Abstract
Microtubules govern actin network remodeling in a wide range of biological processes, yet the mechanisms underlying this cytoskeletal crosstalk have remained obscure. Here we used single-molecule fluorescence microscopy to show that the microtubule plus-end associated protein CLIP-170 binds tightly to formins to accelerate actin filament elongation. Furthermore, we observed mDia1 dimers and CLIP-170 dimers co-tracking growing filament ends for minutes. CLIP-170-mDia1 complexes promoted actin polymerization approximately 18 times faster than free barbed end growth, while simultaneously enhancing protection from capping protein. We used a microtubule-actin dynamics co-reconstitution system to observe CLIP-170-mDia1 complexes being recruited to growing microtubule ends by EB1. The complexes triggered rapid growth of actin filaments that remained attached to the microtubule surface. These activities of CLIP-170 were required in primary neurons for normal dendritic morphology. Thus, our results reveal a cellular mechanism whereby growing microtubule plus-ends direct rapid actin assembly.
Tight coordination between the microtubule (MT) and actin cytoskeletons is required for fundamental processes such as directed cell migration, neuronal arborization, and phagocytosis (1–3). In many of these settings, the growth of MT plus-ends into actin-rich cortical regions triggers changes in actin assembly-based functions (4, 5). In plant cells, following washout of the actin depolymerizing drug Latrunculin B (LatB), all new actin polymerization occurs from MT plus-ends (6). In fission yeast, MT plus-ends direct formin-mediated actin cable assembly during “new end take off” (7, 8). In other systems, MT plus-ends play an instrumental role in steering actin-based motility of neuronal growth cones during neurite outgrowth and axonal guidance (9–11). Thus, it has long been hypothesized that molecular cues associated with MT plus-ends directly govern localized actin assembly; however, no such mechanism has been defined.
Formins directly stimulate both the nucleation and elongation phases of actin filament assembly, and perform essential roles in constructing diverse cellular actin structures, including actin cables, stress fibers, filopodia, phagocytic cups, and cytokinetic rings (3). The dimeric formin homology 2 (FH2) domain nucleates actin filaments and remains processively attached to their growing barbed-ends as they elongate, while the adjacent FH1 domains recruit profilin-bound actin monomers to accelerate elongation (12, 13). The mammalian formin mDia1 supports one of the fastest known rates of actin filament elongation, at 55 subunits s−1 μM−1 (13).
CLIP-170 localizes to MT plus-ends in cells via interactions with the MT end-binding protein EB1, but also to actin-rich cortical zones (5, 14–19). In addition, CLIP-170 binds and colocalizes with mDia1 in macrophages, where both proteins are required for phagocytosis (5), suggesting that their in vivo functions are tied together. The budding yeast protein Smy1 directly inhibits the yeast formin Bnr1, targeting its FH2 domain (20) via ‘formin elongation effector domain’ (FEED) motifs (21). Here we found a FEED-like sequence in human CLIP-170, located in its central coiled-coil-rich region (Fig. 1A & Fig. S1A). We thus decided to purify full-length human CLIP-170 and test its possible effects on mDia1-mediated actin assembly using total internal reflection fluorescence (TIRF) microscopy.
Fig. 1. CLIP-170 accelerates formin-mediated actin filament elongation.
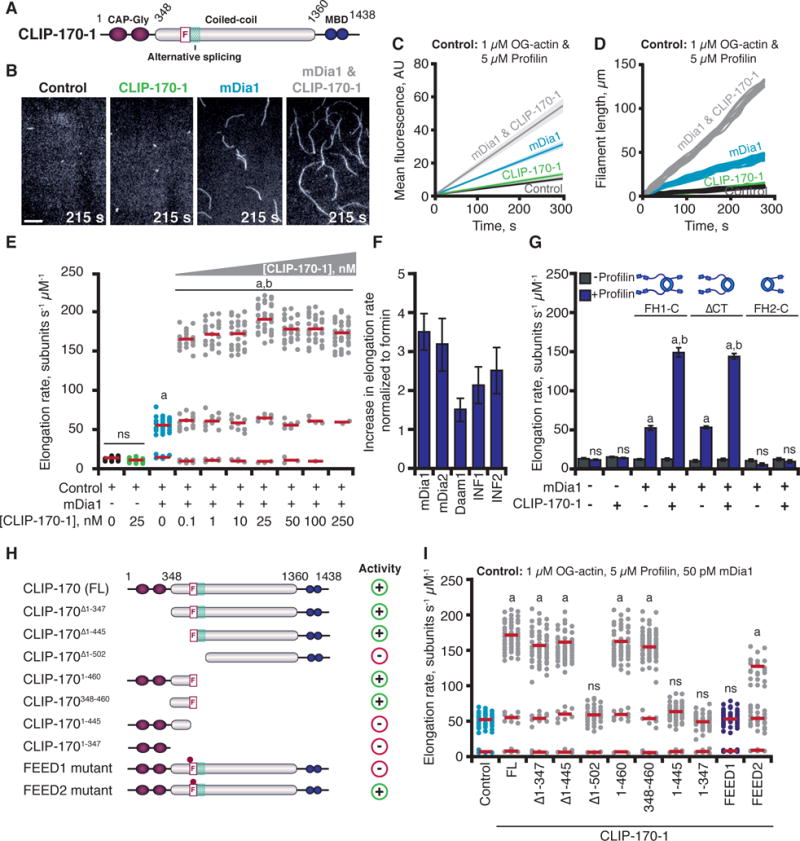
(A) CLIP-170 domains. F, FEED sequence; MBD, metal-binding domain. (B) Images from TIRF assays containing: 50 pM mDia1 and/or 25 nM CLIP-170-1, 215 s after initiation of actin assembly. Scale bar, 20 μm. (C) Kinetics of total actin polymer mass (fluorescence intensity) accumulation averaged from multiple fields of view (n ≥ 3). Linear fits plotted with 95% confidence intervals shaded. (D) Representative filament length traces (10 per condition) from TIRF movies. (E) Distributions of elongation rates from TIRF reactions as in (B) for different concentrations of CLIP-170-1. Distributions are shown from one of three independent experiments (n = 50 each). Red bars show mean elongation rates for subpopulations, measured from all filaments in three separate experiments (n = 150). (F) Fold increase in mean formin-mediated elongation rate stimulated by 25 nM CLIP-170-1. Error bars, SE. (G) Mean elongation rates from TIRF reactions containing different mDia1 constructs +/− profilin. (H) CLIP-170-1 constructs that enhance (+) or fail to enhance (−) the rate of mDia1-mediated elongation. FL, full-length; F, FEED sequence; teal box, alternatively spliced region, dots; FEED1 (445VEEE/AAAA448), FEED2 (450ITKGDLE/AAAAAAA456) mutants. (I) Distributions of elongation rates measured as in (E) for different CLIP-170-1 constructs. Reactions contain 1 μM G-actin (10% OG-labeled; 0.2% biotin-actin), ± 5 μM profilin, ± 50 pM mDia1, 50 pM mDia2, 50 pM Daam1, 100 nM INF1, or 100 nM INF2, ± 25 nM full-length CLIP-170-1 (wild type, FEED1 mutant, or FEED2 mutant). Statistical differences in (E, G, & I): ns, not significantly different from control; a, compared to control (actin and profilin) (p < 0.05); b, compared to formin control (actin, profilin, and formin) (p < 0.05).
Mammalian CLIP-170 is expressed as three alternatively spliced isoforms (CLIP-170-1, CLIP-170-2, and CLIP-170-3) (Fig. S1A). All splicing occurs in a specific region of CLIP-170 (457–502), and the FEED motif, present in all three isoforms, borders the spliced region (Fig. S1A). We purified all three isoforms of CLIP-170 as full-length proteins and tested their effects on mDia1 (FH1-FH2-tail)-mediated actin assembly. CLIP-170 isoforms had no effect on actin assembly alone, but each one dramatically accelerated the rate of mDia1-mediated actin filament elongation (Fig. 1B–D, 1E, S1B–E; Movies S1 & S2), while showing statistically insignificant effects on formin-mediated actin nucleation (Fig. S1F). A closer examination of the distribution of filament elongation rates in reactions containing both mDia1 and CLIP-170 revealed three subpopulations with distinct rates (Fig. 1E & Movie S2). The slowest had a mean elongation rate of 10.1 ± 1.3 (SE) subunits s−1 μM−1, similar to the rate of free barbed-end growth (22). The second subpopulation had a faster rate of 58.8 ± 8.8 subunits s−1 μM−1, similar to filament elongation mediated by mDia1 alone (Fig. 1E & Movie S2). The third was substantially accelerated, elongating at 179.1 ± 39.1 subunits s−1 μM−1, or approximately 18 times faster than free barbed-end growth in the absence of formins. This accelerated elongation occurred exclusively in reactions containing mDia1 and CLIP-170, and was observed at different actin concentrations (Fig. S2). The fraction of filaments in the reaction undergoing accelerated elongation scaled with increasing concentrations of CLIP-170 (Fig. 1E). Because 25 nM CLIP-170 supported the strongest effects, this concentration was used for all further tests. CLIP-170 also increased the elongation rates of filaments assembled by four other formins (mDia2, Daam1, INF1, and INF2) (Fig. 1F & Fig. S3). Thus, the regulatory effects of CLIP-170 extend to multiple formins, not just mDia1.
Accelerated elongation required profilin and the FH1 and FH2 domains of mDia1 (Fig. 1G). These are precisely the same requirements for mDia1-accelerated elongation without CLIP-170 (Fig. 1G) (13, 23). Thus, CLIP-170 may be functioning by further improving the mechanism of elongation already used by formins. We mapped the activities in CLIP-170 to specific domains (Fig. 1H & 1I). Because all three CLIP-170 isoforms had similar effects on mDia1, we mapped activities in the longest isoform, CLIP-170-1. All CLIP-170-1 fragments containing the FEED sequence supported accelerated elongation, and a short fragment (348–460) encompassing the FEED sequence was sufficient, whereas the N-terminal MT- and EB1-binding region (1–347) (15) was dispensable. Next, we generated two mutant alleles (alanine substitutions) in the FEED sequence of full-length CLIP-170-1 (Fig. S1A); the FEED1 mutant abolished accelerated elongation, and the FEED2 mutant reduced the rate of accelerated elongation (Fig. 1I). Thus, CLIP-170-mediated effects on mDia1 are FEED-dependent.
A number of possible mechanisms could explain the effects of CLIP-170 on mDia1, including formation of stable or transient CLIP-170-mDia1 complexes at growing barbed-ends of filaments. To address this, we used multi-wavelength single molecule TIRF microscopy to directly observe fluorescently labeled SNAP-tagged CLIP-170-1 and mDia1 molecules interacting with actin filaments during their assembly. 649-mDia1 and untagged mDia1 are dimeric and have indistinguishable activities (24). Step photobleaching analysis showed that 549-CLIP-170-1 molecules were also predominantly dimers (Fig. S4), as expected (15). 649-mDia1 molecules processively tracked the barbed-ends of filaments (Fig. 2A & Movie S3, left panel) (24), and 549-CLIP-170-1 showed rare interactions with filaments. However, in reactions containing both 649-mDia1 and 549-CLIP-170-1, they tracked the growing barbed-end together (Fig. 2A–E, & Movie S3, right panel). Importantly, 549-CLIP-170-1 and untagged CLIP-170-1 stimulated indistinguishable rates of formin-dependent accelerated elongation (Fig. 2B).
Fig. 2. CLIP-170 and mDia1 form a barbed-end tracking complex that accelerates actin filament elongation.
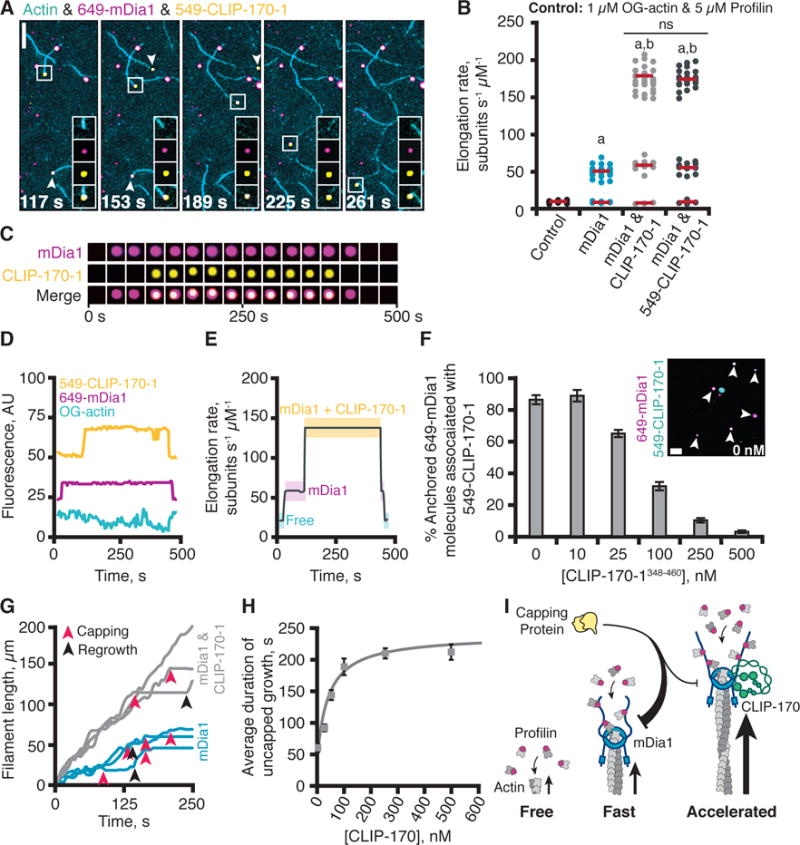
(A) Representative time points from a single molecule TIRF experiment. Reactions contain 1 μM G-actin (10% OG-labeled; 0.2% biotin-actin), 50 pM 649-mDia1, and 25 nM 549-CLIP-170-1. A barbed-end is highlighted in each frame (boxed), with insets showing individual and merged channels. Arrows show two additional growing barbed-ends with 649-mDia1 and 549-CLIP-170-1 associated. Scale bar, 10 μm. Insets 5 × 5 μm. (B) Effects of 25 nM CLIP-170-1 or 549-CLIP-170-1 on rate of mDia1-mediated actin filament elongation. Reactions as in (A). Statistical differences: ns, not significantly different from control; a, compared to control (actin and profilin) (p < 0.05); b, compared to formin control (actin, profilin, and formin) (p < 0.05). (C) Formation and dissociation of a CLIP-170-mDia1 complex at a barbed-end. (D) Fluorescence intensity profiles for each channel showing formation and dissociation of the CLIP-170-mDia1 complex in (C). (E) Elongation rates correlate with arrival and dissociation of mDia1 and/or CLIP-170 at the barbed-end. (F) Single-molecule colocalization of anchored 649-mDia1 and soluble full-length 549-CLIP-170-1 at different concentrations of unlabeled competitor fragment CLIP-170-1348–460. Data averaged from 3 fields of view in each of 3 independent experiments. Error bars, SE. Inset shows a representative field of view from a reaction with no competitor. Scale bar, 5 μm. (G) Representative filament traces from TIRF movies, conditions as in (A) except for the addition of 3 nM CP. Capping events (red arrows) and regrowth events (black arrows) are highlighted. (H) CLIP-170-1 enhances the duration of mDia1-mediated elongation in the presence of CP. Error bars, SE. (I) Cartoon of CLIP-170 joining mDia1 at barbed-end and increasing rate of elongation and duration of growth in the presence of CP.
Accelerated elongation occurred only in instances where 649-mDia1 and 549-CLIP-170-1 were observed together at the barbed-end (300 out of 300 events) (Fig. 2B–E & Movie S4). In our 3 min observation window, CLIP-170 dissociation events were rare (only 2 out of 100 barbed-ends tracked), suggesting that CLIP-170-1-mDia1 barbed-end tracking complexes are long-lived and highly processive. In a few instances, we observed formation and dissolution of the mDia1-CLIP-170 complex (Fig. 2D & 2E). For the filament shown, it initially grew at the free barbed-end rate (with no mDia1 or CLIP-170-1 associated), then upon association of 649-mDia1 growth transitioned to a faster rate, and subsequently when 549-CLIP-170-1 joined the formin on the barbed-end, growth immediately jumped to the further enhanced rate. After several minutes, 549-CLIP-170-1 dissociated, and growth fell back to the mDia1-supported rate, and then when 649-mDia1 dissociated, growth returned to the free barbed-end rate. Thus, when CLIP-170 joins mDia1 at the filament end, it forms a processive barbed-end tracking complex that enhances elongation. Single molecule experiments also showed that CLIP-170-1 binds anchored mDia1 even in the absence of actin, and that binding is competitively displaced by excess CLIP-170-1 fragment (348–460) (Fig. 2F).
A hallmark of formins is their ability to protect growing barbed-ends from capping protein (CP) (25–27). To test whether CLIP-170 influences this function of mDia1 (26, 28), we spiked in different concentrations of CP early in bulk assays. CP slowed mDia1-mediated actin assembly in a dose-dependent manner (Fig. S5A). These results are consistent with recent studies showing that CP and mDia1 can interact with barbed-ends simultaneously, and each catalyze dissociation of the other (29). CLIP-170-1 attenuated the effects of CP on mDia1 in these assays (Fig. S5B). In TIRF microscopy assays, CLIP-170-1 increased the average elongation rate of mDia1-nucleated filaments in the presence of CP (Fig. S5C). Similar effects were observed for each CLIP-170-1 construct that supported accelerated elongation (Fig. S5D). CLIP-170-1 also increased by up to 4-fold the duration of accelerated growth in the presence of CP (Fig. 2G, 2H, & S5C). CP effects were sometimes reversed, as indicated by an abrupt return to growth at mDia1-supported rates, but these events were rare (n < 10 for each condition out of 300 capping events analyzed). On the other hand, CLIP-170 did not affect the average duration of capping before resumed growth significantly (Fig. S5E). Thus, CLIP-170 not only increases the rate of mDia1-mediated actin filament elongation by 3–4 fold but also increases the duration of growth in the presence of CP by 4-fold (see model, Fig. 2I).
CLIP-170 and mDia1 each bind MTs and EB1 (17, 30–33). We thus investigated CLIP-170 and mDia1 interactions, alone and together, with dynamic MTs in the presence and absence of EB1 (Fig. 3A). 549-CLIP-170-1 interacted with MT sides, and was recruited to growing MT ends by EB1 (Fig. 3A, S6A, & S6B; Movie S5), as expected (17). 488-mDia1 bound to MT sides both in the presence and absence of EB1, but was not be recruited to MT ends (Fig. 3A, S6C, & S6D; Movie S6). In the absence of EB1, 488-mDia1 colocalized with 549-CLIP-170-1 on MT sides (Fig. 3B; Movie S6). However, in the presence of both EB1 and 549-CLIP-170-1, 488-mDia1 was recruited to MT plus-ends (Fig. 3C; Movie S6). These observations demonstrate a hierarchal recruitment scheme, in which EB1 recruits CLIP-170, which in turn recruits mDia1 to growing MT ends.
Fig. 3. CLIP-170-mDia1 complexes are recruited to MT plus-ends by EB1 and stimulate actin polymerization from the MT surface.
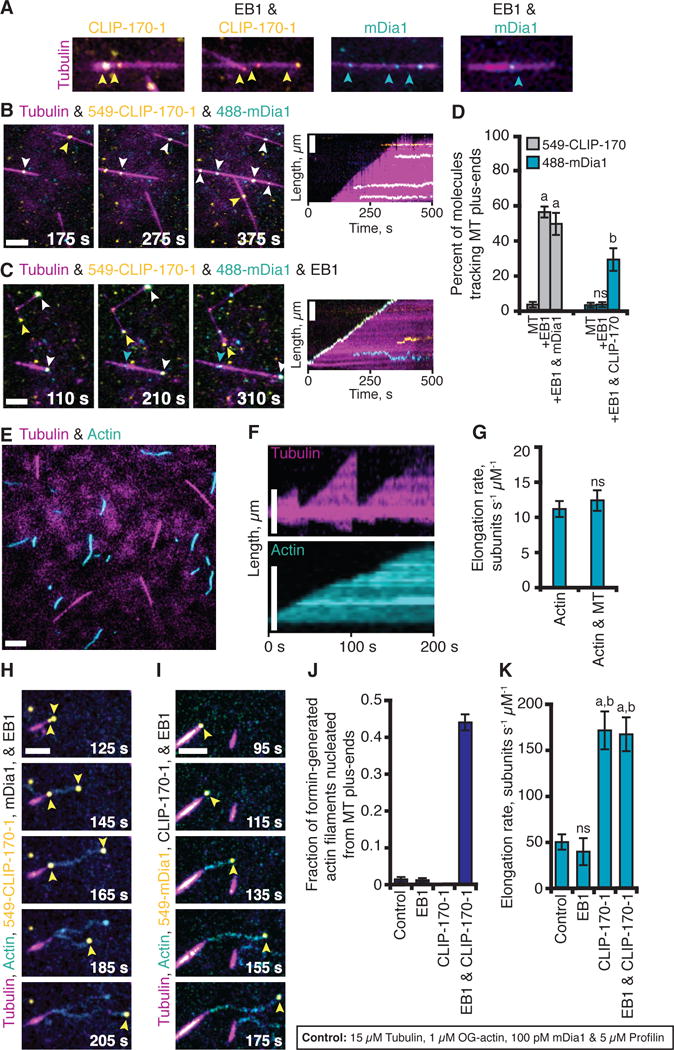
(A) TIRF movies show that 549-CLIP-170-1 binds MT sides, and tracks MT plus-ends only in the presence of EB1 (17). 488-mDia1 binds MT sides, with or without EB1, and does not track MT plus-ends. (B-C) 549-CLIP-170-1 and 488-mDia1 colocalize on MT sides (B), and track MT plus-ends together specifically in the presence of EB1 (C). Reactions in A-C contain 15 μM tubulin (30% AlexaFluor649 labeled), biotinylated GMP-CPP MT seeds, and variable components: 25 nM 549-CLIP-170-1, 100 pM 488-mDia1, and 500 nM EB1. (D) Percentage of molecules tracking MT plus-ends from reactions in (A–C); data averaged from 3 experiments (n > 100). Statistical differences: ns, not different from control; a, compared to CLIP-170-1 (p < 0.05); b, compared to mDia1 (p < 0.05). (E) Co-reconstitution of MTs undergoing dynamic instability and actin filaments polymerizing. Reactions contain all the same components as in (A–C) plus 1 μM G-actin (10% OG-labeled; 0.2% biotin-actin) and 5 μM profilin. (F) Kymographs of MT and actin dynamics. (G) Rate of actin filament elongation does not change significantly in the presence and absence of MTs; data averaged from 3 experiments (n = 50). (H) With the addition of unlabeled EB1 and mDia1, 549-CLIP-170-1 molecules (yellow arrows) were recruited to MT plus-ends, where they triggered assembly of actin filaments that grew at the accelerated rate. (I) Similar observations as in (H) except using 549-mDia1 (yellow arrows) and unlabeled CLIP-170-1. (J) Fraction of formin-generated actin filaments that grew from MT plus-ends in reactions as in (H & I); data from 3 experiments (n = 50). (K) Actin elongation rates from reactions as in (H & I); data from 3 experiments (n = 50). Statistical differences: ns, not different from control; a, compared to actin alone or control (p < 0.05); b, compared to EB1 control (p < 0.05). Error bars in all panels, SE.
We developed a co-reconstitution TIRF system that enabled imaging of MTs undergoing dynamic instability and actin filaments polymerizing simultaneously (Fig. 3E, 3F, & 3G; Movie S7). In the presence of EB1, CLIP-170, and mDia1 (but not in reactions lacking any of these components), we observed striking coordination of growing MT ends with formation of rapidly polymerizing actin filaments. With unlabeled EB1 and mDia1, 549-CLIP-170-1 molecules were dynamically recruited to growing MT plus-ends, where they triggered formation of actin filaments that polymerized from the MT surface (Fig. 3H; Movie S8). Similar observations were made with unlabeled EB1 and CLIP-170-1, and 549-mDia1 (Fig. 3I; Movie S9). About half of the actin filaments assembled by mDia1 originated from MT plus-ends (Fig. 3J), and in each case, 549-CLIP-170-1 or 549-mDia1 tracked the growing barbed-ends, which polymerized at the accelerated rate away from the MT (Fig. 3K). Actin filament pointed ends remained attached to the MT surface on average for 45 ± 10.3 s, until they spontaneously detached or were released by a MT catastrophe event.
To test the importance of these activities in a more physiological setting, we used a short hairpin RNA (shCLIP-170) to interfere with expression of all three CLIP-170 isoforms in rat primary cortical neurons (Fig. 4A & 4B). Dendritic elaboration in these neurons depends on tight coordination between the MT and actin cytoskeletons (11, 16, 34). Depletion of CLIP-170 (validated in N2A cells; Fig. S7) led to dramatically reduced complexity of neuronal processes, as previously reported (16). This phenotype was rescued by a full-length wild type CLIP-170-1 construct resistant to silencing (Fig. 4A & 4B), but not the FEED1 mutant (Fig. 4A & 4B). Conversely, increased expression of wild type CLIP-170-1 over endogenous CLIP-170 led to elevated dendritic complexity as previously shown (16), whereas mutant CLIP-170-1 did not (Fig. 4C & 4D). In N2A cells, wild type and FEED1 mutant CLIP-170-1 were expressed at similar levels (Fig. S7). Further, live imaging showed that they localized to MT plus-ends similarly (Fig. 4E & 4F; Fig. S8A–C; Movie S10) and that the mutant did not alter MT dynamics (Fig. 4G; Fig. S8D & S8E; Movies S10 & S11). Together, these data suggest that CLIP-170 interactions with formins play an important role in coordinating MT and actin dynamics to regulate neuronal process formation.
Figure 4. CLIP-170 interactions with formins promote dendritic branching in primary neurons.
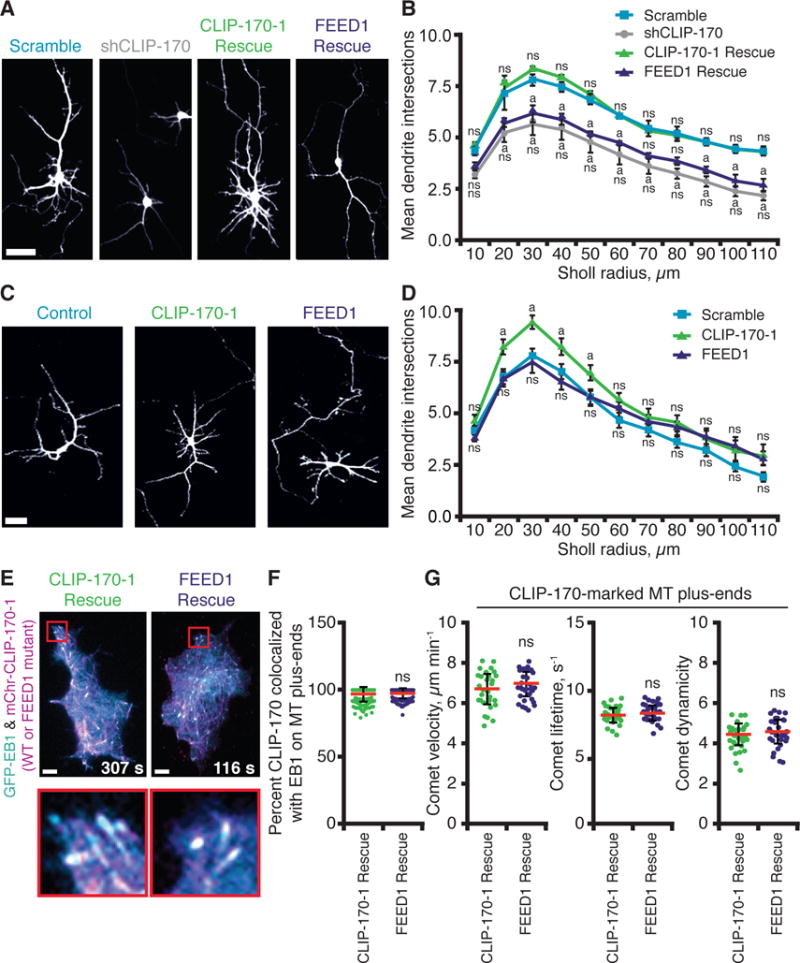
(A) Representative images from 4–5 DIV rat cortical neurons co-transfected with pCMV-GFP and pSUPER containing control (scrambled) shRNA or shRNA against CLIP-170 (shCLIP-170) (16), with or without plasmids expressing shRNA-resistant full-length CLIP-170-1 rescue (wild type or FEED1 mutant). Scale bar, 5 μm. (B) Quantification of dendritic branching complexity by Sholl analysis (n = 90–120 neurons, from two or more independent experiments). Statistical differences: ns, not significantly different from control; a, compared to scramble shRNA control (p < 0.05). There was never a significant difference between the FEED1 rescue the shCLIP-170 knockdown alone. Error bars, SE. (C) Representative images from rat cortical neurons as in (A) expressing full-length CLIP-170-1 (wild type or FEED1 mutant) without silencing endogenous CLIP-170. Scale bar, 5 μm. (D) Quantification of dendritic branching complexity by Sholl analysis as in (B), except data are from n = 60 neurons in two independent experiments. Statistical differences: ns, not significantly different from control; a, compared to scramble shRNA control (p < 0.05). Error bars, SE. (E) Localization of mCherry-CLIP-170-1 (wild type or FEED1 mutant) rescue constructs in N2A cells in which endogenous CLIP-170 was silenced. Scale bars, 10 μm. (F) Quantification of mCherry-CLIP-170-1 colocalization with EB1-GFP at MT plus-ends from cells as in (E). Error bars, SD. (G) Quantitative tracking of mCherry-CLIP-170-1 (wild type or FEED1 mutant) comets on growing MT plus-ends. Analysis includes: comet velocity (or MT growth rate), comet lifetime (MT growth duration), and comet ‘dynamicity’ (a general readout of MT dynamics; (36)). Error bars, SD.
Here we have shown that CLIP-170 tightly interacts with formins to dramatically increase both the rate of actin filament elongation and the duration of elongation in the presence of CP. Further, we have demonstrated that CLIP-170 is part of a mechanism that enables growing MT plus-ends to trigger rapid assembly of actin filaments in vitro, thus directly linking MT and actin dynamics. This mechanism is consistent in a physiological setting, where EB1 and CLIP-170 colocalize on MT plus ends (Fig. 4C & 4D; Fig. S8A–C), and with previous reports showing that growing MT plus-ends survey the actin-rich cortex (10), and that ~10% of mDia1 puncta in cells colocalize with MT plus-ends (31). In neurons, we found that CLIP-170 interactions with formins are required for proper dendritic branching. Similar mechanisms may explain the co-localization and co-functioning of CLIP-170 and mDia1 in phagocytic cup formation (5), and reduced actin-based protrusive activity in neuronal growth cones after CLIP-170 silencing (18, 19, 34, 35).
Supplementary Material
Acknowledgments
We are grateful to S. Paradis for guidance on experiments using neurons, D. Breitsprecher for pioneering the MT-actin co-reconstitution system, J. Gelles for guidance on single molecule analysis, S. Jansen for guidance with cell culture, H. Higgs for providing INF1 and INF2 proteins, and L. Cassimeris for providing pGFP-EB1. This research was supported by a grant from the NIH (GM083137) to B.L.G. J.L.H-R was supported in part by a fellowship from the Leukemia and Lymphoma Society and in part by a training grant from the NIH (T32NS007292).
Footnotes
Author contributions:
J.L.H-R. and B.L.G. designed the experiments and wrote the manuscript; J.L.H-R performed the experiments and data analysis. J.A.E. performed data analysis. A.R. built reagents and performed preliminary experiments. K.K. performed overexpression analysis in neurons and assisted in other neuronal work.
The authors declare no conflict of interest. Supplementary materials contain additional data.
References & Notes
- 1.Forscher P, Smith SJ. Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J Cell Biol. 1988;107:1505–1516. doi: 10.1083/jcb.107.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodriguez OC, et al. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat Cell Biol. 2003;5:599–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- 3.Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- 4.Svitkina TM, et al. Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewkowicz E, et al. The microtubule-binding protein CLIP-170 coordinates mDia1 and actin reorganization during CR3-mediated phagocytosis. J Cell Biol. 2008;183:1287–1298. doi: 10.1083/jcb.200807023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sampathkumar A, et al. Live cell imaging reveals structural associations between the actin and microtubule cytoskeleton in Arabidopsis. Plant Cell. 2011;23:2302–2313. doi: 10.1105/tpc.111.087940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin SG, McDonald WH, Yates JR, Chang F. Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev Cell. 2005;8:479–491. doi: 10.1016/j.devcel.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Martin SG, Rincón SA, Basu R, Pérez P, Chang F. Regulation of the formin for3p by cdc42p and bud6p. Mol Biol Cell. 2007;18:4155–4167. doi: 10.1091/mbc.E07-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suter DM, Forscher P. Substrate-cytoskeletal coupling as a mechanism for the regulation of growth cone motility and guidance. J Neurobiol. 2000;44:97–113. [PubMed] [Google Scholar]
- 10.Salmon WC, Adams MC, Waterman-Storer CM. Dual-wavelength fluorescent speckle microscopy reveals coupling of microtubule and actin movements in migrating cells. J Cell Biol. 2002;158:31–37. doi: 10.1083/jcb.200203022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coles CH, Bradke F. Coordinating neuronal actin-microtubule dynamics. Curr Biol. 2015;25:R677–691. doi: 10.1016/j.cub.2015.06.020. [DOI] [PubMed] [Google Scholar]
- 12.Chesarone M, Gould CJ, Moseley JB, Goode BL. Displacement of Formins from growing barbed ends by Bud14 is critical for actin cable architecture and function. Dev Cell. 2009;16:292–302. doi: 10.1016/j.devcel.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 14.Akhmanova A, et al. CLASPs are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104:923–935. doi: 10.1016/s0092-8674(01)00288-4. [DOI] [PubMed] [Google Scholar]
- 15.Slep KC, Vale RD. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170 and EB1. Mol Cell. 2007;27:976–991. doi: 10.1016/j.molcel.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neukirchen D, Bradke F. Cytoplasmic linker proteins regulate neuronal polarization through microtubule and growth cone dynamics. J Neurosci. 2011;31:1528–1538. doi: 10.1523/JNEUROSCI.3983-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dixit R, et al. Microtubule plus-end tracking by CLIP-170 requires EB1. Proc Natl Acad Sci USA. 2009;106:492–497. doi: 10.1073/pnas.0807614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weng JH, et al. Pregnenolone activates CLIP-170 to promote microtubule growth and cell migration. Nat Chem Biol. 2013;9:636–642. doi: 10.1038/nchembio.1321. [DOI] [PubMed] [Google Scholar]
- 19.Beaven R, et al. Drosophila CLIP-190 and mammalian CLIP-170 display reduced microtubule plus end association in the nervous system. Mol Biol Cell. 2015;8:1491–1508. doi: 10.1091/mbc.E14-06-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chesarone-Cataldo M, et al. The myosin passenger protein Smy1 controls actin cable structure and dynamics by acting as a formin damper. Dev Cell. 2011;21:217–230. doi: 10.1016/j.devcel.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eskin JA, Rankova A, Johnston AB, Alioto SL, Goode BL. Common formin-regulating sequences in Smy1 and Bud14 are required for the control of actin cable assembly in vivo. Mol Biol Cell. 27:828–837. doi: 10.1091/mbc.E15-09-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gould CJ, et al. The formin DAD domain plays dual roles in autoinhibition and actin nucleation. Curr Biol. 2011;21:384–390. doi: 10.1016/j.cub.2011.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breitsprecher D, et al. Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science. 2012;336:1164–1168. doi: 10.1126/science.1218062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moseley JB, et al. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol Biol Cell. 2004;15:896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zigmond SH, et al. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr Biol. 2003;13:1820–1823. doi: 10.1016/j.cub.2003.09.057. [DOI] [PubMed] [Google Scholar]
- 27.Harris ES, Li F, Higgs HN. The mouse formin, FRLα, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J Biol Chem. 2004;279:20076–20087. doi: 10.1074/jbc.M312718200. [DOI] [PubMed] [Google Scholar]
- 28.Wear MA, Cooper JA. Capping Protein: New insights into mechanism and regulation. Trends Biochem Sci. 2004;29:418–428. doi: 10.1016/j.tibs.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Bombardier JP, et al. Single-molecule visualization of a formin-capping protein “decision complex” at the actin filament barbed end. Nat Commun. 2015;6:8707. doi: 10.1038/ncomms9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pierre P, Scheel J, Rickard JE, Kreis TE. CLIP-170 links endocytic vesicles to microtubules. Cell. 1992;70:887–900. doi: 10.1016/0092-8674(92)90240-d. [DOI] [PubMed] [Google Scholar]
- 31.Wen Y, et al. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol. 2004;6:820–830. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- 32.Gaillard J, et al. Differential interactions of the formins INF2, mDia1, and mDia2 with microtubules. Mol Biol Cell. 2011;22:4575–4587. doi: 10.1091/mbc.E11-07-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartolini F, et al. The formin mDia2 stabilizes microtubules independently of its actin nucleation activity. J Cell Biol. 2008;181:523–536. doi: 10.1083/jcb.200709029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swiech L, et al. CLIP-170 and IQGAP1 cooperatively regulate dendrite morphology. J Neurosci. 2011;31:4555–4568. doi: 10.1523/JNEUROSCI.6582-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaiser DA, Vinson VK, Murphy DB, Pollard TD. Profilin is predominantly associated with monomeric actin in Acanthamoeba. J Cell Sci. 1999;112:3779–3790. doi: 10.1242/jcs.112.21.3779. [DOI] [PubMed] [Google Scholar]
- 36.Erck C, et al. A vital role of tubulin-tyrosine-ligase for neuronal organization. Proc Natl Acad Sci USA. 2005;102:7853–7858. doi: 10.1073/pnas.0409626102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Applegate KT, et al. plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. J Struct Biol. 2011;176:168–184. doi: 10.1016/j.jsb.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moseley JB, Maiti S, Goode BL. Formin proteins: purification and measurement of effects on actin assembly. Methods Enzymol. 2006;406:215–234. doi: 10.1016/S0076-6879(06)06016-2. [DOI] [PubMed] [Google Scholar]
- 39.Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 40.Cooper JA, Blum JD, Pollard TD. Acanthamoeba castellanii capping protein: Properties, mechanism of action, immunologic cross-reactivity, and localization. J Cell Biol. 1984;99:217–225. doi: 10.1083/jcb.99.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castoldi M, Popov AV. Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr Purif. 2003;32:83–88. doi: 10.1016/S1046-5928(03)00218-3. [DOI] [PubMed] [Google Scholar]
- 42.Groen AC, Ngyuen PA, Field CM, Ishihara K, Mitchison TJ. Glycogen-supplemented mitotic cytosol for analyzing Xenopus egg microtubule organization. Methods Enzymol. 2014;540:417–433. doi: 10.1016/B978-0-12-397924-7.00023-6. [DOI] [PubMed] [Google Scholar]
- 43.Maiti S, et al. Structure and activity of full-length formin mDia1. Cytoskeleton. 2012;69:393–405. doi: 10.1002/cm.21033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amatruda JF, Cooper JA. Purification, characterization, and immunofluorescence localization of Saccharomyces cerevisiae capping protein. J Cell Biol. 1992;117:1067–1076. doi: 10.1083/jcb.117.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 47.Waterman-Storer CM, Salmon ED. How microtubules get fluorescent speckles. Biophys J. 1998;75:2059–2069. doi: 10.1016/S0006-3495(98)77648-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghiretti AE, Paradis S. The GTPase Rem2 regulates synapse development and dendritic morphology. Dev Neurobiol. 2011;71:374–389. doi: 10.1002/dneu.20868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xia Z, Dudek H, Miranti CK, Greenberg ME. Calcium influx via the NMDA receptor induces immediate early gene transcription by a MAP kinase/ERK-dependent mechanism. J Neurosci. 1996;16:5425–5436. doi: 10.1523/JNEUROSCI.16-17-05425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


