Abstract
Renal ischemia-reperfusion injury is a leading cause of acute kidney injury; the pathogenesis of which remains poorly understood and effective therapies still lacking. Here we tested whether microRNAs, identified as critical regulators of cell health and disease, are involved in this process. We found that miR-17-5p was significantly up-regulated during renal ischemia-reperfusion injury in mice and during hypoxia in cultured renal tubular cells. In cultured cells, MiR-17-5p directly inhibited the expression of death receptor 6 (DR6) and attenuated apoptosis during hypoxia. Blockade of miR-17-5p abolished the suppression of DR6 and facilitated caspase activation and apoptosis. In vivo, an miR-17-5p mimic suppressed DR6 expression and protected against renal ischemia-reperfusion injury. We further verified that miR-17-5p induction during renal ischemia-reperfusion injury was dependent on p53. Inhibition of p53 with pifithrin-α or a dominant-negative mutant led to the repression of miR-17-5p expression under hypoxia in vitro. Moreover, miR-17-5p induction during renal ischemia-reperfusion injury was attenuated in proximal tubule p53 knockout mice, supporting the role of p53 in miR-17-5p induction in vivo. Thus, p53/miR-17-5p/DR6 is a new protective pathway in renal ischemia-reperfusion injury and may be targeted for the prevention and treatment of ischemic acute kidney injury.
Keywords: acute kidney injury, ischemia-reperfusion injury, microRNA, death receptor
Introduction
Renal ischemia-reperfusion injury (IRI) is one of main causes of acute kidney injury (AKI).1, 2 For example, it is unavoidable in patients who undergo major cardiac events (such as infarction or surgery) or kidney transplantation. Renal IRI is associated with high rates of mortality and end-stage kidney failure and notably, even if patients recover from initial injury, renal IRI may have lasting effects including the development of chronic kidney disease (CKD).3 For decades, numerous studies have investigated the molecular and cellular mechanisms of renal IRI and have suggested a variety of pathophysiological changes, including tubular or epithelial cell injury, microvascular dysfunction, and inflammation. However, the mechanistic understanding is incomplete and no therapies are available for effective treatment of IRI-associated kidney injury and renal failure.
MicroRNAs (miRNAs) are recently discovered, critical regulators of gene expression with approximately 21–25 nucleotides in length.4–6 They target downstream genes by binding to the 3′untranslated region (3′UTR) of mRNAs resulting in the repression of translation. Given this critical role, miRNAs not only effect on physiological cellular behavior, but also contribute to the development of diseases in various pathological conditions. In kidneys, miRNAs play important roles in renal development and physiological maintenance and, as a result, the ablation of Dicer (a key enzyme for miRNA production) in specific kidney cell types led to renal dysfunction and renal diseases.7–11 In 2010, we established a conditional Dicer knockout mouse model, in which Dicer was ablated specifically from kidney proximal tubular cells by using PEPCK-Cre .12 Because PEPCK-Cre is expressed for Dicer knockout at 2–3 weeks post-natally, these mice did not have renal development defects and showed normal renal function and histology under control conditions. However, these mice were remarkably resistant to renal IRI, suggesting an important role of miRNAs in the pathogenesis. By microarray analysis, we further identified 23 miRNAs with significant changes in expression during renal IRI.12 More recent studies have investigated the role and regulation of some of these miRNAs in renal IRI, including miR-494, -21, -126, -687, -150, and -489.13–17 Notably, while some of the miRNAs mediate kidney injury during IRI, others may play protective roles.18–20 Accordingly, investigation of these miRNAs may suggest therapeutic strategies for IRI-related kidney diseases. In this study, we specifically examined the role and regulation of miR-17-5p in renal IRI. We verified miR-17-5p induction during renal IRI. The induction was mediated by p53 and, following induction, this microRNA may repress death receptor 6 (DR6) to protect kidney cells and tissues from injury.
Results
miR-17-5p is up-regulated during renal IRI in vivo and hypoxia in vitro
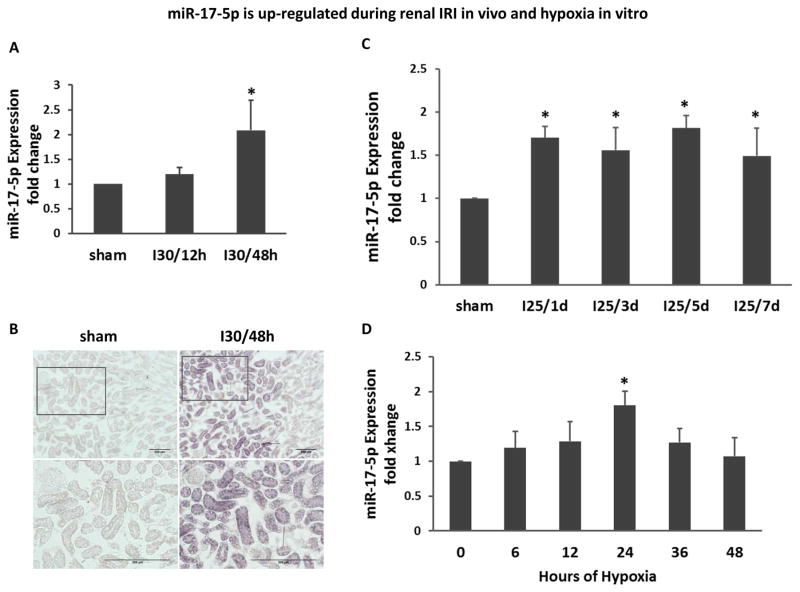
Severe and moderate renal IRI was induced in mice respectively by 30 and 25 minutes of bilateral renal ischemia followed by reperfusion. miR-17-5p in renal cortical tissues was determined by quantitative real time PCR using highly specific Taqman probes. In the severe IRI model, miR-17-5p was marginally induced after 12 hours of reperfusion (I30/12h), but significantly induced after 48 hours of reperfusion (I30/48h) (Figure 1A). To localize miR-17-5p induction, we conducted in situ hybridization. As shown in Figure 1B, miR-17-5p signal was weak in Sham- control kidneys, but it was markedly induced in kidney cortical tissues after 30 minutes of ischemia and 48 hours of reperfusion and the induction was mainly in proximal tubules. In the moderate IRI model, an increase of miR-17-5p was detected at 1 day of reperfusion (I25/1d) and this increase was maintained during the whole observation period of a week (Figure 1C). In cultured rat kidney proximal tubular cells (RPTC), hypoxia (1% oxygen) induced marginal increases in miR-17-5p at 6–12 hours, but the increase became significant at 24 hours of hypoxic incubation (Figure 1D). These results demonstrated the miR-17-5p induction during renal IRI in kidney tissues and hypoxia in cultured tubular cells.
Figure 1. miR-17-5p is up-regulated during renal IRI in vivo and hypoxia in vitro.
(A) miR-17-5p induction in severe renal IRI. Total RNA samples were extracted from kidney cortex of C57BL/6 mice that had been subjected to 30 minutes of bilateral renal ischemia with 12 hours (I30/12h) or 48 hours (I30/48h) of reperfusion, or sham surgery (sham). MiR-17-5p level was analyzed by quantitative real-time PCR that showed a significant up-regulation at I30/48h. Data were shown as mean±SD (n=3), *P<0.05 versus sham. (B) In situ hybridization analysis for miR-17-5p. C57BL/6 mice were subjected to 30 minutes of bilateral renal ischemia with 48 hours of reperfusion (I30/48h) or sham surgery as control to collect frozen sections of kidney tissues. The tissues were analyzed using double-digoxigenin-labeled specific miR-17-5p probe for In situ hybridization. Representative images of each group with 100× and 200× were shown to indicate marked increases of miR-17-5p in renal tubules. (C) miR-17-5p induction in moderate renal IRI. Total RNA samples were extracted from kidney cortex of C57BL/6 mice that had been subjected to 25 minutes of bilateral renal ischemia with reperfusion for 1 day (I35/1d), 3 days (I25/3d), 5 days (I25/5d), and 7 days (I25/7d), or sham surgery (sham). Data were shown as mean±SD (n=3), *P<0.05 versus sham. (D) miR-17-5p induction in cultured renal proximal tubular cells (RPTC) under hypoxia. RPTC cells were incubated under normoxia or hypoxia (1% oxygen) for 6–48 hours. Total RNA samples were extracted for quantitative real time PCR showing the significant induction of miR-17-5p at 24 hours after hypoxia treatment. Data were shown as mean±SD (n=3), *P<0.05 versus 0 hour.
Effects of anti-miR-17-5p and miR-17-5p mimic on tubular cell apoptosis during hypoxia
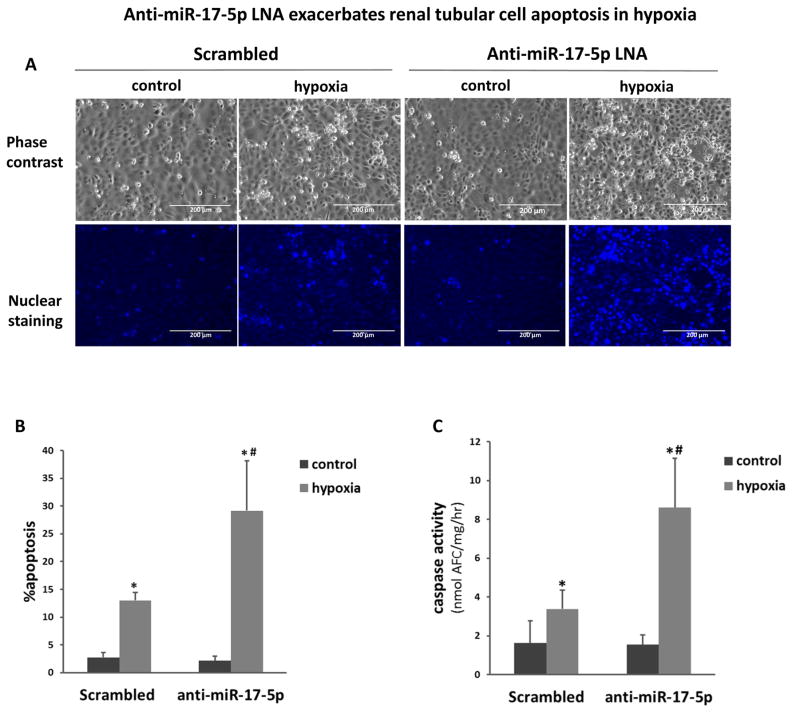
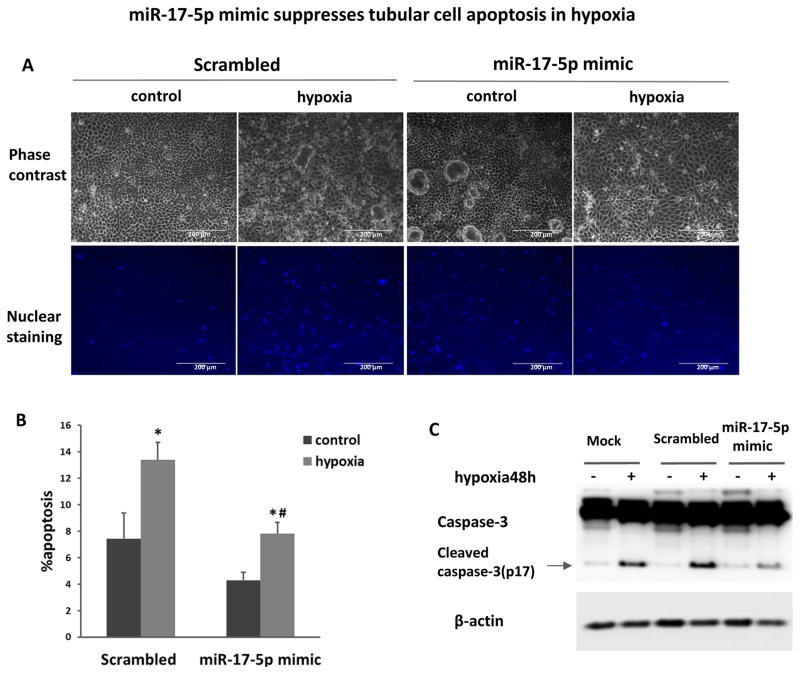
To investigate the pathophysiological role of miR-17-5p, we initially examined the effect of anti-miR-17-5p LNA on hypoxic injury of RPTC cells. Hypoxic incubation resulted in a significant increase of apoptosis in RPTC transfected with scrambled sequence LNA (Figure 2A). Notably, transfection with anti-miR-17-5p LNA almost doubled the percentage of apoptotic cells during hypoxia treatment (Figure 2A, 2B). These morphological evaluations were verified by the measurement of caspase activity (Figure 2C). In contrast, the transfection of miR-17-5p mimic significantly reduced apoptosis during hypoxia as assessed by both morphological assays (Figure 3A, 3B) and immunoblot analysis of active caspase-3 (Figure 3C). Altogether, the results suggested that miR-17-5p induced in renal IRI may act as a protective factor against tubular cell injury and tissue damage.
Figure 2. Anti-miR-17-5p LNA exacerbates renal tubular cell injury under hypoxia.
RPTC cells were transfected with 200nM scramble sequence or anti-miR-17-5p LNA and then incubated in normoxia (control) or hypoxia (1% oxygen) for 48 hours. (A) Representative images to show the cell morphology and nuclear staining of RPTC cells (scale bar is 200 μm). (B) Percentage of RPTC apoptosis by morphological evaluation. Data were expressed as mean±SD (n=6). (C) Enzymatic assay of caspase activity. Data were expressed as mean±SD (n=3), *P<0.05 versus normoxia control, #P<0.05 versus scrambled LNA transfection with hypoxia.
Figure 3. miR-17-5p mimic suppresses tubular cell apoptosis in hypoxia.
RPTC cells were transfected with 200nM miR-17-5p mimic or scrambled oligonucleotides, and then incubated in normoxia (control) or hypoxia (1% oxygen) for 48h. (A) Representative images to show the morphology and nuclear staining of RPTC (scale bar is 200 μm). (B) Percentage of RPTC apoptosis by morphology evaluation. Data were expressed as mean±SD (n=4), *P<0.05 versus normoxia control, #P<0.05 versus scrambled transfection with hypoxia. (C) Immunoblot analysis of caspase-3 cleavage. Whole cell lysate was collected from RPTC cells with miR-17-5p mimic or scrambled oligonucleotides or reagent alone (mock) transfection under normoxia or hypoxia condition for immunoblotting. Caspase-3 cleavage was significantly inhibited by miR-17-5p transfection. β-actin was used as internal control.
MiR-17-5p mimic protects against renal IRI
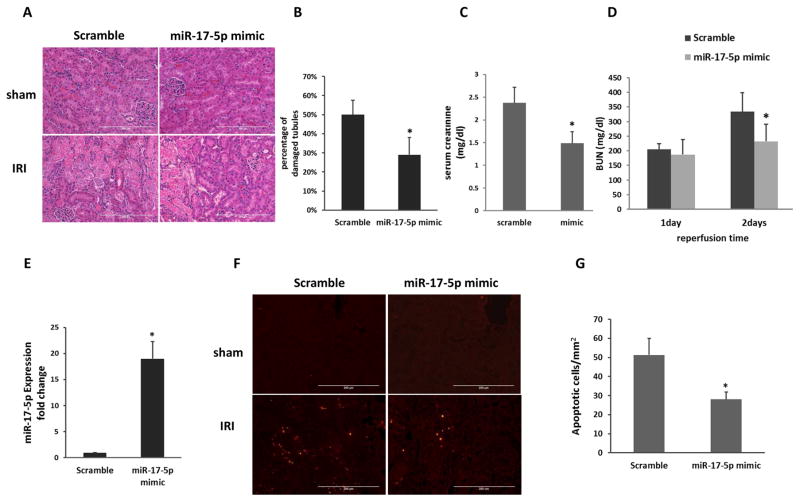
We further examined the effect of miR-17-5p mimic in vivo. Mice were injected with miR-17-5p mimic or scramble sequence oligo, and then subjected to sham or bilateral renal IRI. Compared to scramble oligo group, the mice treated with miR-17-5p showed significantly lower BUN and serum creatinine (Figure 4A, 4B). We further examined renal tissue damage by hematoxylin and eosin (HE) staining. As shown in Figure 4D and 4E, the kidneys from sham surgery group showed healthy histology. Notably, there was severe kidney injury with about 40% tubular damage in scramble oligo-treated mice followed by ischemia 30 minutes and 48 hours of reperfusion. In contrast, miR-17-5p mimic injection significantly reduced the tubular damage to about 20%. Following IRI, these mice also showed significantly less apoptosis (28/mm2) than the scramble control mice (51/mm2) (Figure 4F, 4G). These results suggested that miR-17-5p may play a protective role in ischemic AKI.
Figure 4. miR-17-5p mimic protects against renal IRI.
C57BL/6 mice were injected with scramble oligo or miR-17-5p mimic, and then subjected to 30 minutes of bilateral renal ischemia followed by 48 hours of reperfusion. (A) BUN. (B) Serum creatinine at 48 hours of reperfusion (n=5). *P<0.05 versus scramble oligo group. (C) Representative images of hematoxylin and eosin staining to show kidney tissue damage. Scale bar, 200 μm. (D) Percentage of tubular damage after 30 minutes of ischemia with 48 hours of reperfusion (n=5). *P<0.05 versus scramble oligo group. (E) Representative images of TUNEL staining to show apoptosis in kidney tissues. Scale bar, 200μm. (F) Apoptotic cell numbers per mm square of kidney tissue (n=5). *P<0.05 versus scramble oligo group.
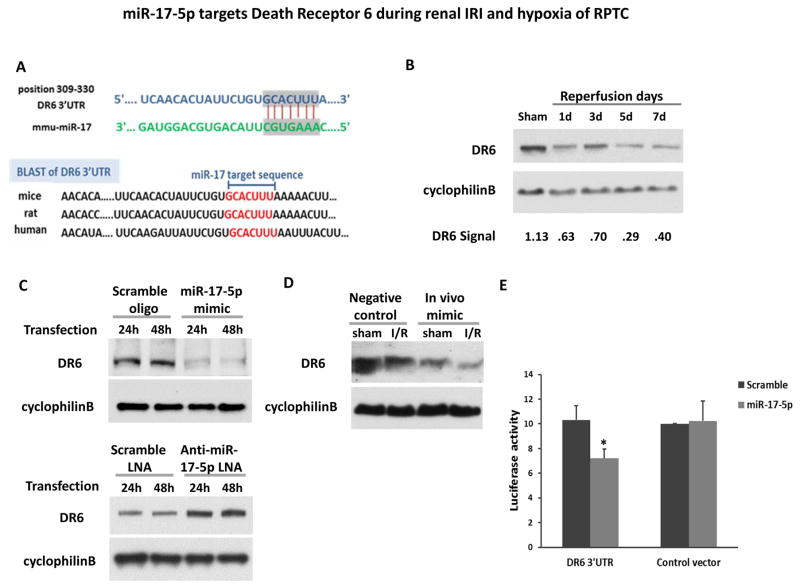
miR-17-5p targets Death Receptor 6 during renal IRI and hypoxia of RPTC
How does miR-17-5p protect? The key to this question is to identify the responsible target gene(s) of this microRNA. Depending on the cellular context, miR-17-5p may target a variety of downstream genes by binding to the 3′UTR of their mRNAs 21. We first searched two microRNA target analysis databases (miRanda and TargetScan), which predicted several hundred putative target genes of miR-17-5p. Among these genes, DR6, also known as tumor necrosis factor receptor superfamily member 21 (TNFRSF21), was consistently predicted as a candidate target of miR-17-5p. Further bioinformatics analysis showed that the miR-17-5p binding site in the 3′UTR of DR6 was conserved across species (Figure 5A). Experimentally, DR6 expression significantly decreased in kidney cortical tissues after 25 minutes of ischemia with 1–7 days of reperfusion (Figure 5B), where miR-17-5p was induced (Figure 1B). In RPTC cells, DR6 expression was suppressed specifically by the transfection of miR-17-5p mimic, whereas anti-miR-17-5p LNA increased DR6 expression (Figure 5C). In vivo, we treated mice with miR-17-5p mimic for 24 hours, which suppressed DR6 expression (Figure 5D). To further determine if miR-17-5p directly targets the 3′UTR of DR6, we conducted miRNA Target Luciferase Reporter assay. To this end, we prepared a construct that contained the putative miR-17-5p targeting site of DR6 3′UTR sequence after the luciferase reporter gene (Luciferase-DR6-3′UTR). HEK cells were co-transfected with the Luciferase-DR6-3′-UTR construct or Luciferase-DR6-3′-empty vector along with miR-17-5p mimic or scrambled-sequence oligonucleotide. miR-17-5p mimic significantly reduced luciferase expression in Luciferase-miR-17-DR6 transfected cells but not the scrambled-sequence oligonucleotide (Figure 5E), indicating that the miR-17-5p directly targeted DR6-3′-UTR. Collectively, these results suggested that miR-17-5p may attenuate the ischemic renal cell death by targeting or repressing DR6.
Figure 5. miR-17-5p targets Death Receptor 6 during renal IRI and hypoxia of RPTC.
(A) The conserved, putative miR-17-5p binding sequence in 3′UTR of DR6 mRNA. (B) Immunoblot analysis of DR6 in mice. C57BL/6 mice were subjected to sham operation or 25 minutes of bilateral kidney ischemia followed by 1, 3, 5, 7 days of reperfusion. Kidney cortical tissues were extracted to analyze DR6 by immunoblotting. Cyclophilin B was used as internal control. For quantification, the DR6 bands were analyzed by densitometry and expressed as the ratio over cyclophilin B. (C) Immunoblot analysis of DR6 in renal proximal tubular cells. RPTC cells were transfected with miR-17-5p mimic or anti-miR-17-5p LNA; the scrambled RNA oligo or LNA was used as control, respectively. The whole cell lysates were collected after transfection for 24 hours or 48 hours for immunoblot analysis of DR6 and cyclophilin B as internal control. . (D) Immunoblot analysis of DR6 in mice injected with miR-17-5p mimic or negative control oligo. The treated mice were subjected to sham surgery or 30 minutes of bilateral renal ischemia and 48 hours of reperfusion. Kidney cortical tissue protein lysate was extracted to analyze DR6 expression by immunoblotting. CyclophilinB level was used as internal control. (E) Target of DR6 3′-UTR by miR-17-5p. The putative miR-17-5p binding sequence was inserted into pMIR-REPORT plasmid and then transfected into HEK cells. The cells were co-transfected along with miR-17-5p mimic or scrambled miRNA. β-gal reporter plasmid was also transfected for normalization. The luciferase activity was measured to show the inhibition effect of miR-17-5p. Data were expressed as mean±SD (n=5), *P<0.05 versus control empty vector co-transfected with scrambled sequence.
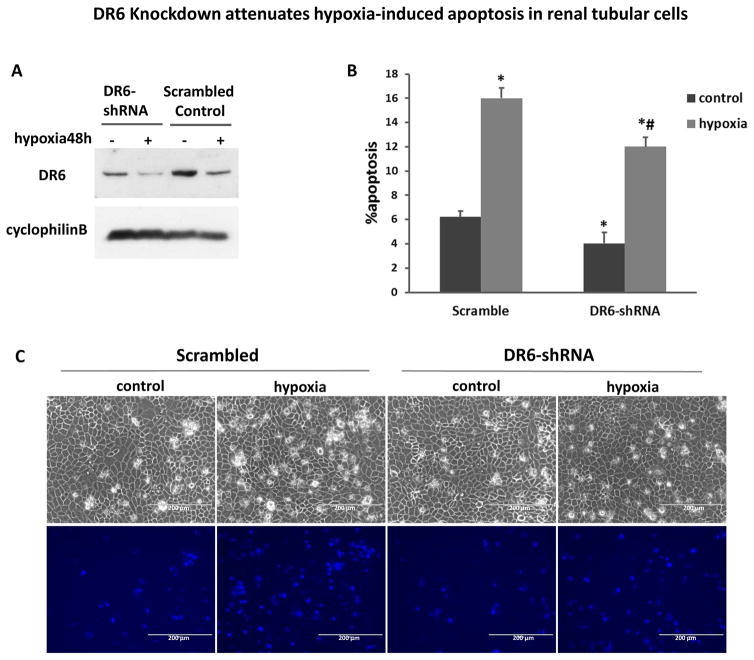
To test the role of DR6 in miR-17-5p-mediated renal protection, we transfected shRNAs into RPTCs to knockdown DR6. Compared with scramble shRNA, DR6-shRNA transfection significantly decreased DR6 expression (Figure 6A). After 48hours of hypoxia (1% oxygen) treatment, ~16 % of apoptosis occurred in scrambled oligo transfected cells, whereas less than 12% in DR6 knockdown cells (Figure 6B). Representative recording of cell morphology further verified that less apoptosis developed following hypoxia in DR6-shRNA cells (Figure 6C). This result supported a pro-apoptotic role of DR6 in renal tubular cells during hypoxic/ischemic injury. This observation also suggested that miR-17-5p may protect kidney cells and tissues by repressing DR6.
Figure 6. DR6 knockdown attenuates hypoxia-induced apoptosis in renal tubular cells.
(A) Immunoblot to show DR6 down-regulation by shRNA. RPTC cells were stably transfected with DR6-shRNA or scrambled control plasmids. The whole cell lysates were analyzed for DR6 by immunoblotting with cyclophilin B as internal control. (B), (C) Both DR6 knockdown and scrambled control RPTC cells were incubated in normoxia (control) or hypoxia (1% oxygen) for 48 hours. The percentage of apoptosis was evaluated morphologically (B). Data were expressed as mean±SD (n=3), *P<0.05 versus normoxia control, #P<0.05 versus scrambled control transfection. (C) Representative images to show the cell morphology and nuclear staining (scale bar-200 μm).
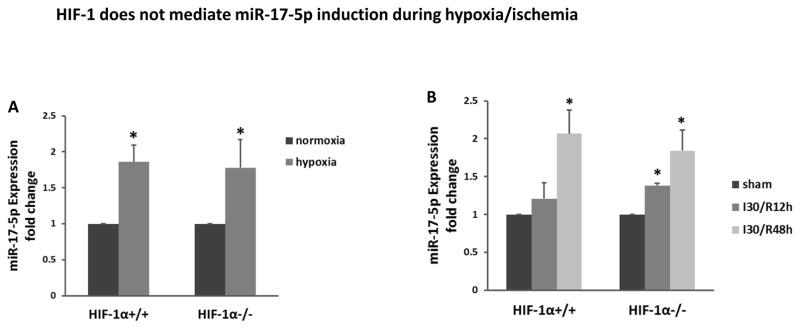
HIF-1 does not mediate miR-17-5p induction during hypoxia/ischemia
After showing DR6 as a down-stream target gene of miR-17-5p (Figures 5, 6), we would like to understand the up-stream mechanism that accounts miR-17-5p induction in renal IRI. HIF-1 is considered to be the “master” transcription factor that regulates gene expression under conditions of hypoxia and ischemia.22 In addition, HIF-1 has been reported to regulate miR-17 in leukemic cells.23 Thus, we hypothesized that HIF-1 mediated miR-17-5p induction during renal IRI and hypoxia of RPTC cells. To test this possibility, we initially compared miR-17-5p induction by hypoxia in wild-type and HIF-1α-null mouse embryonic fibroblasts (MEFs). As shown in Figure 7A, miR-17-5p expression increased in both wild-type and HIF-1α-null cells, negating a role of HIF-1 in hypoxic induction of miR-17-5p. To further test this in vivo, we took advantage of the proximal tubule specific HIF-1α knockout mouse model that was established in our recent work.15, 17 The rationale was that miR-17-5p was mainly induced in kidney proximal tubules (Figure 1B). As shown in Figure 7B, miR-17-5p was induced in both wild-type and proximal tubule-HIF-1α-null mice, irrespective of the HIF-1 status. Thus it was concluded that HIF-1 was not the key transcription factor responsible for miR-17-5p induction by hypoxia in vitro and renal IRI in vivo.
Figure 7. HIF-1 does not mediate miR-17-5p induction during hypoxia/ischemia.
(A) miR-17-5p expression induced by hypoxia in both wild-type (WT) and HIF-1α–deficient MEF cells. The cells were incubated in hypoxia or normoxia condition for 24 hours to isolate RNA for real-time PCR analysis of miR-17-5p. Data were expressed as mean±SD (n=3), *P<0.05 versus normoxia control. (B) miR-17-5p induction in kidney cortex by renal ischemia-reperfusion in proximal tubule HIF-1α knockout (PT-HIF-1α KO) mice and their wild-type littermates. The mice were subjected to bilateral renal ischemia 30 minutes followed by reperfusion of 12 or 48 hours. RNA samples were extracted from the kidney cortical tissues for quantitative real-time PCR analysis of miR-17-5p. All data were expressed as mean±SD (n=3). *P<0.05 versus sham control.
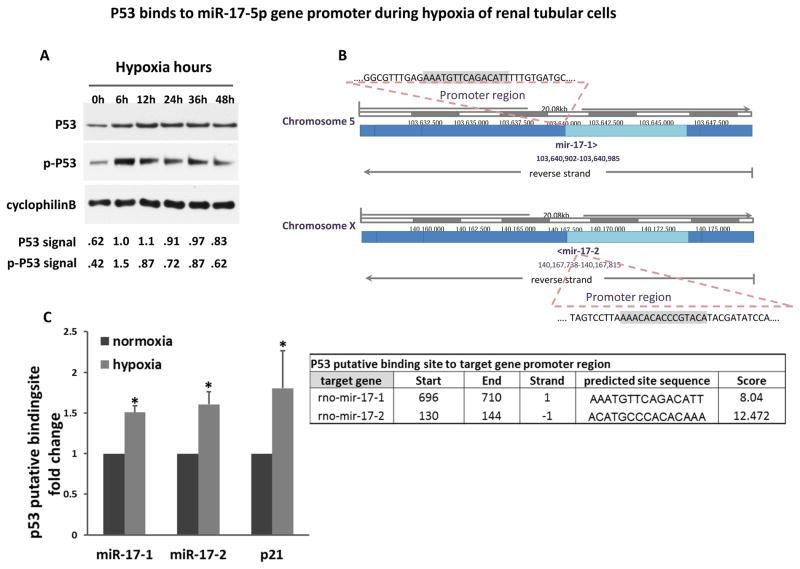
P53 binds to miR-17-5p gene promoter during hypoxia of renal tubular cells
P53 plays an important role in renal IRI.24, 25 Such a conclusion was further established recently by using renal tubule-specific p53-knockout (PT-p53-KO) mouse models.26, 27 In these studies, we further identified multiple genes that are regulated by p53 in renal IRI and may contribute to the pathogenesis.26 Is miR-17-5p subjected to p53 regulation? To address this, we fist verified that p53 was induced or activated by hypoxia (1% oxygen) in vitro in RPTC cells. As shown in Figure 8A, both total and phosphorylated p53 were induced by 6–48 hours of hypoxia. We further used the JASPAR CORE 2016 database to conduct bioinformatics analysis of the promoter region of rat miR-17-5p gene. Interestingly, there appeared to be two miR-17 genes (miR-17-1 and miR-17-2) and both genes contained a putative p53-binding site in their promoter regions (Figure 8B). Experimentally, we analyzed the direct binding of p53 to the miR-17-1 and miR-17-2 promoter regions by chromatin immunoprecipitation (ChIP) assay. As a positive control, p21 (known p53 target gene) was used. As shown in Figure 8C, both predicted p53 binding sites showed significantly more p53 binding in hypoxia-treated RPTC cells. The results implicate that p53 may mediate miR-17-5p expression in hypoxia by direct transcriptional activation.
Figure 8. P53 binds to miR-17-5p gene promoter during hypoxia of renal tubular cells.
(A) P53 activation during hypoxic injury. RPTC cell lysates were collected at 0 – 48 hours of incubation in hypoxia (1% oxygen) for immunoblot analysis of p53 and phospho-p53 (serine-15). Cyclophilin B was used as internal control. For quantification, p53 and p-p53 bands were analyzed by densitometry and expressed as the ratio over cyclophilin B. (B) The putative p53 binding sequence in miR-17 promoter region. (C) P53 binds to miR-17-5p promoter DNA during hypoxia. PRTC cells were incubated under normoxia or hypoxia for 24 hours to collect the chromatin for immunoprecipitation of p53. The immunoprecipated samples were subjected to real-time PCR analysis of miR-17-1 and miR-17-2 promoter sequences. The p53 binding site in p21 promoter was used as positive control. Data were expressed as mean±SD (n=3), *P<0.05 versus normoxia.
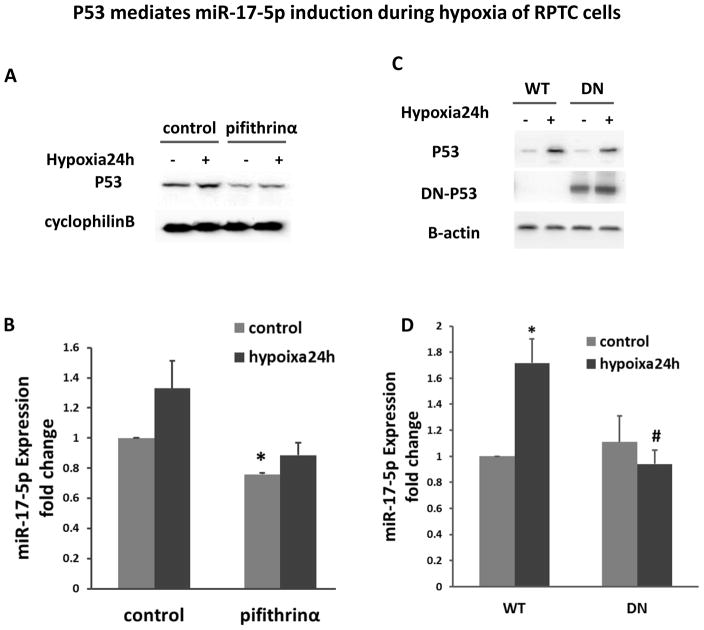
P53 mediates miR-17-5p induction during hypoxia of RPTC cells
To determine if p53 plays a role in miR-17-5p induction during renal hypoxia and IRI, we initially tested the effect of pifithrin-α, a pharmacological inhibitor of p53. We first verified the inhibitory effect of pifithrin-α on p53 in RPTC cells (Figure 9A). Further real-time PCR analysis showed that pifithrin-α blocked miR-17-5p induction during hypoxia of RPTC cells (Figure 9B). To confirm this observation, we examined the stable RPTC cell line transfected with p53 dominant-negative mutant (DN-p53). As shown in Figure 9D, 24 hours of hypoxia treatment resulted in obviously miR-17-5p increase in wild-type (WT) cells, but the induction was clearly blocked in DN-p53 cells (Figure 9C, 9D).
Figure 9. p53 mediates miR-17-5p induction during hypoxia of RPTC cells.
(A), (B) RPTC cells were cultured under normoxia or hypoxia condition with/without pifithrinα. (A) Effect of pifithrinα on p53. The whole cell lysates were collected from RPTC cells and analyzed by immunoblotting with cyclophilin B as internal control. (B) Effect of pifithrinα on miR-17-5p expression. The total RNA samples were extracted from RPTC cells for real-time PCR analysis of miR-17-5p. Data were presented as fold change mean±SD (n=3), *P<0.05 versus normoxia control. (C), (D) RPTC cells with p53 dominant negative mutant (DN) stable expression or the wild type control (WT) were incubated under normoxia or hypoxia. (C) Immunoblot analysis of p53 and dominant negative-p53 (DN-P53). The whole cell lysates were collected for immunoblotting to detect total p53 and DN-p53 by using specific antibody to HA tag. β-actin was used as internal control. (D) Effect of DN-p53 on miR-17-5p. The total RNA samples were extracted for real-time PCR analysis of miR-17-5p to compare the fold change after 24 hours of hypoxia treatment. Data are expressed as mean±SD (n=3), *P<0.05 versus normoxia control, # P<0.05 versus WT hypoxia treatment.
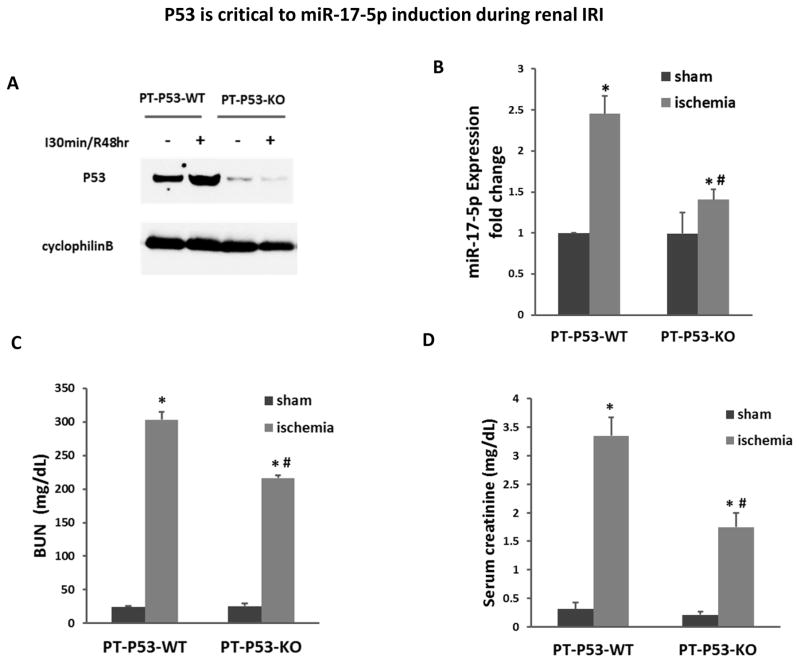
P53 is critical to miR-17-5p induction during renal IRI
Finally, we determined if miR-17-5p induction in renal IRI depends on p53. For this purpose, we used the conditional knockout mouse model from our recent study, which had specific p53 ablation from proximal tubules (PT-P53-KO).26 As expected, compared to wild-type littermates (PT-P53-WT), PT-P53-KO mice showed marked lower p53 in kidney cortical tissues regardless of renal IRI (Figure 10A). Importantly, 30 minutes of bilateral renal ischemia with 48hours of reperfusion led to 2.5 fold increase in miR-17-5p, which was reduced to less than 1.5 fold in PT-P53-KO tissues (Figure 10B). As before,26 PT-P53-KO mice were partially protected from renal IRI as shown by the measurements of BUN and serum creatinine (Figure 10C, 10D).
Figure 10. P53 is critical to miR-17-5p induction during renal IRI.
(A) Proximal tubule p53 wild-type (PT-P53-WT) and proximal tubule p53 knockout (PT-P53-KO) mice were subjected to bilateral renal ischemia for 30 minutes followed by 48 hours of reperfusion. Whole tissue lysates from kidney cortex were prepared for immunoblot analysis of p53. (B) Total RNA samples extracted from kidney cortex were prepared for real-time PCR analysis of miR-17-5p expression. The results show that miR-17-5p expression was significantly induced during renal IRI and this induction was suppressed in proximal tubule p53 knockout mice. (C) BUN of PT-P53-WT and PT-P53-KO mice. (D) Serum creatinine of PT-P53-WT and PT-P53-KO mice. *p<0.05 vs. PT-P53-WT control; # p<0.05 vs. PT-P53-WT Ischemia.
Discussion
MiRNAs are important post-transcriptional regulators of gene expression which mediate various cellular activities and processes of homeostasis by repressing target genes. In the current study, we provide the first evidence of the regulation and functional role of miR-17-5p in ischemic AKI. MiR-17-5p was up-regulated both in vivo in ischemia-reperfused kidneys and in vitro during hypoxic incubation of cultured renal proximal tubular cells. The miR-17-5p induction was shown to be mediated directly by p53, and not by HIF-1. Upon induction, miR-17-5p may play a cytoprotective role for renal cell survival via the suppression of DR6.
miR-17, encoded by the miR-17-92 family cluster, has been reported to increase in response to ischemia and hypoxia in cardiovascular system.28, 29 We and others also demonstrated miR-17 up-regulation in ischemic AKI;15, 30 however, it was unclear if miR-17-5p or miR-17-3p or both are induced. During miRNA biogenesis, both strands (5p, 3p) are produced and it is generally believed that one strand acts as the guidance strand to guide the target mRNA into the RNA-induced silencing complex (RISC), whereas the other (passenger) strand is degraded. Nonetheless, in some conditions, both strands are stable and functional to target different mRNAs. To verify the induction of miR-17 and identify its functional form in ischemic AKI, we conducted quantitative real time PCR using specific Taqman probes to miR-17-5a and -3a. The analysis revealed the induction of both miR-17-5p and miR-17-3p during renal IRI (Figure 1 and data not shown). Further functional study showed that inhibition of miR-17-5p by LNA increased apoptosis during hypoxic incubation (Figure 2), whereas miR-17-3p-LNA had no effect (not shown). Therefore, in the current study, we mainly focused on the regulation and function of miR-17-5p in kidney IRI. Functionally, anit-miR-17-5p-LNA increased apoptosis during hypoxic incubation (Figure 2), while miR-17-5p mimic suppressed apoptosis, supporting a protective role of miR-17-5p. Consistently, the majority of the studies in cardiovascular systems indicate that miR-17-5p may act as a pro-survival factor.28
In various pathological conditions, miR-17-5p may target a variety of downstream genes, including those in the PTEN pathway, WNT/b-catenin pathway, PI3K/AKT pathway, and MAPK/ERK pathways.31–34 In this study, we verified DR6 as a direct target of miR-17-5p by various analyses, including miRNA target reporter luciferase assay. To our knowledge, this is the first report of DR6 as a target of miR-17-5p. DR6, also known as tumor necrosis factor receptor superfamily member 21 (TNFRSF21, belongs to the second group of death receptor signaling complex. DR6 may trigger either apoptotic or survival signals by the recruitment of different sets of molecules.35 When receptor-interacting protein (RIP) and TNFR-associated death domain protein (FRADD) are recruited to the complex, DR6 along with DR3 may trigger NF-κB signaling that promotes the expression of multiple survival gene.36 On the contrary, when Fas-Associated Death Domain (FADD), procaspase-8/10 and FLICE-inhibitory protein (FLIP) are recruited to form the complex, DR6 may activate the downstream death signaling.37 Recent studies suggested the up-regulated endogenous DR6 during hypoxia could exacerbate neuritis damage and inhibition of DR6 could protect neurons and axons from injury after ischemic stroke.38, 39 Consistently, in our study, shRNA knockdown of DR6 led to the protection of RPTC cells from apoptosis, suggesting that DR6 is pro-death in hypoxic renal tubular cells. Knockdown of DR6 also suppressed the basal level of apoptosis in control cells. This effect in control cells is not surprising because DR6 is a member of TNF death receptor superfamily. In kidneys, it is interesting that the induction of miR-17-5p occurred as early as 1day and lasted until 1 week after IRI, which correlated with DR6 inhibition (Figures 1, 5). The sustained expression changes may promote cell survival against initial injury and may also contribute to the ensuing kidney repair process. However, our study does not exclude the possible involvement of other potential miR-17-5p targets, such as PTEN, janus kinase 1 (JAK1), signal transducer and activator of transcription 3 (STAT3), Thioredoxin-interacting protein (Txnip)/apoptosis signal-regulating kinase 1(ASK-1), which have been reported to function downstream of miR-17-5p in different systems.40–43
HIF-1 and p53 are two of the major transcription factors activated during renal IRI that are responsible for the induction of genes, including miRNAs. While the majority of HIF-1 responsive genes are cytoprotective or adaptive to hypoxic/ischemic stress, the induction of injurious genes via HIF-1 have also been reported.22 HIF-1 transcriptionally regulates miR-17-92 cluster in acute myeloid leukemia (AML).23, 44, 45 However, in our present study HIF-1 deficiency did not affect miR-17-5p expression in either hypoxic renal tubular cells or ischemic kidney tissues, by HIF-1 (Figure 7). Instead, we demonstrated that miR-17-5p induction during hypoxia in vitro and renal IRI in vivo is dependent on p53 (Figures 9, 10). CHIP assay further showed the binding of p53 on the gene promoter of miR-17 (Figure 7). P53 is known as a critical pathogenic factor in renal IRI through its regulation of a myriad of genes involved in tubular cell death, cell cycle arrest, and/or renal inflammation.24–27, 46 By identifying miR-17-5p as a transcriptional target of p53 in renal hypoxia/IRI, our work adds new insights into the understanding of p53 regulation in relevant disease conditions. miR-17 induction in renal IRI was largely, yet not completely, diminished in PT-p53-KO mice (Figure 10B). The residual induction was most likely caused by the incomplete p53 knockout in the mouse model, which was established by crossing PEPCK-Cre mice with p53-floxed mice. PEPCK-Cre was expressed in 70–80% proximal tubular cells and, in turn, resulted in partial gene knockout. The 20–30% cells where p53 was not deleted may still respond to renal IRI by inducing miR-17. Our results suggest the activation of p53/miR-17-5p/DR6 axis as a protective pathway during hypoxic/ischemic renal injury. This is somewhat surprising, because p53 is generally considered to contribute to tubular cell death and tissue damage in renal IRI.24–27 However, in cell biology p53 is a stress responsive transcription factor. Among all the genes regulated by p53, some are pro-death whereas others may pro-survival. For example, p21 is a well-documented gene transcribed via p53 and in kidneys, p21 is induced to protect against acute injury.47 In this study, we have identified miR-17-5p as a new protective gene transcribed via p53. During renal IRI, miR-17-5p is induced in tubular cells via p53 and upon induction, miR-17-5p may repress DR-6 to inhibit apoptosis and promote cell survival during hypoxic/ischemic kidney injury.
Material and Methods
Animals and Renal Ischemia-Reperfusion
Proximal tubule-specific p53 or HIF-1α knockout mouse models were established by crossing p53flox/flox or HIF1α flox/flox mice with PEPCK-Cre mice as described in our previous work.15, 26 The transgenic mice and normal C57BL/6 mice were housed and experiments conducted at Charlie Norwood Veterans Affairs Medical Center following a protocol approved by the Institutional Animal Care and Use Committee (IACUC). Renal IRI was induced by bilateral clamping renal pedicles, the clamps were released for reperfusion as detailed recently.15 Sham animals were operated to expose the renal pedicles without clamping.
In vivo miRNA Mimic Delivery
miRNA mimic (or scramble sequence oligos) for in vivo use was delivered by following the protocol of the Invivofectamine 2.0 kit from Invitrogen. Briefly, 500μL of Invivofectamine-mimic complex was prepared by mixing miRNA mimic/scramble oligo, complexation buffer and Invivofectamine at the ratio of 1:1:2. The mixture was incubated at 50°C for 30 minutes and then mixed with 14 volumes of PBS. The mixture was finally concentrated with Millipore centrifugal filter device by 4,000g centrifugation at 4°C for 4 0 minutes. For in vivo test, the Invivofectamine-mimic complex was injected to the mouse 24 hours before ischemic surgery via tail vein at 20–40 μL/second at a total injection volume of 10 μL/g body weight.
Cell Lines and Hypoxia Treatment
The rat kidney proximal tubular cell (RPTC) line was originally obtained from Dr. Ulrich Hopfer at Case Western Reserve University. Wild-type (WT) and dominant-negative mutant (DN-p53) RPTC were prepared in our previous study.48 Wild-type (HIF-1α+/+) and HIF-1α–null mouse embryonic fibroblasts (MEFs) were prepared as previously.15 To establish DR6 knockdown stable RPTC cells, DR6-shRNA was purchased from Origene Technologies (Rockville, MD) to transfect into RPTCs by using Lipofectamine LTX (Life Technologies, Grand Island, NY), and then cells were selected by puromycin for two weeks. The knockdown effect was confirmed by immunoblotting using anti-DR6 antibody (Santa Cruz Biotechnology, TX). For hypoxia treatment, cells were plated overnight and then changed for pre-equilibrated medium incubated in hypoxia chamber with 1% oxygen for experimental time periods.
Morphological Analysis of Apoptosis
Apoptosis was examined morphologically as described previously15. Briefly, cell nuclei was stained with 10μg/ml of Hoechst 33342 for 2–3 minutes, the typical apoptotic cells including cellular shrinkage and nuclear condensation and fragmentation were monitored by phase-contrast and fluorescence microscopy. For quantification of apoptosis, three fields with approximately 200 cells of each dish were checked to estimate the percentage of apoptosis, and each experiment was repeated for three times.
Analysis of Renal Function and Histology
Renal function was monitored by measuring blood urea nitrogen (BUN) and serum creatinine as previously49, 50. Briefly, blood samples were collected and centrifuged to separate serum. The BUN and serum creatinine were measured by using the commercial kits from Biotron Diagnostics Inc (Hemet, CA) and Stanbio Laboratory (Boerne, TX), respectively. Kidney tissue damage and apoptosis were examined by Hematoxylin-eosin (H&E) and Terminal transferase-dUTP nick-end labeling (TUNEL) assay as described in our recent work50–52. Briefly, tissues were fixed with 4% paraformaldehyde and embedded in paraffin. Each sample was sectioned at 4μm and deparaffinized with standard protocol, then stained with standard H&E or TUNEL procedures. Renal tubular damage was indicated by tubular dilation, brush border loss, protein/cell cast formation, and tubular degeneration. Samples were blind evaluated to determine the percentage of tubular damage. For apoptosis evaluation, 10–20 fields were randomly selected in each sample to count positive staining cells using an Axioplan2 fluorescence microscope.
Real-time PCR Analysis of miRNA
Total RNAs were extracted from cells and kidney tissues by using the mirVana kit (Ambion, Austin, TX). The miRNA Reverse Transcription kit (Applied Biosystems) was used for reverse transcription of RNA into cDNA. Then Taqman probes were used for real-time quantitative PCR by using Taqman miRNA assay kit (Applied Biosystems). Fold change were quantified using 2−ΔΔCt values.
In Situ Hybridization (ISH) of miRNA
In situ hybridization was performed by following the standard protocol of the IsHyb ISH kit from BioChain. Briefly, 7μm frozen sections of kidney tissue were prepared and fixed with 4% paraformaldehyde at room temperature for 20 minutes. The sections were de-proteinated with 10μg/ml proteinase-K at 37°C for 10 minutes. The sections were pre-hybridized in prehybridization solution for 3 hours at 76°C. The hybridization was then performed at 76°C overnight with 50nM miRCURY LNA detection probe sequence as 5′–3′/5DigN/TAC CTG CAC TGT AAG CAC TTTG (Exiqon). After stringent washes with 2× SSC,1× SSC at 73°C for 10 minutes, and twice of 0.2×SSC at 37°C for 20 minute s, the sections were incubated with blocking solution at room temperature for 1 hour and further incubated with anti-DIG alkaline phosphatase conjugated antibody (1:100 in blocking solution) at 4°C overnight. The sections were washed with 1×PBS and alkaline phosphatase buffer, and then the detection was performed by using NBT/BCIP solution.
ChIP Assay of P53 Binding to miR-17-5p Promoter
ChIP assay was conducted to analyze the binding of p53 to miR-17-5p promoter region by using R&D Systems assay kit (Minneapolis, MN). Briefly, cultured cells were fixed with 1% formaldehyde to collect cell lysate and shear chromatin by sonication for immunoprecipitation with anti-p53 antibody. The resultant of immunoprecipitation was subjected to real-time PCR for amplification of putative p53 binding sequence and positive control gene p21 by using specifically designed primers.
Luciferase Reporter Assay
The synthesized forward and reverse sequences, including miR-17-5p-binding site in DR6 3′UTR, were annealed and then inserted into pMIR- REPORT luciferase miRNA Expression Reporter Vector (Applied Biosystems) between HindIII and Spel sites. HEK293 cells were co-transfected with the subcloned construct or non-targeting control vector along with miR-17-5p mimic or scrambled-sequence oligonucleotide. A pMIR- REPORT β-gal designed plasmid was used for transfection normalization. Cell lysate was collected after 24hours of transfection to proceed to luciferase assay measurement.
Bioinformatics Analysis of Regulatory Element and Target of miRNA
Two algorithms were used to predict targets of miR-17-5p computationally, including miRanda 2010 database (http://www.microrna.org/microrna/searchMirnas) and Targetscan 7.0 (http://www.targetscan.org/). JASPAR CORE 2016 (http://jaspar.genereg.net/) was used to analyze the regulatory element of miR-17-5p host gene promoter region to identify the putative p53-binding sites.
Immunoblot Analysis
Immunoblot analysis was carried out following a standard protocol. Briefly, the whole cell lysate was collected by using 2%SDS buffer. The same amount (30μg) of protein samples were resolved by SDS-PAGE gel and then transferred to PVDF membrane. The blots were blocked in 5% non-fat milk for 1hour before incubated in primary and secondary antibodies subsequently. The antibodies were purchased from the following sources: anti-DR6 from Santa Cruz Biotechnology; anti-p53, anti–phospho-p53 (serine-15) and anti-caspase-3 from Cell Signaling Technology; anti-β-actin and cyclophilin B from Sigma-Aldrich.
Statistical Analysis
Quantitative values were expressed as mean ± standard deviation (SD) (n≥3). Statistical differences were determined using the Student’s t-test with Microsoft EXCEL 2010. P<0.05 was considered statistically significant.
Acknowledgments
This work was supported in part by the grants from National Natural Science Foundation of China (81430017), Major Fundamental Research Program of Shanghai Committee of Science and Technology (12DJ1400300), the Key Projects in the National Science & Technology Pillar Program in the Twelfth Five-year Plan Period (Grant No. 2011BAl10B07), American Heart Association (12SDG8270002), the National Institutes of Health and Department of Veterans Administration of USA.
Footnotes
Disclosure
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bonventre JV, Yang L. Cellular pathophysiology of ischemic acute kidney injury. J Clin Invest. 2011;121:4210–4221. doi: 10.1172/JCI45161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharfuddin AA, Molitoris BA. Pathophysiology of ischemic acute kidney injury. Nat Rev Nephrol. 2011;7:189–200. doi: 10.1038/nrneph.2011.16. [DOI] [PubMed] [Google Scholar]
- 3.Venkatachalam MA, Weinberg JM, Kriz W, et al. Failed Tubule Recovery, AKI-CKD Transition, and Kidney Disease Progression. J Am Soc Nephrol. 2015;26:1765–1776. doi: 10.1681/ASN.2015010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lorenzen JM, Haller H, Thum T. MicroRNAs as mediators and therapeutic targets in chronic kidney disease. Nat Rev Nephrol. 2011;7:286–294. doi: 10.1038/nrneph.2011.26. [DOI] [PubMed] [Google Scholar]
- 6.Bhatt K, Mi QS, Dong Z. microRNAs in kidneys: biogenesis, regulation, and pathophysiological roles. Am J Physiol Renal Physiol. 2011;300:F602–610. doi: 10.1152/ajprenal.00727.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harvey SJ, Jarad G, Cunningham J, et al. Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J Am Soc Nephrol. 2008;19:2150–2158. doi: 10.1681/ASN.2008020233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho J, Ng KH, Rosen S, et al. Podocyte-specific loss of functional microRNAs leads to rapid glomerular and tubular injury. J Am Soc Nephrol. 2008;19:2069–2075. doi: 10.1681/ASN.2008020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagalakshmi VK, Ren Q, Pugh MM, et al. Dicer regulates the development of nephrogenic and ureteric compartments in the mammalian kidney. Kidney Int. 2011;79:317–330. doi: 10.1038/ki.2010.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel V, Hajarnis S, Williams D, et al. MicroRNAs regulate renal tubule maturation through modulation of Pkd1. J Am Soc Nephrol. 2012;23:1941–1948. doi: 10.1681/ASN.2012030321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sequeira-Lopez ML, Weatherford ET, Borges GR, et al. The microRNA-processing enzyme dicer maintains juxtaglomerular cells. J Am Soc Nephrol. 2010;21:460–467. doi: 10.1681/ASN.2009090964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei Q, Bhatt K, He HZ, et al. Targeted deletion of Dicer from proximal tubules protects against renal ischemia-reperfusion injury. J Am Soc Nephrol. 2010;21:756–761. doi: 10.1681/ASN.2009070718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu H, Jiang W, Xi X, et al. MicroRNA-21 attenuates renal ischemia reperfusion injury via targeting caspase signaling in mice. Am J Nephrol. 2014;40:215–223. doi: 10.1159/000368202. [DOI] [PubMed] [Google Scholar]
- 14.Bijkerk R, van Solingen C, de Boer HC, et al. Silencing of miRNA-126 in kidney ischemia reperfusion is associated with elevated SDF-1 levels and mobilization of Sca-1+/Lin- progenitor cells. Microrna. 2014;3:144–149. doi: 10.2174/2211536604666150121000340. [DOI] [PubMed] [Google Scholar]
- 15.Bhatt K, Wei Q, Pabla N, et al. MicroRNA-687 Induced by Hypoxia-Inducible Factor-1 Targets Phosphatase and Tensin Homolog in Renal Ischemia-Reperfusion Injury. J Am Soc Nephrol. 2015 doi: 10.1681/ASN.2014050463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ranganathan P, Jayakumar C, Tang Y, et al. MicroRNA-150 deletion in mice protects kidney from myocardial infarction-induced acute kidney injury. Am J Physiol Renal Physiol. 2015;309:F551–558. doi: 10.1152/ajprenal.00076.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei Q, Liu Y, Liu P, et al. MicroRNA-489 is induced via HIF-1 to protect against kidney ischemia. J Am Soc Nephrol. 2016 doi: 10.1681/ASN.2015080870. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang IK, Sun KT, Tsai TH, et al. MiR-20a-5p mediates hypoxia-induced autophagy by targeting ATG16L1 in ischemic kidney injury. Life Sci. 2015;136:133–141. doi: 10.1016/j.lfs.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Bijkerk R, van Solingen C, de Boer HC, et al. Hematopoietic microRNA-126 protects against renal ischemia/reperfusion injury by promoting vascular integrity. J Am Soc Nephrol. 2014;25:1710–1722. doi: 10.1681/ASN.2013060640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu X, Kriegel AJ, Liu Y, et al. Delayed ischemic preconditioning contributes to renal protection by upregulation of miR-21. Kidney Int. 2012;82:1167–1175. doi: 10.1038/ki.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marrone AK, Stolz DB, Bastacky SI, et al. MicroRNA-17~92 is required for nephrogenesis and renal function. J Am Soc Nephrol. 2014;25:1440–1452. doi: 10.1681/ASN.2013040390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semenza GL. Hypoxia-inducible factor 1 and cardiovascular disease. Annu Rev Physiol. 2014;76:39–56. doi: 10.1146/annurev-physiol-021113-170322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He M, Wang QY, Yin QQ, et al. HIF-1alpha downregulates miR-17/20a directly targeting p21 and STAT3: a role in myeloid leukemic cell differentiation. Cell Death Differ. 2013;20:408–418. doi: 10.1038/cdd.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly KJ, Plotkin Z, Vulgamott SL, et al. P53 mediates the apoptotic response to GTP depletion after renal ischemia-reperfusion: protective role of a p53 inhibitor. J Am Soc Nephrol. 2003;14:128–138. doi: 10.1097/01.asn.0000040596.23073.01. [DOI] [PubMed] [Google Scholar]
- 25.Molitoris BA, Dagher PC, Sandoval RM, et al. siRNA targeted to p53 attenuates ischemic and cisplatin-induced acute kidney injury. J Am Soc Nephrol. 2009;20:1754–1764. doi: 10.1681/ASN.2008111204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang D, Liu Y, Wei Q, et al. Tubular p53 regulates multiple genes to mediate AKI. J Am Soc Nephrol. 2014;25:2278–2289. doi: 10.1681/ASN.2013080902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ying Y, Kim J, Westphal SN, et al. Targeted deletion of p53 in the proximal tubule prevents ischemic renal injury. J Am Soc Nephrol. 2014;25:2707–2716. doi: 10.1681/ASN.2013121270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du W, Pan Z, Chen X, et al. By targeting Stat3 microRNA-17-5p promotes cardiomyocyte apoptosis in response to ischemia followed by reperfusion. Cell Physiol Biochem. 2014;34:955–965. doi: 10.1159/000366312. [DOI] [PubMed] [Google Scholar]
- 29.Zhou M, Cai J, Tang Y, et al. MiR-17-92 cluster is a novel regulatory gene of cardiac ischemic/reperfusion injury. Med Hypotheses. 2013;81:108–110. doi: 10.1016/j.mehy.2013.03.043. [DOI] [PubMed] [Google Scholar]
- 30.Kaucsar T, Revesz C, Godo M, et al. Activation of the miR-17 family and miR-21 during murine kidney ischemia-reperfusion injury. Nucleic Acid Ther. 2013;23:344–354. doi: 10.1089/nat.2013.0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang Y, Xu C, Fu Y. MicroRNA-17-5p induces drug resistance and invasion of ovarian carcinoma cells by targeting PTEN signaling. J Biol Res (Thessalon) 2015;22:12. doi: 10.1186/s40709-015-0035-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu F, Lu Z, Huang K, et al. MicroRNA-17-5p-activated Wnt/beta-catenin pathway contributes to the progression of liver fibrosis. Oncotarget. 2016;7:81–93. doi: 10.18632/oncotarget.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao E, Jiang C, Ji M, et al. The miRNA-17 approximately 92 cluster mediates chemoresistance and enhances tumor growth in mantle cell lymphoma via PI3K/AKT pathway activation. Leukemia. 2012;26:1064–1072. doi: 10.1038/leu.2011.305. [DOI] [PubMed] [Google Scholar]
- 34.Oeztuerk-Winder F, Guinot A, Ochalek A, et al. Regulation of human lung alveolar multipotent cells by a novel p38alpha MAPK/miR-17-92 axis. EMBO J. 2012;31:3431–3441. doi: 10.1038/emboj.2012.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavrik I, Golks A, Krammer PH. Death receptor signaling. J Cell Sci. 2005;118:265–267. doi: 10.1242/jcs.01610. [DOI] [PubMed] [Google Scholar]
- 36.Karin M, Lin A. NF-kappaB at the crossroads of life and death. Nat Immunol. 2002;3:221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- 37.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 38.Nikolaev A, McLaughlin T, O’Leary DD, et al. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 2009;457:981–989. doi: 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Mi S, Lee X, Hu Y, et al. Death receptor 6 negatively regulates oligodendrocyte survival, maturation and myelination. Nat Med. 2011;17:816–821. doi: 10.1038/nm.2373. [DOI] [PubMed] [Google Scholar]
- 40.Tokudome T, Sasaki A, Tsuji M, et al. Reduced PTEN expression and overexpression of miR-17-5p, -19a-3p, -19b-3p, -21-5p, -130b-3p, -221-3p and -222-3p by glioblastoma stem-like cells following irradiation. Oncol Lett. 2015;10:2269–2272. doi: 10.3892/ol.2015.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katz G, Pobezinsky LA, Jeurling S, et al. T cell receptor stimulation impairs IL-7 receptor signaling by inducing expression of the microRNA miR-17 to target Janus kinase 1. Sci Signal. 2014;7:ra83. doi: 10.1126/scisignal.2005221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar R, Sahu SK, Kumar M, et al. MicroRNA 17-5p regulates autophagy in Mycobacterium tuberculosis-infected macrophages by targeting Mcl-1 and STAT3. Cell Microbiol. 2015 doi: 10.1111/cmi.12540. [DOI] [PubMed] [Google Scholar]
- 43.Dong D, Fu N, Yang P. MiR-17 down-regulation by high glucose stabilizes Thioredoxin-interacting protein and removes thioredoxin inhibition on ASK1 leading to apoptosis. Toxicol Sci. 2015 doi: 10.1093/toxsci/kfv313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haase VH. Mechanisms of hypoxia responses in renal tissue. J Am Soc Nephrol. 2013;24:537–541. doi: 10.1681/ASN.2012080855. [DOI] [PubMed] [Google Scholar]
- 45.Semenza GL. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu Rev Pathol. 2014;9:47–71. doi: 10.1146/annurev-pathol-012513-104720. [DOI] [PubMed] [Google Scholar]
- 46.Sutton TA, Hato T, Mai E, et al. p53 is renoprotective after ischemic kidney injury by reducing inflammation. J Am Soc Nephrol. 2013;24:113–124. doi: 10.1681/ASN.2012050469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Megyesi J, Safirstein RL, Price PM. Induction of p21WAF1/CIP1/SDI1 in kidney tubule cells affects the course of cisplatin-induced acute renal failure. J Clin Invest. 1998;101:777–782. doi: 10.1172/JCI1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang M, Yi X, Hsu S, et al. Role of p53 in cisplatin-induced tubular cell apoptosis: dependence on p53 transcriptional activity. Am J Physiol Renal Physiol. 2004;287:F1140–1147. doi: 10.1152/ajprenal.00262.2004. [DOI] [PubMed] [Google Scholar]
- 49.Wei Q, Dong G, Franklin J, et al. The pathological role of Bax in cisplatin nephrotoxicity. Kidney Int. 2007;72:53–62. doi: 10.1038/sj.ki.5002256. [DOI] [PubMed] [Google Scholar]
- 50.Wei Q, Dong G, Yang T, et al. Activation and involvement of p53 in cisplatin-induced nephrotoxicity. Am J Physiol Renal Physiol. 2007;293:F1282–1291. doi: 10.1152/ajprenal.00230.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brooks C, Wei Q, Cho SG, et al. Regulation of mitochondrial dynamics in acute kidney injury in cell culture and rodent models. J Clin Invest. 2009;119:1275–1285. doi: 10.1172/JCI37829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei Q, Hill WD, Su Y, et al. Heme oxygenase-1 induction contributes to renoprotection by G-CSF during rhabdomyolysis-associated acute kidney injury. Am J Physiol Renal Physiol. 2011;301:F162–170. doi: 10.1152/ajprenal.00438.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]