Abstract
Objective
The authors aimed to explore the relationship between the expression/polymorphisms of TLR-9 and susceptibility to colon cancer development in the Saudi Arabian population.
Methods
In total, blood samples from 115 patients with colon cancer and 102 participants without colon cancer were analyzed in this study. Three single-nucleotide polymorphisms (SNPs) were selected from the TLR-9 gene, including two sites within the TLR-9 gene’s promoter region (rs352144 and rs187084) and one site in a TLR-9 intron region (rs5743839). Odds ratios (ORs) and 95% confidence intervals (CIs) were computed from logistic regression models after adjusting for age, gender, and tumor localization. To investigate the differential expression of TLR-9 in colon cancer, TLR-9 expression was evaluated using quantitative real-time reverse transcription polymerase chain reaction on 40 matched normal and colon tissues.
Results
The authors found that TLR-9 expression was decreased in colon cancer tissues as compared with that in normal tissues. Moreover, significant associations between the TLR-9 rs187084 SNP and colon cancer risk were observed in female patients only. In rs187084, the T allele had a significantly lower frequency (2.8 times) in female cancer patients than in controls (0.27 vs 0.41). The TLR-9 rs352139 and rs352144 SNPs were significantly associated with colon cancer development when the tumor was located in the rectal area.
Conclusion
The findings support the hypothesis that TLR-9 has an anticancer role in colon cancer development. Furthermore, genetic variation may influence colon cancer development, and SNPs in TLR-9 could serve as biomarkers for decision making in the treatment of females with rectal cancer.
Keywords: Innate, immunity, TLR polymorphisms, rs187084, rs352139, rs352144
Introduction
The innate immune system is the first line of defense against pathogens and tissue injury.1 The human immune system is a well-coordinated network of cells, organs, and glands that protects the body from foreign invaders. Therefore, an optimized immune system is the key to health and longevity, and the immune system plays a crucial role in preventing cancer. Toll-like receptors (TLRs) appear to be involved in the first line of defense against invading pathogens, initiating inflammatory responses and playing a key role in immune cell regulation, survival, and proliferation. Through their role in the immune system, TLRs are potential tools for curing and preventing cancer. It is estimated that between 10 and 13 TLR families (named TLR-1 to TLR-13)2–5 are activated by various ligands expressed in different types of immune cells. These receptors are located on the cell surface, and TLR-3, TLR-7, TLR-8, and TLR-9 are localized to the endosomal/lysosomal compartment in most mammalian species. Human TLR-9, which has been mapped to chromosome 3p21.3,2 spans ~5 kb and contains two exons. The TLR-9 gene encodes a protein of 1032 amino acids2 and is preferentially expressed by B cells and plasmacytoid dendritic cells.6 TLR-9 is one of the most important receptors for the initiation of innate immune responses against intracellular pathogens. Unlike other products of the TLR gene family, which are membrane-bound pattern recognition receptors, TLR-9 is localized on the endoplasmic reticulum membrane (in the resting state) or on the endosomal/lysosomal membrane (after ligand stimulation and trafficking);7,8 however, TLR-9 interacts with unmethylated CpG DNA from bacteria and some viruses.9,10 Alternatively, TLR-9 functions through the MyD88-dependent pathway, leading to nuclear factor-kappa-B (NF-κB) activation, cytokine secretion, and inflammatory response.11,12 To date, TLR-9 is the only TLR for which a systemically administered specific agonist has shown substantial evidence of anticancer activity in human clinical trials.13 Several studies have shown that TLR-9 engagement on cluster of differentiation 4-positive (CD4+) T cells can enhance their survival and therefore potentiate anticancer responses by prolonging T-cell activity.14 Previous studies have indicated that TLR-9 polymorphisms may be associated with the risk of developing several types of cancers, including bladder cancer,15 prostate cancer,16–18 acute lymphoblastic leukemia,19 hepatocellular carcinoma,20 gastric cancer,21,22 cervical cancer,23 Hodgkin’s lymphoma,24 breast cancer,25 Burkitt’s lymphoma,26 non-Hodgkin’s lymphoma,27 endometrial cancer,28 esophageal cancer,23 and lymphoma.24,27,29 However, the results are inconsistent and inconclusive. The link between TLR-9 polymorphisms and cancer was specifically investigated in the context of chronic inflammation, which is thought to increase the risk of cancer.30 Most previous studies have focused on three common single-nucleotide polymorphisms (SNPs), ie, rs352140 (C/T), rs5743836 (T/C), and rs187084 (C/T) (also referred to as 2848C/T, 1237T/C, and 1486C/T, respectively); however, the results of these studies were inconsistent.30
In this study, the authors hypothesized that TLR SNPs may affect cancer through inappropriate TLR signaling, causing downstream elevations in proinflammatory cytokine levels, which in turn promotes cancer. In the present study we have investigated the relationship between TLR-9 expression/polymorphism and susceptibility to colorectal cancer development in the Saudi Arabian population.
Subjects and methods
Study population
A population-based, case–control study that included 115 patients with colon cancer and 102 healthy controls was conducted in the Kingdom of Saudi Arabia (KSA). The participants were recruited from King Khalid University Hospital in Riyadh, KSA, and the study was approved by the ethics committee at King Khalid University Hospital in Riyadh, KSA (project E-12-596, 12/3352/IRB). All questionnaire data and samples (tissues and blood) were collected during the initial recruitment of both the cases and controls. Informed written consent and a self-administered questionnaire regarding sociodemographic character (eg, age, family history of cancer, etc), lifestyle (eg, smoking habits and alcohol intake), and personal medical history were collected from all the participants. Cases and controls were frequency-matched by age and gender. The clinico-pathological characteristics of the patients, including age, family history, smoking habits, stage of colon cancer, medications, and presence of other diseases, were collected and compared with those of controls. Patients in the study population ranged in age from 45 to 88 years, with mean ages ± SD of 57.04±14.37 years in patients with colon cancer and 56.51±15.70 years in controls. Among the colon cancer patients, the authors selected 66 males (57.4%) and 49 females (42.6%); among the healthy controls, the authors had 60 males (58.8%) and 42 females (41.2%), as shown in Table 1. Age and gender matched controls were used. All the control subjects were healthy without any health problems. Patients were divided into 2 groups based on the location of the tumor, the controls were chosen so that the tumor tissue and matching control tissue were from the same area, ie, the normal tissues were sampled from a very small area near the tumor. The tissue samples were used for RNA extraction. Blood samples were stored in ethylenediaminetetraacetic acid (EDTA)-containing tubes at −80°C until genomic DNA extraction.
Table 1.
Clinical characteristics of the subjects selected for genotyping
| Characteristics | Cancer (n=115) | Control (n=102) |
|---|---|---|
| Gender | ||
| Male | 66 | 60 |
| Female | 49 | 42 |
| Age (years) | ||
| Male | 55.50±13.94 | 53.24±17.4 |
| Female | 56.95±13.70 | 50.78±15.21 |
| Localization | ||
| Colon | 76 | – |
| Rectum | 39 | – |
| Therapy | ||
| Chemotherapy | ||
| Yes | 3 | – |
| No | 112 | – |
| Radiotherapy | ||
| Yes | 5 | – |
| No | 110 | – |
DNA extraction
Sample collection and storage were performed as previously described.31 Genomic DNA was extracted from whole blood using a QIAmp kit (QIAmp DNA Blood Mini Kit; Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. Briefly, 200–300 μL blood stored in an EDTA tube at −80°C was equilibrated at room temperature, mixed with protease and lysis buffer, and then incubated at 56°C for 10 min. Next, 100% ethanol was added, and the mixture was spun through the column. The column membrane was washed, and the DNA was eluted with 100 μL elution buffer (AE). The extracted DNA concentration was determined using a NanoDrop8000 spectrophotometer (Thermo Scientific). The DNA purity was evaluated by determining standard A260/A280 and A260/A230 ratios.
Total RNA isolation
Total RNA was extracted from 40 colon cancer tissues and 40 matched normal colon tissues using an AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s protocol. The isolated RNA concentration, purity, and quality were determined using an Agilent 2100 Bioanalyzer system and Agilent Small RNA analysis kit (Agilent Technologies, Waldbronn, Germany) according to the manufacturer’s instructions.
Complementary DNA (cDNA) synthesis
As described by Semlali et al,32,33 RNA (1 μg of each sample) was reverse-transcribed into cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Warrington, PA, USA). The preparation conditions of the cDNA templates for polymerase chain reaction (PCR) were as follows: 10 minutes at 25°C, 2 hours at 37°C, and 5 minutes at 85°C. The synthesized cDNA was stored at −20°C or 4°C for immediate use in subsequent PCRs.
Quantitative real-time PCR (qPCR)
qPCR was performed using a SYBR® Green PCR Supermix (Applied Biosystems) and specific primers. The primers for TLR-9 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) control were as follows: TLR-9: (forward) 5′-GGACCTCTGGTACTGCTTCCA-3′ and (reverse) 5′-AAGCTCGTTGTACACCCAGTCT-3′; GAPDH: (forward) 5′-GGTATCGTCGAAGGACTCATGAC-3′ and (reverse) 5′-ATGCCAGTGAGCTTCCCGTTCAGC-3′. All reactions were performed on a 7500 Real-Time PCR System (Applied Biosystems) with an annealing temperature of 60°C for both TLR-9 and GAPDH genes. Cycle thresholds (Cts) were automatically determined using Applied Biosystems software, with each reaction performed in triplicate. Results were analyzed using the 2−∆∆Ct (Livak) relative expression method.
Genotyping
TLR-9 SNPs (rs187084, rs352139, and rs352144) were genotyped using a TaqMan allelic discrimination assay, as previously described.34 The additional information of the SNPs is shown in Table S1. From each sample, 10–20 ng DNA was used per reaction with 5.6 μL of 2× Universal Master Mix and 200 nM primers. All genotypes were determined by an end-point reading on an ABI 7500 real-time PCR machine. The primer and probe mixtures were purchased from Applied Biosystem’s “Assays-on-Demand™” service. Five percent of the samples were randomly selected and subjected to repeat analyses as a quality control measure for verifying genotyping procedures.
Statistical analyses
Genotypic and allelic frequencies were computed and checked for deviation with the Hardy–Weinberg equilibrium (https://ihg.helmholtz-muenchen.de/cgi-bin/hw/hwa1.pl). Case–control and other genetic comparisons were performed using the chi-square test with Yates’s correction, and allelic odds ratios (ORs) with 95% confidence intervals (CIs) were calculated with Fisher’s exact tests (two-tailed). Statistical analysis was performed using the Statistical Package for the Social Sciences (SPSS) 16.0 software for Windows. The authors considered P-values less than 0.05 to be significant. Linkage disequilibrium analysis was conducted using Haploview (v5.0).
Results
Analysis of clinical data parameters
A total of 115 colon cancer cases and 102 healthy controls were included in the study. The clinical characteristics of the patients, including age, gender, nationality, and location of the tumor, were collected and compared between colon cancer patients and controls (Table 1). Non-Saudi Arabian patients or patients with a family history of smoking were excluded from this study. The age of patients in the study population ranged from 45 to 88 years, with mean ages ± SD of 56.04±14.37 years in patients with colon cancer and 52.84±15.88 years in control patients. There were no significant differences in the ages of participants in the two groups. The male-to-female ratio was not significantly different between cases and controls (66/49 for patients with colorectal cancer; 60/42 for controls).
TLR-9 expression in colon cancer cells and colon cancer tissues
To determine whether there was a link between TLR-9 expression and colon cancer, the authors analyzed the expression of TLR-9 in 10 matched tissues (colon cancer and matched normal tissues) using RT-qPCR to compare TLR-9 mRNA levels in normal and colon cancer tissues. Analysis of the expression data revealed that TLR-9 mRNA levels were significantly (P<0.0001) decreased in colon cancer tissues (1.02±0.02 folds in normal colon tissue versus 0.44±0.02 in colon cancer tissue; Figure 1).
Figure 1.
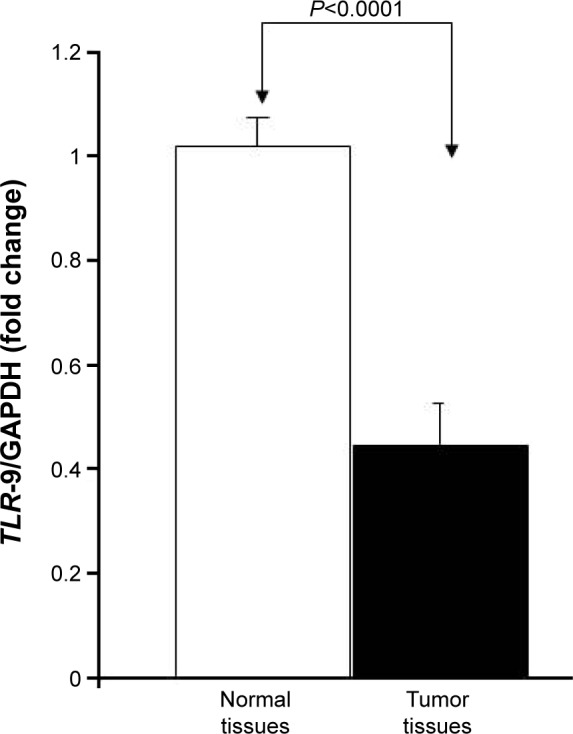
TLR-9 mRNA expression in colon cancer cells and colon cancer tissues.
Notes: Total RNA of tissues was extracted from matching normal and colon cancer tissues, reverse-transcribed into cDNA, and then used to measure TLR-9 mRNA expression with specific primers. TLR-9 expression in colon cancer tissues and matching control tissues is shown as mean ± SD.
Abbreviations: cDNA, complementary DNA; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
TLR-9 polymorphism frequencies and susceptibility to colon cancer development in Saudi Arabian patients
No statistically significant differences were found between colon cancer cases and controls (Table 2). The TLR-9 rs187084 SNP is located in the TLR-9 gene promoter region. The frequencies of CC, CT, and TT genotypes in the TLR-9 rs187084 control group were 35% (34/102), 53% (53/102), and 12% (12/102), respectively, whereas those in the case group were 43% (47/115), 43% (47/115), and 14% (15/115), respectively. No significant differences in TLR-9 rs187084 frequencies were found between the case and control groups (P>0.01). Compared with the CC genotype, carriers of the CT + TT genotype were less susceptible to colon cancer (OR =0.69, 95% CI: 0.39–1.21; P=0.19; Table 2). The same observations were made for TLR-9 rs352139 and TLR-9 rs352144 (Table 2).
Table 2.
Genotype frequencies of TLR-9 gene polymorphisms in colorectal cancer and controls
| Gene | SNP ID | Genotype | Case, n (%) | Control, n (%) | OR | 95% CI | χ2 value | P-value |
|---|---|---|---|---|---|---|---|---|
| TLR-9 | rs187084 | CC | 47 (0.43) | 34 (0.35) | Ref | |||
| CT | 47 (0.43) | 53 (0.53) | 0.642 | 0.355–1.158 | 2.18 | 0.13991 | ||
| TT | 15 (0.14) | 12 (0.12) | 0.904 | 0.376–2.176 | 0.05 | 0.82221 | ||
| CT + TT | 62 (0.57) | 65 (0.65) | 0.690 | 0.393–1.210 | 1.68 | 0.19487 | ||
| C | 141 (0.65) | 121 (0.61) | Ref | |||||
| T | 77 (0.35) | 77 (0.39) | 0.858 | 0.576–1.278 | 0.57 | 0.45166 | ||
| rs352139 | AA | 33 (0.29) | 31 (0.30) | Ref | ||||
| AG | 55 (0.49) | 55 (0.54) | 0.939 | 0.507–1.740 | 0.04 | 0.84243 | ||
| GG | 26 (0.23) | 16 (0.16) | 1.527 | 0.691–3.372 | 1.10 | 0.29448 | ||
| AG + GG | 81 (0.71) | 71 (0.70) | 1.072 | 0.597–1.923 | 0.05 | 0.81642 | ||
| A | 121 (0.54) | 117 (0.57) | Ref | |||||
| G | 107 (0.48) | 87 (0.43) | 1.189 | 0.813–1.740 | 0.80 | 0.37163 | ||
| rs352144 | AA | 2 (0.02) | 0 (0) | Ref | ||||
| AC | 5 (0.04) | 7 (0.07) | 0.147 | 0.006–3.706 | 2.33 | 0.12663 | ||
| CC | 110 (0.94) | 94 (0.93) | 0.234 | 0.011–4.932 | 1.70 | 0.19294 | ||
| AC + CC | 115 (0.98) | 101 (1) | 0.228 | 0.011–4.796 | 1.74 | 0.18682 | ||
| A | 9 (0.04) | 7 (0.03) | Ref | |||||
| C | 225 (0.96) | 195 (0.97) | 0.897 | 0.328–2.455 | 0.04 | 0.83298 |
Abbreviations: CI, confidence interval; OR, odds ratio; Ref, reference; SNP, single-nucleotide polymorphism.
Moreover, out of the three SNPs analyzed, all the three SNPs were in linkage disequilibrium in both cases (r2≥0.80) and controls (r2≥0.80) (Figure 2).
Figure 2.
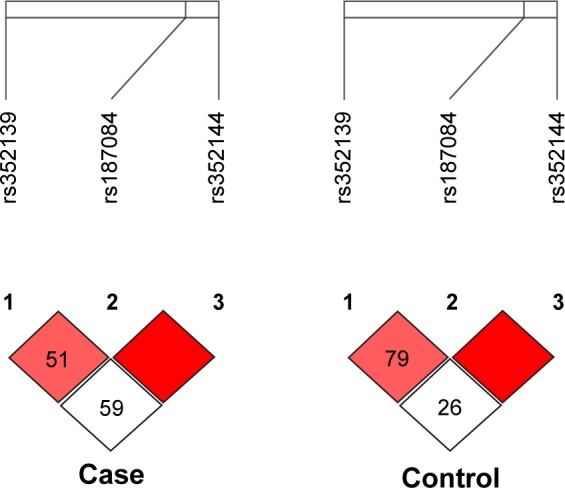
LD of the studied TLR-9 loci rs187084, rs352139, and rs352144 in colorectal cancer case and control groups.
Abbreviation: LD, linkage disequilibrium.
Association between the genotype frequencies of TLR-9 gene polymorphisms and clinical characteristics
Analysis of the TLR-9 genotype distribution for correlations with age, gender, and tumor localization revealed that there were no significant differences (P>0.05) between cases and controls for both subpopulations of patients aged ≤52 and ≥52 years for all three TLR-9 SNPs (rs187084, rs352139, and rs352144; Table 3A and B). The median age of onset in the colon cancer group was 56 years, and the median age of participants in the control group was 52 years. To evaluate the association between colon cancer risk and individual SNPs based on a patient’s gender, the genotype distributions according to gender in the colon cancer group were compared with those of the control subjects (Table 4A and B). Interestingly, in the female group (46 patients and 41 controls), the T allele of SNP rs187084 had a significantly lower frequency (2.8 times) in patients than in the controls (0.27 vs 0.41, OR: 0.527, 95% CI: 0.279–0.995; P=0.04). However, no associations were found for the genotype frequencies of TLR-9 rs352139 and rs352144 in affected and unaffected females (P>0.05; Table 4B). In contrast, all three SNPs from TLR-9 were not associated with colorectal cancer in males (Table 4A). To investigate the possible association between TLR-9 genotype polymorphism frequencies and tumor localization, the authors compared TLR-9 SNP variations in patients with colon cancer when tumors were located in the rectum or in the colon area with variations in control patients. Significant difference was not observed in all three SNPs in patients with tumors located in colon area (Table 5A). However, associations of TLR-9 rs352139 and rs352144, but not TLR-9 rs187084, were found in tumors in the rectal area (Table 5B). In patients with tumors in the rectal area, no significant differences in the frequencies of TLR-9 rs187084 were found between the case and control groups (P>0.05). The frequencies of the AA, AG, and GG genotypes in TLR-9 rs352139 in the control group were 30% (31/102), 54% (55/102), and 16% (16/102), respectively, whereas those in the case group were 18% (6/34), 50% (17/34), and 32% (11/34), respectively (Table 5B). The frequencies of the AA, AC, and CC genotypes in TLR-9 rs352144 in the control group were 0% (0/102), 7% (7/102), and 93% (94/102), respectively, whereas those in the case group were 3% (1/36), 3% (1/36), and 94% (33/36), respectively (Table 5B).
Table 3A.
Genotype frequencies of TLR-9 gene polymorphism in patients younger than 57 years with colorectal cancer
| Gene | SNP ID | Genotype | Case, n (Frequency in %) |
Control, n (Frequency in %) |
OR | 95% CI | χ2 value | P-value |
|---|---|---|---|---|---|---|---|---|
| TLR-9 | rs187084 | CC | 22 (0.42) | 16 (0.27) | Ref | |||
| CT | 23 (0.43) | 35 (0.58) | 0.478 | 0.208–1.098 | 3.07 | 0.07989 | ||
| TT | 8 (0.15) | 9 (0.15) | 0.646 | 0.205–2.041 | 0.56 | 0.45578 | ||
| CT + TT | 31 (0.58) | 44 (0.85) | 0.512 | 0.232–1.130 | 2.78 | 0.09558 | ||
| C | 67 (0.63) | 67 (0.56) | Ref | |||||
| T | 39 (0.37) | 53 (0.44) | 0.736 | 0.431–1.256 | 1.27 | 0.26013 | ||
| rs352139 | AA | 20 (0.36) | 24 (0.39) | Ref | ||||
| AG | 27 (0.48) | 28 (0.45) | 1.157 | 0.523–2.562 | 0.13 | 0.71882 | ||
| GG | 9 (0.16) | 10 (0.16) | 1.080 | 0.367–3.175 | 0.02 | 0.88876 | ||
| AG + GG | 36 (0.64) | 38 (0.61) | 1.137 | 0.538–2.403 | 0.11 | 0.73687 | ||
| A | 67 (0.60) | 76 (0.61) | Ref | |||||
| G | 45 (0.40) | 48 (0.39) | 1.063 | 0.630–1.794 | 0.05 | 0.81763 | ||
| rs352144 | AA | 2 (0.03) | 0 (0) | Ref | ||||
| AC | 4 (0.07) | 4 (0.06) | 0.200 | 0.007–5.453 | 1.67 | 0.19671 | ||
| CC | 53 (0.90) | 58 (0.94) | 0.183 | 0.009–3.897 | 2.15 | 0.14284 | ||
| AC + CC | 57 (0.97) | 62 (1) | 0.184 | 0.009–3.914 | 2.14 | 0.14378 | ||
| A | 8 (0.07) | 4 (0.03) | Ref | |||||
| C | 110 (0.93) | 120 (0.97) | 0.458 | 0.134–1.565 | 1.62 | 0.20304 |
Abbreviations: CI, confidence interval; OR, odds ratio; Ref, reference; SNP, single-nucleotide polymorphism.
Table 3B.
Genotype frequencies of TLR-9 gene polymorphism in patients older than 57 years with colorectal cancer
| Gene | SNP ID | Genotype | Case, n (Frequency in %) |
Control, n (Frequency in %) |
OR | 95% CI | χ2 value | P-value |
|---|---|---|---|---|---|---|---|---|
| TLR-9 | rs187084 | CC | 25 (0.45) | 18 (0.46) | Ref | |||
| CT | 24 (0.43) | 18 (0.46) | 0.960 | 0.406–2.270 | 0.01 | 0.92592 | ||
| TT | 7 (0.12) | 3 (0.08) | 1.680 | 0.382–7.395 | 0.48 | 0.48975 | ||
| CT + TT | 31 (0.55) | 21 (0.54) | 1.063 | 0.468–2.415 | 0.02 | 0.88428 | ||
| C | 74 (0.66) | 54 (0.69) | Ref | |||||
| T | 38 (0.34) | 24 (0.31) | 1.155 | 0.622–2.147 | 0.21 | 0.64774 | ||
| rs352139 | AA | 13 (0.22) | 7 (0.17) | Ref | ||||
| AG | 28 (0.48) | 27 (0.68) | 0.558 | 0.193–1.612 | 1.18 | 0.27836 | ||
| GG | 17 (0.30) | 6 (0.15) | 1.526 | 0.413–5.642 | 0.40 | 0.52559 | ||
| AG + GG | 45 (0.78) | 33 (0.83) | 0.734 | 0.264–2.042 | 0.35 | 0.55304 | ||
| A | 54 (0.47) | 41 (0.51) | Ref | |||||
| G | 62 (0.53) | 39 (0.49) | 1.207 | 0.682–2.135 | 0.42 | 0.51772 | ||
| rs352144 | AA | 0 (0) | 0 (0) | Ref | ||||
| AC | 1 (0.02) | 3 (0.08) | 0.429 | 0.005–33.596 | – | 1.000 | ||
| CC | 57 (0.98) | 36 (0.92) | 1.575 | 0.031–81.149 | – | 1.000 | ||
| AC + CC | 58 (1) | 39 (1) | 1.481 | 0.029–76.201 | – | 1.000 | ||
| A | 1 (0.01) | 3 (0.04) | Ref | |||||
| C | 115 (0.99) | 75 (0.96) | 4.600 | 0.470–45.053 | 2.06 | 0.30785 |
Abbreviations: CI, confidence interval; OR, odds ratio; Ref, reference; SNP, single-nucleotide polymorphism.
Table 4A.
Genotype frequencies of TLR-9 gene polymorphism in male patients with colorectal cancer
| Gene | SNP ID | Genotype | Case, n (Frequency in %) |
Control, n (Frequency in %) |
OR | 95% CI | χ2 value | P-value |
|---|---|---|---|---|---|---|---|---|
| TLR-9 | rs187084 | CC | 22 (0.35) | 19 (0.33) | Ref | |||
| CT | 30 (0.48) | 35 (0.62) | 0.740 | 0.338–1.621 | 0.57 | 0.45162 | ||
| TT | 11 (0.17) | 3 (0.05) | 3.167 | 0.768–13.05 | 2.70 | 0.10042 | ||
| CT + TT | 41 (0.65) | 38 (0.67) | 0.932 | 0.438–1.985 | 0.03 | 0.85473 | ||
| C | 74 (0.59) | 73 (0.64) | Ref | |||||
| T | 52 (0.41) | 41 (0.36) | 1.251 | 0.743–2.108 | 0.71 | 0.39956 | ||
| rs352139 | AA | 19 (0.30) | 15 (0.25) | Ref | ||||
| AG | 31 (0.48) | 34 (0.58) | 0.720 | 0.313–1.657 | 0.60 | 0.43896 | ||
| GG | 14 (0.22) | 10 (0.17) | 1.105 | 0.384–3.180 | 0.03 | 0.85273 | ||
| AG + GG | 45 (0.70) | 44 (0.75) | 0.807 | 0.365–1.787 | 0.28 | 0.59734 | ||
| A | 69 (0.54) | 64 (0.54) | Ref | |||||
| G | 59 (0.46) | 54 (0.46) | 1.013 | 0.613–1.674 | 0 | 0.95849 | ||
| rs352144 | AA | 2 (0.03) | 0 (0) | Ref | ||||
| AC | 1 (0.01) | 3 (0.05) | 0.086 | 0.002–3.101 | 3.00 | 0.08326 | ||
| CC | 64 (0.96) | 55 (0.95) | 0.232 | 0.011–4.945 | 1.69 | 0.19299 | ||
| AC + CC | 65 (0.97) | 58 (1) | 0.224 | 0.011–4.760 | 1.76 | 0.18469 | ||
| A | 5 (0.04) | 3 (0.03) | Ref | |||||
| C | 129 (0.96) | 113 (0.97) | 0.685 | 0.160–2.930 | 0.26 | 0.72813 |
Abbreviations: CI, confidence interval; OR, odds ratio; Ref, reference; SNP, single-nucleotide polymorphism.
Table 4B.
Genotype frequencies of TLR-9 gene polymorphism in female patients with colorectal cancer
| Gene | SNP ID | Genotype | Case, n (Frequency in %) |
Control, n (Frequency in %) |
OR | 95% CI | χ2 value | P-value |
|---|---|---|---|---|---|---|---|---|
| TLR-9 | rs187084 | CC | 25 (0.54) | 15 (0.37) | Ref | |||
| CT | 17 (0.37) | 18 (0.44) | 0.567 | 0.225–1.424 | 1.47 | 0.22539 | ||
| TT | 4 (0.09) | 8 (0.19) | 0.300 | 0.077–1.169 | 3.18 | 0.07439 | ||
| CT + TT | 21 (0.46) | 26 (0.63) | 0.485 | 0.205–1.146 | 2.75 | 0.09703 | ||
| C | 67 (0.73) | 48 (0.59) | Ref | |||||
| T | 25 (0.27) | 34 (0.41) | 0.527 | 0.279–0.995 | 3.95 | 0.04686 | ||
| rs352139 | AA | 14 (0.28) | 15 (0.36) | Ref | ||||
| AG | 24 (0.48) | 21 (0.50) | 1.224 | 0.481–3.118 | 0.18 | 0.67089 | ||
| GG | 12 (0.24) | 6 (0.14) | 2.143 | 0.632–7.266 | 1.52 | 0.21765 | ||
| AG + GG | 36 (0.72) | 27 (0.64) | 1.429 | 0.591–3.454 | 0.63 | 0.42761 | ||
| A | 52 (0.52) | 51 (0.61) | Ref | |||||
| G | 48 (0.48) | 33 (0.39) | 1.427 | 0.792–2.568 | 1.41 | 0.23559 | ||
| rs352144 | AA | 0 (0) | 0 (0) | Ref | ||||
| AC | 4 (0.08) | 4 (0.1) | 1.000 | 0.016–62.3 | – | 1.000 | ||
| CC | 46 (0.92) | 38 (0.9) | 1.208 | 0.023–62.29 | – | 1.000 | ||
| AC + CC | 50 (1) | 42 (1) | 1.188 | 0.023–61.16 | – | 1.000 | ||
| A | 4 (0.04) | 4 (0.05) | Ref | |||||
| C | 96 (0.96) | 80 (0.95) | 1.200 | 0.291–4.951 | 0.06 | 1.000 |
Note: Bold values are statistically significantly different.
Abbreviations: CI, confidence interval; OR, odds ratio; Ref, reference; SNP, single-nucleotide polymorphism.
Table 5A.
Genotype frequencies of TLR-9 gene polymorphisms in colorectal tumors located in the colon area
| Gene | SNP ID | Genotype | Case, n (Frequency in %) |
Control, n (Frequency in %) |
OR | 95% CI | χ2 value | P-value |
|---|---|---|---|---|---|---|---|---|
| TLR-9 | rs187084 | CC | 30 (0.45) | 34 (0.35) | Ref | |||
| CT | 27 (0.41) | 53 (0.53) | 0.577 | 0.294–1.134 | 2.56 | 0.10952 | ||
| TT | 9 (0.14) | 12 (0.12) | 0.850 | 0.315–2.297 | 0.10 | 0.74850 | ||
| CT + TT | 36 (0.55) | 65 (0.65) | 0.628 | 0.332–1.188 | 2.06 | 0.15130 | ||
| C | 87 (0.66) | 121 (0.61) | Ref | |||||
| T | 45 (0.34) | 77 (0.39) | 0.813 | 0.513–1.287 | 0.78 | 0.37640 | ||
| rs352139 | AA | 23 (0.34) | 31 (0.30) | Ref | ||||
| AG | 32 (0.47) | 55 (0.54) | 0.784 | 0.392–1.569 | 0.47 | 0.49166 | ||
| GG | 13 (0.19) | 16 (0.16) | 1.095 | 0.441–2.718 | 0.04 | 0.84470 | ||
| AG + GG | 45 (0.66) | 71 (0.70) | 0.854 | 0.443–1.646 | 0.22 | 0.63780 | ||
| A | 78 (0.57) | 117 (0.57) | Ref | |||||
| G | 58 (0.43) | 87 (0.43) | 1.000 | 0.645–1.551 | 0 | 1.000 | ||
| rs352144 | AA | 0 (0) | 0 (0) | Ref | ||||
| AC | 4 (0.06) | 7 (0.07) | 0.600 | 0.01–35.86 | 0.001 | 1.000 | ||
| CC | 65 (0.94) | 94 (0.93) | 0.693 | 0.014–35.37 | 0.001 | 1.000 | ||
| AC + CC | 69 (1) | 101 (1) | 0.685 | 0.013–34.92 | 0 | 1.000 | ||
| A | 4 (0.03) | 7 (0.03) | Ref | |||||
| C | 134 (0.97) | 195 (0.97) | 1.203 | 0.345–4.189 | 0.08 | 1.000 |
Abbreviations: CI, confidence interval; OR, odds ratio; Ref, reference; SNP, single-nucleotide polymorphism.
Table 5B.
Genotype frequencies of TLR-9 gene polymorphisms in colorectal cancer tumors located in rectal area
| Gene | SNP ID | Genotype | Case, n (Frequency in %) |
Control, n (Frequency in %) |
OR | 95% CI | χ2 value | P-value |
|---|---|---|---|---|---|---|---|---|
| TLR-9 | rs187084 | CC | 14 (0.42) | 34 (0.35) | Ref | |||
| CT | 15 (0.46) | 53 (0.53) | 0.687 | 0.295–1.602 | 0.76 | 0.38390 | ||
| TT | 4 (0.12) | 12 (0.12) | 0.810 | 0.223–2.945 | 0.10 | 0.74819 | ||
| CT + TT | 19 (0.58) | 65 (0.65) | 0.710 | 0.317–1.588 | 0.70 | 0.40332 | ||
| C | 43 (0.65) | 121 (0.61) | Ref | |||||
| T | 23 (0.35) | 77 (0.39) | 0.841 | 0.470–1.503 | 0.34 | 0.55786 | ||
| rs352139 | AA | 6 (0.18) | 31 (0.30) | Ref | ||||
| AG | 17 (0.50) | 55 (0.54) | 1.597 | 0.570–4.471 | 0.80 | 0.37026 | ||
| GG | 11 (0.32) | 16 (0.16) | 3.552 | 1.11–11.37 | 4.81 | 0.02825 | ||
| AG + GG | 28 (0.82) | 71 (0.70) | 2.038 | 0.767–5.415 | 2.09 | 0.14812 | ||
| A | 29 (0.43) | 117 (0.57) | Ref | |||||
| G | 39 (0.57) | 87 (0.43) | 1.809 | 1.038–3.150 | 4.44 | 0.03519 | ||
| rs352144 | AA | 1 (0.03) | 0 (0) | Ref | ||||
| AC | 1 (0.03) | 7 (0.07) | 0.067 | 0.002–2.563 | 3.94 | 0.04722 | ||
| CC | 33 (0.94) | 94 (0.93) | 0.118 | 0.005–2.972 | 2.79 | 0.09506 | ||
| AC + CC | 34 (0.97) | 101 (1) | 0.113 | 0.005–2.847 | 2.91 | 0.08819 | ||
| A | 3 (0.04) | 7 (0.03) | Ref | |||||
| C | 67 (0.96) | 195 (0.97) | 0.802 | 0.202–3.189 | 0.10 | 0.73581 |
Note: Bold values are statistically significantly different.
Abbreviations: CI, confidence interval; OR, odds ratio; Ref, reference; SNP, single-nucleotide polymorphism.
Discussion
In this study, the authors demonstrated that TLR-9 mRNA expression was decreased in colon cancer tissues compared with that in normal tissues, suggesting that TLR-9 may have an anticancer role in colon cancer development. These data support the development of TLR-9 agonists (synthetic oligodeoxynucleotides [ODNs]) for cancer treatment. Baines and Celis35 used a murine cervical carcinoma model, and mice with established subcutaneous tumors that were treated with CpG ODNs injected at a distant site exhibited significant regression. Moreover, CpG ODN-treated mice exhibited significantly improved survival (P<0.001) compared with control mice.35 Tuomela et al36 discovered that low tumor TLR-9 expression is associated with significantly shortened disease-specific survival in patients with triple-negative, but not with estrogen receptor-positive (ER+), breast cancers.
The authors also demonstrated a significant association between the TLR-9 rs187084 SNP and colon cancer risk in females, which is closely linked with the female sex hormones. Previous studies have described the role of estrogen and progesterone in protecting against colon cancer in females.37–40 There is evidence that gender influences the clinical course of colorectal cancer and can also affect disease severity; indeed, females tend to exhibit a benign course more frequently than males. Moreover, males with colorectal cancer tend to have a poorer prognosis. The exact mechanisms through which TLR-9 polymorphisms contribute to cancer pathogenesis are unknown. Based on information from the National Center for Biotechnology Information SNP database, the TLR-9 rs187084 polymorphism in the TLR-9 gene is intronic and is most likely not a true disease-causing variant but could reflect the effects of a functional variant located elsewhere in the TLR-9 gene. Previous studies have shown that C genotype frequencies are closely associated with decreased TLR-9 transcription activity when compared with the T genotype. Some introns may contain regulatory sequences or even encode proteins or RNAs with regulatory functions. Additionally, the authors found significant associations between TLR-9 rs352139/rs352144 SNPs and rectal area localization, but not colon area localization. The TLR-9 rs352139 and rs352144 SNPs are located in the promoter region and most likely affect TLR-9 expression. It is possible that the intron and upstream variant of the TLR-9 gene play an important role in regulating the TLR-mediated immunologic response. However, it is not clear how this intronic SNP (rs352144) reduces TLR-9 expression. It is possible that this SNP influences signaling by creating an alternative splicing site, thus affecting the mRNA transcription and the final protein product. Mutations in the promoter region are predicted to affect the expression/stability of the regulatory regions (ie, the 5′- and 3′-untranslated regions [UTRs]), explaining why the expression levels of these TLR-9 proteins were decreased in colon cancer tissues compared with those in normal tissues. The 3′-UTR contains several types of regulatory elements, including binding sites for microRNAs (miRNAs) and AU-rich elements, which are known to regulate gene expression. Moreover, miRNAs can bind to mRNAs and control protein abundance by suppressing translation or marking mRNAs for degradation.41 Li et al42 reported that rs5743305 is located in the promoter region, 1 kb upstream of exon 1 in the TLR-3 gene, and may influence the transcriptional activity of TLR-3.
Genetic variations play crucial roles in phenotypic variation and disease susceptibility by affecting gene expression. Recent studies by the group43 have integrated a number of resources and technologies to assess several aspects of genome variation affecting gene expression. Additionally, recent findings have suggested that protein divergence between species strongly correlates with expression divergence between species.44,45
Results of this study showed that the TLR-9 rs187084, rs352139, and rs352144 genotypes in the TLR-9 gene were associated with colon cancer risk, suggesting that TLR-9 genetic polymorphisms could serve as biomarkers for decision making in the treatment of females with rectal cancer. Further studies are necessary to investigate the potential mechanisms through which TLR SNPs may mediate pathogenesis and the functional aspects of these TLR-9 SNP polymorphisms in colon cancer.
Supplementary material
Table S1.
Characteristics of selected polymorphisms in TLR-9
| SNP ID | Chromosome/position | Nucleotide change | Position | Region | Minor allele frequency (%)
|
|
|---|---|---|---|---|---|---|
| Cases | Controls | |||||
| rs187084 | 3/52227015 | C>T | 4149 | Promoter | 77 (0.35) | 77 (0.39) |
| rs352139 | 3/52216841 | A>G | 6808 | Intron | 107 (0.48) | 87 (0.43) |
| rs352144 | 3/52219937 | A>C | 3712 | Upstream variant | 225 (0.96) | 195 (0.97) |
Abbreviation: SNP, single-nucleotide polymorphism.
Acknowledgments
This project was supported by the Research Group Program (number RGP-VPP-260) of the Kingdom of Saudi Arabia.
Abbreviations
- SNP
single-nucleotide polymorphism
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- OR
odds ratio
- CI
confidence interval
- Ct
cycle threshold
- EDTA
ethylenediaminetetraacetic acid
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2(8):675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 2.Du X, Poltorak A, Wei Y, Beutler B. Three novel mammalian toll-like receptors: gene structure, expression, and evolution. Eur Cytokine Netw. 2000;11(3):362–371. [PubMed] [Google Scholar]
- 3.Chuang TH, Ulevitch RJ. Cloning and characterization of a sub-family of human toll-like receptors: hTLR7, hTLR8 and hTLR9. Eur Cytokine Netw. 2000;11(3):372–378. [PubMed] [Google Scholar]
- 4.Tabeta K, Georgel P, Janssen E, et al. Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc Natl Acad Sci U S A. 2004;101(10):3516–3521. doi: 10.1073/pnas.0400525101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumagai Y, Takeuchi O, Akira S. Pathogen recognition by innate receptors. J Infect Chemother. 2008;14(2):86–92. doi: 10.1007/s10156-008-0596-1. [DOI] [PubMed] [Google Scholar]
- 6.Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5(10):987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 7.Kutikhin AG. Association of polymorphisms in TLR genes and in genes of the toll-like receptor signaling pathway with cancer risk. Hum Immunol. 2011;72(11):1095–1116. doi: 10.1016/j.humimm.2011.07.307. [DOI] [PubMed] [Google Scholar]
- 8.Chang ZL. Important aspects of toll-like receptors, ligands and their signaling pathways. Inflamm Res. 2010;59(10):791–808. doi: 10.1007/s00011-010-0208-2. [DOI] [PubMed] [Google Scholar]
- 9.Rakoff-Nahoum S, Medzhitov R. Toll-like receptors and cancer. Nat Rev Cancer. 2009;9(1):57–63. doi: 10.1038/nrc2541. [DOI] [PubMed] [Google Scholar]
- 10.So EY, Ouchi T. The application of toll like receptors for cancer therapy. Int J Biol Sci. 2010;6(7):675–681. doi: 10.7150/ijbs.6.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pandey S, Agrawal DK. Immunobiology of toll-like receptors: emerging trends. Immunol Cell Biol. 2006;84(4):333–341. doi: 10.1111/j.1440-1711.2006.01444.x. [DOI] [PubMed] [Google Scholar]
- 12.Pandey S, Mittal B, Srivastava M, et al. Evaluation of toll-like receptors 3 (c.1377C/T) and 9 (G2848A) gene polymorphisms in cervical cancer susceptibility. Mol Biol Rep. 2011;38(7):4715–4721. doi: 10.1007/s11033-010-0607-z. [DOI] [PubMed] [Google Scholar]
- 13.O’Neill LA, Bryant CE, Doyle SL. Therapeutic targeting of toll-like receptors for infectious and inflammatory diseases and cancer. Pharmacol Rev. 2009;61(2):177–197. doi: 10.1124/pr.109.001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahman AH, Taylor DK, Turka LA. The contribution of direct TLR signaling to T cell responses. Immunol Res. 2009;45(1):25–36. doi: 10.1007/s12026-009-8113-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh V, Srivastava N, Kapoor R, Mittal RD. Single-nucleotide polymorphisms in genes encoding toll-like receptor-2, -3, -4, and -9 in a case–control study with bladder cancer susceptibility in a North Indian population. Arch Med Res. 2013;44(1):54–61. doi: 10.1016/j.arcmed.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 16.Mandal RK, George GP, Mittal RD. Association of toll-like receptor (TLR) 2, 3 and 9 genes polymorphism with prostate cancer risk in North Indian population. Mol Biol Rep. 2012;39(7):7263–7269. doi: 10.1007/s11033-012-1556-5. [DOI] [PubMed] [Google Scholar]
- 17.Zheng SL, Augustsson-Balter K, Chang B, et al. Sequence variants of toll-like receptor 4 are associated with prostate cancer risk: results from the cancer prostate in Sweden study. Cancer Res. 2004;64(8):2918–2922. doi: 10.1158/0008-5472.can-03-3280. [DOI] [PubMed] [Google Scholar]
- 18.De Marzo AM, Platz EA, Sutcliffe S, et al. Inflammation in prostate carcinogenesis. Nat Rev Cancer. 2007;7(4):256–269. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miedema KG, Tissing WJ, Te Poele EM, et al. Polymorphisms in the TLR6 gene associated with the inverse association between childhood acute lymphoblastic leukemia and atopic disease. Leukemia. 2012;26(6):1203–1210. doi: 10.1038/leu.2011.341. [DOI] [PubMed] [Google Scholar]
- 20.Xie JJ, Jiang SY, Shi MM, et al. The association between toll-like receptor 2 single-nucleotide polymorphisms and hepatocellular carcinoma susceptibility. BMC Cancer. 2012;12:57. doi: 10.1186/1471-2407-12-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kupcinskas J, Wex T, Bornschein J, et al. Lack of association between gene polymorphisms of angiotensin converting enzyme, Nod-like receptor 1, toll-like receptor 4 and Fas/Fasl with the presence of Helicobacter pylori-induced premalignant gastric lesions and gastric cancer in Caucasians. Helicobacter. 2011;12:112. doi: 10.1186/1471-2350-12-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hold GL, Rabkin CS, Gammon MD, et al. CD14-159C/T and TLR9-1237T/C polymorphisms are not associated with gastric cancer risk in Caucasian populations. Eur J Cancer Prev. 2009;18(2):117–119. doi: 10.1097/CEJ.0b013e3283101292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roszak A, Lianeri M, Sowinska A, Jagodzinski PP. Erratum to: involvement of toll-like receptor 9 polymorphism in cervical cancer development (vol 39, p. 8425, 2012) Mol Biol Rep. 2013;40(1):731. doi: 10.1007/s11033-012-1695-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mollaki V, Georgiadis T, Tassidou A, et al. Polymorphisms and haplotypes in TLR9 and MYD88 are associated with the development of Hodgkin’s lymphoma: a candidate-gene association study. J Hum Genet. 2009;54(11):655–659. doi: 10.1038/jhg.2009.90. [DOI] [PubMed] [Google Scholar]
- 25.Etokebe GE, Knezevic J, Petricevic B, Pavelic J, Vrbanec D, Dembic Z. Single-nucleotide polymorphisms in genes encoding toll-like receptor-2,-3,-4, and -9 in case-control study with breast cancer. Genet Test Mol Biomarkers. 2009;13(6):729–734. doi: 10.1089/gtmb.2009.0045. [DOI] [PubMed] [Google Scholar]
- 26.Noack J, Jordi M, Zauner L, et al. TLR9 agonists induced cell death in Burkitt’s lymphoma cells is variable and influenced by TLR9 polymorphism. Cell Death Dis. 2012;3:e323. doi: 10.1038/cddis.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carvalho A, Cunha C, Almeida AJ, et al. The rs5743836 polymorphism in TLR9 confers a population-based increased risk of non-Hodgkin lymphoma. Genes Immun. 2012;13(2):197–201. doi: 10.1038/gene.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashton KA, Proietto A, Otton G, et al. Toll-like receptor (TLR) and nucleosome-binding oligomerization domain (NOD) gene polymorphisms and endometrial cancer risk. BMC Cancer. 2010;10:382. doi: 10.1186/1471-2407-10-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rahman HA, Khorshied MM, Khorshid OM, Mahgoub SM. Toll-like receptor 2 and 9 genetic polymorphisms and the susceptibility to B cell non-Hodgkin lymphoma in Egypt. Ann Hematol. 2014;93(11):1859–1865. doi: 10.1007/s00277-014-2131-z. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L, Qin H, Guan X, Zhang K, Liu Z. The TLR9 gene polymorphisms and the risk of cancer: evidence from a meta-analysis. PloS one. 2013;8(8):e71785. doi: 10.1371/journal.pone.0071785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Semlali A, Reddy Parine N, Arafah M, et al. Expression and polymorphism of toll-like receptor 4 and effect on NF-kappaB mediated inflammation in colon cancer patients. PLoS One. 2016;11(1):e0146333. doi: 10.1371/journal.pone.0146333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Semlali A, Jacques E, Plante S, et al. TGF-beta suppresses EGF-induced MAPK signaling and proliferation in asthmatic epithelial cells. Am J Respir Cell Mol Biol. 2008;38(2):202–208. doi: 10.1165/rcmb.2007-0031OC. [DOI] [PubMed] [Google Scholar]
- 33.Semlali A, Leung KP, Curt S, Rouabhia M. Antimicrobial decapeptide KSL-W attenuates Candida albicans virulence by modulating its effects on toll-like receptor, human beta-defensin, and cytokine expression by engineered human oral mucosa. Peptides. 2011;32(5):859–867. doi: 10.1016/j.peptides.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 34.Alanazi M, Pathan AA, Abduljaleel Z, et al. Association between PARP-1 V762A polymorphism and breast cancer susceptibility in Saudi population. PLoS One. 2013;8(12):e85541. doi: 10.1371/journal.pone.0085541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baines J, Celis E. Immune-mediated tumor regression induced by CpG-containing oligodeoxynucleotides. Clin Cancer Res. 2003;9(7):2693–2700. [PubMed] [Google Scholar]
- 36.Tuomela J, Sandholm J, Karihtala P, et al. Low TLR9 expression defines an aggressive subtype of triple-negative breast cancer. Breast Cancer Res Treat. 2012;135(2):481–493. doi: 10.1007/s10549-012-2181-7. [DOI] [PubMed] [Google Scholar]
- 37.Marino M. Xenoestrogens challenge 17β-estradiol protective effects in colon cancer. World J Gastrointest Oncol. 2014;6(3):67–73. doi: 10.4251/wjgo.v6.i3.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barzi A, Lenz AM, Labonte MJ, Lenz HJ. Molecular pathways: estrogen pathway in colorectal cancer. Clin Cancer Res. 2013;19(21):5842–5848. doi: 10.1158/1078-0432.CCR-13-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.al-Azzawi F, Wahab M. Estrogen and colon cancer: current issues. Climacteric. 2002;5(1):3–14. [PubMed] [Google Scholar]
- 40.Hendrickse CW, Jones CE, Donovan IA, Neoptolemos JP, Baker PR. Oestrogen and progesterone receptors in colorectal cancer and human colonic cancer cell lines. Br J Surg. 1993;80(5):636–640. doi: 10.1002/bjs.1800800531. [DOI] [PubMed] [Google Scholar]
- 41.Valencia-Sanchez MA, Liu JD, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20(5):515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 42.Li H, Zhao H, Yu J, et al. Increased prevalence of regulatory T cells in the lung cancer microenvironment: a role of thymic stromal lymphopoietin. Cancer Immunol Immunother. 2011;60(11):1587–1596. doi: 10.1007/s00262-011-1059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Semlali A, Al Amri A, Azzi A, et al. Expression and new exon mutations of the human beta defensins and their association on colon cancer development. PLoS One. 2015;10(6):e0126868. doi: 10.1371/journal.pone.0126868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lemos B, Bettencourt BR, Meiklejohn CD, Hartl DL. Evolution of proteins and gene expression levels are coupled in Drosophila and are independently associated with mRNA abundance, protein length, and number of protein-protein interactions. Mol Biol Evol. 2005;22(5):1345–1354. doi: 10.1093/molbev/msi122. [DOI] [PubMed] [Google Scholar]
- 45.Nuzhdin SV, Wayne ML, Harmon KL, McIntyre LM. Common pattern of evolution of gene expression level and protein sequence in Drosophila. Mol Biol Evol. 2004;21(7):1308–1317. doi: 10.1093/molbev/msh128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Characteristics of selected polymorphisms in TLR-9
| SNP ID | Chromosome/position | Nucleotide change | Position | Region | Minor allele frequency (%)
|
|
|---|---|---|---|---|---|---|
| Cases | Controls | |||||
| rs187084 | 3/52227015 | C>T | 4149 | Promoter | 77 (0.35) | 77 (0.39) |
| rs352139 | 3/52216841 | A>G | 6808 | Intron | 107 (0.48) | 87 (0.43) |
| rs352144 | 3/52219937 | A>C | 3712 | Upstream variant | 225 (0.96) | 195 (0.97) |
Abbreviation: SNP, single-nucleotide polymorphism.


