Abstract
Selective 2′‐hydroxyl acylation analyzed by primer extension (SHAPE) provides information on RNA structure at single‐nucleotide resolution. It is most often used in conjunction with RNA secondary structure prediction algorithms as a probabilistic or thermodynamic restraint. With the recent advent of ultra‐high‐throughput approaches for collecting SHAPE data, the applications of this technology are extending beyond structure prediction. In this review, we discuss recent applications of SHAPE data in the transcriptomic context and how this new experimental paradigm is changing our understanding of these experiments and RNA folding in general. SHAPE experiments probe both the secondary and tertiary structure of an RNA, suggesting that model‐free approaches for within and comparative RNA structure analysis can provide significant structural insight without the need for a full structural model. New methods incorporating SHAPE at different nucleotide resolutions are required to parse these transcriptomic data sets to transcend secondary structure modeling with global structural metrics. These ‘multiscale’ approaches provide deeper insights into RNA global structure, evolution, and function in the cell. WIREs RNA 2017, 8:e1374. doi: 10.1002/wrna.1374
For further resources related to this article, please visit the WIREs website.
INTRODUCTION
As more than a mere messenger of information from DNA to protein, RNA molecules adopt structures that have functional biological roles.1, 2, 3, 4, 5, 6 Traditionally, secondary structure predictions of RNA molecules from a single sequence minimize a free energy function to obtain a structural model of intramolecular base pairing,7, 8, 9, 10 or sample suboptimal structures from the partition function.11, 12 Evolutionary covariation, where two nucleotides vary in sequence across related RNAs while preserving base pairing ability, is particularly powerful for predicting structures, especially for bacterial and archaeal RNAs or for highly conserved RNA structures.13, 14, 15 Structure prediction algorithms, however, only predict about half of all base pairs that occur in an RNA,16, 17 and some alignments may not have enough information for covariation analysis.18 For these reasons, an experiment capable of rapidly probing RNA structure is particularly appealing.
Multiple quantitative methods to determine RNA structure experimentally are now available, such as selective 2′‐hydroxyl acylation analyzed by primer extension (SHAPE),19 quantitative dimethyl sulfate (DMS) modification,20 and parallel analysis of RNA structure (PARS).5, 21 Each of these methods provides information on the conformational flexibility of nucleotides in an RNA, either through chemical modification (SHAPE and DMS) or through enzymatic probing (PARS). These quantitative probing techniques present a new paradigm in understanding RNA structure on a transcript‐wide or greater scale.
The most common use for SHAPE data is as a restraint (also referred to as a soft constraint) in secondary structure prediction algorithms.18, 22, 23, 24 The incorporation of SHAPE data into structure prediction algorithms refines the probable structure space of an RNA molecule and greatly improves predictions to approximately 90% accuracy.22, 25 Recently, SHAPE data collected in an ultra‐high‐throughput manner is increasingly used in a model‐free approach as an additional feature for evolutionary analysis. In this review, we discuss these diverse approaches to applying SHAPE data to understand RNA structure. As next‐generation sequencing now allows for the rapid quantification of structure probing,5, 25, 26, 27 the future of SHAPE will involve signal processing techniques to understand a transcript's structure on multiple scales. The speed at which these technologies now provide us with structure information will allow for efficient and accurate analysis of comparative RNA structure.
FIRST‐LEVEL APPROACHES TO INTERPRETING SHAPE EXPERIMENTS
SHAPE uses the reactivity of the 2′‐OH of an RNA molecule to understand the structure of that RNA.19, 28, 29, 30, 31 An electrophile, typically 1M7 or NMIA, covalently bonds with the 2′‐O to form an adduct; this reaction occurs preferentially at conformationally flexible, or unpaired, nucleotides.28, 29 The signal is then ‘read’ by reverse transcription. In most protocols, modified nucleotides block the reverse transcriptase causing it to fall off the transcript. Nucleotides that are more reactive will generate more stops, which, as the experiments are traditionally performed with single‐hit kinetics, indicates the relative frequency of adduct formation.30 The relative rates of adduct formation are then normalized to find the SHAPE profile for that RNA, providing information on the reactivity of each nucleotide.19, 32
In general, positions in a SHAPE profile with high reactivities are more likely to be unpaired, and positions with low reactivities are more likely to be paired.22 Because the SHAPE reagent can react at any nucleobase, the SHAPE profile provides high‐resolution structural information.33 Indeed, differences in SHAPE profiles of two sequence variants indicate that an RNA is a riboSNitch, where a single nucleotide variant changes the structure of the RNA.34, 35, 36 Thus, the SHAPE profile alone encodes information on an RNA's structure.
Recently whole‐transcriptome probing methods use the power of next‐generation sequencing with SHAPE structural profiling for an ultra‐high‐throughput way of probing RNA structure, such as the techniques in vivo click SHAPE (icSHAPE)37, 38 and SHAPE‐Seq,26, 39, 40 both of which utilize reverse transcription stops. Of particular interest, SHAPE with mutational profiling (SHAPE‐MaP) uses modified reverse transcription conditions to induce mutations at positions with 2′ adducts, rather than causing a reverse transcription stop.25 This new technique thus allows for high‐resolution quantification of RNA structure for whole transcripts at every nucleotide position, without concerns about signal decay or adaptor ligation bias.41, 42, 43
What Is SHAPE Really Saying?
SHAPE is a powerful technique for probing the structure of an RNA but the results of a SHAPE experiment are not directly interpretable. Although SHAPE reactivities generally correspond to pairing state, the relationship between SHAPE and frequency of base pairing is not linear. Both Cordero et al.44 and Sükösd et al.45 found that the SHAPE reactivities of paired and unpaired nucleotides fall into two different probability distributions, although the two studies found different distributions of SHAPE reactivities for unpaired nucleotides (Figure 1(a)). While high (over 1.0) SHAPE reactivities generally only occur at unpaired nucleotides, lower SHAPE reactivities frequently correspond to both paired and unpaired nucleotides. Other structural factors such as base stacking may result in an unpaired nucleotide having a low SHAPE reactivity.46 Consequently, the SHAPE reactivity alone is not always predictive of whether a base is paired or unpaired.
Figure 1.
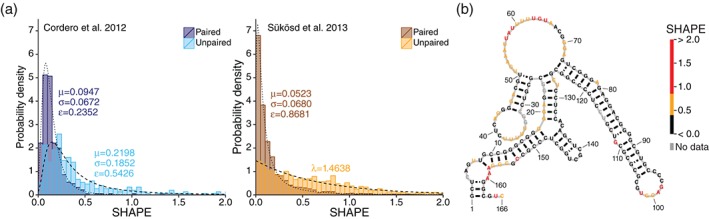
SHAPE reactivities distinguish between paired and unpaired nucleotides. (a) Distributions of SHAPE reactivities for paired and unpaired nucleotides recreated from Cordero et al.44 (left) and Sükösd et al.45 (right; data from Deigan et al.22). SHAPE reactivities for paired nucleotides follow a generalized extreme value distribution in both data sets. SHAPE reactivities for unpaired nucleotides follow a generalized extreme value distribution in Cordero et al. and an exponential distribution in Sükösd et al. In both data sets, the distributions for paired and unpaired nucleotides differ, signifying that SHAPE reactivities are drawn from multiple probability distributions. (b) Example of RNA structure (the RB1 5′ UTR from Kutchko et al.18) overlaid with SHAPE data. Red: high SHAPE; orange: medium SHAPE; black: low SHAPE; gray: no data. Most paired positions have low SHAPE and many unpaired positions have high SHAPE, but SHAPE does not completely distinguish between paired and unpaired nucleotides, in part because this RNA forms multiple conformations.
The distinction that SHAPE provides between paired and unpaired nucleotides is different between the data sets by Cordero et al.44 (Figure 1(a); left) and Deigan et al.22 (shown in Sükösd et al., Figure 1(a); right). The SHAPE data from Deigan et al. has a much larger difference between its paired and unpaired distributions. Thus, the strength of the SHAPE signal depends on the RNA and the experimental conditions, and is not merely a direct measurement of structure.42, 47 These factors are an important and often underappreciated aspect of the experiment. The SHAPE reactivity profile depends on the overall fold of the molecule being probed. As such, experimental conditions can significantly alter the profile, and any comparative analysis of SHAPE reactivity should carefully consider experimental conditions.
SHAPE and Structure Prediction
Besides reporting a likelihood of base pairing for any given nucleotide, SHAPE data also refine secondary structure predictions of an RNA as an energetic restraint. This method was first introduced by Deigan et al.,22 where an additional energy term to the nearest neighbor rules incorporates SHAPE reactivity as an additional pseudo‐free energy term.48 A high SHAPE reactivity makes that nucleotide less likely to base pair, while a low SHAPE reactivity makes base pairing more favorable. The SHAPE reactivities thus refine the structural space to find a structure or structures compatible with the SHAPE data (Figure 1(b)). As RNAs often adopt multiple structural conformations and as SHAPE is a measurement over the entire structural ensemble, the SHAPE data may not match a single structure but instead represent the average reactivity over the ensemble.
Still, projecting SHAPE reactivities onto a single structure, as is the case in Figure 1(b), is the most common approach for visualizing SHAPE data. Although one structure may appear to agree with the data (as in Figure 1(b)), there are often many other structures in the suboptimal ensemble that appear to agree with the data just as well. In fact, in the case of the RB1 5′ UTR illustrated in Figure 1(b), three alternative conformations are all compatible with the data.18 Nonetheless, there is value in visualizing structural models with SHAPE data projected as in Figure 1(b), and these approaches to interpreting SHAPE data will likely remain popular.
The most fitting way of incorporating SHAPE data into structure prediction is a subject of recent debate and thoroughly reviewed in Eddy 2014.49 These methods all attempt to use SHAPE data to constrain structure prediction of that RNA, based on the likelihood of each nucleotide being paired. These methods treat SHAPE measurements in a variety of ways: as a free energy term,22 as a prior probability,50 or as a likelihood of base pairing.24, 51 While each method of incorporating SHAPE into prediction greatly improves the accuracy of the structure predictions, they all perform similarly to each other, with accuracies up to 90–95%.22, 24, 25, 44
When given a SHAPE reactivity, it is impossible to know whether that base is paired or unpaired, especially at lower reactivities. Using structural probing as a restraint in structure prediction provides more information on the structure, with approximately the same accuracy over a wide range of parameters regardless of the method used.22, 24, 25, 44, 50, 51 The next challenge in SHAPE processing is not optimizing these parameters further, but instead evaluating broader structural characteristics that the SHAPE signal can inform us of. Given that the use of SHAPE in structure prediction was very extensively reviewed recently by Eddy 2014,49 we instead decided to focus on recent developments in the analysis of SHAPE signal that attempt to transcend the interpretation of the data as merely informative of a single secondary structure.
RECENT APPLICATIONS OF THE SHAPE SIGNAL
While SHAPE is certainly useful in refining RNA secondary structure prediction, it also provides additional information when analyzed at different scales, or when used in conjunction with other metrics such as evolutionary data. Here, we will discuss recent extensions of SHAPE reactivities beyond structure prediction.
Zooming Out: Regional View of SHAPE Reactivities
At a single‐nucleotide resolution, the traditional conceptualization of SHAPE data is that each reactivity provides a likelihood of that base being paired or unpaired (Figure 2(a)). However, the proportions of unreactive (low‐SHAPE) and reactive (high‐SHAPE) nucleotides are not consistent across larger RNAs because while certain regions within a transcript fold into a well‐defined structure, other regions may adopt multiple distinct structures with similar free energies.
Figure 2.
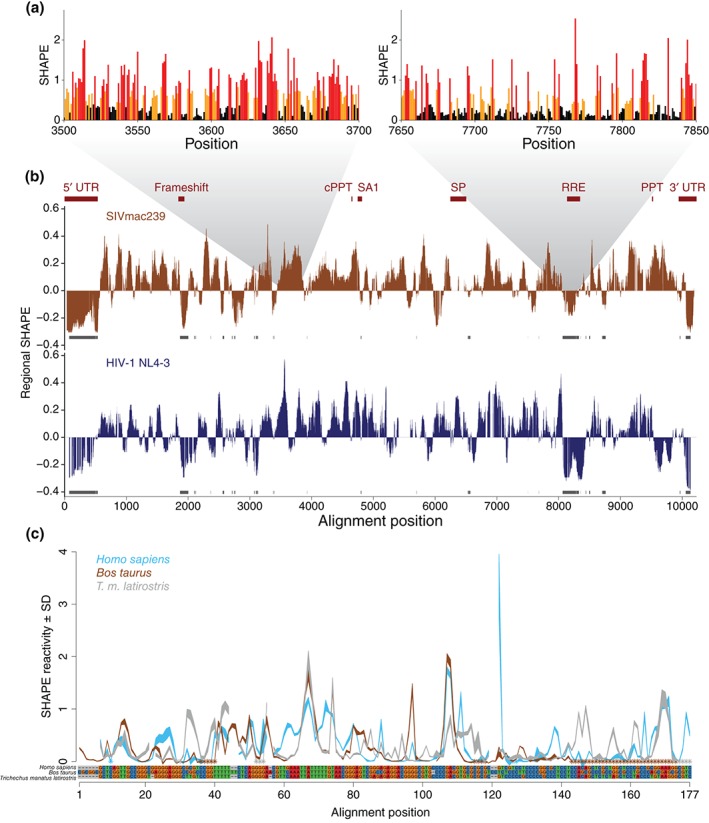
SHAPE data for related RNAs follow similar but not identical patterns. (a) Examples of SHAPE data from high‐SHAPE regions (left) and low‐SHAPE regions (right) from the SIVmac239 SHAPE data from Pollom et al.52 High SHAPE nucleotides are indicated in red, medium in orange, and low in black. Although both profiles have low‐ and high‐SHAPE nucleotides, the frequencies of each are distinct between the two regions. (b) SHAPE data for SIVmac239 (top) and HIV‐1 (bottom) aligned genomes. SHAPE data are from Pollom et al., annotations are from Pollom et al. and the Los Alamos HIV database (http://www.hiv.lanl.gov/), and the sequences were aligned using MAFFT.53 Regional SHAPE represents the windowed median SHAPE over a 75‐nt window, with respect to the global median SHAPE value for each transcript. Values above the line are regionally unstructured, and below the line are regionally structured. Alignment regions where both viruses are regionally structured are annotated in gray. These regions correspond to known structural elements of the SIVmac virus (above, red). (c) SHAPE reactivities aligned by sequence for the RB1 5′ UTR in human (blue), cow (brown), and manatee (gray) from Kutchko et al.18 The SHAPE profiles for each species are similar across most of the UTR. (Reprinted with permission from Ref 18. Copyright 2015 Kutchko et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society).
It is important to note that a majority of transcribed RNAs are long, ranging up to several kilobases.54, 55 The longest transcripts are often RNA virus genomes, such as the well‐studied HIV genome. To quantify these different SHAPE patterns, the median SHAPE reactivity over windows ranging from 50 to 75 nucleotides is a novel way to identify and visualize structured regions in a transcript.25, 52 Although SHAPE reactivities are at single‐nucleotide resolution, averaging reactivities over many nucleotides reveals structured and unstructured regions of an RNA. Such ‘multiscale’ level analysis of these very large transcripts (often greater than 10 kb) provides a different picture of RNA structure than SHAPE at single‐nucleotide resolution (Box 1). Of course, differences in the window size will change the scale at which we are understanding an RNA's structure, and the choice of window size to date has remained largely empirical. There are significant opportunities for algorithm development using SHAPE data at different scales.
BOX 1. NEW INTERPRETATIONS OF SHAPE FOR NEXT‐GENERATION PROBING.
Traditionally, researchers have used SHAPE to understand the structure of a single RNA. Recently, though, developments such as SHAPE‐Seq, icSHAPE, and SHAPE‐MaP now make SHAPE structural probing a next‐generation technology. Now, SHAPE has more applications than just as a measure of whether a single nucleotide is paired or unpaired—it can indicate structuredness at multiple resolutions. SHAPE is now one of multiple metrics—including base pairing probability, Shannon entropy, and nucleotide content—for interpreting both local and broad structure of an RNA. The future of SHAPE as a genome‐wide (for viruses) or transcriptome‐wide (for bacteria and eukaryotes) measurement technique relies on signal processing of this large‐scale data.
Pollom et al.52 compares the windowed SHAPE reactivities of both simian immunodeficiency virus (SIV) and HIV‐1 (Figure 2(b)). Regions where both viruses have low median SHAPE correspond to known RNA structural elements such as the 5′ UTR, the gag‐pro‐pol frameshift, and the Rev response element. Other unannotated regions where both viruses have low median SHAPE reactivities are good targets for identifying new functional RNA elements. This windowed picture of SHAPE reactivity provides structural information—identifying regions of the RNA genomes that have well‐defined structures—that is difficult to see at single‐nucleotide resolution. Median SHAPE helps characterize structure across the transcript, demonstrating that SHAPE profiles have relevance beyond single nucleotide measurements and structure prediction algorithms. Future methods of processing these large SHAPE profiles, using this multiscale approach, may uncover more higher‐order RNA features. This approach also identifies unstructured regions, which may turn out to be just as functional as structure.18
SHAPE Supplements Weak Evolutionary Signals
The traditional method of identifying conserved RNA structures is through mutual information or covariation of nucleotide positions.13, 14, 56 Although covariance is extremely useful for highly conserved RNA structures such as the RNA component of the ribosome,15, 57 the covariance signal may be weak or nonexistent in other eukaryotic RNAs, making it difficult to identify conserved RNA structures in these organisms.18 SHAPE represents a new method to help strengthen the evolutionary signal by finding support for conserved RNA structures independent of structure prediction.
Kutchko et al.18 looked at the structure of the 5′ UTR of RB1, the transcript that codes for the tumor suppressor Retinoblastoma protein, in three different species. Qualitatively, SHAPE data for the three different UTRs have regions of striking similarity (Figure 2(c)). As in reference by Pollom et al.,52 SHAPE data—even in the absence of structure prediction—can indicate structural similarity between different sequences. In addition, SHAPE data can provide insight into observed evolutionary patterns. Watts et al.23 used SHAPE to find that hypervariable regions of the HIV‐1 genome are highly unstructured and insulate low‐SHAPE, highly structured helices. When used in this manner, SHAPE adds a layer of context to evolutionary patterns (Box 2).
BOX 2. MEASURING RNA STRUCTURE CONSERVATION WITH SHAPE.
As SHAPE is a model‐free measure of RNA structure, correlations between SHAPE profiles can find common structures across homologous transcripts. Formal methods for this kind of analysis, such as using SHAPE as a sequence alignment parameter, will aid in finding conserved and functional RNA structures. The ability of technologies including SHAPE‐Seq, icSHAPE, and SHAPE‐MaP to generate large amounts of structural data very quickly presents a new opportunity to understand the evolution of RNA structure. New multiscale metrics, analyzing SHAPE data at multiple resolutions, can identify conserved structures, or structural ensembles, in RNAs throughout evolution.
SHAPE‐Directed Sequence Alignments Identify Conserved RNA Structures
To extend beyond qualitative analysis, the Weeks lab recently published two papers using SHAPE data as a feature for sequence alignment.58, 59 Their algorithm aligns two SHAPE profiles to minimize the difference in SHAPE reactivity at each position, optionally including sequence as an alignment parameter (Figure 3(a)). The difference in SHAPE reactivity between two sequence positions becomes a function applied to the Gotoh alignment algorithm60 (Figure 3(b)). This novel method uses SHAPE reactivity as an additional feature for understanding evolutionary conservation.
Figure 3.
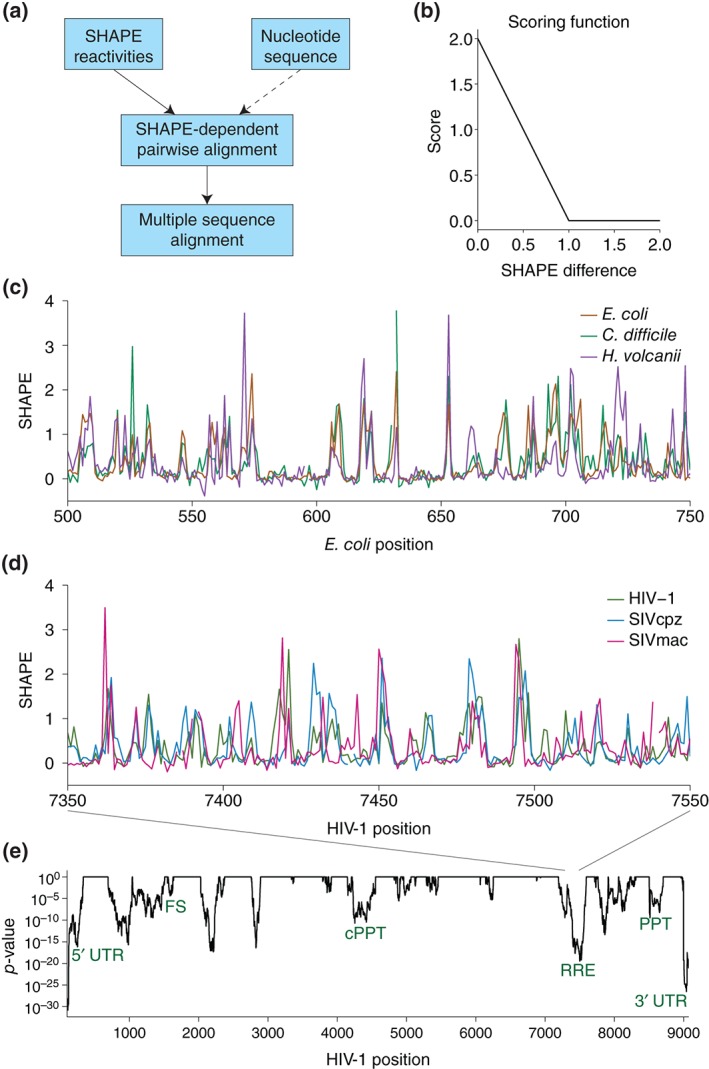
SHAPE data of homologous RNAs can facilitate sequence alignments. (a) Schematic of SHAPE‐directed sequence alignment used in Lavender et al. (1)58 SHAPE reactivities, and optionally nucleotide identity, can be used as parameters in the Gotoh alignment algorithm for pairwise alignment. Pairwise alignments can then be combined to create a SHAPE‐informed multiple sequence alignment. (b) Scoring function from Lavender et al. (1) used to align SHAPE reactivities for two sequences. More similar SHAPE reactivities have a higher score for alignment. If SHAPE reactivities differ by more than 1, the scoring function treats them as unrelated. (c) SHAPE‐only alignment of a section from three 16S rRNA sequences. Data from Lavender et al. (1) The aligned SHAPE reactivities are very similar, reflecting structural homology. (d) SHAPE‐directed multiple sequence alignment of three lentivirus sequences. The aligned SHAPE profiles of each virus have similar patterns. (e) Significance of the correlation of SHAPE between the three viruses, determined by linear regression. Lower p‐values indicate more similarity in SHAPE profiles. The SHAPE profiles have regions of significant correlation across the alignment, particularly in regions known to have functional RNA structures. (Reprinted with permission from Ref 58 Copyright 2015 Lavender et al.; PLOS Computational Biology and Ref 59 Copyright 2015 Lavender et al.; PLoS Computational Biology).
As a proof of concept, they used the method to align ribosomal RNA sequences from Escherichia coli, Clostridium difficile, and Haloferax volcanii with only SHAPE data, not taking sequence into account58 (Figure 3(c)). It should be noted that aligning these distantly related rRNAs based on sequence alone is quite challenging.58 The rRNAs aligned with SHAPE data alone produce an alignment with similar accuracy to the sequence‐only alignment, and using both sequence and SHAPE data reproduce the gold standard, manually curated alignment, suggesting that SHAPE data captures evolutionary patterns in RNA structure.
To extend this method further, they aligned three lentivirus genomes (HIV‐1, SIVcpz, and SIVmac) using sequence and SHAPE together59 (Figure 3(d)). Some regions in viral genomes have very robust structures, although unlike rRNA, viral RNAs also have many unstructured or less‐structured regions. The application of SHAPE‐directed sequence alignments to viral RNAs is thus an important test of this new alignment method. From the aligned SHAPE data, they used a linear regression model to find regions where the SHAPE profiles are correlated between the three viruses (Figure 3(e)). Known RNA functional elements have significant correlations between the SHAPE profiles (Figure 3(e); green), indicating that SHAPE correlation is a signal for structural homology.
SHAPE‐directed sequence alignments are therefore applicable to more divergent RNAs, but most useful for conserved RNA elements where the SHAPE profiles are similar. Systematically using SHAPE as an alignment parameter is a novel approach to identifying conservation of RNA structure, and future extensions of this technique may involve quantifying structural divergence and similarity using SHAPE data.
SHAPE Facilitates Discovery of the Conservation of Multiple RNA Structures
The SHAPE signal does not measure the conformation of a single molecule but instead represents the average reactivities over all copies of that RNA transcript. Indeed the reaction occurs in bulk, and as such the reactivity at any given nucleotide is an ensemble average. As many RNA structures, including riboswitches, adopt multiple conformations,2, 61 it is critical to consider the entire structural ensemble of a transcript when interpreting the SHAPE profile. We can therefore compute the SHAPE‐directed partition function, which models the entire structural ensemble informed by SHAPE data11, 22 and allows us to identify the presence of multiple structural conformations.
As comparative structure analysis now incorporates SHAPE data as additional information, the next step in this direction is using SHAPE to compare multiple structural conformations of an RNA. Our laboratory recently showed that multiple conformations of a purine riboswitch are conserved in sequence62 (Figure 4(a)). However, the covariation signal is much weaker in eukaryotic RNAs than bacterial or archaeal. SHAPE data thus provide us with more information, helping us identify the conservation of multiple structures.
Figure 4.
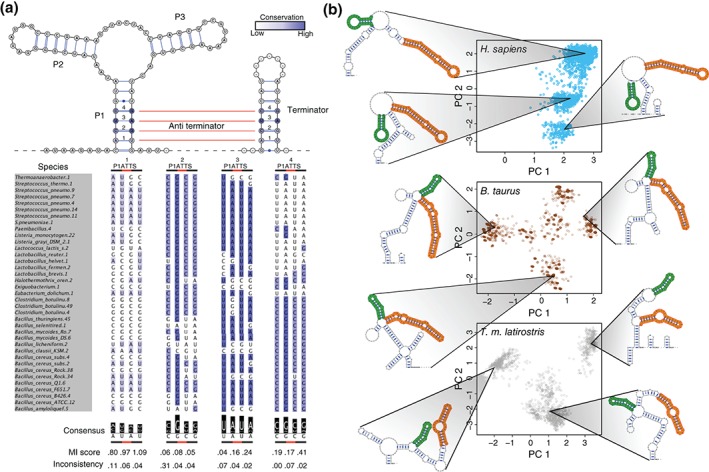
SHAPE reactivities can help identify conservation of multiple RNA structures. (a) Figure from Ritz et al.62 Multiple conformations of an RNA are evolutionarily conserved. Top: Purine riboswitch consensus structure with the anti‐terminator pairs in red lines. Both the P1/terminator conformation and the anti‐terminator conformation must be conserved. Bottom: Evolutionary analysis of bases involved in the P1, terminator, and anti‐terminator stems. Blue indicates conservation of each base. Mutual information shows that the pairs involved in the anti‐terminator stem preserve their ability to base pair, but also to pair with their partners in the P1 and terminator stems. (Reprinted with permission from Ref 62. Copyright 2013 Ritz et al.; PLOS Computational Biology) (b) The 5′ UTR of RB1 forms multiple distinct structures in humans (blue), cow (brown), and manatee (gray), with SHAPE‐directed Boltzmann sampled structures indicated by blue dots. Structures and arc diagrams on the side show representative structures from each conformation. Green and orange stems are conserved across all three organisms. Thus, SHAPE‐directed structure prediction allows us to confidently identify the conservation of multiple RNA structures. (Reprinted with permission from Ref 18. Copyright 2015 Kutchko et al.; Cold Spring Harbor Laboratory Press).
To incorporate SHAPE into conservation analysis, we used SHAPE to probe the 5′ UTR of the human RB1 transcript.18 As SHAPE data greatly improve the accuracy of RNA secondary structure prediction22, 25 we can be confident in our subsequent computational analysis. Using SHAPE‐directed Boltzmann sampling,10, 22, 63 we found that the 5′ UTR adopts three distinct conformations (Figure 4(b)). We applied the same analysis to the homologous UTRs in Bos taurus and Trichechus manatus latirostris and found that they also adopt multiple, similar conformations to the UTR in humans (Figure 4(c)). Here, in conjunction with structure prediction, SHAPE helps uncover the conservation of multiple structures in an RNA. As whole‐transcriptome probing technologies improve, we anticipate further discovery of this type of structural conservation and we encourage others investigating RNA structure to also investigate the conservation of multiple conformations as an important functional consideration.
THE FUTURE OF SHAPE IN THE NEXT‐GENERATION SEQUENCING ERA
The recent developments of high‐throughput structure probing technologies such as SHAPE‐Seq,39 icSHAPE,31 and SHAPE‐MaP25 utilize next‐generation sequencing to quickly obtain the SHAPE profile of one or more RNAs, making SHAPE now a next‐generation probing technology. With the advent of next‐generation sequencing, probing experiments are much faster and more efficient than in the past, and public databases such as the RNA Mapping Database64 allow for large‐scale comparisons of SHAPE data. Several studies recently applied next‐generation sequencing to analyze RNA structure at the whole transcriptome level.5, 27, 36, 65, 66 With this unprecedented amount of RNA structural probing data, the next steps in computational analysis of RNA structure will incorporate methods beyond structure prediction into read alignment, motif analysis, and signal processing.
In this new era of ‘next‐generation structural probing,’ the SHAPE signal is now one of multiple available features to inform us about an RNA. SHAPE represents a model‐free approach to RNA structure analysis and provides information at multiple levels, from single nucleotides to 50‐nt windows to whole transcripts. This multiscale approach to SHAPE structural probing gives us a wealth of information on the structure of a transcript, and combined with other methods such as Shannon entropy25, 67, 68 or mutual information,14 provides us with great detail on the transcript's structure.
Using SHAPE at this whole‐transcriptome, multiscale level will facilitate the application of signal processing methods to these SHAPE profiles of interest and uncover RNA structural features. The use of Fourier transforms to detect periodicity in (aggregated) SHAPE data is the first example of applying signal processing to SHAPE.27, 69 As new model‐free methods are applied to SHAPE profiles, we will gain detailed information on RNA structure at a whole‐transcriptome level and likely discover new levels of structural conservation.
In addition, these new high‐throughput probing technologies allow us to generate SHAPE data for related RNAs very quickly, facilitating the comparison of SHAPE data and structural preservation between homologous sequences. As with HIV and SIV genomes,59 correlations of SHAPE data will identify regions with conserved RNA structure. We live in exciting times for RNA structural biology as these next‐generation structural probing techniques are being developed, refined, and applied to large data sets. The next few years will involve a huge expansion of information regarding the conservation of RNA structure.1 2
ACKNOWLEDGMENTS
We would like to thank Sean Eddy for sharing data and advice regarding the SHAPE distributions in Figure 1(a), and the Weeks lab for sharing code to plot the structure in Figure 1(b). This work is supported by U.S. National Institutes of Health grants NHLBI R01 HL111527, NIGMS R01 GM101237, and NHGRI HG008133 to A.L., and the National Science Foundation Graduate Research Fellowship under grant number DGE‐1144081 to K.M.K.
Conflict of interest: The authors have declared no conflicts of interest for this article.
REFERENCES
- 1. Sharp PA. The centrality of RNA. Cell 2009, 136:577–580. doi:10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 2. Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr Opin Struct Biol 2005, 15:342–348. doi:10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 3. Chen C, Zhang H, Broitman SL, Reiche M, Farrell I, Cooperman BS, Goldman YE. Dynamics of translation by single ribosomes through mRNA secondary structures. Nat Struct Mol Biol 2013, 20:582–588. doi:10.1038/nsmb.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Heaphy S, Dingwall C, Ernberg I, Gait MJ, Green SM, Kern J, Lowe AD, Singh M, Skinner MA. HIV‐1 regulator of virion expression (Rev) protein binds to an RNA stem‐loop structure located within the Rev response element region. Cell 1990, 60:685–693. doi:10.1016/0092-8674(90)90671-Z. [DOI] [PubMed] [Google Scholar]
- 5. Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. Genome‐wide measurement of RNA secondary structure in yeast. Nature 2010, 467:103–107. doi:10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Solem AC, Halvorsen M, Ramos SBV, Laederach A. The potential of the riboSNitch in personalized medicine. Wiley Interdiscip Rev RNA 2015, 6:517–532. doi:10.1002/wrna.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nussinov R, Jacobson A. Fast algorithm for predicting the secondary structure of single‐stranded RNA. Proc Natl Acad Sci USA 1980, 77:6309–6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res 1981, 9:133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zuker M, Sankoff D. RNA secondary structures and their prediction. Bull Math Biol 1984, 46:591–621. [Google Scholar]
- 10. Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci USA 2004, 101:7287–7292. doi:10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McCaskill JS. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers 1990, 29:1105–1119. doi:10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- 12. Ding Y, Lawrence CE. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res 2003, 31:7280–7301. doi:10.1093/nar/gkg938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hofacker IL, Fekete M, Stadler PF. Secondary structure prediction for aligned RNA sequences. J Mol Biol 2002, 319:1059–1066. doi:10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 14. Freyhult E, Moulton V, Gardner P. Predicting RNA structure using mutual information. Appl Bioinformatics 2005, 4:53–59. [DOI] [PubMed] [Google Scholar]
- 15. Gutell R, Larsen N, Woese C. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol Rev 1994, 58:10–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gardner PP, Giegerich R. A comprehensive comparison of comparative RNA structure prediction approaches. BMC Bioinformatics 2004, 5:140. doi:10.1186/1471-2105-5-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mathews DH, Moss WN, Turner DH. Folding and finding RNA secondary structure. Cold Spring Harb Perspect Biol 2010, 2:a003665. doi:10.1101/cshperspect.a003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kutchko KM, Sanders W, Ziehr B, Phillips G, Solem A, Halvorsen M, Weeks KM, Moorman N, Laederach A. Multiple conformations are a conserved and regulatory feature of the RB1 5′ UTR. RNA 2015, 21:1–12. doi:10.1261/rna.049221.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2’‐hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc 2005, 127:4223–4231. doi:10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 20. Tijerina P, Mohr S, Russell R. DMS footprinting of structured RNAs and RNA‐protein complexes. Nat Protoc 2007, 2:2608–2623. doi:10.1038/nprot.2007.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Corley M, Solem A, Qu K, Chang HY, Laederach A. Detecting riboSNitches with RNA folding algorithms: a genome‐wide benchmark. Nucleic Acids Res 2015, 43:1859–1868. doi:10.1093/nar/gkv010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deigan K, Li T, Mathews D, Weeks K. Accurate SHAPE‐directed RNA structure determination. Proc Natl Acad Sci 2009, 106:97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV‐1 RNA genome. Nature 2009, 460:711–716. doi:10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zarringhalam K, Meyer MM, Dotu I, Chuang JH, Clote P. Integrating chemical footprinting data into RNA secondary structure prediction. PLoS One 2012, 7:e45160. doi:10.1371/journal.pone.0045160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Siegfried NA, Busan S, Rice GM, Nelson JAE, Weeks KM. RNA motif discovery by SHAPE and mutational profiling (SHAPE‐MaP). Nat Methods 2014, 11:959–965. doi:10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mortimer SA, Trapnell C, Aviran S, Pachter L, Lucks JB. SHAPE–Seq: high‐throughput RNA structure analysis. Curr Protoc Chem Biol 2012, 4:275–297. doi:10.1002/9780470559277.ch120019. [DOI] [PubMed] [Google Scholar]
- 27. Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. In vivo genome‐wide profiling of RNA secondary structure reveals novel regulatory features. Nature 2014, 505:696–700. doi:10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 28. Mortimer SA, Weeks KM. A fast‐acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc 2007, 129:4144–4145. doi:10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 29. Wilkinson KA, Merino EJ, Weeks KM. Selective 2’‐hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 2006, 1:1610–1616. doi:10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 30. McGinnis JL, Duncan CDS, Weeks KM. High‐throughput SHAPE and hydroxyl radical analysis of RNA structure and ribonucleoprotein assembly. Methods Enzymol 2009, 468:67–89. doi:10.1016/S0076-6879(09)68004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Spitale RC, Flynn RA, Torre EA, Kool ET, Chang HY. RNA structural analysis by evolving SHAPE chemistry. Wiley Interdiscip Rev RNA 2014, 5:867–881. doi:10.1002/wrna.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High‐throughput SHAPE analysis reveals structures in HIV‐1 genomic RNA strongly conserved across distinct biological states. PLoS Biol 2008, 6:e96. doi:10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wilkinson K, Vasa S, Deigan K. Influence of nucleotide identity on ribose 2’‐hydroxyl reactivity in RNA. RNA 2009, 15:1314–1321. doi:10.1261/rna.1536209.structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Halvorsen M, Martin JS, Broadaway S, Laederach A. Disease‐associated mutations that alter the RNA structural ensemble. PLoS Genet 2010, 6:e1001074. doi:10.1371/journal.pgen.1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ritz J, Martin JS, Laederach A. Evaluating our ability to predict the structural disruption of RNA by SNPs. BMC Genomics 2012, 13:S6. doi:10.1186/1471-2164-13-S4-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, Ouyang Z, Zhang J, Spitale RC, Snyder MP, Segal E, et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature 2014, 505:706–709. doi:10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung J‐W, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature 2015, 519:486–490. doi:10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Flynn RA, Zhang QC, Spitale RC, Lee B, Mumbach MR, Chang HY. Transcriptome‐wide interrogation of RNA secondary structure in living cells with icSHAPE. Nat Protoc 2016, 11:273–290. doi:10.1038/nprot.2016.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Aviran S, Trapnell C, Lucks JB, Mortimer SA, Luo S, Schroth GP, Doudna JA, Arkin AP, Pachter L. Modeling and automation of sequencing‐based characterization of RNA structure. Proc Natl Acad Sci USA 2011, 108:11069–11074. doi:10.1073/pnas.1106541108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Loughrey D, Watters KE, Settle AH, Lucks JB. SHAPE‐Seq 2.0: systematic optimization and extension of high‐throughput chemical probing of RNA secondary structure with next generation sequencing. Nucleic Acids Res 2014, 42:e165. doi:10.1093/nar/gku909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pelechano V, Wei W, Steinmetz LM. Widespread co‐translational RNA decay reveals ribosome dynamics. Cell 2015, 161:1400–1412. doi:10.1016/j.cell.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Leonard CW, Hajdin CE, Karabiber F, Mathews DH, Favorov OV, Dokholyan NV, Weeks KM. Principles for understanding the accuracy of SHAPE‐directed RNA structure modeling. Biochemistry 2013, 52:596–599. doi:10.1021/bi300756s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Aviran S, Pachter L. Rational experiment design for sequencing‐based RNA structure mapping. RNA 2014, 20:1864–1877. doi:10.1261/rna.043844.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cordero P, Kladwang W, VanLang C, Das R. Quantitative dimethyl sulfate mapping for automated RNA secondary structure inference. Biochemistry 2012, 51:3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sükösd Z, Swenson MS, Kjems J, Heitsch CE. Evaluating the accuracy of SHAPE‐directed RNA secondary structure predictions. Nucleic Acids Res 2013, 41:2807–2816. doi:10.1093/nar/gks1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bindewald E, Wendeler M, Legiewicz M, Bona MK, Wang Y, Pritt MJ, Le Grice SFJ, Shapiro BA. Correlating SHAPE signatures with three‐dimensional RNA structures. RNA 2011, 17:1688–1696. doi:10.1261/rna.2640111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kladwang W, VanLang CC, Cordero P, Das R. Understanding the errors of SHAPE‐directed RNA structure modeling. Biochemistry 2011, 50:8049–8056. doi:10.1021/bi200524n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xia T, SantaLucia J, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest‐neighbor model for formation of RNA duplexes with Watson‐Crick base pairs. Biochemistry 1998, 37:14719–14735. doi:10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 49. Eddy SR. Computational analysis of conserved RNA secondary structure in transcriptomes and genomes. Annu Rev Biophys 2014, 43:433–456. doi:10.1146/annurev-biophys-051013-022950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Quarrier S, Martin JS, Davis‐Neulander L, Beauregard A, Laederach A. Evaluation of the information content of RNA structure mapping data for secondary structure prediction. RNA 2010, 16:1108–1117. doi:10.1261/rna.1988510.experimental. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Washietl S, Hofacker IL, Stadler PF, Kellis M. RNA folding with soft constraints: reconciliation of probing data and thermodynamic secondary structure prediction. Nucleic Acids Res 2012, 40:4261–4272. doi:10.1093/nar/gks009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pollom E, Dang KK, Potter EL, Gorelick RJ, Burch CL, Weeks KM, Swanstrom R. Comparison of SIV and HIV‐1 genomic RNA structures reveals impact of sequence evolution on conserved and non‐conserved structural motifs. PLoS Pathog 2013, 9:e1003294. doi:10.1371/journal.ppat.1003294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013, 30:772–780. doi:10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ramsköld D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol 2009, 5:1–11. doi:10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lackey L, McArthur E, Laederach A. Increased transcript complexity in genes associated with chronic obstructive pulmonary disease. PLoS One 2015, 10:1–25. doi:10.1371/journal.pone.0140885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nawrocki EP, Eddy SR. Computational identification of functional RNA homologs in metagenomic data. RNA Biol 2013, 10:1170–1179. doi:10.4161/rna.25038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gutell RR, Lee JC, Cannone JJ. The accuracy of ribosomal RNA comparative structure models. Curr Opin Struct Biol 2002, 12:301–310. doi:10.1016/S0959-440X(02)00339-1. [DOI] [PubMed] [Google Scholar]
- 58. Lavender CA, Lorenz R, Zhang G, Tamayo R, Hofacker IL, Weeks KM. Model‐free RNA sequence and structure alignment informed by SHAPE probing reveals a conserved alternate secondary structure for 16S rRNA. PLoS Comput Biol 2015, 11:e1004126. doi:10.1371/journal.pcbi.1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lavender CA, Gorelick RJ, Weeks KM. Structure‐based alignment and consensus secondary structures for three HIV‐related RNA genomes. PLoS Comput Biol 2015, 11:e1004230. doi:10.1371/journal.pcbi.1004230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gotoh O. An improved algorithm for matching biological sequences. J Mol Biol 1982, 162:705–708. [DOI] [PubMed] [Google Scholar]
- 61. Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu Rev Biophys 2008, 37:117–133. doi:10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 62. Ritz J, Martin JS, Laederach A. Evolutionary evidence for alternative structure in RNA sequence co‐variation. PLoS Comput Biol 2013, 9:e1003152. doi:10.1371/journal.pcbi.1003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 2010, 11:129. doi:10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cordero P, Lucks JB, Das R. An RNA mapping data base for curating RNA structure mapping experiments. Bioinformatics 2012, 28:3006–3008. doi:10.1093/bioinformatics/bts554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Underwood JG, Uzilov AV, Katzman S, Onodera CS, Mainzer JE, Mathews DH, Lowe TM, Salama SR, Haussler D. FragSeq: transcriptome‐wide RNA structure probing using high‐throughput sequencing. Nat Methods 2010, 7:995–1001. doi:10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome‐wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 2014, 505:701–705. doi:10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shannon C. A mathematical theory of communiction. Bell Syst Tech J 1948, 27:379–423. [Google Scholar]
- 68. Huynen M, Gutell R, Konings D. Assessing the reliability of RNA folding using statistical mechanics. J Mol Biol 1997, 267:1104–1112. doi:10.1006/jmbi.1997.0889. [DOI] [PubMed] [Google Scholar]
- 69. Del Campo C, Bartholomäus A, Fedyunin I, Ignatova Z. Secondary structure across the bacterial transcriptome reveals versatile roles in mRNA regulation and function. PLoS Genet 2015, 11:1–23. doi:10.1371/journal.pgen.1005613. [DOI] [PMC free article] [PubMed] [Google Scholar]


