Abstract
Background
Schistosomiasis remains one of the most common endemic parasitic diseases affecting over 230 million people worlwide. Schistosoma mansoni is the main species causing intestinal and hepatic schistosomiasis and the fresh water pulmonate snails of the genus Biomphalaria are best known for their role as intermediate hosts of the parasite. The development of new molecular monitoring assays for large-scale screening of snails from transmission sites to detect the presence of schistosomes is an important point to consider for snail control interventions related to schistosomiasis elimination. Our work was focussed on developing and evaluating a new LAMP assay combined with a simple DNA extraction method to detect S. mansoni in experimentally infected snails as a diagnostic tool for field conditions.
Methodology/Principal findings
A LAMP assay using a set of six primers targeting a sequence of S. mansoni ribosomal intergenic spacer 28S-18S rRNA was designed. The detection limit of the LAMP assay was 0.1 fg of S. mansoni DNA at 63°C for 50 minutes. LAMP was evaluated by examining S. mansoni DNA in B. glabrata snails experimentally exposed to miracidia at different times post-exposure: early prepatent period (before cercarial shedding), light infections (snails exposed to a low number of miracidia) and detection of infected snails in pooled samples (within a group of uninfected snails). DNA for LAMP assays was obtained by using a commercial DNA extraction kit or a simple heat NaOH extraction method. We detected S. mansoni DNA in all groups of snails by using no complicated requirement procedure for DNA obtaining.
Conclusions/Significance
Our LAMP assay, named Biompha-LAMP, is specific, sensitive, rapid and potentially adaptable as a cost-effective method for screening of intermediate hosts infected with S. mansoni in both individual snails and pooled samples. The assay could be suitable for large-scale field surveys for schistosomes control campaigns in endemic areas.
Author Summary
Schistosoma mansoni is the main species causing intestinal and hepatic schistosomiasis worldwide and the snails of the genus Biomphalaria are best known for their role as intermediate hosts of the parasite. Molecular xenomonitoring for large-scale screening of snails from transmission sites to detect the presence of schistosomes is an important point to consider for snail control interventions related to schistosomiasis elimination. In our study, we have developed a new simple rapid LAMP assay to detect S. mansoni in Biomphalaria glabrata snails under different situations of infection: early prepatent period, light or low-grade infections and in snails pooled samples. Besides, a simple and rapid method for DNA extraction from snails' tissues was successfully used. This LAMP assay (named Biompha-LAMP) could be potentially useful for large-scale screening in searching infected snails with S. mansoni in field applicable conditions.
Introduction
Human schistosomiasis continues to be one of the most important neglected tropical diseases affecting over 230 million people worldwide. Schistosoma mansoni is the main species causing hepatic and intestinal schistosomiasis in Sub-Saharan Africa and solely in South America [1–4]. The fresh water snails of genus Biomphalaria act as the parasite´s intermediate host which are able to produce a constant output of hundreds or even thousands of cercariae for months [5]. The cercarial emission from infected snails is the route of infection for humans including those who may have been successfully treated in a control program. By themselves, preventive chemotherapy campaigns using mass drug administration have shown not to limit transmission in high-risk areas [6, 7]. The distribution and prevalence of the disease are determined, to a large extent, by the presence or absence of Biomphalaria snails [8]. In addition to health education, safe water supplies, adequate sanitation and environmental management, a snail control would also reduce transmission of human infection and is necessary for a schistosomiasis comprehensive control program [7, 9]. Among different known monitoring approaches for surveillance of active sites for snail-to-human transmission, the detection of cercarial shedding by infected snails after exposure of the specimens to light during 1-24h has been the most traditionally and widely method used [10]. This method has significant limitations to detect the parasite, especially during the prepatent period of snail infections (non-shedding), in low-grade infections and also due to the aborted development of schistosomes in snails [11]. The dissection of snails to detect sporocysts of schistosomes during the prepatent period is a hard task and often unsuccessful because of its tiny size and also, the lack of experienced personnel for accurate identification of infection [12]. Besides, differentiation in cercariae morphology between S. mansoni and other trematodes species sometimes may be difficult [13]. All this produces an underestimation of the true prevalence and incidence of infection by schistosomes in snail populations.
To overcome these limitations in detecting infected snails, molecular xenomonitoring (the detection of parasite DNA or RNA in snails using molecular-based assays) is a great alternative allowing analysis of pooled snails samples and also offering greater efficiency and sensitivity than dissection of snail tissues, especially when large numbers of specimens must be examined. In recent years, several molecular monitoring polymerase chain reaction (PCR)-based assays have been developed for S. mansoni detection in snails, such us conventional PCR [14, 15], nested-PCR [16], multiplex-PCR [17] and real time-PCR [18]. All these studies have demonstrated better results in detecting the parasite than conventional methods but the lack of resources is a major barrier to apply in endemic countries for schistosomiasis because of the highly techniques requirements and skilled personnel. As a potential alternative for molecular xenomonitoring snail sampling adaptable to field conditions could be the loop-mediated isothermal amplification (LAMP) assay [19], a powerful simple and rapid nucleic acid amplification technique with a wide range of possible applications including point-of-care testing in resource-poor settings (such in developing countries) and rapid testing of environmental samples [20].
Several LAMP-based assays have already been reported for the detection of schistosomal DNA in samples from animals in laboratory settings, such as S. japonicum in rabbits [21, 22] or S. mansoni in mice [23, 24] as well as from both human urine and serum samples for detection of S. haematobium [25] and S. japonicum [26], respectively. Additionally, other LAMP assays have been described in order to provide a rapid and effective method to detect schistosomal DNA in field-collected intermediate host snails, including S. japonicum [27], S. haematobium and S. mansoni [10] and potentially later adaptation in a large-scale screening of snails pooled samples to be used as method for snails control [28, 29]. The "development of inexpensive, field-applicable diagnostic assays for the large-scale screening of individual or pooled snails from transmission sites to detect the presence of schistosomes" has been listed as an important point to consider in an agenda for snail control interventions related to schistosomiasis elimination [30].
However, for S. mansoni, no LAMP assay has been evaluated yet to. In our study, we have developed a new simple rapid LAMP assay to detect S. mansoni in Biomphalaria glabrata snails under different situations of infection: early prepatent period, light or low-grade infections and in snails pooled samples. Besides, a simple, rapid and economic method for DNA extraction from snails´ tissues was successfully used. The LAMP assay presented here could be potentially useful for large-scale screening in searching infected snails with S. mansoni in field applicable conditions.
Methods
Ethics statement
Animal procedures complied with the Spanish (Real Decreto RD53/2013) and the European Union (European Directive 2010/63/EU) regulations on animal experimentation for the protection and human use of laboratory animals. Experiments were conducted at the accredited Animal Experimentation Facility of the University of Salamanca (Register number: PAE/SA/001). Procedures were approved by the Ethics Committee of the University of Salamanca (protocol approval number 48531).
S. mansoni maintenance and snails infections
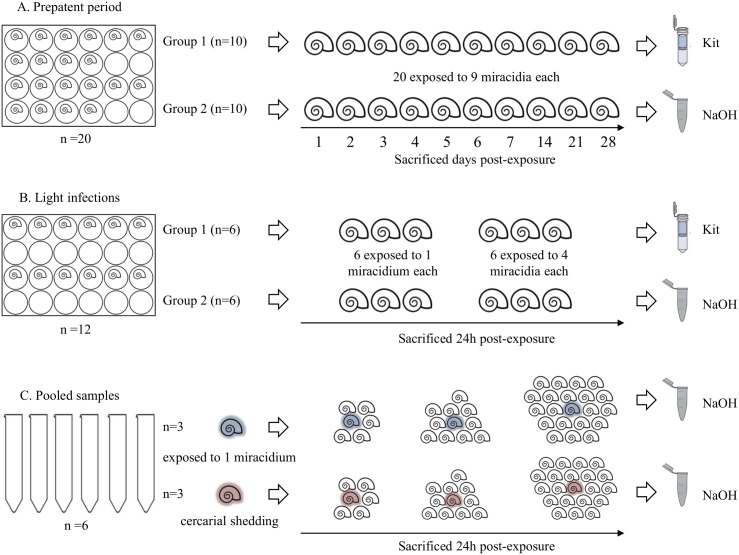
S. mansoni (LE strain) was maintained routinely by passage through Biomphalaria glabrata snails and 4-to-6-week old male CD1 mice (Charles River, Criffa S.A., Barcelona, Spain) at University of Salamanca. Eight weeks after infection mice were humanely euthanized by intraperitoneal injection of sodium pentobarbital (60 mg/kg) plus heparin (2 IU/mL) and the liver was removed and minced to obtain eggs. Purified eggs were put into water to hatch the miracidia for experimental infection of snails. Snails were exposed individually to 9 miracidia in 6-well plates. After 30–40 days, cercariae were shed from infected snails by exposure to light within 60 min at room temperature. Using this routine procedure, a number of B. glabrata snails were exposed to different numbers of miracidia in order to detect subsequently S. mansoni DNA at different times post-exposure (p.e.) simulating different conditions such as: i) detection of infected snails in the early prepatent period (before cercarial shedding), ii) detection of light infections (snails exposed to a low number of miracidia) and iii) detection of infected snails in pooled samples (within a broad group of snails), as described below. A scheme of the different snails infections carried out in the study is showed in fig 1.
Fig 1. Scheme of experimentally infected snails in this study.
(A) Prepatent period. (B) Light infections. (C) Pooled samples. Kit and NaOH, indicate the commercial kit or the heat NaOH extraction method for DNA obtaining, respectively. Number or snails used in each infection, exposition of snails to miracidium/miracidia, snails showing cercarial shedding and sacrificed days post-exposure are indicated by a text in figure.
Prepatent period (Fig 1A)
A total of twenty snails (n = 20) were individually placed into a 24-well polystyrene plate and exposed to 9 miracidia each; next, they were divided into two groups of 10 snails: group 1 and group 2. A specimen from each group was sacrificed every day during the first week (days 1–7), and later on day 14, 21 and 28 p.e.. After crushing, soft tissues from snails belonging to group 1 were immediately extracted from the shells by using a fine needle and storage individually at -20°C until DNA extraction. Snails from group 2 were sacrificed by immersion in pure ethanol and then preserved for a week until extraction of the soft tissues from the shells by using a fine needle; then, the excess ethanol solution was removed by drying on filter paper and promptly processed for DNA obtaining.
Light infections (Fig 1B)
A total of twelve snails (n = 12) were also individually placed into a 24-well polystyrene plate and divided into two groups (group 1 and group 2) of 6 specimens each. Three snails of each group of 6 specimens were exposed to a single miracidium and the other 3 were exposed to 4 miracidia. All specimens were sacrificed at 24 h p.e.. Snails from group 1 were crushed and soft tissues were immediately extracted from the shells by using a fine needle and preserved at -20°C until DNA extraction. Snails from group 2 were sacrificed by immersion in ethanol and subsequently preserved for a week until extraction of the soft tissues from the shells by using a fine needle; then, the excess ethanol solution was removed by drying on filter paper and promptly processed for DNA obtaining.
Pooled samples (Fig 1C)
A total of 6 snails -3 previously exposed to a single miracidium and 3 previously exposed to 9 miracidia and also showing cercarial shedding after 40 days p.e.- were placed individually into 50 mL sterile conical tubes together with 5, 10 and 20 uninfected snails, respectively. After 24h, all the pooled samples were crushed for soft tissues extraction by using a fine needle and promptly processed.
DNA obtaining
Snails and miracidia DNA extraction
DNA from snails for LAMP assays was obtained by using two different methods: i) a commercial DNA extraction kit (NucleoSpin Tissue; Macherey-Nagel, Germany) for snails' tissues preserved at -20°C following the manufacturers' instructions and, ii) a heat sodium hydroxide (NaOH) extraction method [28] for snails´ tissues preserved in absolute ethanol. Briefly, in the heat NaOH extraction method, for processing individual snails a volume of 200 μL of a 50 mM NaOH solution was added and then heated at 95°C for 30 min. Subsequently, the tubes were centrifugated at 5000 rpm for 5 min and a volume of 50 μL of supernatant was recovered in a new clean tube and mixed with an equal volume of a 1 M Tris-HCl (pH 8.0) solution. When preparing pooled samples for DNA extraction, a greater volume of NaOH was used (10 mL instead 200 μL) due to the larger amount of snails' tissues to be digested. Each new solution thus obtained was stored at -20°C until further use as template in LAMP assays. DNA from two separate miracidia to be used as template control for LAMP assays was obtained by the two different mentioned methods. DNA from 2 uninfected snails and 2 snails with confirmed cercarial shedding to be used as additional negative and positive controls, respectively, were also obtained by the two different DNA extraction methods used.
Parasites DNA extraction
DNA from S. mansoni frozen adult worms available in our laboratory was extracted using the NucleoSpin Tissue kit (Macherey-Nagel, Germany) according to the manufacturers' instructions and prepared to a final concentration of 0.5 ng/μL. Then, DNA was 10-fold serially diluted (ranging from 0.05 ng/μL to 0.5 atg/μL) and stored at -20°C until further use. DNA thus prepared was used as a template control in LAMP reactions as well as for assessing sensitivity.
To determine the specificity of LAMP assay, DNA from other several trematodes requiring snails as intermediate hosts in their life cycle with vertebrates were used as heterogeneous control samples, including Schistosoma haematobium and S. intercalatum (affecting people), Schistosoma bovis and Dicrocoelium dendriticum (affecting cattle) and Schistosoma japonicum and Fasciola hepatica (affecting people and/or cattle). These DNA samples were also diluted to a final concentration of 0.5 ng/μL and kept frozen until use.
S. mansoni LAMP primer design
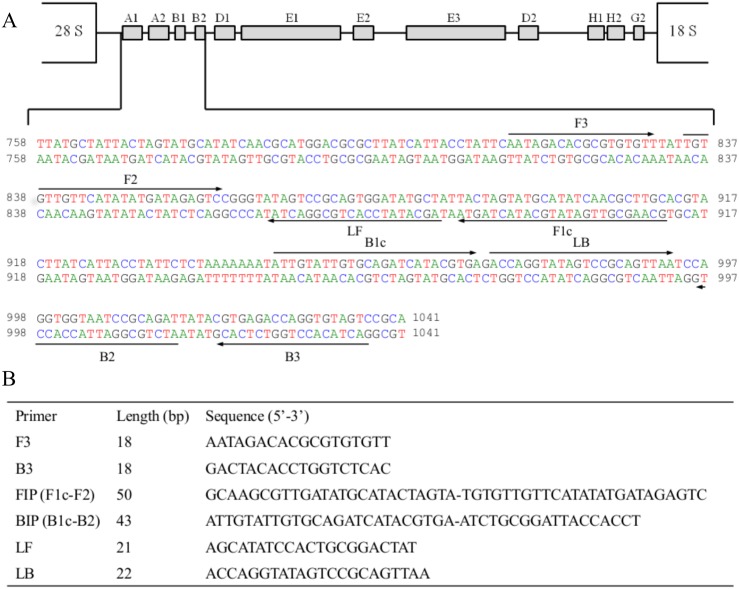
A 3022 base pair (bp) sequence corresponding to the ribosomal intergenic spacer 28S-18S ribosomal RNA gene [31] was retrieved from GenBank (Accesion no. AJ223842) for further studies. Specificity for S. mansoni was tested in silico through BLAST alignment analysis [32], as well as searches and comparisons in available online genomes databases for Schistosoma spp. (e.g. SchistoDB; http://schistodb.net/schisto/). A 284 bp unique region for S. mansoni was selected and used for LAMP primer design using the Primer Explorer v4 software (http://primerexplorer.jp/e/). A set of six primers -including a forward outer primer (F3), a reverse outer primer (B3), a forward inner primer (FIP), a backward inner primer (BIP), a loop forward primer (LF) and a loop backward primer (LB)- was selected based on the criteria described in “A guide to LAMP primer designing” (http://primerexplorer.jp/e/v4_manual/index.html). LAMP primers sequences and their positions in the selected target for S. mansoni are shown in fig 2.
Fig 2. LAMP primer set targeting the selected sequence (GenBank Accesion. No. AJ223842) for ribosomal intergenic spacer 28S-18S ribosomal RNA gene S. mansoni DNA region amplification.
(A) The location of the LAMP primers within the selected sequence is shown. Arrows indicate the direction of extension. (B) Sequence of LAMP primers: F3, forward outer primer; B3, reverse outer primer; FIP, forward inner primer (comprising F1c and F2 sequences); BIP, reverse inner primer (comprising B1c and B2 sequences); LF, loop forward primer; LB, loop reverse primer.
PCR using outer primers F3 and B3
The outer LAMP primer pair (F3 and B3) was initially tested to verify the correct amplification of the selected target of S. mansoni DNA by a touchdown PCR (TD-PCR). Briefly, the PCR F3-B3 assay was conducted in 25 μL reaction mixture containing 2.5 μL of 10x buffer, 1.5 μL of 25 mmol/L MgCl2, 2.5 μL of 2.5 mmol/L dNTPs, 0.5 μL of 100 pmol/L F3 and B3, 2 U Taq-polymerase and 2 μL (10 ng) of DNA template. Initial denaturation was conducted at 94°C for 1 min, followed by a touchdown program for 15 cycles with successive annealing temperature decrements of 1.0°C (from 57°C to 52°C) every 2 cycles. Subsequently, the specificity and sensitivity of PCR F3-B3 were tested using 2 μL of heterogeneous DNA samples included in the study and 2 μL of S. mansoni DNA 10-fold serially diluted, respectively, prepared as mentioned above. Negative controls (ultrapure water) were always also included. The PCR products (3–5 μL) were subjected to 1.5% agarose gel electrophoresis stained with ethidium bromide and visualized under UV light.
Setting up LAMP assay
The LAMP reactions were carried out in a final volume of 25 μL containing 1.6 μM of each of the FIP and BIP primers, 0.2 μM of the F3 and B3 primers, 0.4 μM of the LF and LB primers, 1x Isothermal Amplification Buffer -20 mM Tris-HCl (pH 8.8), 50 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Tween20- (New England Biolabs, UK), 1 M betaine, 6 mM supplementary MgSO4 and 8 U of Bst 2.0 DNA polymerase with 2 μL of template DNA. To establish the optimal reaction time for LAMP assay amplifying the minimum amount of S. mansoni DNA using the set of six primers, three different assays were carried out adding different amounts of S. mansoni DNA (1 ng, 1 pg and 1 fg, respectively) and varying the incubation time at 63°C for 10 min, 20 min, 30 min, 40 min, 50 min and 60 min, followed by 5–10 min to 80°C to terminate the reaction. The optimal reaction time was determinate and used in all the following tests. The amplification results were visually detected by adding 2 μL of 1:10 diluted 10.000X concentration fluorescent dye SYBR Green I (Invitrogen) an also on a 1.5% agarose gel electrophoresis stained with ethidium bromide. LAMP sensitivity and specificity were determinate using genomic DNA from S. mansoni 10-fold serially diluted and other heterogeneous DNA samples from other parasites, respectively, as mentioned above.
Results
PCR F3-B3 sensitivity and specificity
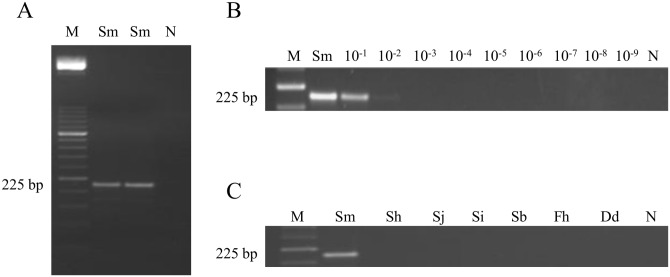
The in silico 225 bp expected amplicon was successfully amplified when using PCR F3-B3 (Fig 3A). The minimum amount of DNA detectable was 0.01 ng (Fig 3B). When DNA samples from other parasites included in the study were subjected to this PCR assay, amplicons were never obtained (Fig 3C).
Fig 3. PCR verification, detection limit and specificity using outer primers F3 and B3.
(A) PCR verification of expected 225 bp target length amplicon. Lane M, 50 bp DNA ladder (Molecular weight marker XIII, Roche); lanes Sm, S. mansoni DNA (1 ng); lane N, negative control (no DNA template). (B) Detection limit of PCR. Lane M, 50 bp DNA ladder (Molecular weight marker XIII, Roche); lane Sm: S. mansoni DNA (1 ng); lanes 10−1–10−9: 10-fold serially dilutions of S. mansoni DNA; lane N, negative control (no DNA template). (C) Specificity of PCR. Lane M, 50 bp DNA ladder (Molecular weight marker XIII, Roche); lanes Sm, Sh, Sj, Si, Sb, Fh, Dd, S. mansoni, S. haematobium, S. japonicum, S. intercalatum, S. bovis, Fasciola hepatica and Dicrocoelium dendriticum DNA samples (1 ng/each), respectively; lane N, negative control (no DNA template).
Setting up the LAMP assay: Biompha-LAMP
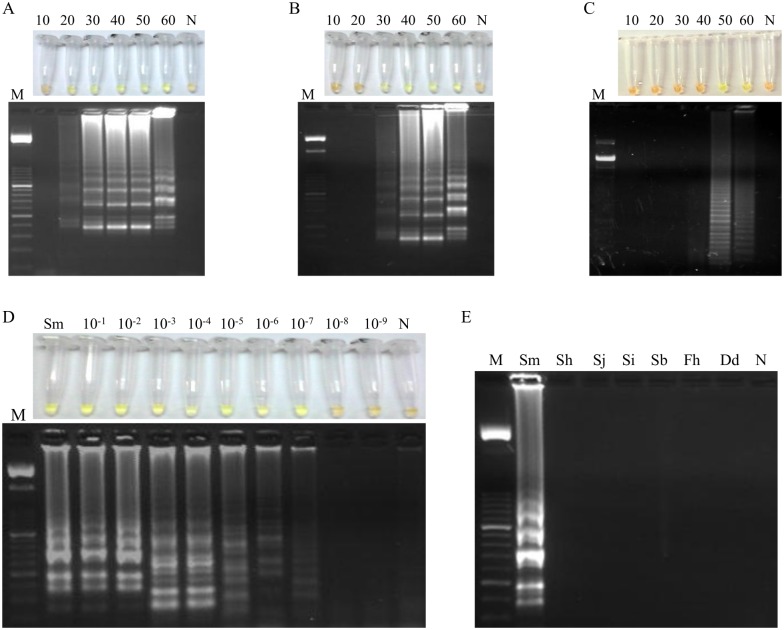
When using 1 ng (Fig 4A), 1 pg (Fig 4B) or 1 fg (Fig 4C) of S. mansoni DNA as template for LAMP assay at 63°C, we obtained positive results as soon as 20 min, 30 min and 50 min, respectively. All amplification results were clearly visualized by naked eye after adding the fluorescent dye as well as on agarose gel electrophoresis showing the typical ladder-like pattern. Afterwards, we evaluated the sensitivity of the LAMP assay at 63°C for 50 min by using S. mansoni DNA 10-fold serially diluted. The limit of detection was 0.1 fg (Fig 4D), showing that LAMP assay is 105 fold higher than PCR F3-B3. Regarding specificity, the LAMP assay was positive only for S. mansoni and no positive DNA products were observed when other species were used as templates (Fig 4E). Thereby, the LAMP assay at 63°C for 50 min was set up as the most suitable to test all the DNA samples from B. glabrata snails included in the study and hereinafter was namely Biompha-LAMP.
Fig 4. Setting up LAMP assay.
LAMP amplification results using (A) 1 ng, (B) 1 pg and (C) 1 fg of S. mansoni DNA obtained at different incubation times (10, 20, 30, 40, 50 and 60 min) tested in a heating block by the addition of SYBR Green I (top) or by visualization on agarose gel (bottom). Lanes M, 50 bp DNA ladder (Molecular weight marker XIII, Roche); lanes N: negative control (no DNA template). (D) Sensitivity assessment performed with LAMP at 63°C for 50 min using serial dilutions of S. mansoni genomic DNA. Lane M: 50 bp DNA ladder (Molecular weight marker XIII, Roche); lane Sm: genomic DNA from S. mansoni (1 ng); lanes 10−1–10−9: 10-fold serially dilutions; lane N: negative control (no DNA template). (E) Specificity of the LAMP assay for S. mansoni. A ladder of multiple bands of different sizes could be only observed in S. mansoni DNA sample. Lane M, 50 bp DNA ladder (Molecular weight marker XIII, Roche); lanes Sm, Sh, Sj, Si, Sb, Fh and Dd, S. mansoni, S. haematobium, S. japonicum, S. intercalatum, S. bovis, Fasciola hepatica and Dicrocoelium dendriticum DNA samples (1 ng/each), respectively; lane N, negative control (no DNA template).
Application of LAMP in snails samples: Biompha-LAMP analysis
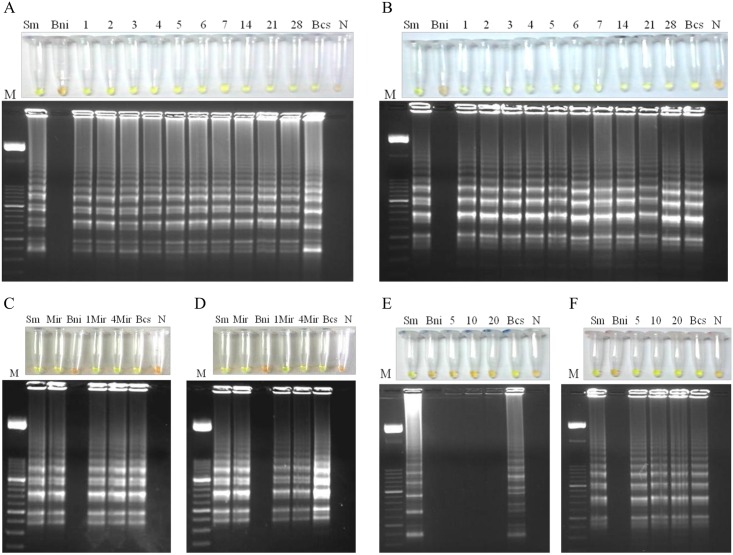
The results obtained in Biompha-LAMP assays to detect S. mansoni DNA in snail samples from the different experimental snails infections carried out in the study are showed in fig 5. We detected S. mansoni DNA in all infected snails tested before cercarial shedding at different days p.e. regardless of the method used for DNA extraction, thus is, the commercial kit (Fig 5A) or the heat NaOH extraction method (Fig 5B). We also obtained Biompha-LAMP positive results in those snails previously exposed to a low number of miracidia (one or four) using both commercial kit or the heat NaOH extraction method for DNA obtaining (Fig 5C and 5D, respectively). We did not obtain positive results in pooled samples containing snails previously exposed to a single miracidium and processed by the heat NaOH extraction method (Fig 5E). However, we obtained Biompha-LAMP positive results in pooled samples containing snails with confirmed cercarial shedding (Fig 5F). In all Biompha-LAMP assays, positive controls included (DNA from S. mansoni adult worms, miracidia or infected snails) showed an amplification product whereas negative controls (distilled water as template or DNA from uninfected snails) never amplified.
Fig 5. Application of LAMP in snails samples: Biompha-LAMP analysis.
(A) and (B) Analysis of snails before cercarial shedding at different days post-exposure to 9 miracidia each using a commercial kit or the heat NaOH extraction method for DNA obtaining, respectively. Lanes M, 50 bp DNA ladder (Molecular weight marker XIII, Roche); lane Sm, Schistosoma mansoni DNA (1 ng); lanes Bni, Biomphalaria glabrata DNA non infected; Lanes 1–7, 14, 21 and 28, days post-exposure to miracidia; lanes Bcs, Biomphalaria glabrata DNA with cercarial shedding; lanes N: negative control (no DNA template). (C) and (D) Analysis of snails exposed to one or four miracidia at 24h post-exposure using a commercial kit or the heat NaOH extraction method, respectively. Lanes M, 50 bp DNA ladder (Molecular weight marker XIII, Roche); lanes Sm, Schistosoma mansoni DNA (1 ng); lanes Mir; DNA obtained from one miracidium; lanes Bni, Biomphalaria glabrata DNA non infected; lanes 1Mir and 4Mir, DNA obtained from snails exposed to one or 4 miracidia, respectively; lanes Bcs, Biomphalaria glabrata DNA with cercarial shedding; lanes N: negative control (no DNA template). (E) and (F) Analysis of pooled samples containing snails previously exposed to 1 miracidium or with confirmed cercarial shedding, respectively, using the heat NaOH extraction method. Lanes M, 50 bp DNA ladder (Molecular weight marker XIII, Roche); lanes Sm, Schistosoma mansoni DNA (1 ng); lanes Bni, Biomphalaria glabrata DNA non infected; lanes 5, 10 and 20, DNA obtained from pooled samples containing one snail exposed to 1 miracidium or with confirmed cercarial shedding together with 5, 10 or 20 uninfected snails; lanes Bcs, Biomphalaria glabrata DNA with cercarial shedding; lanes N: negative control (no DNA template).
Discussion
B. glabrata, as an intermediate snail host for S. mansoni, plays a crucial role in both multiplication and transmission of schistosomes. Thus, snail control interventions are considered a priority and still needed for the interruption of schistosomiasis transmission. The early detection of prepatently infected B. glabrata snails using simple, sensitive and inexpensive molecular methods to detect S. mansoni DNA sequences in snails would be very helpful in rapid evaluation of the potential risk of transmission in suspected areas of schistosomiasis and would also provide for more effective disease control measures. In this sense, LAMP assay has become a most suitable tool than PCR-based methods for rapid molecular monitoring of vectors [33] and intermediate snails hosts of several parasites, including schistosomes [34–36] because of its operational simplicity, less time-consuming and versatility of visual detection readout options for field application [20].
In this work, we have developed and evaluated a rapid, sensitive and specific LAMP assay combined with a simple and economic DNA extraction method to detect experimentally infected snails with S. mansoni. This methodology could be potentially suitable for monitoring of infected snails in endemic areas of schistosomiasis with basic laboratory facilities.
To design the specific set of six primers for our LAMP assay, an intergenic spacer (IGS) of the large subunit (28S) ribosomal RNA gene was selected [31]. Ribosomal genes within Schistosoma species are known to be multi-copy (over 80–137 copies are estimated) and tandemly repeated within S. mansoni genome [37]. Therefore, using a repetitive selected portion of the genome as target for amplification might greatly increase LAMP sensitivity. Additionally, the ribosomal IGS frequently contain specific sequence motifs, thus allowing differentiation of Schistosoma species and also avoiding cross-reactions with other target organisms such as the intermediate snail hosts. Furthermore, a section of the ribosomal IGS of both S. haematobium and S. mansoni has previously been successfully used for molecular detection of schistosomes DNA in freshwater snails by using either RT-PCR or oligochromatographic dipstick assay (PCR-OC) [18]. Nevertheless, in terms of potential use of this new LAMP assay in field conditions, an additional validation using other DNA samples from other Biomphalaria species and also other Schistosoma species should be formerly tested.
After verifying the operation, sensitivity and specificity of PCR F3-B3 in amplification of the in silico expected fragment of 225 bp of S. mansoni IGS 28S rRNA gene, we attempted to set up the best conditions for primers set operation in the LAMP reaction. The design of our LAMP assay included a pair of loop primers (LF and LB), which it has been reported to accelerate the LAMP reaction speed and then reducing the reaction time to about 30 min [38]. When testing different reaction times, we obtained amplification of 1 ng and 1 pg of S. mansoni DNA at 63°C in just only 20 min and 30 min, respectively, as was confirmed by naked eye and electrophoresis. We also obtained amplification of such a small amount as 1 fg of S. mansoni DNA when the reaction was incubated at 63°C for 50 min. According to this result, both this temperature and reaction time were selected to establish the limit of detection of the LAMP assay, which finally resulted in 105 times higher than that obtained by PCR F3-B3 (0.1 fg vs. 10 pg, respectively). Thus, the value of 0.1 fg was considered as the lower limit of the detection threshold of our LAMP assay in detecting S. mansoni genomic DNA. It has been reported that a number of 10 S. mansoni miracidia yield 0.45 ng of genomic DNA [16] and also that S. mansoni genome contains approximately 580 fg of DNA [39]. Then, theoretically our LAMP assay would detect S. mansoni DNA corresponding to less than the equivalent to one single miracidium or a single parasite cell. A high sensitivity has also been previously reported when using other LAMP assays to detect schistosomes in infected snails [10, 29], but a long time of 120 min was required to complete the reaction, whereas our LAMP assay (Biompha-LAMP) takes only 50 min to obtain the same limit of DNA detection.
The applicability and effectiveness of our Biompha-LAMP assay in detecting laboratory-infected snails with S. mansoni could be assessed on a number of Biomphalaria specimens in a very early prepatent period (as soon as one day after miracidial exposure) as well as in low-grade infections (snails infected with only 4 miracidia or even in monomiracidial infections) regardless of the method used for DNA extraction. Our results were consistent with those previously reported in detecting infected snails from prepatent period by molecular methods, such as PCR [14, 16] and other LAMP assays [10, 29]. This feature is of a great value since at 24 hours p.e. sporocysts has not yet undergone germinal cell division [40] and besides that, not all miracidia subsequently complete the infection and develop until cercariae [41]. Thus, Biompha-LAMP could be a good alternative to parasitological methods in detecting trace amounts of S. mansoni DNA present in low-infected snails in low-transmission areas of schistosomiasis. An additional advantage is that not complicate or expensive procedure for snails DNA extraction is required because a simple and economical heat NaOH method is just enough to obtain a quality DNA for LAMP amplification. In this sense, another successful LAMP assays to detect schistosomes DNA without requiring a purified nucleic acid have been already reported [25, 29].
In recent years, the large-scale molecular screening of pooled field-collected snails in transmission areas of schistosomiasis has been reported as a simple and efficient tool for snails surveillance [28, 29]. For potential applications of our Biompha-LAMP in such setting, we tested in laboratory conditions several different size crushed pooled samples. When a snail previously exposed to a single miracidium was placed and crushed together with other 5, 10 or 20 uninfected snails no amplification was obtained. Probably, the greater volume of NaOH used to crush a greater amount of soft tissues for snails DNA extraction diluted in excess DNA concentration to be detected, since a single snail exposed to a single miracidium was previously individually found to be LAMP positive. When testing pooled samples containing a single snail showing cercarial shedding together with other several uninfected snails, a LAMP positive result was always obtained. This is consistent with the fact that a high number of cercariae increase the total amount of S. mansoni DNA in pooled samples.
In conclusion, the current study has demonstrated that our new designed Biompha-LAMP assay is specific, sensitive, rapid and could be a potentially adaptable cost-effective diagnostic method for screening of intermediate hosts infected with S. mansoni in both individual snails and pooled samples. Moreover, the rapidity of the reactions including loop primers shows that Biompha-LAMP is suitable for large-scale field surveys for schistosomes control campaigns in endemic areas.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Health Research Projects: Technological Development Project in Health (grant number DTS16/00207) and Health Research Project (grant number PI16/01784) of funding institution Instituto de Salud Carlos III (http://www.isciii.es/). Unión Europea. Co-financing by Fondo Europeo de Desarrollo Regional (FEDER), “Una manera de hacer Europa”. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Colley DG, Bustinduy AL, Secor WE, King CH. Human Schistosomiasis. Lancet. 2014; 383: 2253–2264. 10.1016/S0140-6736(13)61949-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King CH. Parasites and poverty: the case of schistosomiasis. Acta Trop. 2010; 113: 95–104. 10.1016/j.actatropica.2009.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Utzinger J, Raso G, Brooker S, De Savigny D, Tanner M, Ornbjerg N et al. Schistosomiasis and neglected tropical diseases: towards integrated and sustainable control and a word of caution. Parasitology. 2009; 136: 1859–1874. 10.1017/S0031182009991600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King CH, Dickman K, Tisch DJ. Reassessment of the cost of chronic helmintic infection: a meta-analysis of disability-related outcomes in endemic schistosomiasis. Lancet. 2005; 365: 1561–1569. 10.1016/S0140-6736(05)66457-4 [DOI] [PubMed] [Google Scholar]
- 5.Stensgaard AS, Utzinger J, Vounatsou P, Hürlimann E, Schur N, Saarnak CF, et al. Large-scale determinants of intestinal schistosomiasis and intermediate host snail distribution across Africa: does climate matter? Acta Trop. 2013; 128: 378–390. 10.1016/j.actatropica.2011.11.010 [DOI] [PubMed] [Google Scholar]
- 6.Satayathum SA, Muchiri EM, Ouma JH, Whalen CC, King CH. Factors affecting infection and reinfection with Schistosoma haematobium in coastal Kenya: survival analysis during nine-year, school-based treatment program. Am J Trop Med Hyg. 2006; 75: 83–92. [PMC free article] [PubMed] [Google Scholar]
- 7.King CH, Bertsch D. Historical Perspective: Snail Control to Prevent Schistosomiasis. PLoS Negl Trop Dis. 2015; 9(4): e0003657 10.1371/journal.pntd.0003657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown DS. 1994. Freshwater Snails of Africa and their Medical Importance, 2nd ed Taylor and Francis, London. [Google Scholar]
- 9.World Health Organization. Schistosomiasis: Progress Report 2001–2011 and Strategic plan 2012–2020. 2013.
- 10.Abbasi I, King CH, Muchiri EM, Hamburger J. Detection of Schistosoma mansoni and Schistosoma haematobium DNA by loop-mediated isothermal amplification: identification of infected snails from early prepatency. Am J Trop Med Hyg. 2010; 83: 427–432. 10.4269/ajtmh.2010.09-0764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbosa CS. Methods for malacological work in schistosomiasis. Mem Inst Oswaldo Cruz. 1992; Suppl. IV; 311–312. [Google Scholar]
- 12.Loker ES, Bayne CJ, Buckley PM, Kruse KT. Ultrastructure of encapsulation of Schistosoma mansoni mother sporocysts by hemocytes of juveniles of the 10-R2 strain of Biomphalaria glabrata. J Parasitol. 1982; 68: 84–94. [PubMed] [Google Scholar]
- 13.Favre TC, Bogea THP, Rotenberg L, Silva HS, Peri OS. Carcarial emergence of Schistosoma mansoni from Biomphalaria glabrata and Biomphalaria straminea. Mem Inst Oswaldo Cruz. 1995; 90: 565–567. [DOI] [PubMed] [Google Scholar]
- 14.Hamburger J, He-Na, Xin XY, Ramzy RM, Jourdane J, Ruppel A. A polymerase chain reaction assay for detecting snails infected with bilharzia parasites (Schistosoma mansoni) from very early prepatency. Am J Trop Med Hyg. 1998; 59: 872–876. [DOI] [PubMed] [Google Scholar]
- 15.Jannotti-Passos LK, Vidigal TH, Dias-Neto E, Pena SD, Simpson AJ, Dutra WO et al. PCR amplification of the mitochondrial DNA minisatellite region to detect Schistosoma mansoni infection in Biomphalaria glabrata snails. J Parasitol. 1997; 83: 395–399. [PubMed] [Google Scholar]
- 16.Hanelt B, Adema CM, Mansour MH, Loker ES. Detection of Schistosoma mansoni in Biomphalaria using nested PCR. J Parasitol. 1997; 83: 387–394. [PubMed] [Google Scholar]
- 17.Jannotti-Passos LK, Magalhães KG, Carvalho OS, Vidigal TH. Multiplex PCR for both identification of Brazilian Biomphalaria species (Gastropoda: Planorbidae) and diagnosis of infection by Schistosoma mansoni (Trematoda: Schistosomatidae). J Parasitol. 2006; 92: 401–403. 10.1645/GE-593R1.1 [DOI] [PubMed] [Google Scholar]
- 18.Kane RA, Stothard JR, Rollinson D, Leclipteux T, Evraerts J, Standley CJ et al. Detection and quantification of schistosome DNA in freshwater snails using either fluorescent probes in real-time PCR or oligochromatographic dipstick assays targeting the ribosomal intergenic spacer. Acta Trop. 2013; 128: 241–249. 10.1016/j.actatropica.2011.10.019 [DOI] [PubMed] [Google Scholar]
- 19.Notomi T, Okayama H, Masubuchi H. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000; 28: E63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Notomi T, Mori Y, Tomita N, Kanda H. Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J Microbiol. 2015; 53: 1–5. 10.1007/s12275-015-4656-9 [DOI] [PubMed] [Google Scholar]
- 21.Xu J, Rong R, Zhang HQ, Shi CJ, Zhu XQ, Xia CM. Sensitive and rapid detection of Schistosoma japonicum DNA by loop-mediated isothermal amplification (LAMP). Int J Parasitol. 2009; 40: 327–331. 10.1016/j.ijpara.2009.08.010 [DOI] [PubMed] [Google Scholar]
- 22.Wang C, Chen L, Yin X, Hua W, Hou M, Ji M et al. Application of DNA-based diagnostics in detection of Schistosomal DNA in early infection and after drug treatment. Parasit Vectors. 2011; 4: 164 10.1186/1756-3305-4-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernández-Soto P, Gandasegui Arahuetes J, Sánchez Hernández A, López Abán J, Vicente Santiago B, Muro A. A Loop-Mediated Isothermal Amplification (LAMP) Assay for Early Detection of Schistosoma mansoni in Stool Samples: A Diagnostic Approach in a Murine Model. PLoS Negl Trop Dis. 2014; 8: e3126 10.1371/journal.pntd.0003126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song J, Liu C, Bais S, Mauk MG, Bau HH, Greenberg RM. Molecular Detection of Schistosome Infections with a Disposable Microfluidic Cassette. PLoS Negl Trop Dis. 2015; 9: e0004318 10.1371/journal.pntd.0004318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gandasegui J, Fernández-Soto P, Carranza-Rodríguez C, Pérez-Arellano JL, Vicente B, López-Abán J et al. The Rapid-Heat LAMPellet Method: A Potential Diagnostic Method for Human Urogenital Schistosomiasis. PLoS Negl Trop Dis. 2015; 9: e0003963 10.1371/journal.pntd.0003963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu J, Guan ZX, Zhao B, Wang YY, Cao Y, Zhang HQ et al. DNA Detection of Schistosoma japonicum: Diagnostic Validity of a LAMP Assay for Low-Intensity Infection and Effects of Chemotherapy in Humans. PLoS Negl Trop Dis. 2015; 9: e0003668 10.1371/journal.pntd.0003668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumagai T, Furushima-Shimogawara R, Ohmae H, Wang TP, Lu S, Chen R et al. Detection of early and single infections of Schistosoma japonicum in the intermediate host snail, Oncomelania hupensis, by PCR and loop-mediated isothermal amplification (LAMP) assay. Am J Trop Med Hyg. 2010; 83: 542–548. 10.4269/ajtmh.2010.10-0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tong QB, Chen R, Zhang Y, Yang GJ, Kumagai T, Furushima-Shimogawara R et al. A new surveillance and response tool: Risk map of infected Oncomelania hupensis detected by Loop-mediated isothermal amplification (LAMP) from pooled samples. Acta Trop. 2015; 141: 170–177. 10.1016/j.actatropica.2014.01.006 [DOI] [PubMed] [Google Scholar]
- 29.Hamburger J, Abbasi I, Kariuki C, Wanjala A, Mzungu E, Mungai P et al. Evaluation of loop-mediated isothermal amplification suitable for molecular monitoring of schistosome-infected snails in field laboratories. Am J Trop Med Hyg. 2013; 88: 344–351. 10.4269/ajtmh.2012.12-0208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rollinson D, Knopp S, Levitz S, Stothard JR, Tchuem Tchuenté LA, Garba A et al. Time to set the agenda for schistosomiasis elimination. Acta Trop. 2013; 128:423–440. 10.1016/j.actatropica.2012.04.013 [DOI] [PubMed] [Google Scholar]
- 31.Kane RA, Rollinson D. Comparison of the intergenic spacers and 3' end regions of the large subunit (28S) ribosomal RNA gene from three species of Schistosoma. Parasitology. 1998; 117: 235–242. [DOI] [PubMed] [Google Scholar]
- 32.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990; 5; 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 33.Nzelu CO, Gomez EA, Cáceres AG, Sakurai T, Martini-Robles L, Uezato H, et al. Development of a loop-mediated isothermal amplification method for rapid mass-screening of sand flies for Leishmania infection. Acta Trop. 2014; 132: 1–6. 10.1016/j.actatropica.2013.12.016 [DOI] [PubMed] [Google Scholar]
- 34.Chen R, Tong Q, Zhang Y, Lou D, Kong Q, Lv S et al. Loop-mediated isothermal amplification: rapid detection of Angiostrongylus cantonensis infection in Pomacea canaliculata. Parasite Vector. 2011; 4: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu CY, Song HQ, Zhang RL, Chen MX, Xu MJ, Ai L et al. Specific detection of Angiostrongylus cantonensis in the snail Achatina fulica using a loop-mediated isothermal amplification (LAMP) assay. Mol Cell Probes. 2011; 25:164–167. 10.1016/j.mcp.2011.04.002 [DOI] [PubMed] [Google Scholar]
- 36.Chen Y, Wen T, Lai DH, Wen YZ, Wu ZD, Yang TB et al. Development and evaluation of loop-mediated isothermal amplification (LAMP) for rapid detection of Clonorchis sinensis from its first intermediate hosts, freshwater snails. Parasitology. 2013; 140: 1377–1383. 10.1017/S0031182013000498 [DOI] [PubMed] [Google Scholar]
- 37.Copeland CS, Marz N, Rose D, Hertel J, Brindley PJ, Santana CB et al. (2009) Homology-based annotation of non-coding RNAs in the genomes of Schistosoma mansoni and Schistosoma japonicum. BMC Genomics. 2009; 10: 464 10.1186/1471-2164-10-464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002; 16: 223–229. [DOI] [PubMed] [Google Scholar]
- 39.Gomes AL, Melo FM, Werkhauser RP, Abath FG. Development of a real time polymerase chain reaction for quantitation of Schistosoma mansoni DNA. Mem Inst Oswaldo Cruz. 2006; 101: 133–136. [DOI] [PubMed] [Google Scholar]
- 40.Schutte HJ. Studies on the South African strain of Schistosoma mansoni. part 2: the intra-molluscan larval stages. S Afr J Sci. 1974; 70: 327–346. [Google Scholar]
- 41.Iwanaga Y. Studies On Host-Parasite Relationship Between The Puerto Rican Strain Of Schistosoma mansoni And Biomphalaria Snails. Southeast Asian J Trop Med Public Health. 1994; 25: 509–515. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.