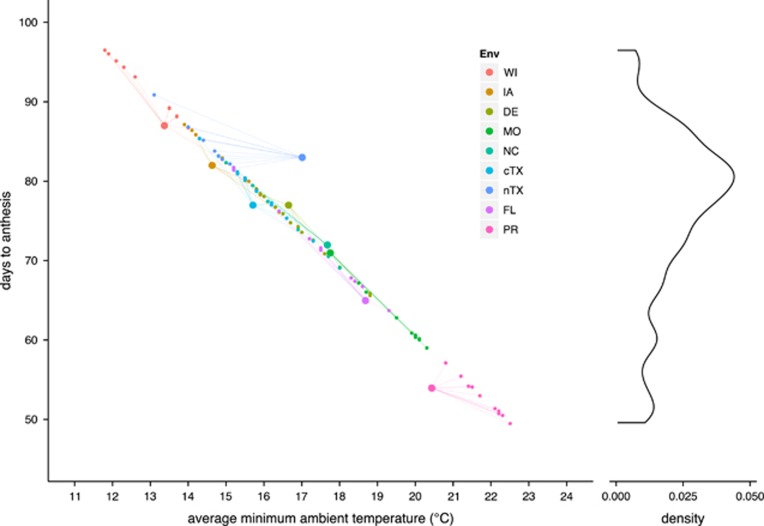
The elastic net models fitted to DTS and DTA environment main effects explained ~98% of the variation. The largest coefficients were negative associations with RHm and the temperature variables (data not shown); however, visual inspection indicated that the relationship with RHm might be unreliable because the association was due to a large deviation in the humidity at one location, Lubbock, TX, which had a much lower RHm than all other environments (Table 1). Excluding RHm, SATn was the next best predictor. Ignoring all other variables, SATn was significantly associated with flowering time 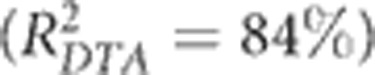 across the 7 °C cline in our MET, and was predictive of a ~4-day increase in flowering time (DTA) with each additional 1 °C in SATn. On the basis of this expectation and historical temperature data from 2001 to 2011, the population was predicted to vary in flowering time (DTA) across years by 8–18 days at a given location (Figure 5).
across the 7 °C cline in our MET, and was predictive of a ~4-day increase in flowering time (DTA) with each additional 1 °C in SATn. On the basis of this expectation and historical temperature data from 2001 to 2011, the population was predicted to vary in flowering time (DTA) across years by 8–18 days at a given location (Figure 5).
Figure 5.
Decadal minimum ambient temperature-associated plasticity in flowering time. The plot to the left shows the observed and predicted relationship between DTA and the average of the minimum ambient temperature. Using the prediction equation 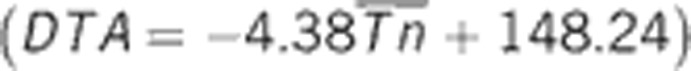 estimated from linear regression of our observed data (large points) on SATn, DTA was predicted (small points) for each environment, using the average minimum ambient temperature
estimated from linear regression of our observed data (large points) on SATn, DTA was predicted (small points) for each environment, using the average minimum ambient temperature  in each year from 2001 to 2011 (averages were calculated based on the environment-specific planting and anthesis dates of our observed data and did not include soil temperature). We acknowledge that some of the prediction is outside the range of the observed data and may be unreliable. The plot to the right shows the marginal density distribution of the predicted DTA values.
in each year from 2001 to 2011 (averages were calculated based on the environment-specific planting and anthesis dates of our observed data and did not include soil temperature). We acknowledge that some of the prediction is outside the range of the observed data and may be unreliable. The plot to the right shows the marginal density distribution of the predicted DTA values.
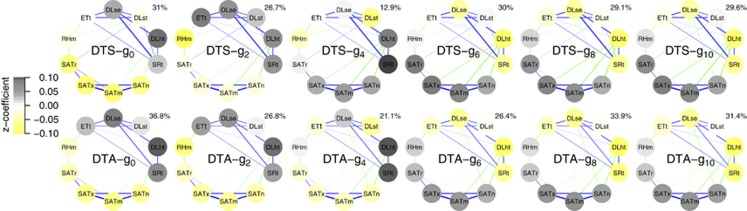
For the analysis of gEI, the environment-specific generation effects estimated by the generation-means model (1) was used for elastic net regression (Figure 6). The percent variation explained by these models was modest, averaging ~30% it is useful to bear in mind that the relatively large amount of unexplained variation could be due to a number of reasons: a low sample size (nine environments), random effects (no true covariate associations), unaccounted for environment variables, opposing fEI effect–environment variable relationships within generations (Supplementary Figure S3) and/or non-linear or interaction effects. The estimated coefficients for the model tended to be largest and exhibited a more consistent direction of change for light (DLme, DLHt and SRt) and temperature variables. The signs of these coefficients switched across g2 to g6, during which the light-related coefficients changed more suddenly than temperature-related variables. Among the light-related variables, DLht was the predictor with the largest coefficient in most generations. The results suggested a transitional adaptation for early flowering time in short-day warm conditions to early flowering time in long-day cool conditions (Figure 6).
Figure 6.
Elastic net estimated effects of environmental variables on generation-by-environment interactions. Network plots are used to represent the magnitude of the estimated effect of each environment variable (node) on flowering time within each generation, the correlation between environment variables (blue line=positive correlation; green line=negative correlation; line thickness represents the relative magnitude of the correlation, with no lines shown for correlations between −0.5 and 0.5), and the percent variation explained by the model (upper right corner of each network). See Materials and methods for a description of the node abbreviations. To allow for comparison among generations and between traits, the dependent variable data were rescaled using a Z-transformation (see Materials and methods).
The statistical approach used above is physiologically naive, with no assumptions about the environmental influence on growth and development embedded in the model. To examine the hypothesis that adaptation to IA was associated with selection for photoperiod insensitivity, we applied a strategy in which the difference in thermal time (GDD) to flowering between long- and short-day environments is used to estimate photoperiod sensitivity—an excess of GDD in long-day environments being indicative of photoperiod sensitivity for maize (Gouesnard et al., 2002; Coles et al., 2010). The fEI effects of GDDTS and GDDTA were correlated with DTS (rGDDTS−DTS=0.93) and DTA (rGDDTA−DTA=0.95), such that adjusting the data for temperature-dependent growth had only a small impact on the variation in genotype-by-environment effects. Nevertheless, g0 flowered later and accumulated significantly more GDDs until flowering in temperate, long-day environments compared with tropical, short-day environments—evidence for long-day sensitivity (Table 2). For the same environmental contrast, g10 exhibited a reverse pattern, flowering earlier and accumulating significantly fewer GDDs until flowering in the temperate, long-day environments (Table 2). The reduction in photoperiod sensitivity in Hallauer's Tusón was supported by comparisons with photoperiod-sensitive and photoperiod-insensitive checks included in the trial (Table 2). Selection appeared to favor the conversion from long-day photoperiod sensitivity in g0 to a requirement for fewer calendar days or GDDs to reach flowering in long-day environments in g10. This interpretation is congruent with the importance of latitude distances in genetic correlations between environments (Figure 4).
Table 2. Mean flowering times for four inbred checks and generations of selection in Tusón in long- and short-day environments.
| Accession |
Calendar days |
Growing degree days |
||||
|---|---|---|---|---|---|---|
| LDa,b | SDa,b | LD-SD | LD | SD | LD-SD | |
| Silkingc | ||||||
| CML254d | 117±3 | 88±0 | 29* | 1760±122 | 1285±22 | 475 |
| Ki14 | 103±3 | 87±1 | 16** | 1515±47 | 1280±32 | 235** |
| g0 | 92±1 | 87±4 | 5 | 1352±22 | 1249±21 | 103* |
| g2 | 83±1 | 81±1 | 2 | 1216±12 | 1181±12 | 35 |
| g4 | 77±0 | 77±0 | 0 | 1123±4 | 1126±2 | −3 |
| g6 | 74±0 | 75±1 | −1 | 1063±6 | 1096±5 | −34* |
| g8 | 72±0 | 74±1 | −2 | 1034±8 | 1081±6 | −47* |
| g10 | 69±1 | 72±2 | −3 | 987±12 | 1044±8 | −58** |
| B97 | 70±1 | 77±4 | −7 | 1011±18 | 1139±54 | −128 |
| B73 | 69±1 | 77±0 | −8** | 990±21 | 1129±3 | −139** |
| Anthesis | ||||||
| CML254d | 109±1 | 88±0 | 21** | 1636±20 | 1279±33 | 357* |
| Ki14 | 87±2 | 82±0 | 5* | 1299±35 | 1198±21 | 101* |
| g0 | 84±0 | 81±1 | 3 | 1255±11 | 1191±8 | 64** |
| g2 | 79±0 | 77±1 | 2 | 1159±5 | 1134±10 | 25 |
| g4 | 75±0 | 74±0 | 1 | 1096±2 | 1091±3 | 5 |
| g6 | 72±0 | 73±1 | −1 | 1041±3 | 1062±4 | −21* |
| g8 | 70±0 | 71±1 | −1 | 1009±5 | 1042±4 | −33** |
| g10 | 68±0 | 70±1 | −2* | 975±7 | 1017±3 | −42** |
| B97 | 68±1 | 72±1 | −4 | 984±19 | 1054±17 | −70* |
| B73 | 69±1 | 75±0 | −6*** | 988±15 | 1110±9 | −122*** |
Abbreviations: LD, long-day environment; SD, short-day environment.
*0.1<P<0.01; **0.01<P<0.001; ***0.001<P<0.0001.
LD environment (WI, IA, DE, MO, NC, nTX); SD environment (FL, PR); cTX was excluded due to uncertainty about LD/SD classification, but the data suggested similarity to a SD environment.
Values correspond to the environment-classified (LD or SD) mean±s.e. of environment-specific effects (calculated using estimates from the generation-means model: for checks: β+ςs=1+κc+κc*ee; for Tusón: β+ςs=2+γg+γg*ee).
No DTS data were available from MO.
No DTS or DTA data were available for CML254 in WI, DE or nTX.
Multivariate responses to selection
Direct conscious selection was applied primarily for early flowering and secondarily for reduced lodging. We used our data set to examine other traits not acknowledged as having been under direct selection: PLH, ERH, TSW and SPL (primary traits) and ASI and PEH (derived traits). Only PLH, ERH, TSW and ASI exhibited significant (P<0.0001) change across generations. For these traits, we tested the null hypothesis that the observed change was greater than the expected correlated change (calculated according to Falconer and Mackay, 1996, p 317) with respect to DTS and DTA (Table 3).
Table 3. Correlated responses to 10 generations of selection for earlier flowering in Tusón, genotypic correlation and heritabilities of unselected traits.
| Trait | ORa | ρORb | Ĥplot2 | σ̂P | DTS(Ĥplot2 : 0.47±0.02) |
DTA(Ĥplot2 : 0.53±0.02) |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| ρ̂Gf(g) | CR | ρ̂OR-CRc | ρ̂Gf(g) | CR | ρ̂OR-CR | |||||
| ASI | −0.47±0.05 | *** | 0.26±0.02 | 2.2 | 0.56±0.05 | −0.94 | NS | 0.11±0.07 | −0.19 | * |
| PLH | −6.11±0.64 | *** | 0.47±0.02 | 18.5 | 0.37±0.06 | −6.93 | NS | 0.34±0.06 | −6.70 | NS |
| ERH | −6.38±0.65 | *** | 0.44±0.02 | 16.3 | 0.41±0.06 | −6.54 | NS | 0.43±0.05 | −7.29 | NS |
| PEH | — | NS | 0.39±0.02 | 13.4 | 0.03±0.07 | — | — | −0.05±0.06 | — | — |
| TSW | −0.17±0.02 | *** | 0.44±0.03 | 1.0 | 0.48±0.06 | −0.49 | NS | 0.37±0.06 | −0.40 | NS |
| SPL | — | NS | 0.44±0.03 | 30.2 | 0.01±0.07 | — | — | −0.00±0.07 | — | — |
Abbreviations: ASI, anthesis-silking interval; ERH, ear height; NS, not significant; PEH, plant height above ear; PLH, plant height; REML, restricted maximum likelihood; SPL, spike length; TSW, tassel weight; *0.1<P<0.01; ***0.001<P<0.0001.
OR: observed response to selection; 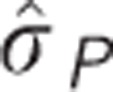 : phenotypic s.d.;
: phenotypic s.d.; 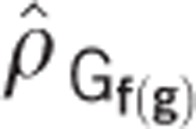 : restricted maximum likelihood estimated genotypic correlation; CR: correlated response to selection.
: restricted maximum likelihood estimated genotypic correlation; CR: correlated response to selection.
P-value based on Wald test; HO: γ>0.
P-value based on t-test with 1 degree of freedom and s.d. equal to that of OR; HO:OR >PR.
The genotypic correlation among families nested within generations, 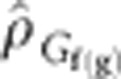 , was estimated between traits. Given that there was random mating among a reasonably large number of selection units (that is, 300–500) per generation,
, was estimated between traits. Given that there was random mating among a reasonably large number of selection units (that is, 300–500) per generation, 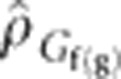 should reflect pleiotropy and disequilibrium at linked loci that started in disequilibrium. The same traits that showed significant change across generations were significantly genetically correlated with DTS or DTA, whereas the traits that did not show significant change across generations were not significantly correlated with DTS or DTA. The direction of observed change was as expected based on the sign of the correlation: early flowering was associated with shorter stature (PLH), lower placement of the primary ear (ERH), smaller tassels (TSW) and a shorter time interval between female and male flowering time (that is, lower values of ASI). These traits exhibited a correlated indirect response to selection that did not differ significantly from expectations (Table 3). The significantly greater response for ASI with respect to DTA could be explained by an indirect response associated with DTS, which was more highly correlated with ASI; no significant difference was found for the observed ASI response with respect to that predicted based on DTS. Therefore, none of the additional traits examined were deduced as having been under direct unconscious selection.
should reflect pleiotropy and disequilibrium at linked loci that started in disequilibrium. The same traits that showed significant change across generations were significantly genetically correlated with DTS or DTA, whereas the traits that did not show significant change across generations were not significantly correlated with DTS or DTA. The direction of observed change was as expected based on the sign of the correlation: early flowering was associated with shorter stature (PLH), lower placement of the primary ear (ERH), smaller tassels (TSW) and a shorter time interval between female and male flowering time (that is, lower values of ASI). These traits exhibited a correlated indirect response to selection that did not differ significantly from expectations (Table 3). The significantly greater response for ASI with respect to DTA could be explained by an indirect response associated with DTS, which was more highly correlated with ASI; no significant difference was found for the observed ASI response with respect to that predicted based on DTS. Therefore, none of the additional traits examined were deduced as having been under direct unconscious selection.
Discussion
The maize landrace Tusón is distributed throughout lowland tropical environments of the Caribbean and Central and South America (Committee on The Preservation of Indigenous Strains of Maize monographs on Races of Maize, 1952). Through a decade of phenotypic mass selection for early flowering (primary selection) and reduced lodging (secondary selection), a population created from multiple accessions of the landrace Tusón was adapted to the temperate summer environment of Ames, IA, by Dr A Hallauer (unpublished data). On the basis of this retrospective examination of phenotypic variation in Hallauer's Tusón across a wide latitudinal range, the population underwent a marked phenotypic transformation, corroborating the inherent diversity within maize and its capacity for environmental adaptation. Among the sample of families from across 10 generations, averaged across nine environments, the timing from planting to silking and anthesis spanned 37 and 27 days, respectively; PLHs ranged from 162 to 267 cm, and ERHs ranged from 73 to 182 cm. The range and magnitude of genotypic variation found here in Hallauer's Tusón exceeded or was comparable to that of the diverse multi-parent NAM population (Buckler et al., 2009; Hung et al., 2012a). The NAM population was designed to maximize the sampling of molecular diversity and has been used to identify numerous loci associated with variation in the aforementioned traits. This indicates that Hallauer's Tusón is genotypically diverse, and that it has good potential for dissecting the genetics of responsive variation for adaptation (Wisser et al., 2011) and to evaluate the goodness-of-fit of realized genetic gain with respect to models describing the genetic architecture of flowering time.
We infer that the genetic basis for adaptation of Hallauer's Tusón is attributable to selection across a fitness landscape structured by evolving genetic architectures of photoperiod-dependent and temperature-dependent flowering time. Relative to other characteristics of maize, variation in photoperiodism is conditioned by a smaller number of loci, with one quantitative trait locus explaining nearly 10% of the variation in photoperiodism (Hung et al., 2012b). This architecture might explain the concomitant, relatively rapid change in the generational mean and variance that initially occurred as the population transitioned toward photoperiod insensitivity (Figure 2; Table 2). The sustained, although subtler, reduction in mean flowering time across generations, coupled with the inflection in generational variance (Figure 2) as the population advanced beyond physiological-model expectations that would implicate photoperiodism (Table 2), suggested an additional adaptive mechanism was at play. Genetically, different allele frequency-dependent models could be invoked to hypothesize on the dynamics of change in this latter phase of selection (for example, with respect to increasing variance: Goodnight, 1988), and ongoing analysis using genotype data will hopefully permit such inference. However, at the level of the whole genotype (individual), variation in temperature-dependent flowering time has been noted for maize across the range of temperatures that occurred in the environments of the present study (Ellis et al., 1992), and analysis of environmental variables associated with gEI in this study revealed adaptation to low temperature as a relevant factor (Figure 6).
A unique aspect of this study was the wide latitudinal range used for replicated evaluation of a genetically defined population. Across these environments, Hallauer's Tusón exhibited large variation in flowering time, the majority of which was associated with the main effects of environment and the main effects of genotype. The overall variation in genotype-by-environment interactions was relatively small, giving rise to a s.d. in flowering time of 1–2 days among families, compared with a 10- and 5-day s.d. associated with environment and genotype main effects, respectively. Nevertheless, changes in genotype-by-environment interactions contributed part of the observed response to selection.
What does this configuration of variation suggest about phenological adaptation of tropical maize and breeding for environmental adaptation? We recognize that the following points are inferred from only a single selected population, so the scope of inference is restricted genetically. Genotypes strongly dictated the relative timing of flowering, giving rise to a high correlation in flowering time among families across a wide geography (that is, the earliest family in one environment was among the earliest set of families in another environment). Yet, the population exhibited a high level of environmental plasticity (variation in environment main effects), such that the absolute timing of flowering between environments varied by as much as 30 days in the MET. The environmental plasticity was directionally associated with the average of the daily minimum temperature from planting to flowering. Our data predict a ~4-day increase in flowering time (DTA) with each additional 1 °C in the average of the minimum temperature, which is remarkably consistent with temperature effects on flowering in other plant species (for example, Fitter et al., 1995; Anderson et al., 2012). Therefore, selection on variation in genotypic main effects to match an environment-specific fitness window, dictated by a temperature-dependent reaction norm (apparently conserved across species), underlies a critical component of flowering time adaptation. On the basis of historical temperature data from 2001 to 2011, we predicted that flowering time could have varied across years by 8–18 days at a given location (Figure 5), suggesting that once a population is deemed adapted in terms of flowering time, interannual temperature differences will still induce substantial variation in flowering time.
The configuration of variation for flowering time offers some insight about the adaptive process underlying phenotypic mass selection for temperate adaptation. Selection was highly effective under a high-G low-gEI configuration, and after 10 generations the population became ‘fully' adapted in terms of flowering time in a temperate environment (referencing the inbred lines B73 and B97 as temperate-adapted checks; Table 2). Given the qualitatively consistent or correlated response to selection across all environments in this study, selection for temperate adaptation seems achievable in practically any environment, with the possible exception of PR where, due to compression of flowering time variation, this selection may not be as effective. This study does not address this conclusion directly although, because a population adapted to only one environment was used. It is not known whether the reduction in fEI variation across the first few generations (Supplementary Figures S2 and S3) would have occurred if selection were performed in another environment. This notion could be addressed by performing parallel selection on a population across a range of environments followed by an evaluation of all of the selected populations in all of the selected environments.
Although the gEI/fEI component of variation was relatively low, it was large enough to suggest that flowering time was partially structured by latitudinal clines in daylength and temperature across the MET environments, as well as to suggest that there was environment-specific adaption possibly restricted to a latitudinal band ranging near IA and DE, with IA having been the location in which selection originally took place. So although it is possible to achieve general adaptation across a wide geography (discussed above), the rate of adaptation will be affected by the relationship between the selection and target environments. Pertaining to Hallauer's Tusón, selecting in another environment besides IA or DE is expected to reduce the IA or DE response to selection for DTS by ~0.5 calendar days or ~10 GDD per generation (excluding PR from consideration, which inflates this effect). This demonstrates that establishing expectations from METs on the distribution of genotype-by-environment interaction in the context of selection can aid in evaluating tradeoffs in choosing environments for phenotypic or genomic selection.
The barrier to temperate adaptation of exotic germplasm is not only flowering time; it is a maladaptive syndrome encompassing a range of phenological, morphological and production characteristics. In the population studied here, it appears that alleles conditioning early flowering time are in coupling phase linkage with alleles conditioning smaller plant architecture and/or the traits are under positive pleiotropic control. Although the data set to which we have applied a statistical approach aiming to deduce the occurrence of direct unconscious selection may be underpowered, failure to reject the null hypothesis is consistent with tightly linked effects found for Ma1 in sorghum (Thurber et al., 2013) and pleiotropic effects found for Ghd7 in rice (Xue et al., 2008). In Hallauer's Tusón, there was secondary selection for reduced lodging, which could also explain some of the changes in PLH and ERH. Nevertheless, PLH and ERH did not exhibit a significant deviation in generational change over the expectation established based on DTS or DTA, the primary trait under selection. Taken together, selection on flowering time alone may be enough to alleviate many of the symptoms of the maladaptive syndrome. In maize, a major source of diversity resides in its tropical environments of origin, whereas a major source of production occurs in its non-native, temperate environments. Selection under one temperate environment may be sufficient to overcome a number of maladaptive symptoms, but the response to selection for adaptation in terms of yield potential will need additional study.
This study highlights the possibility of adapting entirely new maize germplasm pools and putatively unique sources of diversity for US production. This was readily achieved in Hallauer's Tusón by means of conventional phenotypic selection, which today could be sped by the use of marker-assisted selection procedures. This study also contributes as a reference evidencing the potential for genetic adaptation to help mitigate vulnerabilities of crops to the effects of climate change and other stressors (for example, Craufurd and Wheeler, 2009; Butler and Huybers, 2012).
Data archiving
Data available from the Dryad Digital Repository: doi:10.5061/dryad.8f64f.
Acknowledgments
This project was supported by Agriculture and Food Research Initiative Competitive Grant No. 2011-67003-30342 from the USDA National Institute of Food and Agriculture (Agriculture and Natural Resources Science for Climate Variability and Change Program), Hatch Act funds and the US Department of Agriculture-Agricultural Research Service. The investigators also wish to thank students and technical support staff that assisted with collection of this data.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Aitken Y. (1977). Evaluation of maturity genotype-cimate interactions in maize (Zea mays L.). Zeitschrift für Pflanzenzüchtung 78: 216–237. [Google Scholar]
- Anderson E, Brown WL. (1952). The history of the common maize varieties of the United States corn belt. Agric Hist 26: 2–8. [Google Scholar]
- Anderson JT, Inouye DW, McKinney AM, Colautti RI, Mitchell-Olds T. (2012). Phenotypic plasticity and adaptive evolution contribute to advancing flowering phenology in response to climate change. Proc Biol Sci 279: 3843–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apiolaza LA., Gilmour AR, Garrick DJ. (2000). Variance modelling of longitudinal height data from a Pinus radiata progeny test. Can J Res 30: 645–654. [Google Scholar]
- Betŕan FJ, Mayfield K, Isakeit T, Menz M. (2003). Breeding maize exotic germplasm. In: Lamkey KR, Michael L (eds). Plant Breeding: the Arnel R. Hallauer International Symposium. Blackwell Publishing: Oxford, UK. pp 352–367. [Google Scholar]
- Bonhomme R, Derieux M, Edmeades GO. (1994). Flowering of diverse maize cultivars in relation to temperature and photoperiod in multilocation field trials. Crop science 34: 156–164. [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C et al. (2009). The genetic architecture of maize flowering time. Science 325: 714–718. [DOI] [PubMed] [Google Scholar]
- Butler EE, Huybers P: Adaptation of US maize to temperature variations. Nat Clim Change,. (2012) 3: 68–72 [Google Scholar]
- Chardon F, Moreau L, Falque M, Joets J, Decousset L, Murigneux A et al. (2004). Genetic architecture of flowering time in maize as inferred from quantitative trait loci meta-analysis and synteny conservation with the rice genome. Genetics 168: 2169–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles ND, McMullen MD, Balint-Kurti PJ, Pratt RC, Holland JB. (2010). Genetic control of photoperiod sensitivity in maize revealed by joint multiple population analysis. Genetics 184: 799–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Committee on The Preservation of Indigenous Strains of Maize monographs on Races of Maize. (1952–1963). National Academy of Sciences—National Research, Washington, D.C. Pub. N 1136 (Venezuela); Pub. N 510 (Columbia); Pub. N 511 (Central America); Pub. N 593 (Brazil and other Eastern South American Countries); Pub. N 747 (Bolivia); Pub. N 792 (West Indies); Pub. N 847 (Chile); Pub. N 915 (Peru); Pub. N 975 (Ecuador); Pub. N 453 (Cuba); and Races of Maize in Mexico by the Bussey Institute, Harvard University, MA.
- Craufurd PQ, Wheeler TR. (2009). Climate change and the flowering time of annual crops. J Exp Bot 60: 2529–2539. [DOI] [PubMed] [Google Scholar]
- Crossa J, Vargas M, van Eeuwijk F a, Jiang C, Edmeades GO, Hoisington D. (1999). Interpreting genotype × environment interaction in tropical maize using linked molecular markers and environmental covariables. Theor Appl Genet 99: 611–625. [DOI] [PubMed] [Google Scholar]
- Ellis RH, Summerfield RJ, Edmeades GO, Roberts EH. (1992). Photoperiod, temperature, and the interval from sowing to tassel initiation in diverse cultivars of maize. Crop Sci 32: 1225. [Google Scholar]
- Epinat-Le Signor C, Dousse S, Lorgeou J, Denis J-B, Bonhomme R, Carolo P et al. (2001). Interpretation of genotype X environment interactions for early maize hybrids over 12 Years. Crop Sci 41: 663–669. [Google Scholar]
- Falconer DS, Mackay TFC. (1996). Introduction to Quantitative Genetics 4th edn. Pearson Education Limited: Essex, England. [Google Scholar]
- Fitter AH, Fitter RSR, Harris ITB, Williamson MH. (1995). Relationships between first flowering date and temperature in the flora of a locality in central England. Funct Ecol 9: 55–60. [Google Scholar]
- Forsythe WC, Rykiel EJ, Stahl RS, Wu H, Schoolfield RM. (1995). A model comparison for daylength as a function of latitude and day of year. Ecol Modell 80: 87–95. [Google Scholar]
- Garner WW, Allard HA. (1923). Further studies in photoperiodism, the response of the plant to relative length of day and night. J Agric Res XXIII: 871–920. [DOI] [PubMed] [Google Scholar]
- Gilmour AR, Gogel BJ, Cullis BR, Thompson R. (2009). ASReml User Guide Release 3.0.
- Godfray HCJ, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF et al. (2010). Food security: the challenge of feeding 9 billion people. Science 327: 812–818. [DOI] [PubMed] [Google Scholar]
- Goodman MM. (1999). Broadening the genetic diversity in maize breeding by use of exotic germplasm. In: Coors JG, Pandey S (eds). The Genetics and Exploitation of Heterosis in Crops. CSSA: Madison, WI, USA. pp 139–148. [Google Scholar]
- Goodnight CT. (1988). Epistasis and the effect of founder events on the additive genetic variance. Evolution 42: 441–454. [DOI] [PubMed] [Google Scholar]
- Gouesnard B, Rebourg C, Welcker C, Charcosset A. (2002). Analysis of photoperiod sensitivity within a collection of tropical maize. Genet Resour Crop Evol 49: 471–481. [Google Scholar]
- Hallauer AR, Carena MJ, Miranda Filho JB. (2010). Quantitative Genetics in Maize Breeding. Springer. [Google Scholar]
- Holland JB, Goodman MM, Castillo-Gonzalez F. (1996). Identification of agronomically superior latin american maize accessions via multi-stage evaluations. Crop Sci 36: 778–784. [Google Scholar]
- Holland JB, Nyquist WE, Cervantes-Martínez CT. (2003). Estimating and interpreting heritability for plant breeding: an update. In: Janick J (ed). Plant Breeding Reviews Vol 22, John Wiley and Sons Inc.: Hoboken, NJ, USA. pp 9–112. [Google Scholar]
- Holley RN, Goodman MM. (1988). Yield potential of tropical hybrid maize derivatives. Crop Sci 28: 213–218. [Google Scholar]
- Hung H-Y, Browne C, Guill K, Coles N, Eller M, Garcia A et al. The relationship between parental genetic or phenotypic divergence and progeny variation in the maize nested association mapping population. Heredity,. (2012. a) 108: 490–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung H-Y, Shannon LM, Tian F, Bradbury PJ, Chen C, Flint-Garcia SA et al. (2012. b). ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc Natl Acad Sci 109: E1913–E1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C, Müller AE. (2009). Flowering time control and applications in plant breeding. Trends Plant Sci 14: 563–573. [DOI] [PubMed] [Google Scholar]
- Kiniry JR, Ritchie JT, Musser RL. (1983). Dynamic nature of the photoperiod response in maize. Agron J 75: 700–703. [Google Scholar]
- Kottek M, Grieser J, Beck C, Rudolf B, Rubel F. (2006). World Map of the Köppen-Geiger climate classification updated. Meteorol. Zeitschrift 15: 259–263. [Google Scholar]
- Kuleshov NN. (1933). World's diversity of phenotypes of maize. Agron J 25: 688–700. [Google Scholar]
- Liu K, Goodman M, Muse S, Smith JS, Buckler E, Doebley J. (2003). Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics 165: 2117–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malosetti M, Ribaut J-M, van Eeuwijk FA. (2013). The statistical analysis of multi-environment data: modeling genotype-by-environment interaction and its genetic basis. Front Physiol 4: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manel S, Gugerli F, Thuiller W, Alvarez N, Legendre P, Holderegger R et al. (2012). Broad-scale adaptive genetic variation in alpine plants is driven by temperature and precipitation. Mol Ecol 21: 3729–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melhus IE. (1948). Exploring the maize germplasm of the tropics. Rep Hybrid Corn Ind Res Conf 3: 7–19. [Google Scholar]
- Oakey H, Verbyla A, Pitchford W, Cullis B, Kuchel H. (2006). Joint modeling of additive and non-additive genetic line effects in single field trials. Theor Appl Genet 113: 809–819. [DOI] [PubMed] [Google Scholar]
- Orr HA. (2005). The genetic theory of adaptation: a brief history. Nat Rev Genet 6: 119–127. [DOI] [PubMed] [Google Scholar]
- Piepho HP, Williams ER, Fleck M. (2006). A note on the analysis of designed experiments with complex treatment structure. HortScience 41: 446–452. [Google Scholar]
- Pollak L. (1997). United States LAMP Final Report. In: Salhuana W., Sevilla R., Eberhart SA (eds). Latin American Maize Project Final Report. Pioneer Hi-Bred Int. Des Moines, IA, USA. Special Publication. [Google Scholar]
- Pollak LM. (2003). The history and success of the public-private project on germplasm enhancement of maize (GEM). Adv Agron 78: 45–87. [Google Scholar]
- Quinby JR. (1967). The maturity genes of sorghum. Adv Agron 19: 267–305. [Google Scholar]
- Reymond M, Muller B, Charcosset A, Tardieu F. (2003). Combining quantitative trait loci analysis and an ecophysiological model to analyze the genetic variability of the responses of maize leaf growth to temperature and water deficit. Plant Physiol 131: 664–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz Corral JA, Durán Puga N, Sánchez González JDJ, Ron Parra J, González Eguiarte DR, Holland JB et al. (2008). Climatic adaptation and ecological descriptors of 42 mexican maize races. Crop Sci 48: 1502–1512. [Google Scholar]
- Székely GJ, Rizzo ML. (2013). The distance correlation t-test of independence in high dimension. J Multivar Anal 117: 193–213. [Google Scholar]
- Taiz L, Zeiger E. (2006). Plant Physiology 4th edn. Sinauer Associates Inc.: Sunderland, MA, USA. [Google Scholar]
- Tanksley SD. (1997). Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277: 1063–1066. [DOI] [PubMed] [Google Scholar]
- Tarter JA, Holland JB. (2006). Gains from selection during the development of semiexotic inbred lines from Latin American maize accessions. Maydica 51: 15–23. [Google Scholar]
- Tester M, Langridge P. (2010). Breeding technologies to increase crop production in a changing world. Science 327: 818–822. [DOI] [PubMed] [Google Scholar]
- Thurber CS, Ma JM, Higgins RH, Brown PJ. (2013). Retrospective genomic analysis of sorghum adaptation to temperate-zone grain production. Genome Biol 327: R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer AF. (1999). Background of U.S. hybrid corn. Crop Sci 39: 601–626. [Google Scholar]
- Vigouroux Y, Glaubitz JC, Matsuoka Y, Goodman MM, Sánchez G J, Doebley J. (2008). Population structure and genetic diversity of New World maize races assessed by DNA microsatellites. Am J Bot 95: 1240–1253. [DOI] [PubMed] [Google Scholar]
- Warrington IJ, Kanemasu ET. (1983). Corn growth response to temperature and photoperiod I. seedling emergence, tassel initiation, and anthesis. Agron J 75: 749–754. [Google Scholar]
- Wellhausen EJ. (1965). Exotic germ plasm for improvement of corn belt maize. In: Proceedings of the 20th Annual Hybrid Corn Industry-Research Conference, Chicago, IL, USA. pp 31–45.
- Wisser RJ, Balint-Kurti PJ, Holland JB. (2011). A novel genetic framework for studying response to artificial selection. Plant Genet Resour 9: 281–283. [Google Scholar]
- Wolfinger RD. (1993). Covariance structure selection in general mixed models. Commun Stat Simul Comput 22: 1079–1106. [Google Scholar]
- Wright SI, Bi IV, Schroeder SG, Yamasaki M, Doebley JF, McMullen MD et al. (2005). The effects of artificial selection on the maize genome. Science 308: 1310–1314. [DOI] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40: 761–767. [DOI] [PubMed] [Google Scholar]
- Yano M, Kojima S, Takahashi Y, Hongxuan L, Sasaki T. (2001). Genetic control of flowering time in rice, a short-day plant. Plant Physiol 127: 1425–1429. [PMC free article] [PubMed] [Google Scholar]
- Zou H, Hastie T. (2005). Regularization and variable selection via the elastic net. J R Stat Soc Ser B 67: 301–320. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.