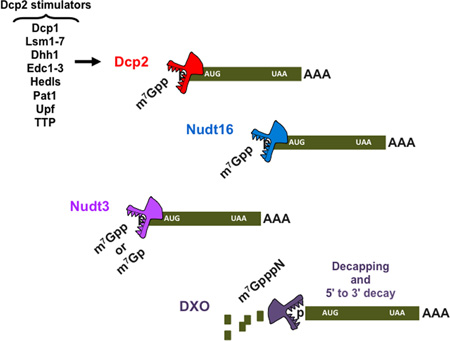
Abstract
Removal of the 5′ end cap is a critical determinant controlling mRNA stability and efficient gene expression. Removal of the cap is exquisitely controlled by multiple direct and indirect regulators that influence association with the cap and the catalytic step. A subset of these factors directly stimulate activity of the decapping enzyme, while others influence remodeling of factors bound to mRNA and indirectly stimulate decapping. Furthermore, the components of the general decapping machinery can also be recruited by mRNA-specific regulatory proteins to activate decapping. The Nudix hydrolase, Dcp2, identified as a first decapping enzyme, cleaves capped mRNA and initiates 5′ to 3′ degradation. Extensive studies on Dcp2 led to broad understanding of its activity and the regulation of transcript specific decapping and decay. Interestingly, seven additional Nudix proteins possess intrinsic decapping activity in vitro and at least two, Nudt16 and Nudt3, are decapping enzymes that regulate mRNA stability in cells. Furthermore, a new class of decapping proteins within the DXO family preferentially function on incompletely capped mRNAs. Importantly, it is now evident that each of the characterized decapping enzymes predominantly modulates only a subset of mRNAs, suggesting the existence of multiple decapping enzymes functioning in distinct cellular pathways.
Keywords: decapping, mRNA decay, Nudix proteins, Dcp2, Nudt3, Nudt16, DXO family
Graphical Abstract
Eukaryotic transcripts are capped at their 5′ ends by a 5′-5′ triphosphate bridged 7-methylguanosine to the first transcribed nucleotide of the mRNA chain1. The cap fulfills a variety of functions in all aspects of mRNA metabolism - synthesis, nucleo-cytoplasmic transport, translation, silencing and turnover2. The presence of the 5′ cap structure increases both the accuracy and efficiency of pre-mRNA splicing3, 4 and participates in mRNA processing and exports5. In the cytosol, the cap is specifically recognized by the translation initiation factor eIF4E and required for efficient mRNA translation6. The cap is also an important determinant of mRNA stability where capped mRNAs are more stable than uncapped transcripts7. The cap protects the RNA from 5′-3′ exonuclease decay8, 9 and its removal is a critical regulatory event that influences transcript expression10 and affects overall protein synthesis within the cell.
Dcp2 was initially discovered in yeast through a genetic screen11 and subsequently identified as the first eukaryotic decapping enzyme12–14. For almost a decade since its discovery, Dcp2 has been assumed to be the only enzyme responsible for the majority of eukaryotic mRNA decapping. Interestingly, the findings in recent years have demonstrated that at least three additional enzymes are also capable of removing the cap from an mRNA in cells15–17. Moreover, it is now evident that each decapping enzyme regulates only a small fraction of cellular transcripts suggesting that a number of distinct decapping complexes exist, each with their own substrate specificities and modulatory pathways. In this review, we will initially summarize the properties that confer transcript specificity to the extensively characterized Dcp2 protein and expand to our current understanding of additional decapping enzymes, Nud16 and Nudt3 as well as the DXO family of proteins that hydrolyze incompletely capped RNAs.
Regulators of Dcp2 decapping
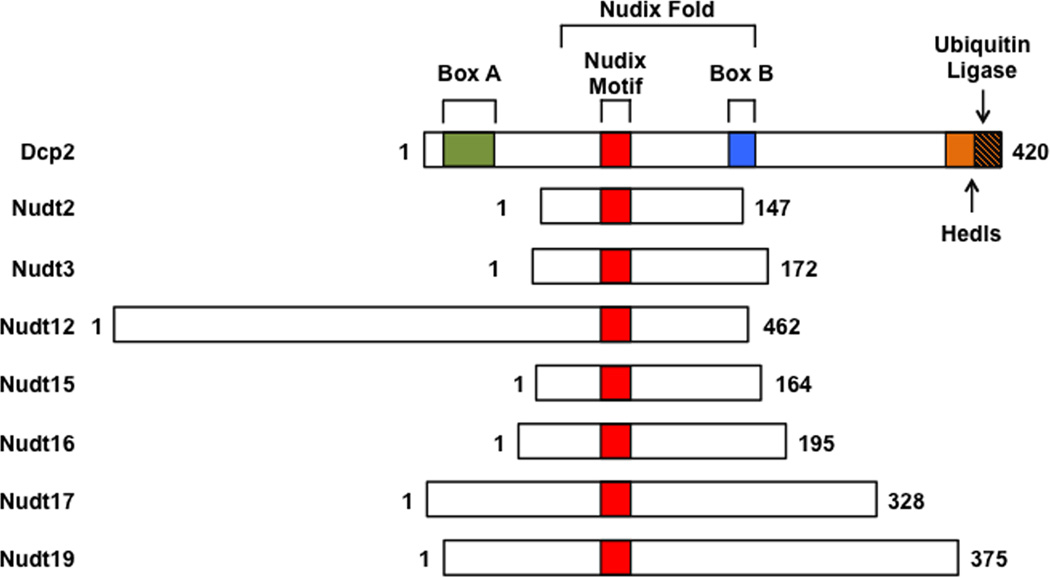
Dcp2 (Nudt20) is a member of the evolutionary conserved Nudix hydrolase superfamily of proteins that are known to catalyze hydrolysis of a wide range of small nucleotide substrates composed of a nucleoside diphosphate linked to another moiety X (Nudix)18. They contain a conserved Nudix motif with the consensus sequence GX5EX7REUXEEXGU (where U represents a hydrophobic residue, and X represents any amino acid), which forms part of the versatile catalytic site for diphosphate hydrolysis18 (Figure 1). The catalytic site within the Nudix motif of Dcp2 contains three conserved glutamate residues that coordinate a divalent cation essential for cap hydrolysis19, 20. Dcp2 also possesses a conserved region known as Box A and Box B (Figure 1). Box A is important for catalytic fidelity of decapping20 and at least in yeast, for an interaction with the decapping activator Dcp1p21 (Figure 2). Box B is required for the RNA binding property of Dcp2 and its removal decreases both RNA-binding and decapping activity in vitro20. Dcp2 cleaves between the α- and β-phosphate moieties of the cap generating m7GDP and 5′-end monophosphate RNA12, 14 (Figure 3). Importantly, the RNA body contributes to the Dcp2 substrate specificity20 and both the cap and an oligonucleotide chain of approximately 25 nucleotides are required for the enzyme activity19. For more extensive reviews on Dcp2 structure and its interaction with the cap see22–24.
Figure 1. Schematic representation of the Nudix proteins with decapping activity.
Nudix proteins with decapping activity are aligned relative to their Nudix motif. The human Dcp2 protein is schematically depicted at the top with the evolutionarily conserved domains, with Nudix Motif (red), Box A (green), Box B (blue) and regulatory domains encompassing the binding sites for Hedls (orange) and ubiquitin ligases (orange with stripes).
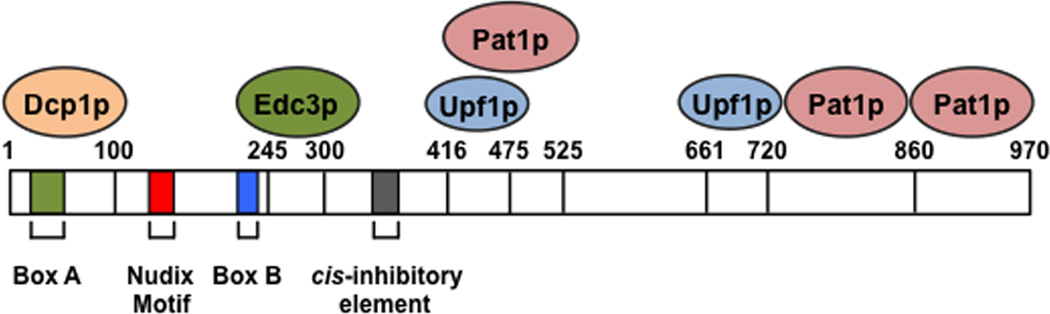
Figure 2. Schematic representation of the yeast Dcp2p decapping protein and its interacting partners.
S. cerevisiae Dcp2p is schematically depicted with Nudix Motif (red), Box A (green), Box B (blue), cis-inhibitory element (grey) and binding sites for Dcp1p, Edc3p, Pat1p and Upf1p interacting proteins denoted along with their corresponding amino acid regions numbered.
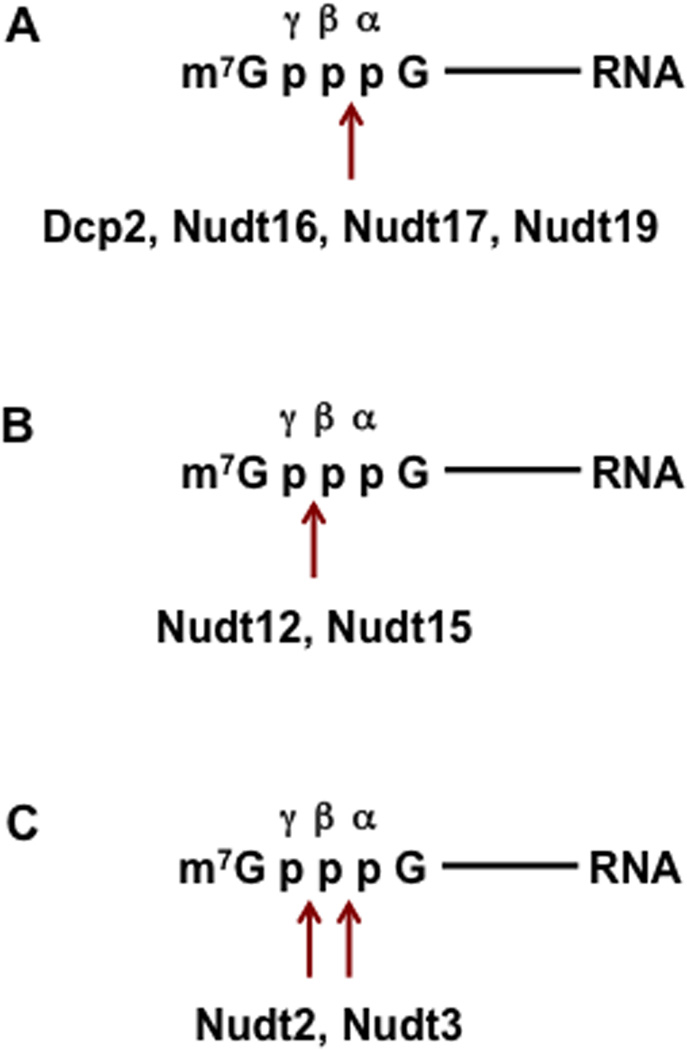
Figure 3. A summary of the predominant cleavage site(s) within the cap triphosphate bridge for Nudix proteins with decapping activity.
The position of the α, β and γ phosphates are shown with the cleave sites indicated by the arrow
In yeast, decapping is carried out by a single enzyme composed of a catalytic subunit (Dcp2p) and a regulatory subunit (Dcp1p) and facilitated by decapping activators25 (Figure 2). Based on the mechanistic models that rely on in vitro enzymatic assays, protein-protein interaction studies and structural analyses, the activators mainly function to promote decapping by inducing conformational changes in Dcp2p required for the decapping reaction26, 27. Structural analyses of a ternary complex consisting of Dcp2p-Dcp1p with the decapping enhancer Edc1p reveals the requirement of a rotation of the Dcp2p catalytic domain in the presence of Dcp1p and Edc1p to position Dcp2p and Dcp1p residues to bind the capped mRNA substrate explaining the decapping enhancement by Dcp1p and Edc1p28. Conserved from yeast to humans, Pat1p, Dhh1p, Edc3p and the Lsm1–7 complex activate general decapping whereas Upf1p, Upf2p and Upf3p are NMD-specific regulators. Edc1p and Edc2p decapping stimulators are present exclusively in yeast19. In metazoans, the Hedls protein (also known as Edc4 or Ge-1) enhances Dcp2 decapping in vitro by promoting a stable interaction of Dcp1 with Dcp229, 30. Pat1p, initially identified in yeast as a decapping activator31, interacts directly with Dcp1p32 to stimulate decapping33. Pat1p associates with the Lsm1p-7p protein complex and binds selectively to deadenylated mRNAs to promote decapping. The Dhh1p decapping activator (known as DDX6 or Rck/p54 in human and Me31B in Drosophila) was also initially identified in yeast where its depletion results in the accumulation of capped mRNAs34–36. Mammalian Dhh1 interacts with Dcp2, Edc3 and Pat1 decapping components to stimulate decapping32, 37, 38. Importantly, a point mutation in the human Edc3 that compromises its ability to activate Dcp2 decapping in vitro was identified in a family with cognitive disability39, indicating that Edc3 and Dcp2 contribute to neural functions.
There are at least two categories of decapping stimulatory proteins, general activators and transcript specific activators. In Drosophila S2 cells, Dcp1 and Edc4 regulate overlapping mRNAs indicating a general decapping stimulatory function, whereas Edc3 targets a group of transcripts distinct from Dcp1 and Edc440. Moreover, positive regulation of decapping can be mediated through interaction of the Dcp2 C-terminal domain with specific decapping activators that control substrate specificity and trigger Dcp2 enzyme activation (Figure 2). In S. cerevisiae, Edc3p specifically promotes decapping of the Rps28 mRNA41. Rps28p protein recognizes a conserved hairpin structure in the 3′ UTR of Rps28 mRNA and recruits Edc3p. The decapping is stimulated by an interaction of Edc3p with Dcp1p and Dcp2p34, 42, 43. Pat1p and Upf1p can also bind both their respective substrate mRNAs and distinct sites in Dcp2p44–47 (Figure 2), implying that these decapping activators directly control the targeting or substrate specificity of the decapping enzyme.
In addition to positive mediators of Dcp2p, Dcp2p is also subjected to a surprising negative regulatory circuit by a cis-inhibitory element in its C-terminus47 (Figure 2). Removal of the inhibitory element results in constitutive activation of Dcp2p decapping and loss of substrate mRNA specificity. In the presence of the inhibitory element, Edc3p is an obligate cofactor for Dcp2p-mediated YRA1 pre-mRNA decay. However, when the inhibitory element is removed, YRA1 pre-mRNA degradation could bypass its requirement for Edc3p. The negative element appears to inhibit the enzymatic or substrate-binding activity of Dcp2p whereas the positive elements promote the binding of specific decapping activators such as Edc3p, Pat1p, and Upf1p to Dcp2p to alleviate the inhibitory activity and promote formation of distinct decapping complexes.
mRNA cis-elements and RNA binding protein-mediated recruitment of Dcp2
Dcp2 is an RNA-binding protein that selectively binds a subset of mRNAs containing a stem-loop structure within 10 bases of the 5´ cap48. The structure, termed the Dcp2 binding and decapping element, was initially identified in the mRNAs including the mRNA encoding human exosome component, Rrp41 and promotes preferential Dcp2-directed decapping on a heterologous mRNA48. Importantly, the secondary structure, but not primary sequence, is critical for Dcp2 recognition and decapping49. A genome-wide analysis predicts several hundred human mRNAs could harbor a similar stem loop structure at their 5′ end.
In addition to direct RNA recruitment, cis elements can also serve as a platform to recruit RNA binding proteins and activate decapping. The AU-rich element (ARE) located in the 3′ UTRs of many mRNAs encoding transcription factors, cytokines and proto-oncogenes can recruit ARE-binding proteins and subject the mRNA to rapid decapping and degradation50. One well-characterized ARE-binding protein is tristetraprolin (TTP), which recruits Edc3 and Dcp251 and promotes ARE-dependent Dcp2 decapping activity in vitro29. Knockdown of Dcp2 reduces 5′-decay of c-fos ARE-containing mRNAs52 while overexpression of Dcp2 results in accelerated degradation of an mRNA containing the GM-CSF ARE29.
The Lsm1p-7p-Pat1p complex is important for the recruitment of the decapping complex to deadenylated mRNA and activation of decapping53. The purified Lsm1–7-Pat1p complex from yeast has an inherent ability to distinguish between oligoadenylated and polyadenylated mRNAs and binds preferentially to deadenylated transcripts containing a U-tract at their 3′-end44. In mammalian cells, a U-tract at the 3′-end of a generic RNA triggers Dcp2-mediated decapping in an Lsm1–7 complex-dependent manner54. Stimulation of decapping by oligouridylation is also observed for an endogenous mRNA. Mammalian histone mRNAs contain a stem-loop structure instead of a poly(A) tail at their 3′-end and undergo tailing by unconventional terminal ribonucleotidyltransferases55. Uridylation induces rapid decay of histone mRNAs through both a Dcp2 dependent 5′-3′ degradation and exosome mediated 3′-5′ degradation55–57. Interestingly, uridylation, which was initially detected at the 3′ ends of miRNA-directed cleavage products in Arabidopsis and mammalian cells58, also occurs on polyadenylated mRNAs and is a conserved feature of mRNA metabolism in eukaryotes59–63.
Dcp2 protein modification and controlled levels as a means to regulate decapping
In addition to the regulation of mRNA decapping by direct or protein-mediated indirect Dcp2 recruitment, modifications of the Dcp2 protein and its expression can also determine the fate of a subset of mRNAs. In particular, the C-terminus of human Dcp2 can act as a regulatory domain to modulate Dcp2 activity (Figure 1) by either promoting decapping complex assembly through Hedls or restricting Dcp2 levels by targeting uncomplexed Dcp2 for ubiquitin-mediated proteasome degradation64. Moreover, Dcp2 levels are limiting relative to Dcp2 decapping stimulatory proteins64 and ~10-fold less abundant then other decapping factors65, indicative of the importance of maintaining its expression in check within a cell.
A role for Dcp2 in autophagy provides another example of regulated Dcp2 decapping by protein modification. A conserved pathway in both yeast and humans negatively regulates autophagy through modulation of the autophagy-related (ATG) mRNA stability in an mTOR-dependent manner66. Under normal growth conditions RCK promotes Dcp2-dependent decapping of the ATG mRNA and subjects it to 5′-3′ degradation. mTOR-associated phosphorylation of Dcp2 is required for transcript decapping and subsequent degradation. Exposure to stress by nutrient deprivation inactivates mTOR and prevents Dcp2 phosphorylation leading to dissociation of RCK from the ATG mRNA. This, in turn, facilitates accumulation of ATG transcripts and induction of autophagy.
Levels of Dcp2 mRNA and protein are responsive to viral challenge in mouse embryonic fibroblast (MEF) cells where Dcp2 levels are elevated67. Importantly, MEF cells deficient in Dcp2 upregulate a group of innate immune response genes involved in type I IFN response67. In particular, the stability of a key transcription factor in the antiviral response, IFN regulatory factor 7 (IRF-7) mRNA is regulated by Dcp2 decapping. MEF cells defective in Dcp2 expression are more resistant to viral challenge than wild type MEF cells indicating Dcp2 may be involved in curtailing the innate immune response following activation67. Induction of Dcp2 expression is also important in the modulation of inflammatory response genes through the celiac disease-associated lnc13 RNA. In celiac patient macrophages, or lipopolysaccharide induced macrophages, Dcp2 levels are transcriptionally induced by nuclear factor kappa B (NF-κB) which initiates decay of lnc13 RNA68. Lnc13 represses the expression of a subset of inflammatory genes and reduction of lnc13 levels relieves the repression thus promoting the disease associated inflammatory response68. Whether regulation of Dcp2 decapping can be exploited for the amelioration of celiac disease remains to be determined.
Mammalian cells contain multiple mRNA decapping enzymes
Since its discovery, Dcp2 has been postulated as the major decapping enzyme in eukaryotic cells. However, recent studies have demonstrated that Dcp2 is not the default all encompassing decapping enzyme in multicellular organisms, but is one member of a growing list of decapping enzymes. Several lines of evidence were critical for the postulate that multiple decapping enzymes likely exist in mammals. First, Dcp2 is an RNA binding protein that must bind to the 5′ end of an RNA to recognize and hydrolyze the cap14, 19, 20. As an RNA-binding protein, it would be expected to bind different RNA sequences/structures with different affinities and thus preferentially hydrolyze different mRNAs. This hypothesis was substantiated by the demonstration that Dcp2 can selectively regulate a subset of mRNAs possessing a Dcp2 element at their 5′ end48, 49. Second, Dcp2 is not ubiquitously expressed in all mouse tissues. Of the eight mouse tissues tested, Dcp2 protein is highly expressed in brain and testes, detected in lung and spleen but devoid in liver, muscle, heart and kidney15. Moreover, Dcp2 is developmentally regulated. In two adult tissues that do not express detectable levels of Dcp2 protein, heart and liver, robust Dcp2 expression is detected in mouse embryos and the levels decrease to undetectable upon birth15. In contrast, Dcp2 can be detected in the brain throughout development. Most significantly, MEF cell lines either containing or lacking measurable Dcp2 expression are equally competent to decap exogenous RNA substrates introduced into these cells15 indicating that decapping proteins other than Dcp2 are also functional in mammalian cells.
Nudt16 is an mRNA decapping enzyme
The first direct demonstration that mammalian mRNA 5′-end degradation is more complex and extends beyond just Dcp2-directed decapping came with the finding that Nudt16 can function as a decapping enzyme in cells. Nudt16 is a nucleolar U8 snoRNA binding protein69 that possesses decapping activity and was initially proposed to be a U8 snoRNA specific nucleolar RNA decapping enzymes69, 70. More recently analysis of mouse Nudt16 uncovered it is ubiquitously expressed across mouse tissues, predominantly expressed in the cytoplasm and is involved in mRNA decapping15, 71.
Consistent with the premise of multiple mRNA decapping enzymes that each function on a subset of mRNAs, Dcp2 and Nudt16 uniquely influence the stability of only a subset of mRNA transcripts. Global profiling of mRNA in Nudt16 knockdown cells following one and four-hour transcriptional arrest identified 40 and 134 mRNAs, respectively whose stabilities increase15. Importantly mRNAs responsive to Nudt16 are minimally responsive to Dcp2 and vice versa. Furthermore, it appears Dcp2 preferentially functions on more labile mRNAs since a greater number of mRNAs are altered at the one-hour transcriptional arrest while Nudt16 may function on more long lived mRNAs that are apparent after four hours of transcriptional arrest15. The two decapping enzymes also appear to differentially function in different decay mechanisms, where nonsense-mediated mRNA decay predominantly utilizes Dcp2, while degradation of ARE containing mRNAs and miRNA-mediated silencing can use either Dcp2 or Nudt1672. Decapping, at least in mammals, appears to involve multiple enzymes that catalyze hydrolysis of the mRNA cap and each preferentially function on distinct subsets of transcripts.
Multiple Nudix proteins can decap RNA in vitro
There are 22 Nudix proteins in the human genome, with each protein reported to hydrolyze distinct and overlapping substrates including dNTPs, diadenosine polyphosphates, nucleotide sugars73 and in the case of Dcp2 (Nudt20) and Nudt16, capped mRNAs74. Six additional Nudix proteins (Nudt2, Nudt3, Nudt12, Nudt15, Nudt16, Nudt17 and Nudt19 shown schematically in Figure 1) have the capacity to decap RNA in vitro74. Similar to Dcp2 and Nudt16, Nudt17 and Nudt19 primarily cleave the pyrophosphate linkage between the α-and β-phosphates to generate m7GDP + pRNA (Figure 3A). Nudt12 and Nudt15 hydrolyze the β-γ-phosphate bond to generate m7GMP + ppRNA products (Figure 3B). Nudt2 and Nudt3 cleave between the α-β- or the β-γ-phosphates with comparable activity (Figure 3C)74. The function of ppRNA in cells is currently unknown. Since all known 5´ to 3´ exoribonucleases utilize a 5´ end monophosphorylated RNA substrate, ppRNAs are unlikely to be direct targets for 5´ end decay. Alternatively, they may be substrates for cytoplasmic recapping that utilizes ppRNA75. The reported accumulation of uncapped and untranslated mRNA76 may be constituted by decapping generated ppRNA maintained in a silenced state prior to restoration into the translating pool by cytoplasmic recapping.
With the exception of Nudt12 and Nudt16, all the decapping competent Nudix proteins only hydrolyze a cap that is at the 5′ end of an RNA and not free cap structure74. The requirement of an RNA for the majority of the Nudix decapping enzymes suggests that like Dcp2, they are also RNA-binding proteins and likely selectively function on specific mRNAs as is also the case for Nudt1615 and Nudt3 (17 and below). In addition to RNA decapping, Nudt12 and Nudt16 can hydrolyze m7GpppG cap structure in vitro74 and Nudt16 can also hydrolyze inosine diphosphate (IDP) and its cognate deoxyribose (dIDP) into IMP and Pi77, 78. Cap structure decapping is a well-characterized function of the scavenger decapping protein, DcpS79–81. DcpS is a member of the histidine triad family of proteins80 that cleaves m7GpppG to m7Gp + ppG. The Fragile Histidine Triad (FHIT) protein can also hydrolyze cap structure in vitro82. However, the significance of Nudt12, Nudt16 and FHIT in hydrolyzing cap structure in cells remains an open question considering all three enzymes have several orders of magnitude lower cap structure decapping activity than does DcpS (74 and unpublished).
Surprisingly, decapping is not restricted to eukaryotic proteins. The bacterial Nudix protein RppH, which accelerates decay of E. coli mRNA decay by converting their triphosphorylated 5′ ends to monophosphorylated form83, can hydrolyze m7G capped RNA exclusively between the α-and β-phosphates to release m7GDP + pRNA74. Similar to the eukaryotic decapping enzymes above, it requires a cap substrate linked to an RNA moiety and does not function on cap structure alone. The significance of this observation is currently unclear since prokaryotes lack an m7G cap. However, the recent demonstration that bacterial mRNAs can possess a nicotinamide adenine dinucleotide (NAD) on their 5′ end and the ability of a bacterial Nudix protein, NudC, to hydrolyze NAD at the 5´ end of an RNA84 suggests there might yet exist additional 5′ end modifications in prokaryotes that are targeted by RppH. Moreover, the RppH decapping activity also indicates the ability of Nudix proteins to hydrolyze a triphosphorylated nucleoside at the 5′ end evolved prior to eukaryotic decapping.
Nudt3 is an mRNA decapping enzyme
Nudt3 is a cytoplasmic protein that, along with Nudt4, Nudt10, and Nudt11, belongs to the diphosphoinositol polyphosphate phosphohydrolase (DIPP) family of proteins and can hydrolyze diphosphoinositol polyphosphates (DIPs) and ApnA dinucleotides73. However, only Nudt3 contains decapping activity in vitro74 and in cells17. Moreover, the S. cerevisiae ortholog, Ddp1p, contains robust decapping activity in vitro74 suggesting that like Nudt3, Ddp1p may be a decapping enzyme in yeast.
A genome-wide RNA-seq analysis performed in MCF-7 cells with reduced levels of Nudt3 reveals that, similarly to Dcp2 and Nudt16, steady state levels of a small subset of RNAs are altered as a function of Nudt3 with a disproportional percentage of the responsive mRNAs involved in cell motility17. Intriguingly, many of the mRNAs regulated by Nudt3 are either cell surface receptors or secreted extracellular proteins involved in cell migration, suggesting an important role for Nudt3 in controlling cell motility. Indeed, reduction of Nudt3 levels in MCF7 cells promotes enhanced lamellipodia/filopodia formation and corresponding increase in cell migration17.
Of the four characterized Nudt3 responsive mRNAs involved in cell motility, Nudt3 decapping is directly involved in the stability of integrin β6 and lipocalin-2 mRNAs. A similar alteration in mRNA levels are not observed in cells knocked down for the related Nudt4 protein which demonstrates the changes in integrin β6 and lipocalin-2 mRNAs are not due to DIPP or ApnA dinucleotide hydrolysis activities shared by Nudt3 and Nudt473, but rather due to the unique decapping property of Nudt3. Interestingly, the steady state increase in the remaining two mRNAs, fibronectin and S100A8, following Nudt3 knock down are independent of mRNA stability and likely a consequence of indirect modulation by Nudt3 decapping.
DXO Family of Decapping Enzymes
Historically, the cotranscriptional addition of the N7-methyl guanosine cap to a nascent pre-mRNA was thought to be a default process that always proceeded to completion. This perception was upended by the identification of a novel class of enzymes that specifically recognize and remove incomplete caps at the 5′ end of an mRNA in a capping quality control mechanism (reviewed in85, 86). In S. cerevisiae, Rai1p which exists in a heterodimeric complex with the nuclear 5′-3′ exonuclease Rat187 possesses RNA 5′ pyrophosphohydrolase (PPH) activity, releasing pyrophosphate (PPi) from 5′ triphosphorylated RNA88 (Table 1). Interestingly, Rai1p homologs from Ashbya gossypii, Candida glabrata and Scheffersomyces stipites, remove the entire first nucleotide of an RNA 5′ triphosphate with a 5′ triphosphonucleotide hydrolase (TPH) activity, releasing pppN + RNA from 5′ pppN-RNA89. All the fungal Rai1 proteins tested thus far also cleave incompletely capped GpppN-RNA releasing unmethylated GpppN16 (Table 1), in contrast to the canonical decapping enzymes, which cleave within the pyrophosphate linkage (see above). All the Rai1p activities generate an RNA with a 5′ monophosphate that is subsequently degraded by Rat1p. Furthermore, both Rai1p and Rat1p mutually stimulate each others activity.
Table 1.
Summary of DXO family member biochemical activities
| Activity | PPH | TPH | Decapping | 5’-3’ Exo | |
|---|---|---|---|---|---|
| Substrate | pppRNA | pppRNA | m7GpppRNA | GpppRNA | pRNA |
| Cleavage site | |||||
| Products | PPi + pRNA |
pppN + pRNA |
m7GpppN + pRNA |
GpppN + pRNA |
pN + pRNA |
| A.gossypii Rai1 | − | ++ | ++ | ++ | ++ |
| C.glabrata Rai1 | − | + | − | + | − |
| S.pombe Rai1 | + | − | − | + | − |
| K.lactis Dxo1 | − | − | ++ | ++ | ++ |
| M.musculus DXO | ++ | − | ++ | ++ | ++ |
+ ≤ 50% activity; ++ ≥ 50% activity
Data derived from89
Yeast also contain a second protein with extensive structural identity to Rai1 termed Decapping exonuclease 1(Dxo1p)90. S. cerevisiae Dxo1p is predominantly a cytoplasmic protein91 and similar to Rai1, the K. lactis Dxo1 can preferentially hydrolyze an unmethylated cap from an mRNA, but lacks PPH activity (Table 1). Surprisingly, K. lactis Dxo1p also has intrinsic 5′-3′ exonuclease activity and can single handedly both decap and degrade an incompletely capped RNA90 an outcome analogous to the Rai1p-Rat1p heterodimer within the cytoplasm. Most significantly, aberrantly capped mRNAs accumulate in rai1Δ S. cerevisiae under stress conditions16, or rai1Δ dxo1Δ double disrupted S. cerevisiae, suggesting capping is a regulated process and Rai1p and Dxo1p function in a capping quality control mechanism to ensure 5′ end fidelity.
Mammalian cells appear to have one homolog of Rai1p and Dxo1p, termed DXO (formerly DOM3Z) and it possesses the cumulative actives of both proteins. It contains PPH activity to cleave between the α-β-phosphates of the triphosphate 5′ end, it hydrolyzes unmethylated incompletely capped RNA, and possesses 5′-3′exonuclease activity92 (Table 1). The decapping activity of DXO does not discriminate between methylated and unmethylated caps in vitro. However, cap binding proteins inhibit DXO activity92 and likely protect the mature, methylated cap from DXO-mediated degradation. Surprisingly, DXO preferentially degrades incompletely capped pre-mRNAs in cells suggesting a role in pre-mRNA quality surveillance. Of particular note, incompletely capped pre-mRNAs are impaired in their efficiency to splice internal introns which extends earlier observations that demonstrated a role for the 5′ cap in splicing of the first intron3, 4, 93 and provides a link between 5′ capping quality and subsequent splicing.
Conclusion
Regulation of mRNA 5′ end decapping occurs at multiple levels. First is the availability of the decapping enzyme, whether it is constitutively or temporally expressed and its presence in a particular cell type. Second is its recognition of the mRNA substrate which involves both direct and indirect association with the 5′ end cap and modulatory proteins that influence the access and activity of the enzyme. Studies focused on Dcp2 have lead to an extensive understanding of its activity and function including the surprising preferential targeting of only a small fraction of cellular mRNAs, in particular, mRNAs involved in the innate immune response. More recently, it has become apparent that mammalian cells possess a multitude of decapping enzymes in addition to Dcp2. Of the seven additional Nudix proteins with intrinsic decapping activity, two thus far have been shown to function as decapping enzymes that modulate mRNA stability in cells, Nudt3 and Nudt16. Similar to Dcp2, both do not indiscriminately function on all mRNAs and preferentially target a select subset. Furthermore, a new class of decapping enzymes within the DXO family preferentially function on incompletely capped mRNAs. It is anticipated that the remaining Nudix decapping-competent enzymes are also cellular decapping enzymes that function on select mRNAs with each decapping protein modulating a subset of distinct cellular pathways.
The identification of multiple decapping proteins raises intriguing future areas of inquiries. Considering the number of regulatory proteins involved in Dcp2 decapping (Figure 2), it is tempting to speculate additional confirmed decapping proteins will have both stimulatory and inhibitory proteins that control their activity. Whether each will have unique or overlapping associated proteins remains to be determined. Are the enzymes polysome associated and are they active when polysome bound? Does decay occur in a specific subcellular location or throughout the cytoplasm? These and many other questions can now be addressed with Nudt3 and Nudt16 and others as they are also confirmed as decapping enzymes. The presence of numerous modified nucleotides, such as pseudouridine, 5-methylcytidine, 5-hydrozymethylcytidine, and N6-methyladenosine94–97 in RNA provides further potential avenues of transcript specific degradation by decapping enzymes. In particular, modifications of the first encoded nucleotide following the cap would be predicted to have a disproportional influence. The last one and a half decades since the demonstration of Dcp2 as a decapping enzyme has provided a tremendous wealth of unexpected findings. Nevertheless, it is anticipated that we are only at the beginning phase of understanding all the decapping components that function in cells and the physiological roles that they fulfill.
Acknowledgments
This work was supported by NIH grant GM065007 to MK.
Footnotes
Conflict of Interest statement. The authors have no conflict to declare.
References
- 1.Muthukrishnan S, Both GW, Furuichi Y, Shatkin AJ. 5'-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature. 1975;255:33–37. doi: 10.1038/255033a0. [DOI] [PubMed] [Google Scholar]
- 2.Topisirovic I, Svitkin YV, Sonenberg N, Shatkin AJ. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev RNA. 2011;2:277–298. doi: 10.1002/wrna.52. [DOI] [PubMed] [Google Scholar]
- 3.Konarska MM, Padgett RA, Sharp PA. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984;38:731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- 4.Edery I, Sonenberg N. Cap-dependent RNA splicing in a HeLa nuclear extract. Proc Natl Acad Sci U S A. 1985;82:7590–7594. doi: 10.1073/pnas.82.22.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Visa N, Izaurralde E, Ferreira J, Daneholt B, Mattaj IW. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J Cell Biol. 1996;133:5–14. doi: 10.1083/jcb.133.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonenberg N, Rupprecht KM, Hecht SM, Shatkin AJ. Eukaryotic mRNA cap binding protein: purification by affinity chromatography on sepharose-coupled m7GDP. Proc Natl Acad Sci U S A. 1979;76:4345–4349. doi: 10.1073/pnas.76.9.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furuichi Y, LaFiandra A, Shatkin AJ. 5'-Terminal structure and mRNA stability. Nature. 1977;266:235–239. doi: 10.1038/266235a0. [DOI] [PubMed] [Google Scholar]
- 8.Hsu CL, Stevens A. Yeast cells lacking 5'-->3' exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5' cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sachs AB. Messenger RNA degradation in eukaryotes. Cell. 1993;74:413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- 10.Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunckley T, Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci U S A. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song MG, Li Y, Kiledjian M. Multiple mRNA decapping enzymes in mammalian cells. Mol Cell. 2010;40:423–432. doi: 10.1016/j.molcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiao X, Xiang S, Oh C, Martin CE, Tong L, Kiledjian M. Identification of a quality-control mechanism for mRNA 5'-end capping. Nature. 2010;467:608–611. doi: 10.1038/nature09338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grudzien-Nogalska E, Jiao X, Song MG, Hart RP, Kiledjian M. Nudt3 is an mRNA decapping enzyme that modulates cell migration. RNA. 2016 doi: 10.1261/rna.055699.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bessman MJ, Frick DN, O'Handley SF. The MutT proteins or "Nudix" hydrolases, a family of versatile, widely distributed, "housecleaning" enzymes. J Biol Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 19.Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R. Analysis of recombinant yeast decapping enzyme. RNA. 2003;9:231–238. doi: 10.1261/rna.2151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piccirillo C, Khanna R, Kiledjian M. Functional characterization of the mammalian mRNA decapping enzyme hDcp2. Rna. 2003;9:1138–1147. doi: 10.1261/rna.5690503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.She M, Decker CJ, Svergun DI, Round A, Chen N, Muhlrad D, Parker R, Song H. Structural basis of dcp2 recognition and activation by dcp1. Mol Cell. 2008;29:337–349. doi: 10.1016/j.molcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Kiledjian M. Regulation of mRNA decapping. Wiley Interdiscip Rev RNA. 2010;1:253–265. doi: 10.1002/wrna.15. [DOI] [PubMed] [Google Scholar]
- 23.Ling SH, Qamra R, Song H. Structural and functional insights into eukaryotic mRNA decapping. Wiley Interdiscip Rev RNA. 2011;2:193–208. doi: 10.1002/wrna.44. [DOI] [PubMed] [Google Scholar]
- 24.Arribas-Layton M, Wu D, Lykke-Andersen J, Song H. Structural and functional control of the eukaryotic mRNA decapping machinery. Biochim Biophys Acta. 2013;1829:580–589. doi: 10.1016/j.bbagrm.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker R. RNA degradation in Saccharomyces cerevisae. Genetics. 2012;191:671–702. doi: 10.1534/genetics.111.137265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Floor SN, Jones BN, Hernandez GA, Gross JD. A split active site couples cap recognition by Dcp2 to activation. Nat Struct Mol Biol. 2010;17:1096–1101. doi: 10.1038/nsmb.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borja MS, Piotukh K, Freund C, Gross JD. Dcp1 links coactivators of mRNA decapping to Dcp2 by proline recognition. RNA. 2011;17:278–290. doi: 10.1261/rna.2382011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valkov E, Muthukumar S, Chang CT, Jonas S, Weichenrieder O, Izaurralde E. Structure of the Dcp2-Dcp1 mRNA-decapping complex in the activated conformation. Nat Struct Mol Biol. 2016 doi: 10.1038/nsmb.3232. [DOI] [PubMed] [Google Scholar]
- 29.Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 30.Xu J, Yang JY, Niu QW, Chua NH. Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell. 2006;18:3386–3398. doi: 10.1105/tpc.106.047605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatfield L, Beelman CA, Stevens A, Parker R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nissan T, Rajyaguru P, She M, Song H, Parker R. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol Cell. 2010;39:773–783. doi: 10.1016/j.molcel.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 34.Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischer N, Weis K. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 2002;21:2788–2797. doi: 10.1093/emboj/21.11.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haas G, Braun JE, Igreja C, Tritschler F, Nishihara T, Izaurralde E. HPat provides a link between deadenylation and decapping in metazoa. J Cell Biol. 2010;189:289–302. doi: 10.1083/jcb.200910141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ozgur S, Chekulaeva M, Stoecklin G. Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies. Mol Cell Biol. 2010;30:4308–4323. doi: 10.1128/MCB.00429-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahmed I, Buchert R, Zhou M, Jiao X, Mittal K, Sheikh TI, Scheller U, Vasli N, Rafiq MA, Brohi MQ, et al. Mutations in DCPS and EDC3 in autosomal recessive intellectual disability indicate a crucial role for mRNA decapping in neurodevelopment. Hum Mol Genet. 2015;24:3172–3180. doi: 10.1093/hmg/ddv069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Badis G, Saveanu C, Fromont-Racine M, Jacquier A. Targeted mRNA degradation by deadenylation-independent decapping. Mol Cell. 2004;15:5–15. doi: 10.1016/j.molcel.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 42.Tritschler F, Eulalio A, Truffault V, Hartmann MD, Helms S, Schmidt S, Coles M, Izaurralde E, Weichenrieder O. A divergent Sm fold in EDC3 proteins mediates DCP1 binding and P-body targeting. Mol Cell Biol. 2007;27:8600–8611. doi: 10.1128/MCB.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harigaya Y, Jones BN, Muhlrad D, Gross JD, Parker R. Identification and analysis of the interaction between Edc3 and Dcp2 in Saccharomyces cerevisiae. Mol Cell Biol. 2010;30:1446–1456. doi: 10.1128/MCB.01305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chowdhury A, Mukhopadhyay J, Tharun S. The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA. 2007;13:998–1016. doi: 10.1261/rna.502507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johansson MJ, He F, Spatrick P, Li C, Jacobson A. Association of yeast Upf1p with direct substrates of the NMD pathway. Proc Natl Acad Sci U S A. 2007;104:20872–20877. doi: 10.1073/pnas.0709257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He F, Li C, Roy B, Jacobson A. Yeast Edc3 targets RPS28B mRNA for decapping by binding to a 3' untranslated region decay-inducing regulatory element. Mol Cell Biol. 2014;34:1438–1451. doi: 10.1128/MCB.01584-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He F, Jacobson A. Control of mRNA decapping by positive and negative regulatory elements in the Dcp2 C-terminal domain. RNA. 2015;21:1633–1647. doi: 10.1261/rna.052449.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Song MG, Kiledjian M. Transcript-specific decapping and regulated stability by the human Dcp2 decapping protein. Mol Cell Biol. 2008;28:939–948. doi: 10.1128/MCB.01727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y, Ho ES, Gunderson SI, Kiledjian M. Mutational analysis of a Dcp2-binding element reveals general enhancement of decapping by 5'-end stem-loop structures. Nucleic Acids Res. 2009;37:2227–2237. doi: 10.1093/nar/gkp087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murray EL, Schoenberg DR. A+U-rich instability elements differentially activate 5'-3' and 3'-5' mRNA decay. Mol Cell Biol. 2007;27:2791–2799. doi: 10.1128/MCB.01445-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 54.Song M, Kiledjian M. 3' Terminal oligo U-tract-mediated stimulation of decapping. RNA. 2007;13:2356–2365. doi: 10.1261/rna.765807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5' to 3' and 3' to 5'. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoefig KP, Rath N, Heinz GA, Wolf C, Dameris J, Schepers A, Kremmer E, Ansel KM, Heissmeyer V. Eri1 degrades the stem-loop of oligouridylated histone mRNAs to induce replication-dependent decay. Nat Struct Mol Biol. 2013;20:73–81. doi: 10.1038/nsmb.2450. [DOI] [PubMed] [Google Scholar]
- 57.Slevin MK, Meaux S, Welch JD, Bigler R, Miliani de Marval PL, Su W, Rhoads RE, Prins JF, Marzluff WF. Deep sequencing shows multiple oligouridylations are required for 3' to 5' degradation of histone mRNAs on polyribosomes. Mol Cell. 2014;53:1020–1030. doi: 10.1016/j.molcel.2014.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen B, Goodman HM. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- 59.Chang H, Lim J, Ha M, Kim VN. TAIL-seq: genome-wide determination of poly(A) tail length and 3' end modifications. Mol Cell. 2014;53:1044–1052. doi: 10.1016/j.molcel.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 60.Knusel S, Roditi I. Insights into the regulation of GPEET procyclin during differentiation from early to late procyclic forms of Trypanosoma brucei. Mol Biochem Parasitol. 2013;191:66–74. doi: 10.1016/j.molbiopara.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 61.Rissland OS, Norbury CJ. Decapping is preceded by 3' uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol. 2009;16:616–623. doi: 10.1038/nsmb.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sement FM, Ferrier E, Zuber H, Merret R, Alioua M, Deragon JM, Bousquet-Antonelli C, Lange H, Gagliardi D. Uridylation prevents 3' trimming of oligoadenylated mRNAs. Nucleic Acids Res. 2013;41:7115–7127. doi: 10.1093/nar/gkt465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomas MP, Liu X, Whangbo J, McCrossan G, Sanborn KB, Basar E, Walch M, Lieberman J. Apoptosis Triggers Specific, Rapid, and Global mRNA Decay with 3' Uridylated Intermediates Degraded by DIS3L2. Cell Rep. 2015;11:1079–1089. doi: 10.1016/j.celrep.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Erickson SL, Corpuz EO, Maloy JP, Fillman C, Webb K, Bennett EJ, Lykke-Andersen J. Competition between Decapping Complex Formation and Ubiquitin-Mediated Proteasomal Degradation Controls Human Dcp2 Decapping Activity. Mol Cell Biol. 2015;35:2144–2153. doi: 10.1128/MCB.01517-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Paabo S, Mann M. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hu G, McQuiston T, Bernard A, Park YD, Qiu J, Vural A, Zhang N, Waterman SR, Blewett NH, Myers TG, et al. A conserved mechanism of TOR-dependent RCK-mediated mRNA degradation regulates autophagy. Nat Cell Biol. 2015;17:930–942. doi: 10.1038/ncb3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Y, Dai J, Song M, Fitzgerald-Bocarsly P, Kiledjian M. Dcp2 Decapping Protein Modulates mRNA Stability of the Critical Interferon Regulatory Factor (IRF) IRF-7. Mol Cell Biol. 2012;32:1164–1172. doi: 10.1128/MCB.06328-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Castellanos-Rubio A, Fernandez-Jimenez N, Kratchmarov R, Luo X, Bhagat G, Green PH, Schneider R, Kiledjian M, Bilbao JR, Ghosh S. A long noncoding RNA associated with susceptibility to celiac disease. Science. 2016;352:91–95. doi: 10.1126/science.aad0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ghosh T, Peterson B, Tomasevic N, Peculis BA. Xenopus U8 snoRNA Binding Protein Is a Conserved Nuclear Decapping Enzyme. Mol Cell. 2004;13:817–828. doi: 10.1016/s1097-2765(04)00127-3. [DOI] [PubMed] [Google Scholar]
- 70.Taylor MJ, Peculis BA. Evolutionary conservation supports ancient origin for Nudt16, a nuclear-localized, RNA-binding, RNA-decapping enzyme. Nucleic Acids Res. 2008;36:6021–6034. doi: 10.1093/nar/gkn605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu G, Zhang J, Li Y, Li Z, Zhang N, Xu X, Wang T, Guan Z, Gao GF, Yan J. hNUDT16: a universal decapping enzyme for small nucleolar RNA and cytoplasmic mRNA. Protein Cell. 2011;2:64–73. doi: 10.1007/s13238-011-1009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Y, Song M, Kiledjian M. Differential utilization of decapping enzymes in mammalian mRNA decay pathways. RNA. 2011;17:419–428. doi: 10.1261/rna.2439811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McLennan AG. The Nudix hydrolase superfamily. Cell Mol Life Sci. 2006;63:123–143. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Song MG, Bail S, Kiledjian M. Multiple Nudix family proteins possess mRNA decapping activity. RNA. 2013;19:390–399. doi: 10.1261/rna.037309.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Otsuka Y, Kedersha NL, Schoenberg DR. Identification of a cytoplasmic complex that adds a cap onto 5'-monophosphate RNA. Mol Cell Biol. 2009;29:2155–2167. doi: 10.1128/MCB.01325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mukherjee C, Patil DP, Kennedy BA, Bakthavachalu B, Bundschuh R, Schoenberg DR. Identification of cytoplasmic capping targets reveals a role for cap homeostasis in translation and mRNA stability. Cell Rep. 2012;2:674–684. doi: 10.1016/j.celrep.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Iyama T, Abolhassani N, Tsuchimoto D, Nonaka M, Nakabeppu Y. NUDT16 is a (deoxy)inosine diphosphatase, and its deficiency induces accumulation of single-strand breaks in nuclear DNA and growth arrest. Nucleic Acids Res. 2010;38:4834–4843. doi: 10.1093/nar/gkq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Abolhassani N, Iyama T, Tsuchimoto D, Sakumi K, Ohno M, Behmanesh M, Nakabeppu Y. NUDT16 and ITPA play a dual protective role in maintaining chromosome stability and cell growth by eliminating dIDP/IDP and dITP/ITP from nucleotide pools in mammals. Nucleic Acids Res. 2010;38:2891–2903. doi: 10.1093/nar/gkp1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang Z, Kiledjian M. Functional Link between the Mammalian Exosome and mRNA Decapping. Cell. 2001;107:751–762. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]
- 80.Liu H, Rodgers ND, Jiao X, Kiledjian M. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 2002;21:4699–4708. doi: 10.1093/emboj/cdf448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gu M, Fabrega C, Liu SW, Liu H, Kiledjian M, Lima CD. Insights into the structure, mechanism, and regulation of scavenger mRNA decapping activity. Mol Cell. 2004;14:67–80. doi: 10.1016/s1097-2765(04)00180-7. [DOI] [PubMed] [Google Scholar]
- 82.Taverniti V, Seraphin B. Elimination of cap structures generated by mRNA decay involves the new scavenger mRNA decapping enzyme Aph1/FHIT together with DcpS. Nucleic Acids Res. 2015;43:482–492. doi: 10.1093/nar/gku1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5' pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- 84.Cahova H, Winz ML, Hofer K, Nubel G, Jaschke A. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature. 2015;519:374–377. doi: 10.1038/nature14020. [DOI] [PubMed] [Google Scholar]
- 85.Jurado AR, Tan D, Jiao X, Kiledjian M, Tong L. Structure and function of pre-mRNA 5'-end capping quality control and 3'-end processing. Biochemistry. 2014;53:1882–1898. doi: 10.1021/bi401715v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhai LT, Xiang S. mRNA quality control at the 5' end. J Zhejiang Univ Sci B. 2014;15:438–443. doi: 10.1631/jzus.B1400070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xue Y, Bai X, Lee I, Kallstrom G, Ho J, Brown J, Stevens A, Johnson AW. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol Cell Biol. 2000;20:4006–4015. doi: 10.1128/mcb.20.11.4006-4015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xiang S, Cooper-Morgan A, Jiao X, Kiledjian M, Manley JL, Tong L. Structure and function of the 5'-->3' exoribonuclease Rat1 and its activating partner Rai1. Nature. 2009;458:784–788. doi: 10.1038/nature07731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang VY, Jiao X, Kiledjian M, Tong L. Structural and biochemical studies of the distinct activity profiles of Rai1 enzymes. Nucleic Acids Res. 2015;43:6596–6606. doi: 10.1093/nar/gkv620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chang JH, Jiao X, Chiba K, Oh C, Martin CE, Kiledjian M, Tong L. Dxo1 is a new type of eukaryotic enzyme with both decapping and 5'-3' exoribonuclease activity. Nat Struct Mol Biol. 2012;19:1011–1017. doi: 10.1038/nsmb.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 92.Jiao X, Chang JH, Kilic T, Tong L, Kiledjian M. A mammalian pre-mRNA 5' end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing. Mol Cell. 2013;50:104–115. doi: 10.1016/j.molcel.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 94.Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fu L, Guerrero CR, Zhong N, Amato NJ, Liu Y, Liu S, Cai Q, Ji D, Jin SG, Niedernhofer LJ, et al. Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J Am Chem Soc. 2014;136:11582–11585. doi: 10.1021/ja505305z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]