Abstract
Gene expression is regulated at many levels, including after generation of the primary RNA transcript from DNA but before translation into protein. Such post-translational gene regulation occurs via the action of a multitude of RNA binding proteins and include varied actions from splicing to regulation of association with the translational machinery. Primary evidence that such processes might contribute to disease mechanisms in neurodegenerative disorders comes from the observation of mutations in RNA binding proteins, particularly in diseases in the amyotrophic lateral sclerosis-frontotemporal dementia spectrum and in some forms of ataxia and tremor. The bulk of evidence from recent surveys of the types of RNA species that are affected in these disorders suggests a global deregulation of control rather than a very small number of RNA species, although why some groups of neurons are sensitive to these changes is not well understood. Overall, these data suggest that neurodegeneration can be initiated by mutations in RNA binding proteins and, as a corollary, that neurons are particularly sensitive to loss of control of gene expression at the post-transcriptional level. Such observations have implications not only for understanding the nature of neurodegenerative disorders but also how we might intervene therapeutically in these diseases.
Graphical Abstract
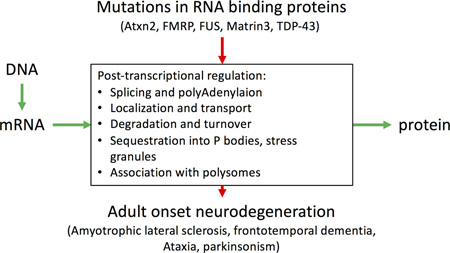
Mutated RNA binding proteins cause neurodegenerative diseases such as ALS by disrupting the information flow from DNA to protein
Introduction
Genetic mechanisms have been shown to influence the lifetime risk of several neurodegenerative diseases. In general terms, there are both variants that are inherited in families [1] and more subtle risk factor variants that act at a population level [2]. Furthermore, there are examples where the same gene is relevant for both inherited and sporadic disease, suggesting that mechanisms in both cases are likely shared [3]. In many cases, common variation around genes that affects risk of sporadic disease is not associated with amino acid changes, but rather variants that change gene regulation, by expression or splicing [4]. This observation suggests that some forms of gene regulation might be relevant to the pathogenesis of several neurodegenerative diseases.
Post-transcriptional gene regulation occurs at the level of RNA, and includes a variety of modifications to the nucleic acid that can be accomplished by RNA binding proteins [5] or by the interactions of non-coding RNAs with mRNAS [6] (Fig. 1). As might therefore be expected, post-transcriptional gene regulation has been considered to influence processes that are relevant to neurodegeneration. One causal way in which this might occur is by mutations in RNA-binding proteins, of which there are several that cause neurodegenerative diseases. However, there are also other, less direct but still critical, mechanisms by which post-transcriptional regulation might contribute to pathways relevant for the development of neuropathology.
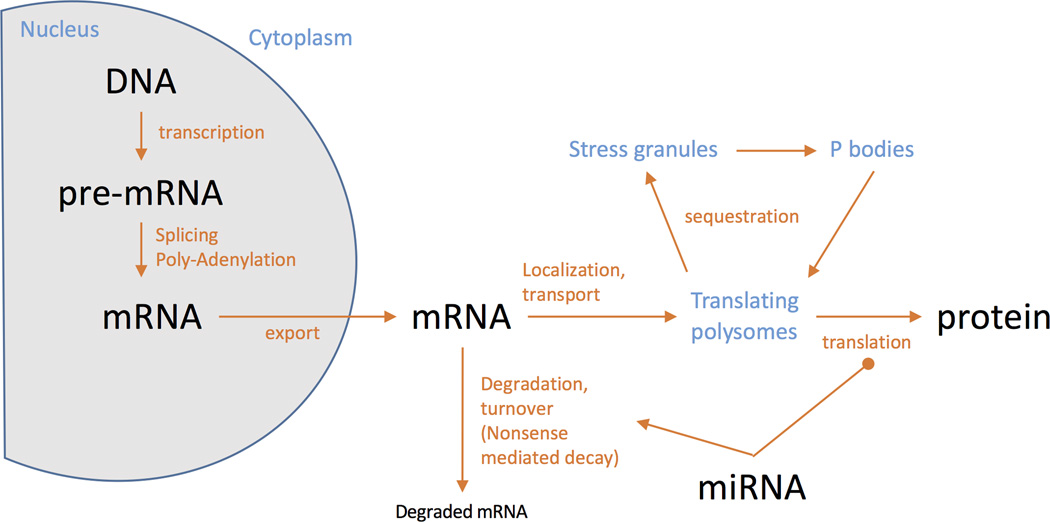
Figure 1. Post-transcriptional gene regulation.
Key aspects of post-transcriptional gene regulation. Although the central dogma of molecular biology, that DNA makes RNA makes protein (biomolecules are shown in black), is generally true and is useful, there are a variety processing steps (red) that control the flow of information between the key events. Additionally, the control of biological information is compartmentalized within the cell, and here subcellular structures are identified in blue. It should be noted that microRNA (miRNA) species interact with mRNA and can either promote degradation or inhibit translation to protein.
In this chapter, I will discuss some of the potential ways in which post-translational gene regulation might contribute to neurodegeneration. While this discussion will not be comprehensive, I will give examples of known mechanisms in specific neurodegenerative diseases and indicate where these might generalize to other disorders. This review will focus on the best studied examples of RNA-binding proteins that have been shown to play causal roles in neurodegeneration. I will use those situations where mutations in the genes encoding for RNA-binding proteins, as causality can be reasonably inferred in those cases. However, it should be noted that similar mechanisms are likely to play roles in other neurological conditions, including sporadic diseases, especially where there is evidence of some misregulation of RNA binding proteins.
Gene mutations in RNA binding proteins in Amyotrophic Lateral Sclerosis and frontotemporal dementia
Amyotrophic Lateral Sclerosis (ALS; also known a motor neurone disease or Lou Gehrig’s disease) is a notably unpleasant disease, mainly because from diagnosis to demise often occurs over a relatively rapid time course, with a range of intervals between 1.5–2 years [7] (see sidebar for description of the disease process). As for many of the diseases discussed here, FTD-ALS has age-dependent prevalence and is generally rare before the age of 50 but much more common in the subsequent two decades of life, with an overall lifetime prevalence of about 4 cases per 100,000 population.
Brain regions affected in various neurodegenerative diseases.
The central nervous system contains a startling variety of different cell types. For neurons, the principal definitions of these cells were first morphological and anatomical then, later, by the types of neurotransmitters produced and by molecular characterization. Neurodegenerative diseases are characterized by the progressive loss of various subtypes of neurons that then result in symptoms in patients that are related to loss of the normal functions of those cells. Although patterns of neuronal loss in different diseases are not absolutely restricted to a single set of cells or neurotransmitter type, the following descriptions outline some of the major changes seen in these disease.
Alzheimer’s disease (AD) is predominantly a diseases of the neocortex, where protein deposits occur in the entorhinal cortex then are found in progressively more brain regions, including for example the hippocampus. Two types of protein deposits are found; extracellular plaques composed of aggregated fragments of the amyloid precursor protein (APP) and intracellular tangles, composed largely of the neuronal microtubule protein, Tau. The progression of AD pathology is mirrored by memory loss and other mood and personality changes. By the end of the disease, the number of neurons that have died is sufficient that there is volumetric loss of the cerebral cortex.
In Amyotrophic lateral sclerosis, a progressive disease course and eventually fatal events arises from death of neurons in the upper motor tracts linking the motor regions of the cerebral cortex to the spinal cord, and lower motor neurons in the ventral horn of the spinal cord that project to the musculature. As these neurons become dysfunctional and drop out, control of the muscles that they innervate is lost, leading to weakness, paralysis and eventually loss of control of breathing when the neurons that innervate the diaphragm are damaged. Genetically, ALS is related to an apparently quite different condition, frontotemporal dementia (FTD) where forebrain neurons are damaged, resulting in variable changes in cognition and personality. Both ALS and FTD are often associated with the cytoplasmic accumulation.
Parkinson’s disease (PD) is a movement disorder characterized clinically by slowed movement, tremor, an unstable posture and gait problems. Many, but not all, of the motor signs of PD relate to the loss of dopamine producing neurons that project from the substantia nigra pars compacta in the midbrain to the caudate and putamen, two parts of the human striatum. There is evidence that the loss of neurons in PD is established very early in the disease course but continues over time to have more and more impact on movement, leading to a need to treat patients usually by replacing dopamine directly or by surgical approaches that restore function of the basal ganglia motor circuit. The main pathology of PD is the deposit of α-synuclein, a small protein that is normally found at synapses but becomes aggregated and deposited in surviving neurons as Lewy bodies and Lewy neurites. Using Lewy bodies as a sensitive marker reveals that there are actually many brain regions affected in PD, resulting in a range of non-motor complications including cognitive changes that are currently difficult to treat.
Spinocerebellar ataxias are a heterogeneous set of conditions where loss of Purkinje cells in the cerebellum, a part of the brain responsible for motor learning and co-ordination, are lost. Damage to Purkinje cells therefore results in uncoordinated movements and speech problems and, like the other diseases discussed here, usually get progressively worse over time.
There are no effective treatments to modify neurodegeneration in FTD-ALS and, given this acute need, there is a substantial literature on trying to understand the causal basis for disease risk in individuals, with the hope that this would then lead to identified pathways that could be tractable for treatment. Although most cases are sporadic in nature, that is to say there is no easily discernible single causes, one successful approach to understanding causality has been to interrogate the genetic basis of ALS [8]. Although frank mutations account for only a minority of the overall cases of ALS, some of the more common variants occur in relatively high percentages of cases. Importantly, it has been observed that even within single families with the same mutation, the actual presentation of disease can be quite variable. Specifically, a relatively common hexanucleotide expansion in the first intron of the c9orf72 gene [9,10], is associated with both ALS and frontotemporal degeneration (FTD, sometimes also called frontotemporal lobar neurodegeneration or FTLD), a condition where dementia is the typical presentation of the disease due to extensive atrophy of the frontal and temporal lobes of the cerebral cortex. These observations have led to the recognition that ALS-FTD represent a clinicopathologcial spectrum of disease [11], perhaps additionally explaining why looking for cause of diseases such as ALS was difficult as families with such mixed phenotypes may not have been considered to have the same underlying disease.
Although there are a variety of genes involved in FTD-ALS, a prominent subset of them are known or suspected RNA binding proteins. Specifically, mutations in fused in TAR-binding protein (TARDBP) [12,13], fused in sarcoma (FUS) [14,15], and matrin-3 (MATR3) [16] all cause ALS and all three protein products have domain structures consistent with RNA binding activity. Interestingly, deposition of the protein product of TARDBP, TDP-43 (also known as Transactive response DNA-binding protein 43), in cases of both FTD and ALS had been identified prior to mutations being reported [17], thus anticipating the genetic link between these two diseases. While TDP-43 is normally a nuclear protein, in ALS-FTD cases it is found to relocalize to the cytosol of vulnerable neuronal types. The accumulation of cytosolic TDP-43 occurs in sporadic ALS-FTD, suggesting a further link between familial and idiopathic forms of these diseases. Two questions are prominent in light of this information; how do mutations affect RNA binding activity of these proteins and which RNAs are misregulated leading to disease?
Mutations in FUS, TDP-43 and MATR3 directly implicate mRNA regulation in the pathobiology of ALS
Each of FUS, TDP43 and MATR3 have one or more RNA recognition motifs (Fig. 2). However, mutations are generally not in these domains but instead are clustered in relatively less structured regions, particularly the glycine-rich regions that are at the C-terminus of TDP-43 and in the center of the protein in FUS. There is an additional cluster of FUS mutations at the C-terminal nuclear localization sequence. For MATR3, mutations are found in regions of the protein that are outside of well-recognized domains. Therefore, it is not immediately certain from examination of the effects of mutations on the primary protein sequence, whether or not RNA binding is actually important for the pathogenic effects of these mutant proteins. To address this question, we might first look at what RNAs are bound to these three proteins, what the proposed effects of binding on RNA regulation are and how mutations influence those functions.
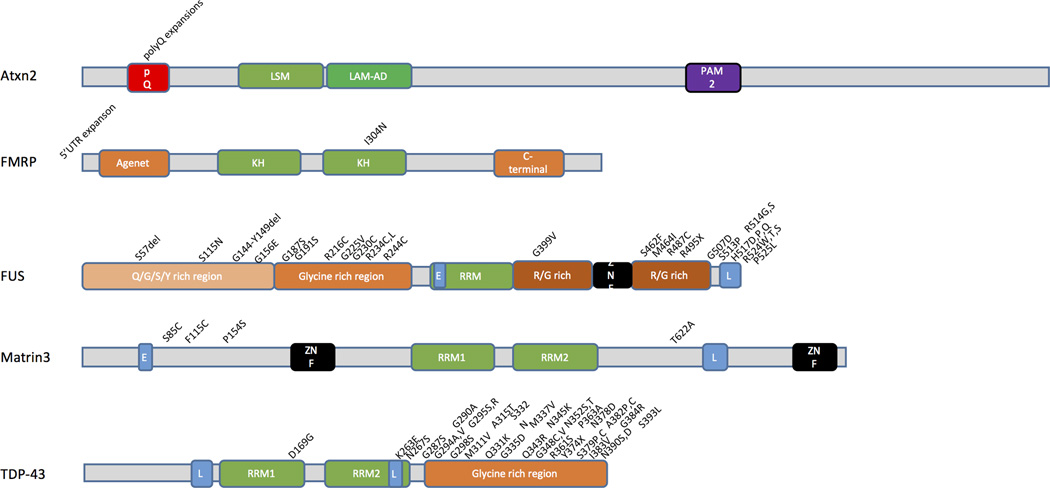
Figure 2. Mutations in RNA binding proteins associated with neurodegeneration.
A variety of proteins associated with adult onset neurodegenerative disease contain RNA binding domains. The putative domain structures of proteins that are mutated in neurodegenerative diseases are shown, in alphabetical order. For each protein, RNA binding domains are in green, nuclear localization (L) or exclusion (E) sequences are in blue, Zinc finger domains are in black and amino acid repeats or amino-acid enriched domains are in shades of red and orange. Above each ideogram are examples of specific mutations found in each gene. Additional abbreviations; KH, heterogeneous nuclear ribonucleoprotein K homology; Lsm, Like Sm; Lsm-AD, Lsm associated domain; PAM2, PABP [poly(A)-binding protein] interacting motif; pQ, poly Q (glutamine); RRM, RNA recognition motif; ZNF, zinc finger. It is worth noting that mutations are only rarely found in the RNA binding motifs themselves but rather in the structural domains that surround them.
RNA binding targets of TDP-43, FUS and MATR3
Both FUS and TDP43 have been shown to bind directly to a series of RNA targets. TDP-43 has been proposed to bind to a large number of RNA targets in different cell and tissue types [18–22]. The binding of TDP-43 to RNA is mediated by stretches of UG in the targets [23]. Poly-A sequences are also found in the lists of TDP-43 targets [24], although whether TDP-43 binds directly to these directly is unclear as TDP-43 can interact with a number of other proteins to access a number of potential sequences [25]. Additionally, binding targets of TDP-43 include microRNA (miRNA) species, again perhaps working together with other proteins, specifically components of the Dicer and Drosha complexes [26]. As has been noted by others [27], the overlap between different surveys of TDP-43 bound RNA or miRNA targets is relatively small although a specific set of RNAs has been confirmed across studies, including TDP-43 itself [28].
FUS was identified as a RNA binding protein many years before mutations were associated with ALS as there is a translocation event in some forms of sarcoma [29]. As for TDP43, there have been several genome-wide surveys of the types and sequences of RNA species bound to FUS that do not overlap substantially [30–32]. Consistent with variation between tissues, some studies have reported that FUS and TDP43 have different RNA binding profiles[21], but other approaches have suggested some overlap in downstream targets in neurons [33,34]. One consistent target is FUS itself, although the mechanisms by which FUS controls its own expression are different from TDP43 [35] Similar to TDP43, FUS may bind to miRNA via the Drosha complex [36].
Matrin-3 also binds to several other RNA binding proteins and high throughput sequencing has identified a series of RNA targets that are bound to these protein complexes [37]. Conversely, Matrin-3 can be isolated from pulldowns of other RNA binding proteins [38] and co-purifies with retina-expressed small RNAs [39]. Finally, like TDP-43, Matrin-3 interacts physically with PTB1, a polypyrimidine binding protein, with which it binds motifs within RNA targets [40].
Based on these data, we can infer that each of the three RNA binding proteins that are mutated in familial FTD-ALS each work within a network of other ribonucleiprotein complexes to bind a large series of both mRNAS and small RNAs, the latter including miRNAs. There is modest evidence of sharing of RNA targets between the different proteins, but given that the lists of potential candidates are very long, this is perhaps to be expected by chance alone. Therefore, the RNA targets are not especially informative as to how mutations in these genes leads to ALS. It is important to therefore consider what the function of RNA binding is and how mutations affect those functions.
Post-transcriptional functions of RNA binding proteins involved in ALS
Again prior to nomination as an FTD-ALS gene, TDP-43 had been proposed to control the splicing of exon 9 of the cystic fibrosis transmembrane receptor (CFTR) gene, an event that occurs in the nucleus [41]. Along the same lines, the binding of TDP-43 to its own mRNA controls splicing of the 3’-UTR region and affects its own hence mRNA stability in a post-transcriptional manner [28,42,43]. This leads to the general concept that TDP-43 plays an important role in RNA splicing. To address this hypothesis, several genome-wide surveys in cells and genetically modified mice have been performed and have identified multiple mRNA species that show aberrant splicing in the absence of TDP-43 [19,20,22,44]. Whether all of these are direct targets of TDP-43 is unclear, although some studies have suggested that the altered splicing events are surrounded by the same UG motifs as recovered in direct binding assays [44].
FUS likewise is involved in pre-mRNA splicing via interactions with small nuclear ribonucleoprotein (snRNP) complexes [45,46] and potentially link these to translation via an interaction with RNA-polymerase II [47]. The motif by which FUS binds to RNA is less well defined that the TDP-43 motif and the protein may bind long introns generally [31]. Therefore, and consistent with the binding studies discussed above, the range of splicing targets affected by FUS is broad and vary between studies perhaps due to differences in expression levels between cells and tissues [21,30,31,33,34]. Matrin-3 has also been proposed to play a broad role in the control of splicing [40].
These results might suggest that deregulated splicing might be a general mechanism by which neurons become vulnerable to neurodegeneration in ALS, perhaps because the brain has a relatively high level of alternate splicing events compared to other tissues [48]. However, all three proteins may also be involved in events other than splicing including regulation of miRNA processing [36,49,50], expression of long non coding RNAs [51], mRNA transport in neurons [52–55] and mRNA translation[56,57]. The multiplicity of these functions have been reviewed elsewhere [27,58], but because there are so many proposed functions, it is not clear as to exactly what level these proteins act to cause disease. An important next consideration, therefore, is how mutations affect protein function and, specifically, whether post-translational regulation is important in disease pathogenesis.
Effects of mutations on post-translational gene regulation
As noted in figure 1, mutations that are associated with FTD-ALS are found in several different domains of FUS, TDP-43 and MATR3 but are generally away from the RNA binding regions. This observation means that it is formally possible that the pathogenic effects of mutations are unrelated to RNA binding potential and, therefore, to post-transcriptional regulation of gene expression. Additionally, the accumulation of insoluble TDP-43 protein in FTD-ALS generally is in the cytoplasm not in the nucleus where this protein normally resides [17]. Therefore, it would be important to distinguish between the possibilities that mutations have a toxic function in the cytoplasm or that lack their normal function in the nucleus.
One way to potentially discriminate between these effects might be to compare knockout and transgenic models for a given protein. Deletion of Tardbp in adult motor neurons results in loss of those cells specifically and ALS-like phenotypes [59], which might suggest that loss of nuclear function is important in maintenance of adult motor neurons. However, precise interpretation of these data is complicated as a generalized knockout in mice results in embryonic lethality [60–62], suggesting that RNA binding of TDP-43 is important for the survival of many cell types.
Conversely, expression of mutant forms of TDP-43 in adult mice results in toxicity towards motor neurons [63]. In general, the accumulation of cytoplasmic aggregates of TDP-43 is not a required event for toxicity. Again, however, clear interpretation here is complicated by the observation that the wild type protein is toxic in a manner that depends on the level of transgene over-expression [63]. Therefore, some of the mechanisms by which toxicity occurs might be related to expression levels while others might relate to the effects of mutations. At least one recent knock-in model has been reported where the expression events may be avoided [64]. This model was reported to have damage to both upper (cortical) and lower (spinal cord) motor neurons, suggesting that at least some effects of TDP-43 mutations are independent of expression levels.
These data support the idea that normal function of TDP-43 is important for toxic effects of mutations, although the possibility that mutations have a neomorphic toxic function is difficult to fully rule out. However, experiments in Drosophila models suggest that deletion of the RRM domain, which should abolish RNA binding, is sufficient to mitigate the toxicity associated with expression of human wild type or mutant TDP-43 [65]. Additionally, manipulation of expression of RNA binding proteins can also block TDP-43 toxicity [66,67]. Finally, transgenic expression of mutant TDP-43 in mice results in altered splicing, consistent with the normal function of wild type protein being altered by mutation [68].
Overall, the balance of evidence suggests that the normal functions of TDP-43 are relevant to the toxicity caused by mutations associated with ALS-FTD. Because TDP-43 has a large number of RNA targets, these observations suggest that mutations will result in a global and generalized effect on post-translational gene regulation.
Along the same lines, mutations in FUS require RNA binding to exert toxic effects [69] and cause a global deregulation of RNA metabolism that can be mitigated by expression of a master regulator of nonsense-mediated decay [70]. Because FUS and TDP-43 may have different effects on distinct sets of RNA targets [21,33] it is unlikely that there are a small number of common downstream targets, although this possibility cannot completely be discounted. Taken together, these data suggest global rather than highly specific events.
Overall, the available data suggests that post-transcriptional control of gene expression by RNA binding proteins is a major mediator of pathogenesis in familial ALS-FTD. Why specific groups of neurons are especially vulnerable to global deregulation of RNA metabolism is unclear at this time and how one might use this information to intervene in disease remains to be clarified.
Post-transcriptional gene regulation in other neurodegenerative diseases
The above discussion clearly identifies post-transcriptional gene regulation by RNA-binding proteins as potentially causal in ALS-FTD. If some groups of neurons are specifically vulnerable to global deregulation of RNA metabolism, a reasonable question is then whether other age-related neurodegenerative disorders are similarly a consequence of altered gene regulation. The following examples are mainly focussed on those diseases where causal mutations in RNA binding proteins have been identified as these represent the strongest and most direct evidence for post-transcriptional gene regulation in each disorder.
Fragile-X and Fragile-X tremor ataxia syndrome
Mutations in the FRM1 (fragile X mental retardation 1) gene cause the developmental disorder fragile X, a relatively common cause of intellectual disability with additional features including autistic-like behaviours [71]. The most common underlying mutation is a triplet repeat expansion in the 5’ untranslated region of a gene on Chromosome X that encodes the FMRP protein [72]. The expanded triplet repeats are heavily methylated and this epigenetic modification of the gene results, in males, in loss of FMRP function. Importantly for the discussion here, FMRP has several recognizable RNA binding domains [73] and binds a variety of mature RNA species to repress translation [74]. Thus, in fragile X syndrome it is likely excessive translation of RNA to protein, an important post-transcriptional gene expression process, that leads to a neurodevelopmental disorder. A point mutation, I304N (Fig. 2) is associated with a severe form of fragile-x syndrome [75] and the mutant protein has diminished RNA binding [76], further supporting the concept that loss of FMRP function is the proximate cause of this disorder. Also, as for TDP-43 and FUS, FMRP may regulate its own mRNA in a post-transcriptional manner [77]
Additionally, carriers of a pre-mutation allele, with a repeat length intermediate between the typical length in the population and the pathogenic expansion found in fragile X, develop a tremor ataxia syndrome later in life [78]. The key distinction between the mechanisms underlying fragile X and FXTAS (fragile X tremor ataxia syndrome) is that in the latter situation, FMR1 is translated to produce RNA with expanded repeats. The mechanism by which this aberrant species induces toxicity is not fully resolved, but one widely studied hypothesis is that the mutant RNA is directly toxic to neurons, potentially by sequestering RNA binding proteins in cytoplasmic foci and thus disrupting overall post-transcriptional control in neurons [79]. Another potential contributor to toxicity comes from direct translation of the repeats themselves to produce small toxic peptides or RAN translation [80]. Which mode of toxicity is dominant in the human condition is still debated, although it is important to note that both RAN translation and RNA toxicity are proposed to occur in a variety of other triplet repeat disorders including a relatively common cause of familial ALS-FTD, hexanucleotide expansions in the c9orf72 gene [81], again suggesting that generalized dysregulation of post-transcriptional events can be toxic to motor neurons.
Complex phenotypes related to ATXN2 mutations
Another example of a triplet repeat expansion is found in the gene ATXN2 where the triplet repeats are part of the gene coding sequence and therefore produce a protein with an increased number of repeating glutamates or polyQ [82]. It is therefore possible that the polyQ expansion in mutant ATXN2 alters its normal function, leading to spinocerebellar ataxia, an adult onset neurological disorder with prominent loss of gait and balance.
As in many of the other examples here, ATXN2 is an RNA binding protein that has been shown to play roles in post-transcriptional regulation of gene expression. Specifically, ATXN2 interacts with a series of other RNA binding proteins and participates in several aspects of post-transcriptional RNA metabolism including translation at polysomes [83] and disruption of cytosolic structures containing RNA, stress granules or P-bodies [84] that may result in protein translation changes as these structures are normally translationally silent.
ATXN2 may also contribute to other neurodegenerative conditions than ataxia. Some ATXN2 mutation carriers have symptoms of parkinsonism, suggesting loss of dopamine neurons in the substantia nigra pars compacta, either alone or in combination with ataxia [85]. These data suggest that several types of neurons are sensitive to deregulation of translational control, which can lead to multiple neuropathologies.
Additionally, the ATXN2 gene has been proposed to be a modifier of risk for ALS [86]. Specifically, intermediate polyQ repeat lengths are proposed to increase lifetime risk of ALS by about 4-fold and ATXN2 acts as a modifier of TDP-43 toxicity in yeast and Drosophila models. The initial genetic observations have been confirmed in multiple studies and extended to show that both a similar effect is seen in both familial and sporadic ALS [87]. While the exact mechanism(s) by which ATXN2 repeats influence ALS risk are not fully elucidated, given that multiple RNA metabolism genes are mutated in ALS and that ATXN2 plays a role in RNA metabolism, it is reasonable to suggest that these data further supports the idea that ALS can be a consequence of deregulated post-transcriptional gene control.
Other common diseases
The above examples are deliberately focussed on situations where gene mutations exert a strong effect on disease risk. However, there are likely to be other mechanisms at play, perhaps most cogently in sporadic diseases. It is therefore appropriate to discuss potential mechanisms in some of the more common neurodegenerative diseases.
Alzheimer’s disease
The principle driving pathogenic pathway, more certainly in familial Alzheimer’s disease (AD) but also likely in sporadic cases, is the generation of amyloid-beta fragments that aggregate to form amyloid plaques in the brain parenchyma. Secondarily, plaques lead to the formation of tau-positive tangles within neurons [88]. While this ‘amyloid cascade’ hypothesis has some limitations [89], particularly with respect to its ability to predict useful drug interventions, it does nonetheless position AD as a post-translational disease, being focussed on protein processing and deposition events.
Despite this overarching theme in AD research, there have been several attempts to understand whether other mechanisms might be in play in the disease, including post-transcriptional gene regulation. Particular attention has been paid to the possibility that microRNAs (miRNA) might be deregulated in the AD brain and affect, particularly, enzymes involved in amyloid processing [90]. Another possible post-transcriptional event relevant to AD is that miRNAs may control cholesterol metabolism in the brain, which is thought to represent an important pathway for control of amyloid production [91]. However, different surveys of miRNA in the AD brain have nominated different candidates that do not overlap between studies [92–95]. Therefore, at this stage it is not clear if there is a particularly robust miRNA signal that differentiates AD from controls, although this is certainly an area of investigation that merits further consideration perhaps with larger sample series.
Parkinson’s disease
Parkinson’s disease (PD) is also usually characterized by the deposition of protein, prominently α-synuclein, in neurons in the form of Lewy bodies and Lewy neurites in various brain regions [96]. Initial surveys of the miRNA complement of PD brains versus controls nominated miR-133b as differential between groups and further claimed that mir-133b and the transcription factor Pitx3 might have inter-dependent effects on dopamine neuron development, setting the PD brain up for subsequent loss of these cells [97]. However, because PD pathology prominently includes loss of dopamine neurons especially as the disease progresses, it is not clear whether the diminishment of mir-133b is a cause or a consequence of neurodegeneration in this region. Subsequent surveys of miRNAs in PD did not identify mir-133b, although other miRNA differences were noted [98]. Therefore, as for AD, whether miRNA-mediated post-transcriptional effects are important in the sporadic PD brain remains ambiguous and larger surveys across multiple affected brain regions in the human disease, ideally at different stages of progression, are warranted.
There are additional proposed events that might link post-transcriptional gene regulation to familial PD. Several years ago, my lab proposed that DJ-1, a rare cause of familial parkinsonism [99], might participate in RNA regulation, consistent with prior nomination of the protein as part of an RNA binding complex [100]. Unfortunately, this result has not yet been widely replicated and it is not clear if there is any direct binding of DJ-1 protein to RNA. Along the same lines, there are claims that LRRK2 (Leucine-rich repeat kinase 2[101]) can bind to the proteins involved in miRNA production and thereby control miRNA regulation [102]. However, again, the primary observations of direct protein interactions have not been widely replicated and it remains to be seen if there is a direct, or perhaps indirect, effect of these mutations on post-transcriptional control.
Conclusion
Overall, the examples given here allow us to confidently conclude that post-transcriptional gene regulation is an important driving event in some forms of neurodegenerative disease. Most convincingly, dominantly inherited forms of ALS-FTD are associated with mutations in several RNA binding proteins, including TDP-43, TLS/FUS and Matrin-3. RNA binding proteins can also be mutated in inherited SCA and the clinical phenotypes of ALS and SCA may overlap, with at least one SCA protein, ATXN2 being a modifier of ALS risk. However, the RNA targets and the functions performed by these RNA binding proteins varies, suggesting a global deregulation of post-transcriptional control rather than a set of very specific RNAs is likely to be detrimental for neurons function and survival in the human brain In contrast, the evidence for post-transcriptional gene regulation in other common neurodegenerative disorders is fragmentary and it is often difficult to discern whether the direction of effect. That is to say whether deregulation of gene expression affects neuronal survival in AD or PD or whether the disease process secondarily affects aspects of gene expression is uncertain. However, given that neurons in general are sensitive to loss of regulation of gene expression at the level of RNA binding proteins suggests that this is a hypothesis worth pursuing, even to exclusion, in multiple neurodegenerative diseases.
Important future directions include resolving the very difficult question of whether specific subsets of RNA species that are affected by loss of RNA binding proteins are responsible for neurodegeneration or whether a more global deregulation of post-transcriptional processes is important in triggering neuronal cell death. The balance of evidence probably supports the latter, but this is an important question for developing therapeutics if there were a small number of key mediators of cell death, then these could be targeted more effectively than a large ensemble of dysregulated genes. Another fundamental remaining question is why, at least in those diseases where there is strongest evidence that RNA binding proteins can drive pathogenesis, some groups of neurons are sensitive to alterations in their function. Perhaps the complexity of post-transcriptional gene regulation is particularly high in neurons, which have specific requirements for alternate splicing and translation at synapses. As for resolving they key pathway targets, answering this fundamental question may be important in the eventual development of novel therapeutics to reverse the course of these diseases.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute on Aging, NIH.
Footnotes
No conflicts of interest.
References
- 1.Hardy J, Orr H. The genetics of neurodegenerative diseases. J. Neurochem. 2006;97(6):1690–1699. doi: 10.1111/j.1471-4159.2006.03979.x. [DOI] [PubMed] [Google Scholar]
- 2.Lill CM, Bertram L. Towards unveiling the genetics of neurodegenerative diseases. Semin. Neurol. 2011;31(5):531–541. doi: 10.1055/s-0031-1299791. [DOI] [PubMed] [Google Scholar]
- 3.Singleton A, Hardy J. A generalizable hypothesis for the genetic architecture of disease: pleomorphic risk loci. Hum. Mol. Genet. 2011;20(R2):R158–R162. doi: 10.1093/hmg/ddr358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tak YG, Farnham PJ. Making sense of GWAS: using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics Chromatin. 2015;8:57. doi: 10.1186/s13072-015-0050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neelamraju Y, Hashemikhabir S, Janga SC. The human RBPome: from genes and proteins to human disease. J. Proteomics. 2015;127(Pt A):61–70. doi: 10.1016/j.jprot.2015.04.031. [DOI] [PubMed] [Google Scholar]
- 6.Im H-I, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012;35(5):325–334. doi: 10.1016/j.tins.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiò A, Logroscino G, Hardiman O, et al. Prognostic factors in ALS: A critical review. Amyotroph. Lateral Scler. Off. Publ. World Fed. Neurol. Res. Group Mot. Neuron Dis. 2009;10(5–6):310–323. doi: 10.3109/17482960802566824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Renton AE, Chiò A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat. Neurosci. 2014;17(1):17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper-Knock J, Shaw PJ, Kirby J. The widening spectrum of C9ORF72-related disease; genotype/phenotype correlations and potential modifiers of clinical phenotype. Acta Neuropathol. (Berl.) 2014;127(3):333–345. doi: 10.1007/s00401-014-1251-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kabashi E, Valdmanis PN, Dion P, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 2008;40(5):572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 13.Sreedharan J, Blair IP, Tripathi VB, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319(5870):1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vance C, Rogelj B, Hortobágyi T, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323(5918):1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwiatkowski TJ, Bosco DA, Leclerc AL, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323(5918):1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 16.Johnson JO, Pioro EP, Boehringer A, et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat. Neurosci. 2014;17(5):664–666. doi: 10.1038/nn.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 18.Sephton CF, Cenik C, Kucukural A, et al. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J. Biol. Chem. 2011;286(2):1204–1215. doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polymenidou M, Lagier-Tourenne C, Hutt KR, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 2011;14(4):459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tollervey JR, Curk T, Rogelj B, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011;14(4):452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colombrita C, Onesto E, Megiorni F, et al. TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells. J. Biol. Chem. 2012;287(19):15635–15647. doi: 10.1074/jbc.M111.333450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao S, Sanelli T, Dib S, et al. RNA targets of TDP-43 identified by UV-CLIP are deregulated in ALS. Mol. Cell. Neurosci. 2011;47(3):167–180. doi: 10.1016/j.mcn.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Bhardwaj A, Myers MP, Buratti E, Baralle FE. Characterizing TDP-43 interaction with its RNA targets. Nucleic Acids Res. 2013;41(9):5062–5074. doi: 10.1093/nar/gkt189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narayanan RK, Mangelsdorf M, Panwar A, et al. Identification of RNA bound to the TDP-43 ribonucleoprotein complex in the adult mouse brain. Amyotroph. Lateral Scler. Front. Degener. 2013;14(4):252–260. doi: 10.3109/21678421.2012.734520. [DOI] [PubMed] [Google Scholar]
- 25.Mohagheghi F, Prudencio M, Stuani C, et al. TDP-43 functions within a network of hnRNP proteins to inhibit the production of a truncated human SORT1 receptor. Hum. Mol. Genet. 2016;25(3):534–545. doi: 10.1093/hmg/ddv491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawahara Y, Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc. Natl. Acad. Sci. U.S.A. 2012;109(9):3347–3352. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ratti A, Buratti E. Physiological Functions and Pathobiology of TDP-43 and FUS/TLS proteins. J. Neurochem. 2016 doi: 10.1111/jnc.13625. [DOI] [PubMed] [Google Scholar]
- 28.Ayala YM, De Conti L, Avendaño-Vázquez SE, et al. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 2011;30(2):277–288. doi: 10.1038/emboj.2010.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crozat A, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363(6430):640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 30.Hoell JI, Larsson E, Runge S, et al. RNA targets of wild-type and mutant FET family proteins. Nat. Struct. Mol. Biol. 2011;18(12):1428–1431. doi: 10.1038/nsmb.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogelj B, Easton LE, Bogu GK, et al. Widespread binding of FUS along nascent RNA regulates alternative splicing in the brain. Sci. Rep. 2012;2:603. doi: 10.1038/srep00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakaya T, Alexiou P, Maragkakis M, et al. FUS regulates genes coding for RNA-binding proteins in neurons by binding to their highly conserved introns. RNA N. Y. N. 2013;19(4):498–509. doi: 10.1261/rna.037804.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagier-Tourenne C, Polymenidou M, Hutt KR, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat. Neurosci. 2012;15(11):1488–1497. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Honda D, Ishigaki S, Iguchi Y, et al. The ALS/FTLD-related RNA-binding proteins TDP-43 and FUS have common downstream RNA targets in cortical neurons. FEBS Open Bio. 2013;4:1–10. doi: 10.1016/j.fob.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Y, Liu S, Liu G, et al. ALS-associated FUS mutations result in compromised FUS alternative splicing and autoregulation. PLoS Genet. 2013;9(10):e1003895. doi: 10.1371/journal.pgen.1003895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morlando M, Dini Modigliani S, Torrelli G, et al. FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO J. 2012;31(24):4502–4510. doi: 10.1038/emboj.2012.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salton M, Elkon R, Borodina T, et al. Matrin 3 binds and stabilizes mRNA. PloS One. 2011;6(8):e23882. doi: 10.1371/journal.pone.0023882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tenzer S, Moro A, Kuharev J, et al. Proteome-wide characterization of the RNA-binding protein RALY-interactome using the in vivo-biotinylation-pulldown-quant (iBioPQ) approach. J. Proteome Res. 2013;12(6):2869–2884. doi: 10.1021/pr400193j. [DOI] [PubMed] [Google Scholar]
- 39.Yamazaki F, Kim HH, Lau P, et al. pY RNA1-s2: a highly retina-enriched small RNA that selectively binds to Matrin 3 (Matr3) PloS One. 2014;9(2):e88217. doi: 10.1371/journal.pone.0088217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coelho MB, Attig J, Bellora N, et al. Nuclear matrix protein Matrin3 regulates alternative splicing and forms overlapping regulatory networks with PTB. EMBO J. 2015;34(5):653–668. doi: 10.15252/embj.201489852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J. Biol. Chem. 2001;276(39):36337–36343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- 42.Avendaño-Vázquez SE, Dhir A, Bembich S, et al. Autoregulation of TDP-43 mRNA levels involves interplay between transcription, splicing, and alternative polyA site selection. Genes Dev. 2012;26(15):1679–1684. doi: 10.1101/gad.194829.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D’Alton S, Altshuler M, Lewis J. Studies of alternative isoforms provide insight into TDP-43 autoregulation and pathogenesis. RNA N. Y. N. 2015;21(8):1419–1432. doi: 10.1261/rna.047647.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ling JP, Pletnikova O, Troncoso JC, Wong PC. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science. 2015;349(6248):650–655. doi: 10.1126/science.aab0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamazaki T, Chen S, Yu Y, et al. FUS-SMN protein interactions link the motor neuron diseases ALS and SMA. Cell Rep. 2012;2(4):799–806. doi: 10.1016/j.celrep.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gerbino V, Carrì MT, Cozzolino M, Achsel T. Mislocalised FUS mutants stall spliceosomal snRNPs in the cytoplasm. Neurobiol. Dis. 2013;55:120–128. doi: 10.1016/j.nbd.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 47.Yu Y, Reed R. FUS functions in coupling transcription to splicing by mediating an interaction between RNAP II and U1 snRNP. Proc. Natl. Acad. Sci. U. S. A. 2015;112(28):8608–8613. doi: 10.1073/pnas.1506282112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5(10):R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Carlo V, Grossi E, Laneve P, et al. TDP-43 regulates the microprocessor complex activity during in vitro neuronal differentiation. Mol. Neurobiol. 2013;48(3):952–963. doi: 10.1007/s12035-013-8564-x. [DOI] [PubMed] [Google Scholar]
- 50.Dini Modigliani S, Morlando M, Errichelli L, et al. An ALS-associated mutation in the FUS 3’-UTR disrupts a microRNA-FUS regulatory circuitry. Nat. Commun. 2014;5:4335. doi: 10.1038/ncomms5335. [DOI] [PubMed] [Google Scholar]
- 51.Lourenco GF, Janitz M, Huang Y, Halliday GM. Long noncoding RNAs in TDP-43 and FUS/TLS-related frontotemporal lobar degeneration (FTLD) Neurobiol. Dis. 2015;82:445–454. doi: 10.1016/j.nbd.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 52.Fallini C, Bassell GJ, Rossoll W. The ALS disease protein TDP-43 is actively transported in motor neuron axons and regulates axon outgrowth. Hum. Mol. Genet. 2012;21(16):3703–3718. doi: 10.1093/hmg/dds205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alami NH, Smith RB, Carrasco MA, et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014;81(3):536–543. doi: 10.1016/j.neuron.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fujii R, Takumi T. TLS facilitates transport of mRNA encoding an actin-stabilizing protein to dendritic spines. J. Cell Sci. 2005;118(Pt 24):5755–5765. doi: 10.1242/jcs.02692. [DOI] [PubMed] [Google Scholar]
- 55.Belly A, Moreau-Gachelin F, Sadoul R, Goldberg Y. Delocalization of the multifunctional RNA splicing factor TLS/FUS in hippocampal neurones: exclusion from the nucleus and accumulation in dendritic granules and spine heads. Neurosci. Lett. 2005;379(3):152–157. doi: 10.1016/j.neulet.2004.12.071. [DOI] [PubMed] [Google Scholar]
- 56.Ishiguro A, Kimura N, Watanabe Y, et al. TDP-43 binds and transports G-quadruplex-containing mRNAs into neurites for local translation. Genes Cells Devoted Mol. Cell. Mech. 2016;21(5):466–481. doi: 10.1111/gtc.12352. [DOI] [PubMed] [Google Scholar]
- 57.Yasuda K, Zhang H, Loiselle D, et al. The RNA-binding protein Fus directs translation of localized mRNAs in APC-RNP granules. J. Cell Biol. 2013;203(5):737–746. doi: 10.1083/jcb.201306058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fiesel FC, Kahle PJ. TDP-43 and FUS/TLS: cellular functions and implications for neurodegeneration. FEBS J. 2011;278(19):3550–3568. doi: 10.1111/j.1742-4658.2011.08258.x. [DOI] [PubMed] [Google Scholar]
- 59.Iguchi Y, Katsuno M, Niwa J, et al. Loss of TDP-43 causes age-dependent progressive motor neuron degeneration. Brain J. Neurol. 2013;136(Pt 5):1371–1382. doi: 10.1093/brain/awt029. [DOI] [PubMed] [Google Scholar]
- 60.Kraemer BC, Schuck T, Wheeler JM, et al. Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol. (Berl.) 2010;119(4):409–419. doi: 10.1007/s00401-010-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sephton CF, Good SK, Atkin S, et al. TDP-43 is a developmentally regulated protein essential for early embryonic development. J. Biol. Chem. 2010;285(9):6826–6834. doi: 10.1074/jbc.M109.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu L-S, Cheng W-C, Hou S-C, et al. TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genes. N. Y. N 2000. 2010;48(1):56–62. doi: 10.1002/dvg.20584. [DOI] [PubMed] [Google Scholar]
- 63.Tsao W, Jeong YH, Lin S, et al. Rodent models of TDP-43: recent advances. Brain Res. 2012;1462:26–39. doi: 10.1016/j.brainres.2012.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stribl C, Samara A, Trümbach D, et al. Mitochondrial dysfunction and decrease in body weight of a transgenic knock-in mouse model for TDP-43. J. Biol. Chem. 2014;289(15):10769–10784. doi: 10.1074/jbc.M113.515940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ihara R, Matsukawa K, Nagata Y, et al. RNA binding mediates neurotoxicity in the transgenic Drosophila model of TDP-43 proteinopathy. Hum. Mol. Genet. 2013;22(22):4474–4484. doi: 10.1093/hmg/ddt296. [DOI] [PubMed] [Google Scholar]
- 66.Chou C-C, Alexeeva OM, Yamada S, et al. PABPN1 suppresses TDP-43 toxicity in ALS disease models. Hum. Mol. Genet. 2015;24(18):5154–5173. doi: 10.1093/hmg/ddv238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coyne AN, Yamada SB, Siddegowda BB, et al. Fragile X protein mitigates TDP-43 toxicity by remodeling RNA granules and restoring translation. Hum. Mol. Genet. 2015;24(24):6886–6898. doi: 10.1093/hmg/ddv389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arnold ES, Ling S-C, Huelga SC, et al. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc. Natl. Acad. Sci. U. S. A. 2013;110(8):E736–E745. doi: 10.1073/pnas.1222809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Daigle JG, Lanson NA, Smith RB, et al. RNA-binding ability of FUS regulates neurodegeneration, cytoplasmic mislocalization and incorporation into stress granules associated with FUS carrying ALS-linked mutations. Hum. Mol. Genet. 2013;22(6):1193–1205. doi: 10.1093/hmg/dds526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barmada SJ, Ju S, Arjun A, et al. Amelioration of toxicity in neuronal models of amyotrophic lateral sclerosis by hUPF1. Proc. Natl. Acad. Sci. U. S. A. 2015;112(25):7821–7826. doi: 10.1073/pnas.1509744112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kidd SA, Lachiewicz A, Barbouth D, et al. Fragile X syndrome: a review of associated medical problems. Pediatrics. 2014;134(5):995–1005. doi: 10.1542/peds.2013-4301. [DOI] [PubMed] [Google Scholar]
- 72.Nelson DL, Orr HT, Warren ST. The unstable repeats--three evolving faces of neurological disease. Neuron. 2013;77(5):825–843. doi: 10.1016/j.neuron.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vasilyev N, Polonskaia A, Darnell JC, et al. Crystal structure reveals specific recognition of a G-quadruplex RNA by a β-turn in the RGG motif of FMRP. Proc. Natl. Acad. Sci. U. S. A. 2015;112(39):E5391–E5400. doi: 10.1073/pnas.1515737112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Darnell JC, Van Driesche SJ, Zhang C, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146(2):247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.De Boulle K, Verkerk AJ, Reyniers E, et al. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat. Genet. 1993;3(1):31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- 76.Siomi H, Choi M, Siomi MC, et al. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell. 1994;77(1):33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 77.Schaeffer C, Bardoni B, Mandel JL, et al. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20(17):4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hagerman PJ, Hagerman RJ. Fragile X-associated tremor/ataxia syndrome. AnnNYAcad. Sci. 2015;1338:58–70. doi: 10.1111/nyas.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mohan A, Goodwin M, Swanson MS. RNA-protein interactions in unstable microsatellite diseases. Brain Res. 2014;1584:3–14. doi: 10.1016/j.brainres.2014.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cleary JD, Ranum LPW. Repeat-associated non-ATG (RAN) translation in neurological disease. Hum. Mol. Genet. 2013;22(R1):R45–R51. doi: 10.1093/hmg/ddt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Edbauer D, Haass C. An amyloid-like cascade hypothesis for C9orf72 ALS/FTD. Curr. Opin. Neurobiol. 2016;36:99–106. doi: 10.1016/j.conb.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 82.Orr HT. Cell biology of spinocerebellar ataxia. J. Cell Biol. 2012;197(2):167–177. doi: 10.1083/jcb.201105092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Satterfield TF, Pallanck LJ. Ataxin-2 and its Drosophila homolog, ATX2, physically assemble with polyribosomes. Hum. Mol. Genet. 2006;15(16):2523–2532. doi: 10.1093/hmg/ddl173. [DOI] [PubMed] [Google Scholar]
- 84.Nonhoff U, Ralser M, Welzel F, et al. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol. Biol. Cell. 2007;18(4):1385–1396. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gwinn-Hardy K, Chen JY, Liu HC, et al. Spinocerebellar ataxia type 2 with parkinsonism in ethnic Chinese. Neurology. 2000;55(6):800–805. doi: 10.1212/wnl.55.6.800. [DOI] [PubMed] [Google Scholar]
- 86.Elden AC, Kim H-J, Hart MP, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466(7310):1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang M-D, Gomes J, Cashman NR, et al. Intermediate CAG repeat expansion in the ATXN2 gene is a unique genetic risk factor for ALS--a systematic review and meta-analysis of observational studies. PloS One. 2014;9(8):e105534. doi: 10.1371/journal.pone.0105534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 2016;8(6):595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Karran E, De Strooper B. The amyloid cascade hypothesis: are we poised for success or failure? J. Neurochem. 2016 doi: 10.1111/jnc.13632. [DOI] [PubMed] [Google Scholar]
- 90.Sun X, Bromley-Brits K, Song W. Regulation of β-site APP-cleaving enzyme 1 gene expression and its role in Alzheimer’s disease. J. Neurochem. 2012;120(Suppl 1):62–70. doi: 10.1111/j.1471-4159.2011.07515.x. [DOI] [PubMed] [Google Scholar]
- 91.Yoon H, Flores LF, Kim J. MicroRNAs in brain cholesterol metabolism and their implications for Alzheimer’s disease. Biochim. Biophys. Acta. 2016 doi: 10.1016/j.bbalip.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nunez-Iglesias J, Liu C-C, Morgan TE, et al. Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer’s disease cortex reveals altered miRNA regulation. PloS One. 2010;5(2):e8898. doi: 10.1371/journal.pone.0008898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hébert SS, Horré K, Nicolaï L, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc. Natl. Acad. Sci. U. S. A. 2008;105(17):6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang W-X, Huang Q, Hu Y, et al. Patterns of microRNA expression in normal and early Alzheimer’s disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. (Berl.) 2011;121(2):193–205. doi: 10.1007/s00401-010-0756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shioya M, Obayashi S, Tabunoki H, et al. Aberrant microRNA expression in the brains of neurodegenerative diseases: miR-29a decreased in Alzheimer disease brains targets neurone navigator 3. Neuropathol. Appl. Neurobiol. 2010;36(4):320–330. doi: 10.1111/j.1365-2990.2010.01076.x. [DOI] [PubMed] [Google Scholar]
- 96.Langston JW, Schüle B, Rees L, et al. Multisystem Lewy body disease and the other parkinsonian disorders. Nat. Genet. 2015;47(12):1378–1384. doi: 10.1038/ng.3454. [DOI] [PubMed] [Google Scholar]
- 97.Kim J, Inoue K, Ishii J, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317(5842):1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Miñones-Moyano E, Porta S, Escaramís G, et al. MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum. Mol. Genet. 2011;20(15):3067–3078. doi: 10.1093/hmg/ddr210. [DOI] [PubMed] [Google Scholar]
- 99.Bonifati V. Autosomal recessive parkinsonism. Parkinsonism Relat. Disord. 2012;18(Suppl 1):S4–S6. doi: 10.1016/S1353-8020(11)70004-9. [DOI] [PubMed] [Google Scholar]
- 100.van der Brug MP, Blackinton J, Chandran J, et al. RNA binding activity of the recessive parkinsonism protein DJ-1 supports involvement in multiple cellular pathways. Proc. Natl. Acad. Sci. U. S. A. 2008;105(29):10244–10249. doi: 10.1073/pnas.0708518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cookson MR. The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson’s disease. Nat. Rev. Neurosci. 2010;11(12):791–797. doi: 10.1038/nrn2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gehrke S, Imai Y, Sokol N, Lu B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466(7306):637–641. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]