Abstract
Extracellular matrix materials such as decellularized cartilage (DCC) and chondroitin sulfate (CS) may be attractive chondrogenic materials for cartilage regeneration. The goal of the current study was to investigate the effects of encapsulation of DCC and CS in homogeneous microsphere-based scaffolds, and to test the hypothesis that encapsulation of these extracellular matrix materials would induce chondrogenesis of rat bone marrow stromal cells. Four different types of homogeneous scaffolds were fabricated from microspheres of poly(D,L-lactic-co-glycolic acid): Blank (poly(D,L-lactic-co-glycolic acid) only; negative control), transforming growth factor-β3 encapsulated (positive control), DCC encapsulated, and CS encapsulated. These scaffolds were then seeded with rat bone marrow stromal cells and cultured for 6 weeks. The DCC and CS encapsulation altered the morphological features of the microspheres, resulting in higher porosities in these groups. Moreover, the mechanical properties of the scaffolds were impacted due to differences in the degree of sintering, with the CS group exhibiting the highest compressive modulus. Biochemical evidence suggested a mitogenic effect of DCC and CS encapsulation on rat bone marrow stromal cells with the matrix synthesis boosted primarily by the inherently present extracellular matrix components. An important finding was that the cell seeded CS and DCC groups at week 6 had up to an order of magnitude higher glycosaminoglycan contents than their acellular counterparts. Gene expression results indicated a suppressive effect of DCC and CS encapsulation on rat bone marrow stromal cell chondrogenesis with differences in gene expression patterns existing between the DCC and CS groups. Overall, DCC and CS were easily included in microsphere-based scaffolds; however, there is a requirement to further refine their concentrations to achieve the differentiation profiles we seek in vitro.
Keywords: Chondroitin sulfate, decellularized cartilage, microsphere-based scaffolds, cartilage regeneration
Introduction
Scaffold-based regenerative strategies for osteochondral tissue that take into consideration physiological and hierarchical variations in properties of native bone and cartilage have been increasingly gaining attention.1–4 Several of these strategies employ extracellular matrix (ECM)-based materials because of their ability to regulate behavior such as migration, proliferation, and differentiation of resident or transplanted cells.5,6 For cartilage regeneration, cartilage matrix has been used as a chondroinductive material because of its potential to retain bioactive molecules to which the regenerating tissue is naturally predisposed to respond.6–9 Moreover, materials like chondroitin sulfate (CS), the major sulfated glycosaminoglycan (GAG) found in the ECM of native cartilage, are used for cartilage regeneration because of their ability to create a favorable microenvironment for cells.10–12
Microsphere-based scaffolds possess an immense potential for musculoskeletal regeneration because of their characteristics like rigidity in shape, ability to provide a porous network, and uniform mechanical properties.13 Additionally, they offer a variety of alternatives in terms of materials for microsphere matrices, and methods for microsphere fabrication and sintering.14–19 We have previously demonstrated that three- dimensional (3D) microsphere-based gradient scaffolds containing gradients of growth factors are capable of directing cell phenotype by influencing them to secrete tissue-specific ECM components to promote osteochondral regeneration.20–23 In addition, we have shown that microsphere-based scaffolds containing gradients of CS and tricalcium phosphate can provide “raw materials” for synthesis of new ECM components, and in combination with growth factors (or alone) can furnish the surrounding progenitor cells with bioactive signals for their differentiation along the chondro- and osteogenic lineages in different regions of the scaffolds.24–26 Furthermore, we recently evaluated the response of decellularized cartilage (DCC) encapsulation in homogeneous microsphere- based scaffolds. The DCC encapsulation at a concentration of 10 wt% evoked a biosynthetic response from the seeded rat bone marrow stromal cells (rBMSCs) with comparable gene expression to cells seeded on transforming growth factor-β3 (TGF-β3) encapsulated scaffolds.9 To establish the benefits of our raw material gradient microsphere-based scaffolds, it is imperative to identify raw materials that are most efficacious in promoting osteogenesis and chondrogenesis. For determining the leading chondrogenic materials, the most rational step would be to evaluate the performance of homogeneous microsphere-based scaffolds incorporating chondrogenic materials in propelling chondrogenesis. Therefore, the objective of this study was to investigate the effects of encapsulating a higher concentration of ECM materials (DCC and CS), compared with what we have previously used, on influencing rBMSC chondrogenesis in homogeneous microsphere- based scaffolds. The results would have implications for identifying raw material concentrations that can then be combined with osteogenic raw materials for use in microsphere-based gradient scaffolds toward osteochondral repair.
In the present study, we investigated whether encapsulated raw materials (DCC and CS) at a higher concentration in poly(D,L-lactic-co-glycolic acid) (PLGA) microsphere-based scaffolds would provide building blocks and drive the differentiation of the seeded cells toward a chondrogenic lineage. Homogeneous microsphere-based scaffolds were fabricated encapsulating DCC and CS (at a concentration of 30 wt%) as chondrogenic raw materials. The response of seeded rBMSCs to the raw materials was evaluated when cultured for 6 weeks in a medium consisting of dissolved factors. We hypothesized that encapsulation of raw materials, DCC or CS, in homogeneous microsphere- based scaffolds would induce chondrogenesis in rBMSCs.
Materials and methods
Materials
All reagents for the decellularization process were purchased from Sigma–Aldrich (St. Louis, MO) unless otherwise noted. PLGA (50:50 lactic acid: glycolic acid ratio, ester end group) with an intrinsic viscosity (i.v.) of 0.37 dL/g, was obtained from Evonik Industries (Essen, Germany). Human TGF-β3 and murine insulin-like growth factor (IGF)-I were obtained from PeproTech, Inc. (Rocky Hill, NJ). Chondroitin sulfate A sodium salt (from bovine trachea) was obtained from Sigma (St. Louis, MO). All other reagents and organic solvents utilized were of cell culture or ACS grade. Two porcine knees obtained from a Berkshire hog (castrated male that was approximately 7–8 months old and weighed 120 kg) were purchased from a local abattoir (Bichelmeyer Meats, Kansas City, KS).
Tissue retrieval and decellularization
Articular cartilage was harvested from hip and knee joint surfaces using scalpels and immediately rinsed in phosphate-buffered saline (PBS). PBS was then drained from the cartilage and the tissue was stored at −20°C. After freezing overnight, the cartilage was thawed and coarsely cryoground with dry ice pellets using a cryogenic tissue grinder (BioSpec Products, Bartlesville, OK). The dry ice was allowed to sublime overnight in the freezer. Decellularization of the cartilage was performed using our previously described protocol.9,27 Coarse-ground cartilage particles were packed into dialysis tubing (3500 MWCO) and stored in hypertonic salt solution (HSS) overnight at room temperature with gentle agitation (70 rpm). The packets were then subjected to 220 rpm agitation with two reciprocating washes, encompassing triton X-100 (0.01% v/v) followed with HSS, to permeabilize intact cellular membranes. The tissue was then treated overnight with benzonase (0.0625 KU mL−1) at 37°C and later treated with sodium-lauroylsarcosine (NLS, 1% v/v) overnight to further lyse cells and denature cellular proteins. After NLS exposure, the tissue was washed with ethanol (40% v/v) at 50 rpm and subjected to organic exchange resins to extract the organic solvents at 65 rpm. Afterward, the tissue was washed in saline-mannitol solution at 50 rpm followed by 2 h of rinsing with DI water at 220 rpm. The tissue was then removed from the packets and was frozen and lyophilized. The DCC particles were further cryoground into a fine powder with a freezer-mill (SPEX SamplePrep, Metuchen, NJ) and then lyophilized. The DCC powder was filtered using a 45 μm mesh (ThermoFisher Scientific, Waltham, MA) to remove large particles and then frozen until use.
Preparation of microspheres
Four different types of microspheres were fabricated for the study: (a) PLGA microspheres (BLANK), (b) TGF-β3 encapsulated in PLGA microspheres (TGF), (c) DCC encapsulated in PLGA microspheres (DCC), and (d) CS encapsulated in PLGA microspheres (CS). For fabricating TGF-β3 encapsulated microspheres, TGF-β3 was first reconstituted in 10 mM citric acid. The reconstituted protein solution was mixed with 20% w/v PLGA dissolved in dichloromethane (DCM) at a loading of 30 ng TGF-β3 per 1.0 mg of PLGA. The final mixture was then sonicated over ice (50% amplitude, 20 s). The DCC and CS encapsulated microspheres were fabricated by adding 6% w/v DCC or 6% w/v CS to 14% w/v PLGA dissolved in DCM, respectively. Using the PLGA-protein and PLGA-DCC/CS emulsions, microspheres were fabricated via our previously reported technology.9,20–26,28–31 In brief, using acoustic excitation produced by an ultrasonic transducer (Branson Ultrasonics, Danbury, CT), regular jet instabilities were created in the polymer stream, thereby creating uniform polymer droplets. An annular carrier non-solvent stream of 0.5% w/v poly(vinyl alcohol) (PVA, 88% hydrolyzed, 25 kDa, Polysciences, Inc., Warrington, PA) in deionized water (DI H2O) carried the droplets (i.e. microspheres) into a beaker containing the non-solvent solution at 0.5% w/v in DI H2O (cold PVA solution in case of DCC microspheres). The microspheres were stirred for 1 h to allow for solvent to evaporate and then filtered, rinsed and stored at −20°C. The microspheres were then lyophilized for 48 h before further use.
Scaffold fabrication
Scaffolds were prepared using our previously established technology.9,20–26,28–30 Briefly, lyophilized microspheres (30–50 mg) were dispersed in DI H2O and loaded into a syringe. The dispersion was then pumped using a programmable syringe pump (PHD 22/2000; Harvard Apparatus, Inc., Holliston, MA) into a cylindrical plastic mold (diameter ~ 4 mm) having a filter at the bottom until a height of about 2 mm was reached. The scaffolds were 3.8–4.0 mm in diameter and around 2 mm in height. The packed microspheres were then sintered with ethanol-acetone (95:5 v/v) for 55 min. The scaffolds were lyophilized for 48 h and sterilized with ethylene oxide for 12 h prior to cell seeding experiments. A total of four different groups were tested in the study and were named according to the composition of microspheres as BLANK, TGF, DCC, and CS.
Cell seeding of scaffolds
rBMSCs were obtained from the femurs of eight young male Sprague–Dawley rats (176–200 g, SASCO) following a University of Kansas approved IACUC protocol (175–08) and cultured in medium consisting of αMEM supplemented with 10% FBS (MSC-Qualified, cat #10437-028) and 1% penicillin–streptomycin (P/S) (all from Thermofisher Scientific, Waltham, MA). When the cells were 80% to 90% confluent, they were trypsinized and re-plated at 7500 cells/cm2. Seeding was performed when cells reached P4. Scaffolds were sterilized using ethylene oxide for 12 h, allowed to ventilate overnight after sterilization, and placed in a 48-well plate. Cells (P4) were resuspended in culture medium at a concentration of approximately 50 million/mL. A total of 25 μL of this cell suspension (~1.25 M cells) was placed directly onto the top of the scaffold, which infiltrated the scaffold via capillary action.25,30 Cells were allowed to attach for 1 h, after which 1 mL of culture medium was added. After 24 h, the culture medium was replaced by 1 mL of differentiation medium consisting of αMEM, 1% P/S, 10% FBS, 50 μg/mL ascorbic acid (Sigma, St. Louis, MO), 40 μg/mL L-proline (Sigma, St. Louis, MO), 100 μM sodium pyruvate (Thermofisher Scientific, Waltham, MA), 100 nM dexamethasone (DEX) (MP Biomedicals, Santa Ana, CA), 1% insulintransferrin-selenium 100X (ITS) (Thermofisher Scientific, Waltham, MA), 1% non-essential amino adds (NEAA) (Thermofisher Scientific, Waltham, MA), 15 mM HEPES buffer (Thermofisher Scientific, Waltham, MA), and 100 ng/mL murine IGF-I. Every 48 h for 6 weeks, three-fourths of the differentiation medium was replaced with fresh medium.
Scanning electron microscopy (SEM) and energy Dispersion Spectroscopy (EDS)
Microspheres and acellular scaffolds were imaged via a Versa 3D Dual Beam (FEI, Hillsboro, OR) scanning electron microscope with a detector for EDS. The microspheres were cryo-fractured using a sharp blade, and the dispersion of TGF-β3, DCC, and CS within the microspheres was further analyzed using EDS at an accelerating voltage of 10 kV. Pixel maps for atomic nitrogen and sulfur were generated using Aztec analysis software (Oxford Instruments, Abingdon, UK). The PLGA (BLANK) microspheres were also imaged to confirm the absence of nitrogen and sulfur in the EDS maps (Supplementary Figure 1).
Mechanical testing
Unconfined compression tests of acellular (i.e. week 0) microsphere-based scaffolds (n = 6) were conducted using a uniaxial testing apparatus (Instron Model 5848, Canton, MA) with a 50 N load cell. A custom-made stainless steel bath and a compression-plate assembly were mounted in the apparatus.32 Cylindrical scaffold samples were compressed to 40% strain at a strain rate of 10%/min in PBS (0.138 M sodium chloride, 0.0027 M potassium chloride) at 37°C. Compressive moduli of elasticity were calculated from the initial linear regions of the stress–strain curves (i.e. at ~5% strain) as described previously.9,25,26,28–30
Porosity measurement
A fluid saturation method as described previously25 was used in this study to calculate the porosities of the scaffolds:
where VB, m, d, h, Ww, WD, and VP are the bulk volume, mass, diameter, height, wet weight, dry weight, and pore volume of the scaffolds, respectively. WWater and ρWater are the weight and density of water, respectively. Briefly, wet and dry weights of scaffolds were recorded after fabrication and porosities were determined by the above-described equations.
Biochemical analyses
Engineered constructs (n = 6) were analyzed for matrix production at 0 (i.e. 24-h post seeding), 3, and 6 weeks. The samples were digested in papain solution consisting of 125 mg/mL papain (from papaya latex), 5 mM N-acetyl cysteine, 5 mM ethylenediaminetetraacetic acid, and 100 mM potassium phosphate buffer (20 mM monobasic potassium phosphate, 79 mM dibasic potassium phosphate) (all reagents from Sigma Aldrich) in DI H2O. Engineered constructs were removed from culture in a sterile manner, placed in microcentrifuge tubes, homogenized with the papain solution (1 mL), and allowed to digest overnight in a 60°C water bath. The digested scaffolds were then centrifuged at 10,000 rpm for 5 min to pellet fragments of polymer and other impurities and stored at −20°C. Later, the supernatant was used to determine DNA, GAG, and hydroxyproline (HYP) contents using the PicoGreen (Molecular Probes, Eugene, OR), dimethylmethylene blue (DMMB) (Biocolor, Newtownabbey, Northern Ireland), and HYP (cat #MAK008, Sigma Aldrich, St. Louis, MO) assays, respectively. The acellular controls from the DCC group were also analyzed for their inherent DNA, HYP and GAG content while the CS group acellular scaffolds were evaluated for their GAG content only at weeks 0, 3, and 6. The DNA content values of the acellular DCC scaffolds were subtracted from the corresponding values of their cellular counterparts at each time point in an effort to distinguish cell proliferation on the cellular DCC scaffolds from the residual DNA present in these scaffolds.
Gene expression analyses
Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) was performed for gene expression analyses in microsphere-based constructs (n = 6) at weeks 0, 1.5, 3, and 6. Certain groups at certain time points (indicated in “Results” section) had no Ct values, indicating that the fluorescence intensities in these samples did not cross the threshold fluorescence. These samples were marked as zero for RNA expression. RNA was isolated and purified using QIAshredders and an RNeasy Kit (Qiagen, Valencia, CA) according to the manufacturer’s guidelines. Isolated RNA was converted to complementary DNA using a TaqMan High Capacity kit (Applied Biosystems, Foster City, CA) in an Eppendorf RealPlex Mastercycler. TaqMan Gene expression assays from Applied Biosystems for appropriate genes (Table 1) were run in the Eppendorf system. A 2−ΔΔCt method was used to evaluate the relative level of expression for each target gene. For quantification, the BLANK constructs at week 0 were designated as the calibrator group and GAPDH expression as the endogenous control.
Table I.
Genes used for RT-qPCR analysis.
| Gene | Symbol | TaqMan Assay ID |
|---|---|---|
| Glyceraldehyde 3-phosphate dehydrogenase | GAPDH | Rn01775763_gl |
| Collagen type I | COLIAI | Rn01463848_ml |
| Collagen type II | COL2AI | Rn01751069_mH |
| Collagen type X | COLI0AI | Rn01408029_gl |
| Aggrecan | ACAN | Rn00573424_ml |
| SRY (sex determining region Y)-box 9 | SOX-9 | Rn01751069_mH |
| Runt-related transcription factor 2 | RUNX2 | Rn01512298_ml |
| Integrin-binding sialoprotein | IBSP | Rn00561414_ml |
Histology and immunohistochemistry (IHC)
At 6 weeks, microsphere-based constructs (n = 3) were soaked in 30% w/v sucrose (Thermofisher Scientific, Waltham, MA) solution in PBS for 24 h. Afterward, the constructs were equilibrated in optimal cutting temperature embedding medium (OCT, Tissue-Tek, Torrance, CA) overnight at 37°C and then frozen at −20°C. In all, 10-μm thick sections were cut using a cryostat (Micron HM-550 OMP, Vista, CA) and stained using hematoxylin (cell nuclei) and eosin (cytoplasm); Masson’s trichrome for collagen, cell nuclei, and cytoplasm; Safranin O for GAGs; and Sudan Black for residual polymer. Acellular constructs (n = 2) at week 6 from the DCC and CS groups were also stained using Safranin O. The sections from cellular constructs were stained for the presence of collagen type I, collagen type II, and aggrecan using IHC. Mouse monoclonal primary antibodies (all from Thermofisher Scientific, Waltham, MA) against collagen type I (1:200 dilution), collagen type II (1:200 dilution), and aggrecan (1:50 dilution) were used for the immunostaining. Following the primary antibody, biotinylated secondary antibody was used followed with the ABC complex (Vector Laboratories, Burlingame, CA). The antibodies were visualized with the diaminobenzidine (DAB) substrate per the manufacturer’s (Vector Laboratories) protocol. Negative controls were also run with the primary antibody omitted. Histological and IHC staining images from the CS constructs could not be obtained as the sections washed off the slides during the procedures of staining, washing, dehydration, and clearing.
Statistical analyses
GraphPad Prism 6 statistical software (GraphPad Software, Inc., La Jolla, CA) was used to compare experimental groups using a one-factor ANOVA (sections Mechanical Testing and Porosity Measurement) or a two-factor ANOVA (sections Biochemical Analyses and Gene Expression Analyses) followed by a Tukey’s post hoc test, where p < 0.05 was considered significant. Additionally, standard box plots were constructed to eliminate outliers. All quantitative results are reported as average ± standard deviation.
Results
Tissue decellularization
Following decellularization and cryo-grinding, the DNA, GAG, and HYP contents were reduced by 44% (p < 0.05), 23% (p < 0.05), and 23% (p < 0.05), respectively.
SEM
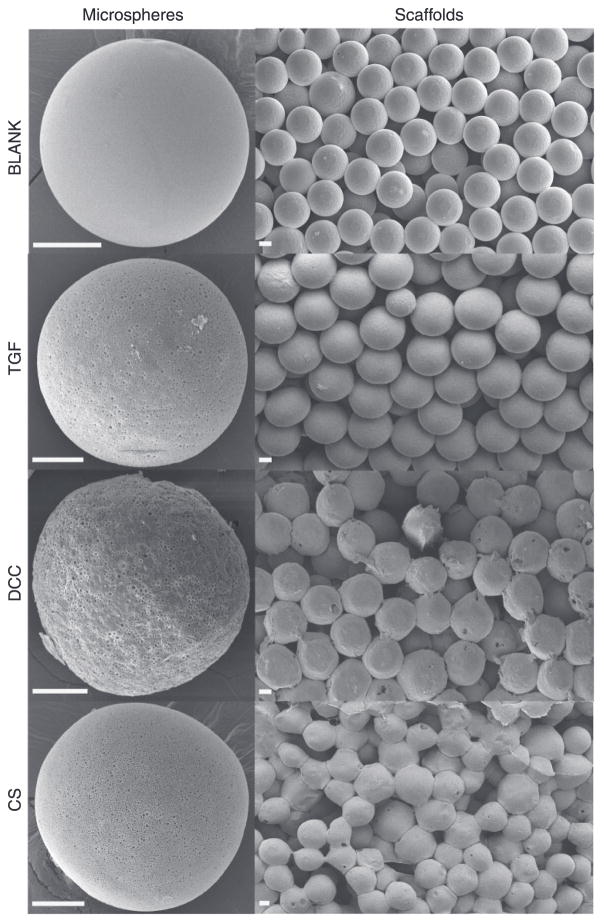
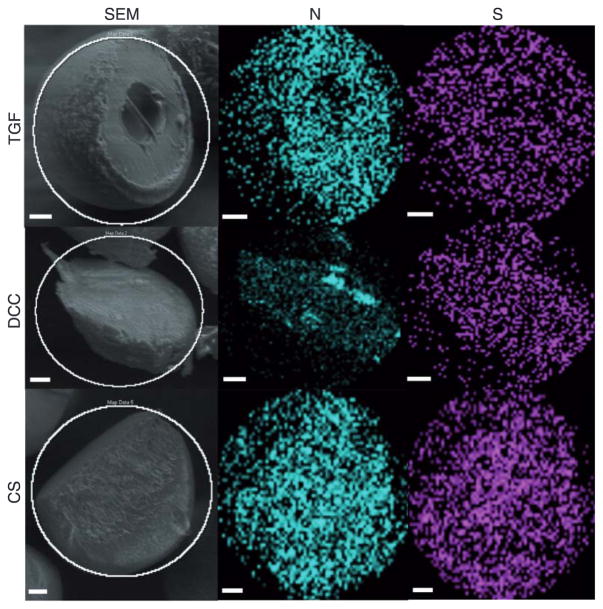
Figure 1 represents the scanning electron micrographs of microspheres and scaffolds from the four different groups. All four types of microspheres had a spherical morphology with the BLANK, TGF, and CS microspheres depicting a smooth surface while the DCC microspheres possessed a rough surface. The microspheres in the TGF, DCC, and CS groups had micron and sub-micron sized pores present throughout the surface while no pores were observed on the surface of the BLANK microspheres. The images of the scaffolds demonstrated the overall porous nature of microsphere-based scaffolds with similar degrees of microsphere sintering (extent of interconnections) among the BLANK, TGF, and DCC groups; however, the microspheres in the CS scaffolds appeared to be fused more with each other than what was observed in the other three groups. Figure 2 illustrates the distribution of atomic nitrogen (N) and sulfur (S) in the interior of TGF, DCC, and CS microspheres. Nitrogen was distributed uniformly within the TGF, DCC, and CS microspheres. Sulfur was observed to be present inside the TGF, DCC, and CS microspheres. The spectral maps for the BLANK microspheres showed that the nitrogen and sulfur were essentially absent from these microspheres (Supplementary Figure 1).
Figure 1.
Scanning electron micrographs of microspheres (left column) and scaffolds (right column). BLANK (PLGA-only), TGF (PLGA with TGF-β3 encapsulated), DCC (PLGA with 30 wt% DCC), and CS (PLGA with 30 wt% CS) microspheres and scaffolds. The images reveal the distinct morphological features of the microspheres and scaffolds; note the porous nature of the surface of the TGF microspheres, rough surface of DCC microspheres, and relatively greater degree of sintering in the CS scaffolds. Scale bars: 50 μm.
Figure 2.
Scanning electron micrographs (left column) and energy dispersive spectral maps (center and right columns) of cryo-fractured microspheres for atomic nitrogen (N) and sulfur (S). TGF (PLGA with TGF-β3 encapsulated), DCC (PLGA with 30 wt% DCC), and CS (PLGA with 30 wt% CS) microspheres. Note the uniform distribution of nitrogen and sulfur in the TGF, DCC, and CS microspheres. Scale bars: 25 μm.
Mechanical testing
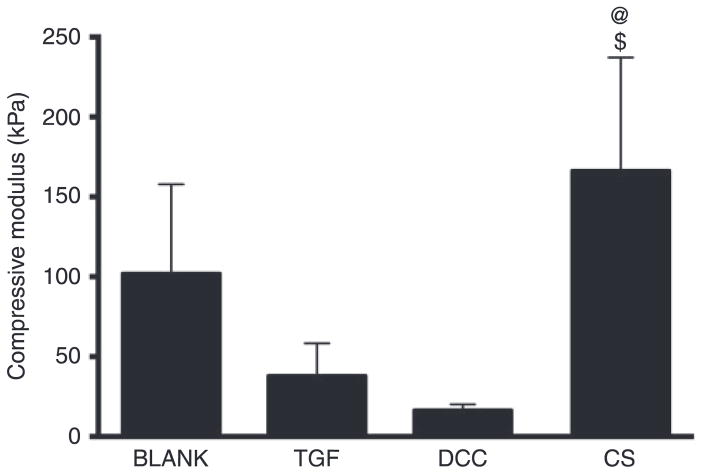
The average compressive moduli for BLANK, TGF, DCC, and CS scaffolds were 102 ± 56 kPa, 38 ± 20 kPa, 16.5 ± 3.7 kPa, and 166 ± 71 kPa, respectively (Figure 3). The compressive modulus for the CS group was 4.4-fold (p < 0.05) and 10-fold (p < 0.05) higher than the moduli of the TGF and DCC groups, respectively. No significant differences were observed in compressive modulus among the BLANK, TGF, and DCC groups.
Figure 3.
Average compressive moduli of elasticity of acellular microsphere-based scaffolds at week 0. All values are expressed as the average + standard deviation (n = 6). The CS group had a significantly higher modulus than the TGF and DCC groups. @significant difference from the TGF group and $significant difference from the DCC group (p < 0.05).
Porosity measurement
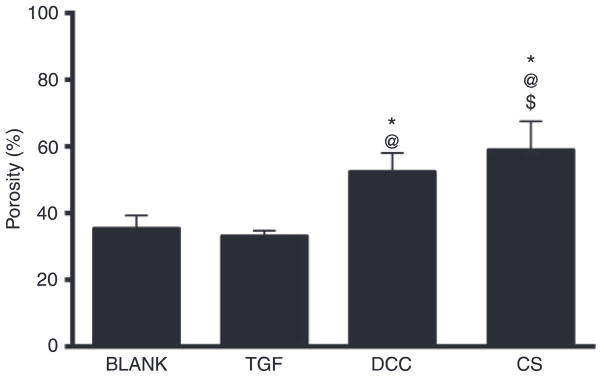
The average porosity of the CS group was 1.7-fold (p < 0.05), 1.8-fold (p < 0.05), and 1.1-fold (p < 0.05) higher than the average porosities of the BLANK, TGF, and DCC groups, respectively (Figure 4). The porosity of the DCC group was 1.5-fold (p < 0.05) and 1.6-fold (p < 0.05) higher than the porosities of the BLANK and TGF groups, respectively. No significant differences in porosities were observed between the BLANK and TGF groups.
Figure 4.
Average porosities of different scaffold groups. All values are expressed as the average + standard deviation (n = 6). Both the DCC and CS groups had higher porosities than the BLANK and TGF groups. *Significant difference from the BLANK group, @significant difference from the TGF group, and $significant difference from the DCC group (p < 0.05).
Biochemical analysis
DNA content
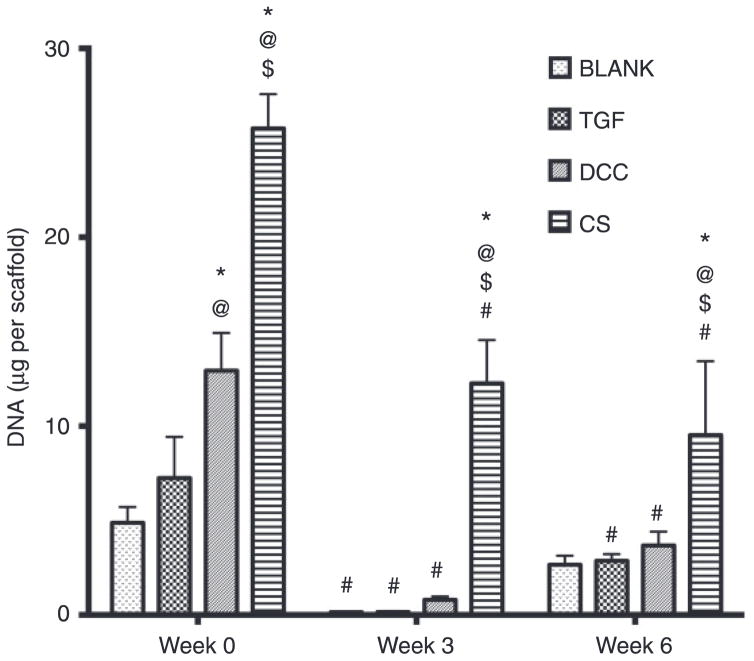
At week 0, the DCC and CS groups outperformed the BLANK group in DNA content with 2.7-fold (p < 0.05) and 5.3-fold (p < 0.05) higher DNA contents, respectively (Figure 5). Additionally, the DCC and CS groups at week 0 had 1.8-fold (p < 0.05) and 3.6-fold (p < 0.05) higher DNA contents than the DNA content in the TGF group, respectively. Moreover, the CS group at week 0 had 2-fold (p < 0.05) higher DNA content than the DCC group. No significant differences in DNA content were observed between the BLANK and TGF groups at week 0. At week 3, the CS group had 88-fold (p < 0.05), 82-fold (p < 0.05), and 15-fold (p < 0.05) more DNA than the BLANK, TGF, and DCC groups, respectively. Similarly, the CS group at week 6 had 3.6-fold (p < 0.05), 3.3-fold (p < 0.05), and 2.6- fold (p < 0.05) more DNA than the BLANK, TGF, and DCC groups, respectively. No significant differences in DNA content were observed among the BLANK, TGF, and DCC groups at weeks 3 or 6. All of the groups showed statistically significant decreases in DNA contents with time. The BLANK group at week 0 had 35-fold (p < 0.05) more DNA than its matching value at week 3. The TGF group at week 0 had 48-fold (p < 0.05) and 2.5-fold (p < 0.05) more DNA than its corresponding values at weeks 3 and 6, respectively. The DCC group at week 0 had 15-fold (p < 0.05) and 3.5-fold (p < 0.05) higher DNA content than at weeks 3 and 6, respectively. Week 0 DNA content in the CS group was 2.1-fold (p < 0.05) and 2.7-fold (p < 0.05) higher than its matching values at weeks 3 and 6, respectively. It must be noted that the values of DNA content obtained in the DCC group represent the amount of DNA present as a result of cell proliferation in these scaffolds. The values do not represent the residual amount of DNA present in these scaffolds, as the leftover DNA from the acellular DCC controls was subtracted at each time point.
Figure 5.
Total DNA content in different scaffold groups at weeks 0, 3, and 6. All values are expressed as the average + standard deviation (n = 6). The CS group had the highest DNA content at all time points by at least a factor of 2. *Significant difference from the BLANK group at same time point, @significant difference from the TGF group at same time point, $significant difference from the DCC group at same time point, and #significant difference from its value at week 0 (p < 0.05).
GAG content
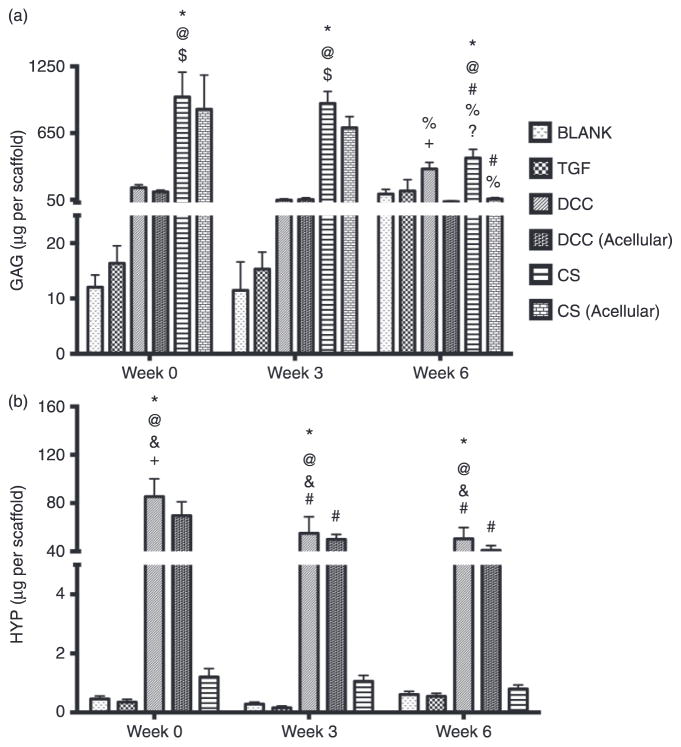
The GAG content in the CS group at week 0 was 81-fold (p < 0.05), 60-fold (p < 0.05), and 6.2-fold (p < 0.05) higher than the GAG contents in the BLANK, TGF, and DCC groups, respectively (Figure 6(a)). Similarly, the GAG content in the CS group at week 3 was 80-fold (p < 0.05), 60-fold (p < 0.05), and 19-fold (p < 0.05) higher than the GAG contents in the BLANK, TGF, and DCC groups, respectively. No significant differences in GAG content were observed among the BLANK, TGF, and DCC groups at weeks 0 and 3. At week 6, the GAG content in the CS group was 4.2-fold (p < 0.05) and 2.6-fold (p < 0.05) higher than the GAG contents in the BLANK and TGF groups, respectively.
Figure 6.
Total GAG content (a) and HYP content (b) in different scaffold groups at weeks 0, 3, and 6. All values are expressed as the average + standard deviation (n = 6). The DCC and CS groups had significantly higher GAG content than their acellular counterparts at week 6. The DCC group at week 0 also had significantly higher HYP content than the DCC (acellular) group. *Significant difference from the BLANK group at same time point, @significant difference from the TGF group at same time point, $significant difference from the DCC group at same time point, + significant difference from the DCC (acellular) group at same time point, ?significant difference from the CS (acellular) group at same time point, #significant difference from its value at week 0, and %significant difference from its value at week 3 (p < 0.05).
Most notably, the CS and DCC groups at week 6 had up to an order of magnitude higher GAG contents than their acellular counterparts. Specifically, the CS group GAG content at week 6 was 7.5-fold (p < 0.05) higher than the GAG content of the CS (acellular) group. Likewise, the DCC group at week 6 had 10-fold (p < 0.05) higher GAG content than the DCC (acellular) group. No significant differences in GAG content were observed between the BLANK and TGF groups at week 6. Only the CS and DCC groups showed statistically significant changes in GAG content over time. The GAG contents in the CS and CS (acellular) groups at week 0 were 2.3-fold (p < 0.05) and 15-fold (p < 0.05) higher than their corresponding values at weeks 6, respectively. Additionally, week 3 GAG amounts in the CS and CS (acellular) groups were 2.2-fold (p < 0.05) and 12-fold (p < 0.05) higher than their matching values at week 6, respectively. On the other hand, the DCC group had 6.9-fold (p < 0.05) more GAG at week 6 than at week 3. No significant differences in GAG content were observed in the BLANK, TGF, and DCC (acellular) groups over time.
HYP content
The HYP content results revealed that the DCC group at week 0 had 185-fold (p < 0.05), 244-fold (p < 0.05), and 71-fold (p < 0.05) higher HYP content than the BLANK, TGF, and CS groups, respectively (Figure 6(b)). In addition, the DCC group at week 0 outperformed the DCC (acellular) with a HYP content that was 1.2-fold (p < 0.05) higher. Week 3 HYP content results showed that the HYP content in the DCC group was 189-fold (p < 0.05), 458-fold (p < 0.05), and 52-fold (p < 0.05) higher than in the BLANK, TGF, and CS groups, respectively. Likewise, the HYP content in the DCC group at week 6 was 83-fold (p < 0.05), 99-fold (p < 0.05), and 62-fold (p < 0.05) higher than in the BLANK, TGF, and CS groups, respectively. No significant differences in HYP content were observed among the BLANK, TGF, and CS groups at weeks 0, 3, or 6. Only the DCC and DCC (acellular) groups showed statistically significant differences in HYP over time. The DCC group at week 0 had 1.6-fold (p < 0.05) 1.7-fold (p < 0.05) higher HYP content than at weeks 3 and 6, respectively. The HYP content in the DCC (acellular) group at week 0 was 1.4-fold (p < 0.05) and 1.7-fold (p < 0.05) higher than its matching values at weeks 3 and 6, respectively.
Gene expression
SOX9 and COL2AI
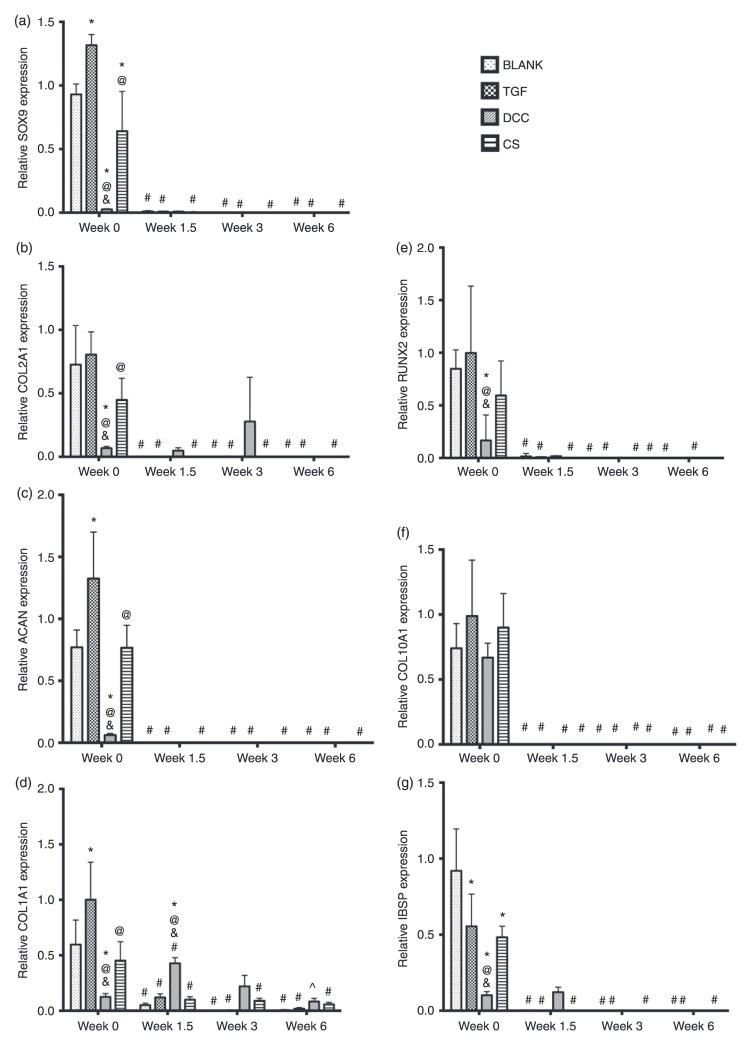
The BLANK group at week 0 had 31-fold (p < 0.05) and 1.1-fold (p < 0.05) higher SOX9 expression than the DCC and CS groups, respectively (Figure 7(a)). The TGF group SOX9 expression at week 0 was 1.4-fold (p < 0.05), 44-fold (p < 0.05), and 1.6-fold (p < 0.05) higher than the SOX9 expression of the BLANK, DCC, and CS groups, respectively. The CS group at week 0 also had 27-fold (p < 0.05) higher SOX9 expression than the DCC group. No significant differences in SOX9 expression were observed among the groups at weeks 1.5, 3, or 6. The SOX9 expression for all of the groups at week 1.5 and beyond was essentially negligible, with the BLANK, TGF, and CS groups showing statistically significant decrease in expression from their corresponding week 0 values.
Figure 7.
Relative gene expression. (a) SOX9 expression, (b) COL2AI expression, (c) ACAN expression, (d) COLIAI expression, (e) RUNX2 expression, (f) COLI0AI expression, and (g) IBSP expression. All values are expressed as the average + standard deviation (n = 6). The TGF positive control group had higher expression whereas the DCC group had lower expression of chondrogenic signals at week 0. *Significant difference from the BLANK group at same time point, @significant difference from the TGF group at same time point, &significant difference from the CS group at same time point, #significant difference from its value at week 0, and ^significant difference from its value at week 1.5 (p < 0.05).
The COL2A1 expression values for the BLANK group at weeks 1.5, 3, and 6; TGF group at weeks 3 and 6; DCC group at weeks 1.5, 3, and 6; and CS group at weeks 1.5, 3, and 6, were marked as zero because the fluorescence intensities in these samples did not cross the threshold fluorescence. The TGF group at week 0 had 11-fold (p < 0.05) and 2.4-fold (p < 0.05) higher COL2A1 expression than the DCC and CS groups, respectively (Figure 7(b)). The BLANK and the CS groups at week 0 had 10-fold (p < 0.05) and 4.9-fold (p < 0.05) higher COL2A1 expression than the DCC group, respectively. No significant differences in COL2A1 expression were observed among the groups at weeks 1.5, 3, or 6. The BLANK, TGF, and CS groups showed statistically significant changes in COL2A1 expression values over time while no significant differences in COL2A1 expression were observed in the DCC group over time. The week 0 COL2A1 expression values for the BLANK, TGF, and CS groups were statistically significantly higher than their corresponding values at weeks 1.5, 3, and 6.
ACAN and COLIAI
The TGF group at week 0 had 1.5-fold (p < 0.05), 20-fold (p < 0.05), and 1.5-fold (p < 0.05) higher ACAN expression than the expression levels of the BLANK, DCC, and CS groups, respectively (Figure 7(c)). Both the BLANK and the CS group at week 0 had 13-fold (p < 0.05) higher ACAN expression than the DCC group at that time point. No significant differences in ACAN expression were observed among the groups at weeks 1.5, 3, or 6. All the groups at week 1.5 and beyond had negligible ACAN expression, with the BLANK, TGF, and CS groups exhibiting statistically significant decrease in expression from their corresponding week 0 values.
The BLANK, TGF, and CS groups at week 0 had 4.6-fold (p < 0.05), 7.7-fold (p < 0.05), and 3.5-fold (p < 0.05) higher COL1A1 expression than the DCC group, respectively (Figure 7(d)). The TGF group at week 0 had 1.7-fold (p < 0.05) and 2.2-fold (p < 0.05) higher COL1A1 expression than the BLANK and CS groups, respectively. At week 1.5, the DCC group out-performed the BLANK, TGF, and CS groups in COL1A1 expression with expression value that was 8.6-fold (p < 0.05), 3.6-fold (p < 0.05), and 4.3-fold (p < 0.05) higher, respectively. No significant differences in COL1A1 expression were observed among the groups at weeks 3 or 6. All of the groups showed statistically significant changes in COL1A1 expression over time. The BLANK group at week 0 had 12-fold (p < 0.05), 300-fold (p < 0.05), and 60-fold (p < 0.05) higher COL1A1 expression than its expression at weeks 1.5, 3, and 6, respectively. The TGF and the CS groups showed a similar pattern to the BLANK group in COL1A1 expression. The TGF group COL1A1 expression at week 0 was 8.3-fold (p < 0.05), 333-fold (p < 0.05), and 100-fold (p < 0.05) higher than at weeks 1.5, 3, and 6, respectively. The CS group at week 0 had 4.5-fold (p < 0.05), 5-fold (p < 0.05), 7.5-fold (p < 0.05) higher COL1A1 expression than at weeks 1.5, 3, and 6, respectively. The COL1A1 expression in the DCC group peaked at week 1.5 with an expression value that was 3.3-fold (p < 0.05) and 5.4-fold (p < 0.05) higher than at weeks 0 and 6, respectively.
RUNX2, COLI0AI, and IBSP
The BLANK, TGF, and CS groups at week 0 had 13-fold (p < 0.05), 15-fold (p < 0.05), and 13-fold (p < 0.05) higher RUNX2 expression than the DCC group, respectively (Figure 7(e)). No significant differences were observed in RUNX2 expression among the BLANK, TGF, and CS groups at week 0. Additionally, no significant differences were observed in RUNX2 expression among all the groups at weeks 1.5, 3, or 6. The RUNX2 expression for all of the groups at week 1.5 and beyond was negligible, with the BLANK, TGF, and CS groups showing statistically significant decrease in expression from their corresponding week 0 values.
The COL10A1 expression values for the BLANK group at weeks 1.5, 3, and 6; TGF group at weeks 3 and 6; DCC group at weeks 1.5, 3, and 6; and CS group at weeks 1.5, 3, and 6, were marked as zero because the fluorescence intensities in these samples did not cross the threshold fluorescence. No significant differences were observed in COL10A1 expression among the groups at any time point (Figure 7(f)). All of the groups had negligible COL10A1 expression at week 1.5 and beyond with statistically significant decrease in expression from their corresponding week 0 values.
The IBSP expression values for the BLANK group at weeks 3 and 6, and the CS group at week 6, were marked as zero because the fluorescence intensities in these samples did not cross the threshold fluorescence. The BLANK group at week 0 had 2.1-fold (p < 0.05), 10-fold (p < 0.05), and 1.9-fold (p < 0.05) higher IBSP expression than the expression values of the TGF, DCC, and CS groups, respectively (Figure 7(g)). The IBSP expression values of the TGF and CS group at week 0 were 4.9-fold (p < 0.05) and 5.3-fold (p < 0.05) higher than the IBSP expression of the DCC group, respectively. No significant differences were observed in IBSP expression among the groups at weeks 1.5, 3, or 6. The BLANK, TGF, and CS groups had negligible IBSP expression at week 1.5 and beyond with statistically significant decrease in expression from their corresponding week 0 values. On the other hand, the DCC group had negligible IBSP expression at week 3 and 6 with no statistically significant differences in its expression values over time.
Histology and IHC
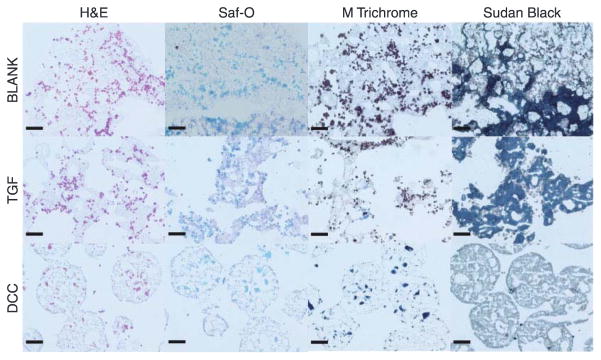
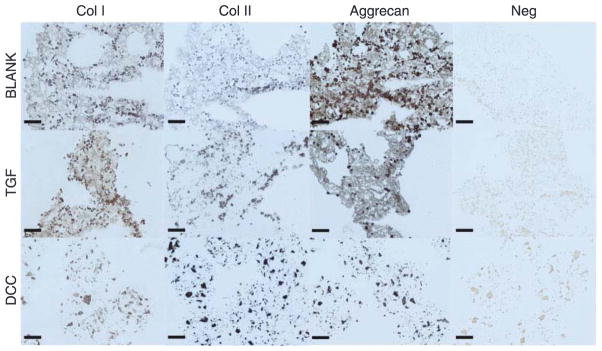
Figure 8 represents the histological images from the BLANK, TGF, and DCC groups at week 6. The sections from the CS scaffolds washed off during the staining process after multiple careful attempts, therefore no histological and IHC images are available from the CS scaffolds. H&E images showed that the cells in the BLANK and TGF groups were present primarily around the periphery of the microspheres, whereas cells in the DCC group were also found to have infiltrated the microspheres. No differences were observed in Safranin O (stains GAGs orange) staining intensities among the groups at week 6. Masson’s trichrome, which stains collagen dark blue, depicted the staining intensities to be greater in the TGF and DCC groups than in the BLANK group. All of the groups stained for Sudan Black, with higher staining intensities in the BLANK and TGF groups than in the DCC group. In addition, the spherical shape of the microspheres was still evident only in the DCC group. Figure 9 depicts the IHC images obtained from the BLANK, TGF, and DCC groups at week 6. All three of the groups stained positively for collagen I, with staining intensities in the BLANK and TGF groups being higher than the intensity in the DCC group. No differences in collagen II staining intensities were observed among the BLANK, TGF, and DCC groups. Aggrecan staining was more intense in the BLANK and TGF groups than in the DCC group. The aggrecan staining in the DCC group appeared to be distributed in clusters within the microspheres themselves, perhaps indicative of the encapsulated DCC itself.
Figure 8.
Histological staining images of cell-seeded microsphere-based constructs at week 6. BLANK, TGF, and DCC scaffolds were stained for H&E, Safranin-O, Masson’s trichrome, and Sudan Black. No images could be obtained from the CS group as the sections washed off the slides during the staining process. The Sudan Black staining intensities for residual polymer were higher in the BLANK and TGF groups compared with the DCC group. Scale bars: 100 μm.
Figure 9.
Immunohistochemical staining images of microsphere-based constructs at week 6. BLANK, TGF, and DCC were stained for collagen I, collagen II, and aggrecan. No images could be obtained from the CS group as the sections washed off from the slides during the staining process. The BLANK and the TGF group stained more intensely for aggrecan than the DCC group. Images of negative controls (primary antibody omitted) are also shown. Scale bars: 100 μm.
Discussion
The current study for the first time compared the effects of encapsulating DCC versus CS in promoting the chondrogenic differentiation of mesenchymal stem cells. DCC or CS could potentially supply the neighboring cells with raw materials (i.e. bioactive signals and building blocks) for differentiation along the chondrogenic lineage.5,6,33,34 Our previous studies have shown that the incorporation of DCC or CS, at concentrations of 10 or 20 wt%, in microsphere-based scaffolds rendered the scaffolds bioactive, which further led to greater cell numbers compared with the “blank” (PLGA-only) controls and also enhanced matrix synthesis by the seeded rBMSCs.9,25,26 In the present study, DCC or CS were incorporated at higher concentrations (30 wt%) in microsphere-based scaffolds than our previous iterations of these scaffolds and the potential of encapsulated DCC or CS were evaluated side-by-side to influence the chondrogenic differentiation of rBMSCs.
The SEM images depicted that all four types of microspheres were uniform in size, with average microsphere diameter ranging between 160 and 180 μm (Supplementary Figure 2). The TGF, DCC, and CS microspheres possessed minute pores on their surface formed perhaps as a result of particulate leaching during solvent evaporation.9,25,26,29 The DCC encapsulation imparted the PLGA microspheres a rough appearance, differing from our prior work where PLGA microspheres encapsulating DCC at a concentration of 10 wt% had a smooth surface.9 The higher concentrations of DCC encapsulated in the current study might have resulted in an uneven surface of the DCC microspheres. The microsphere-based scaffolds from all of the four groups were observed to be porous in nature with interconnected pores, as we have consistently observed in previous work.25,28 Additionally, it was noted that microspheres in the CS scaffolds had a relatively higher degree of sintering than the degree of sintering observed in the other three groups. Failure to observe a similar effect in the DCC group may have indicated differences in partitioning of CS and DCC in the polymeric microspheres that might have contributed to the higher extent of sintering in the CS group. The EDS maps for atomic nitrogen and sulfur demonstrate that TGF, DCC, and CS were uniformly distributed throughout the interior of the microspheres in the corresponding groups with no evidence of agglomeration at any site. The presence of sulfur in the TGF microspheres could be attributed to the cysteine and methionine amino acid residues present in TGF-β3.
In contrast to our previous findings,9,26 it was observed that the compressive modulus of the CS group was significantly higher than the moduli of the TGF and DCC groups. The higher compressive modulus of CS scaffolds may be attributed to the higher degree of microsphere sintering observed in the CS group. The DCC and CS groups also had significantly higher porosities than the control groups. The higher porosities in the DCC and CS groups is likely associated with the presence of minute pores on the surface of the microspheres that imparted them an additional level of microporosity in addition to the macroporosity obtained from the microsphere sintering.25
With regard to biochemical content, the CS groups at all time points had significantly higher cell numbers (i.e. DNA content) than the other three groups. Additionally, it was observed that the DCC group at week 0 had significantly higher cell numbers than controls at that time point. The higher cell numbers in the CS scaffolds is indicative of higher cell proliferation in these scaffolds, as CS is known to have a mitogenic effect on the proliferation of MSCs.10,25,35,36 The higher cell numbers in the DCC scaffolds at week 0 may be attributed to the rough surface of the microspheres that might have promoted initial cell attachment.37,38 We observed a similar phenomenon previously in DCC-coated microspheres where the DCC coated scaffold groups had higher cell numbers at week 0.9 Higher concentrations of DCC used in the current study led to higher amounts of DCC being present on the surface of the microspheres, thus aiding in initial cell attachment by providing additional cell adhesion sites. The CS and DCC groups outperformed the other three groups in GAG and HYP contents at all time points, respectively. Higher amounts of GAG and HYP in the CS and DCC groups was likely ascribed to the inherent GAGs and collagen present in these scaffolds that decreased over time as the scaffolds degraded. It was noted that at week 0, the cell seeded DCC group had a significantly higher HYP content than its acellular counterpart, highlighting the contribution of cell proliferation to matrix synthesis in the DCC scaffolds. A major finding of the study was that the cellular DCC and CS groups at week 6 had significantly higher GAG contents than their acellular equivalents, suggestive of enhanced matrix production and/or retention/incorporation by the seeded cells in the DCC and CS groups. Together, these findings suggest that CS and DCC encapsulation in microsphere-based scaffolds promoted new cartilage-related matrix synthesis, and support our previous findings of a modulatory effect of CS and DCC on rBMSCs.9,25
It must be noted that the expression of the osteogenic markers RUNX2 and IBSP remained low in all of the groups throughout the 6-week culture period, indicating that the rBMSCs did not appreciably differentiate toward the osteogenic lineage in any of the scaffold groups, which might be a limitation with using mesenchymal stem cells that themselves have a propensity for exhibiting a hypertrophic phenotype.39 The gene expression results for SOX9, COL2A1, and ACAN showed that the expression of these genes was largely suppressed in the DCC and CS groups at week 0 compared with the control groups at that time point, with the CS group outperforming the DCC group at week 0, although as expected, the positive control TGF group outperformed the BLANK group in SOX9 and ACAN expression. The lower expression of chondrogenic markers in the DCC and CS groups early on indicated that the DCC and CS inhibited the expression of chondrogenic markers by creating an environment that is already high in cartilage-like ECM components. We previously observed a similar phenomenon in hydroxyapatite (HAp) and tricalcium phosphate (TCP) encapsulated microsphere- based scaffolds where the expression of osteogenic markers in MSCs was largely suppressed due to the presence of inherent minerals in the scaffolds.25,30 Additionally, the chondrogenic gene expression in the DCC group did not increase over time, which was in contrast to the findings of some other groups utilizing cartilage matrix, where chondrogenic gene expression in the cartilage matrix scaffolds either was maintained or increased over time.8,40 Failure to observe up-regulation of the chondrogenic genes at later time points in the DCC group hint that the decellularization process might have impaired some critical cartilage matrix components required for cells to guide them toward a chondrogenic lineage, or perhaps that the encapsulation process itself may have been detrimental to bioactivity.41 Since decellularization can result in changes in cartilage matrix, we believe that the encapsulation of other forms of cartilage matrix34 (e.g. devitalized cartilage, DVC) might enhance the chondroinductivity of microsphere-based scaffolds, which is a matter of further investigation. In our prior work, we have demonstrated the raw materials such as CS in combination with growth factors like TGF-β3 can enhance the secretion of cartilage specific matrix components. Moreover, Almeida et al.42 noticed that a combination of a cartilage-ECM-derived scaffold and stimulation with TGF-β3 can induce chondrogenesis in human fat-pad-derived stem cells, so perhaps encapsulating CS, DCC, or DVC in combination with the growth factor may provide a synergistic effect, thus boosting the chondrogenic potential of microsphere-based scaffolds.
The histological images at week 6 pointed toward higher cell numbers in the BLANK and TGF groups than in the DCC group; however, no significant differences were observed in the DNA content among the three groups at week 6. The cells in the BLANK and TGF groups were found to be predominantly present around the periphery of the microspheres, while cells in the DCC group were also observed within the microsphere matrix, suggesting that the porous nature of DCC microspheres allowed for cell infiltration to occur within the microsphere matrix or perhaps there was residual DNA from the DCC itself. The Safranin- O staining intensities were not different among the BLANK, TGF, and DCC groups, which was consistent with no observed differences in the GAG content among the three groups at that time point. The Masson’s trichrome images were in agreement with our HYP content results, both showing that the DCC group had higher collagen content than the BLANK and TGF groups at week 6. The higher net collagen content in the DCC group was due to the inherent collagen present in the DCC scaffolds as confirmed by the Masson’s trichrome staining images of the acellular DCC scaffolds (Supplementary Figure 3). Sudan Black staining hinted that encapsulation of DCC altered polymer degradation (perhaps accelerating it). The staining intensities for residual polymer were significantly higher in the BLANK and TGF groups than the intensity in the DCC group. PLGA microspheres degrade via bulk erosion where the rate-limiting step is the diffusion of water molecules into the microsphere core. DCC microspheres because of their porous nature may have allowed faster diffusion of water into their core, thereby accelerating the polymer degradation rate relative to the BLANK and TGF groups. The IHC images illustrated that the BLANK and TGF groups stained more intensely for collagen I and aggrecan, which was consistent with the higher gene expression of collagen I and aggrecan in the BLANK and TGF groups than the DCC group at week 0.
In conclusion, the results of the current study demonstrated that encapsulation of DCC and CS altered the morphological and structural properties of both the microspheres and the scaffolds. Moreover, the encapsulation of DCC and CS led to enhanced cell attachment and proliferation on microsphere-based scaffolds thereby, corroborating with our earlier studies suggesting that both DCC and CS were bioactive when incorporated into microsphere-based scaffolds.9,25 By providing an environment rich in GAGs and collagen, the DCC and CS scaffolds initially impeded the chondrogenic gene expression in rBMSCs; however, biochemical evidence suggested of a modulatory effect of DCC and CS on matrix synthesis by rBMSCs. Additionally, the differences highlighted between the DCC and CS groups by the biochemical content analysis and the gene expression patterns hint that rBMSCs responded differently to both DCC and CS encapsulated into the microsphere-based scaffolds. Although the cellular response did not provide compelling evidence of DCC and CS enhancing chondrogenesis in microsphere-based scaffolds, the increased GAG content in these groups relative to acellular controls after 6 weeks was encouraging. There is a need to further refine the technology by using even higher concentrations of CS and DCC, or perhaps different forms of cartilage matrix (e.g. devitalized cartilage), or combinations of these raw materials with TGF-β.
Acknowledgments
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors thank Prem Thapa for his assistance with SEM imaging and EDS mapping.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health (R01 AR056347) and Kansas Bioscience Authority Rising Star Award.
Footnotes
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Di Luca A, Van Blitterswijk C, Moroni L. The osteochondral interface as a gradient tissue: From development to the fabrication of gradient scaffolds for regenerative medicine. Birth Defects Res C: Embryo Today. 2015;105:34–52. doi: 10.1002/bdrc.21092. [DOI] [PubMed] [Google Scholar]
- 2.Shimomura K, Moriguchi Y, Murawski CD, et al. Osteochondral tissue engineering with biphasic scaffold: current strategies and techniques. Tissue Eng B. 2014;20:468–476. doi: 10.1089/ten.TEB.2013.0543. [DOI] [PubMed] [Google Scholar]
- 3.Seo SJ, Mahapatra C, Singh RK, et al. Strategies for osteochondral repair: Focus on scaffolds. J Tissue Eng. 2014;5:1–14. doi: 10.1177/2041731414541850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopa S, Madry H. Bioinspired scaffolds for osteochondral regeneration. Tissue Eng A. 2014;20:2052–2076. doi: 10.1089/ten.tea.2013.0356. [DOI] [PubMed] [Google Scholar]
- 5.Cheng CW, Solorio LD, Alsberg E. Decellularized tissue and cell-derived extracellular matrices as scaffolds for orthopaedic tissue engineering. Biotechnol Adv. 2014;32:462–484. doi: 10.1016/j.biotechadv.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benders KEM, van Weeren PR, Badylak SF, et al. Extracellular matrix scaffolds for cartilage and bone regeneration. TRENDS Biotechnol. 2013;31:171–178. doi: 10.1016/j.tibtech.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Cheng N-C, Estes BT, Young T-H, et al. Engineered cartilage using primary chondrocytes cultured in a porous cartilage-derived matrix. Regen Med. 2011;6:81–93. doi: 10.2217/rme.10.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrigues NW, Little D, Sanchez-Adams J, et al. Electrospun cartilage-derived matrix scaffolds for cartilage tissue engineering. J Biomed Mater Res A. 2014;102:3998–4008. doi: 10.1002/jbm.a.35068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutherland AJ, Detamore MS. Bioactive microsphere-based scaffolds containing decellularized cartilage. Macromol Biosci. 2015;15:979–989. doi: 10.1002/mabi.201400472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawatjui N, Damrongrungruang T, Leeanansaksiri W, et al. Silk fibroin/gelatin–chondroitin sulfate–hyaluronic acid effectively enhances in vitro chondrogenesis of bone marrow mesenchymal stem cells. Mater Sci Eng: C. 2015;52:90–96. doi: 10.1016/j.msec.2015.03.043. [DOI] [PubMed] [Google Scholar]
- 11.Madeira C, Santhagunam A, Salgueiro JB, et al. Advanced cell therapies for articular cartilage regeneration. Trends Biotechnol. 2015;33:35–42. doi: 10.1016/j.tibtech.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Huang Z, Nooeaid P, Kohl B, et al. Chondrogenesis of human bone marrow mesenchymal stromal cells in highly porous alginate-foams supplemented with chondroitin sulfate. Mater Sci Eng: C. 2015;50:160–172. doi: 10.1016/j.msec.2015.01.082. [DOI] [PubMed] [Google Scholar]
- 13.Huang W, Li X, Shi X, et al. Microsphere based scaffolds for bone regenerative applications. Biomater Sci. 2014;2:1145–1153. doi: 10.1039/c4bm00161c. [DOI] [PubMed] [Google Scholar]
- 14.Borden M, Attawia M, Laurencin CT. The sintered microsphere matrix for bone tissue engineering: in vitro osteoconductivity studies. J Biomed Mater Res. 2002;61:421–429. doi: 10.1002/jbm.10201. [DOI] [PubMed] [Google Scholar]
- 15.Brown JL, Nair LS, Laurencin CT. Solvent/non-solvent sintering: a novel route to create porous microsphere scaffolds for tissue regeneration. J Biomed Mater Res B: Appl Biomat. 2008;86:396–406. doi: 10.1002/jbm.b.31033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nukavarapu SP, Kumbar SG, Brown JL, et al. Polyphosphazene/nano-hydroxyapatite composite microsphere scaffolds for bone tissue engineering. Biomacromolecules. 2008;9:1818–1825. doi: 10.1021/bm800031t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown LR, Gombotz WR, Healy MS. Very low temperature casting of controlled release microspheres. US Patent Office; 1991. [Google Scholar]
- 18.Berkland C, Kim K, Pack DW. Fabrication of PLG microspheres with precisely controlled and monodisperse size distributions. J Control Release. 2001;73:59–74. doi: 10.1016/s0168-3659(01)00289-9. [DOI] [PubMed] [Google Scholar]
- 19.Blaker JJ, Knowles JC, Day RM. Novel fabrication techniques to produce microspheres by thermally induced phase separation for tissue engineering and drug delivery. Acta Biomater. 2008;4:264–272. doi: 10.1016/j.actbio.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 20.Dormer NH, Singh M, Zhao L, et al. Osteochondral interface regeneration of the rabbit knee with macroscopic gradients of bioactive signals. J Biomed Mater Res A. 2012;100:162–170. doi: 10.1002/jbm.a.33225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohan N, Dormer NH, Caldwell KL, et al. Continuous gradients of material composition and growth factors for effective regeneration of the osteochondral interface. Tissue Eng A. 2011;17:2845–2855. doi: 10.1089/ten.tea.2011.0135. [DOI] [PubMed] [Google Scholar]
- 22.Dormer NH, Busaidy K, Berkland CJ, et al. Osteochondral interface regeneration of rabbit mandibular condyle with bioactive signal gradients. J Oral Maxillof Surg. 2011;69:e50–e57. doi: 10.1016/j.joms.2010.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dormer NH, Singh M, Wang L, et al. Osteochondral interface tissue engineering using macroscopic gradients of bioactive signals. Ann Biomed Eng. 2010;38:2167–2182. doi: 10.1007/s10439-010-0028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohan N, Gupta V, Sridharan BP, et al. Microsphere- based gradient implants for osteochondral regeneration: a long-term study in sheep. Regen Med. 2015;10:709–728. doi: 10.2217/rme.15.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta V, Mohan N, Berkland C, et al. Microsphere-based scaffolds carrying opposing gradients of chondroitin sulfate and tricalcium phosphate. Front Bioeng Biotechnol. 2015;3:96. doi: 10.3389/fbioe.2015.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohan N, Gupta V, Sridharan B, et al. The potential of encapsulating “raw materials” in 3D osteochondral gradient scaffolds. Biotechnol Bioeng. 2014;111:829–841. doi: 10.1002/bit.25145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sutherland AJ, Beck EC, Dennis SC, et al. Decellularized cartilage may be a chondroinductive material for osteochondral tissue engineering. PLoS ONE. 2015;10:e0121966–13. doi: 10.1371/journal.pone.0121966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh M, Morris CP, Ellis RJ, et al. Microsphere-based seamless scaffolds containing macroscopic gradients of encapsulated factors for tissue engineering. Tissue Eng C-Methods. 2008;14:299–309. doi: 10.1089/ten.tec.2008.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh M, Dormer N, Salash JR, et al. Three-dimensional macroscopic scaffolds with a gradient in stiffness for functional regeneration of interfacial tissues. J Biomed Mater Res A. 2010;94:870–876. doi: 10.1002/jbm.a.32765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dormer NH, Qiu Y, Lydick AM, et al. Osteogenic differentiation of human bone marrow stromal cells in hydroxyapatite-loaded microsphere-based scaffolds. Tissue Eng A. 2012;18:757–767. doi: 10.1089/ten.tea.2011.0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeon JH, Bhamidipati M, Sridharan B, et al. Tailoring of processing parameters for sintering microsphere-based scaffolds with dense-phase carbon dioxide. J Biomed Mater Res B: Appl Biomater. 2013;101:330–337. doi: 10.1002/jbm.b.32843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh M, Detamore MS. Stress relaxation behavior of mandibular condylar cartilage under high-strain compression. J Biomech Eng. 2009;131:061008. doi: 10.1115/1.3118776. [DOI] [PubMed] [Google Scholar]
- 33.Renth AN, Detamore MS. Leveraging “raw materials” as building blocks and bioactive signals in regenerative medicine. Tissue Eng B. 2012;18:341–362. doi: 10.1089/ten.teb.2012.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sutherland AJ, Converse GL, Hopkins RA, et al. The bioactivity of cartilage extracellular matrix in articular cartilage regeneration. Adv Healthcare Mater. 2014;4:29–39. doi: 10.1002/adhm.201400165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan H, Hu Y, Zhang C, et al. Cartilage regeneration using mesenchymal stem cells and a PLGA–gelatin/chondroitin/hyaluronate hybrid scaffold. Biomaterials. 2006;27:4573–4580. doi: 10.1016/j.biomaterials.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 36.Uygun BE, Stojsih SE, Matthew HW. Effects of immobilized glycosaminoglycans on the proliferation and differentiation of mesenchymal stem cells. Tissue Eng A. 2009;15:3499–3512. doi: 10.1089/ten.tea.2008.0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kay S, Thapa A, Haberstroh KM, et al. Nanostructured polymer/nanophase ceramic composites enhance osteoblast and chondrocyte adhesion. Tissue Eng. 2002;8:753–761. doi: 10.1089/10763270260424114. [DOI] [PubMed] [Google Scholar]
- 38.Boyan BD, Hummert TW, Dean DD, et al. Role of material surfaces in regulating bone and cartilage cell response. Biomaterials. 1996;17:137–146. doi: 10.1016/0142-9612(96)85758-9. [DOI] [PubMed] [Google Scholar]
- 39.Somoza RA, Welter JF, Correa D, et al. Chondrogenic differentiation of mesenchymal stem cells: challenges and unfulfilled expectations. Tissue Eng B. 2014;20:596–608. doi: 10.1089/ten.teb.2013.0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moradi A, Ataollahi F, Sayar K, et al. Chondrogenic potential of physically treated bovine cartilage matrix derived porous scaffolds on human dermal fibroblast cells. J Biomed Mater Res A. 2016;104:245–256. doi: 10.1002/jbm.a.35561. [DOI] [PubMed] [Google Scholar]
- 41.Keane TJ, Swinehart IT, Badylak SF. Methods of tissue decellularization used for preparation of biologic scaffolds and in vivo relevance. Methods. 2015;84:25–34. doi: 10.1016/j.ymeth.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 42.Almeida HV, Liu Y, Cunniffe GM, et al. Controlled release of transforming growth factor-β3 from cartilage-extra-cellular-matrix-derived scaffolds to promote chondrogenesis of human-joint-tissue-derived stem cells. Acta Biomater. 2014;10:4400–4409. doi: 10.1016/j.actbio.2014.05.030. [DOI] [PubMed] [Google Scholar]