Abstract
As it is a hard-wired system for responses to microbes, innate immunity is particularly susceptible to classical genetic analysis. Mutations led the way to the discovery of many of the molecular elements of innate immune sensing and signaling pathways. In turn, the need for a faster way to find the molecular causes of mutation-induced phenotypes triggered a huge transformation in forward genetics. During the 1980s and 1990s, many heritable phenotypes were ascribed to mutations through positional cloning. In mice, this required three steps. First, a genetic mapping step was used to show that a given phenotype emanated from a circumscribed region of the genome. Second, a physical mapping step was undertaken, in which all of the region was cloned and its gene content determined. Finally, a concerted search for the mutation was performed. Such projects usually lasted for several years, but could produce breakthroughs in our understanding of biological processes. Publication of the annotated mouse genome sequence in 2002 made physical mapping unnecessary. More recently we devised a new technology for automated genetic mapping, which eliminated both genetic mapping and the search for mutations among candidate genes. The cause of phenotype can now be determined instantaneously. We have created more than 100,000 coding/splicing mutations. And by screening for defects of innate and adaptive immunity we have discovered many “new” proteins needed for innate immune function.
Keywords: forward genetics, genetic mapping, N-ethyl-N-nitrosourea, Toll-like receptor, innate immunity
Forward genetics and its use in studying the innate immune system
Forward genetics operates rather as Gregor Mendel might have hoped it would. One begins with a phenotype and seeks to find its physical cause, embodied in differences at the level of the gene. Classical geneticists, including Mendel and his many successors, have always sought exceptions to the norm and set out to explain them. Morgan (1) and Muller (2) each realized that mutagens could increase the frequency at which variant phenotypes were observed. And this classical approach has not nearly run its course, for in no organism has saturation been achieved where all biological processes of interest are concerned.
In the relatively new era of innate immunity, marked by the discovery of receptors that sense microbial infections of all kinds, mutations have been particularly illuminating. For example, two distantly related strains of mice, C3H/HeJ and C57BL/10ScCr, were each found to be refractory to lipopolysaccharide (LPS) (3;4), apparently as the result of allelic mutations (5). This made it seem certain that a single protein, very likely a receptor capable of recognizing structurally diverse LPS molecules from most Gram-negative organisms, was needed to mount an inflammatory response to infection (6). There was a great sense of urgency about finding this protein. But there was only a phenotype: nothing more. No trace of a difference between either refractory strain and closely related controls could be discerned using the best of biochemical or immunological methods (7;8).
Positional cloning: rigorous and slow
How does one find the cause of a heritable phenotype? At one time, only a biochemical solution to the question could be achieved. The causes of primaquine sensitivity (9), alkaptonuria (10), and sickle cell disease (11;12) were determined prior to the advent of molecular cloning and DNA sequencing technologies. But more often a biochemical solution remained elusive, and with it, the cause of the phenotype itself.
In the 1980s, positional cloning offered a way forward. It was accomplished in three steps (Figure 1). First, high-resolution genetic mapping was necessary to establish a critical region within which the genetic cause of the phenotype was proven to reside. This critical region had to be relatively small (if possible well under one centiMorgan) given the fact that both the number and location of genes in the genome were unknown, and it was impractical to clone large tracts of mammalian DNA. Second, in a physical mapping step, it was necessary to clone all of the critical region as large, overlapping pieces of DNA. Cosmids, yeast artificial chromosomes, and bacterial artificial chromosomes each played a part in this process. When polymerase chain reaction could be used to identify unique sequences near the ends of genomic DNA clones, the search for overlapping pieces of DNA was much facilitated, though still difficult and relatively slow. Third, in a final mutation identification step, it was necessary to determine the gene content of the critical region and find the mutation. Knowing the gene content initially depended on exon trapping, or direct hybridization of cDNA molecules to the DNA of the critical region. Later, as databases of RNA sequences (expressed sequence tags, or ESTs) became comprehensive, it depended on sequencing DNA from the critical region and performing in silico searches for homology with ESTs. Finding the mutation itself depended on sequencing DNA from all candidate genes (whether in humans or in mice), and identifying a mutation that unerringly distinguished the normal from aberrant phenotype.
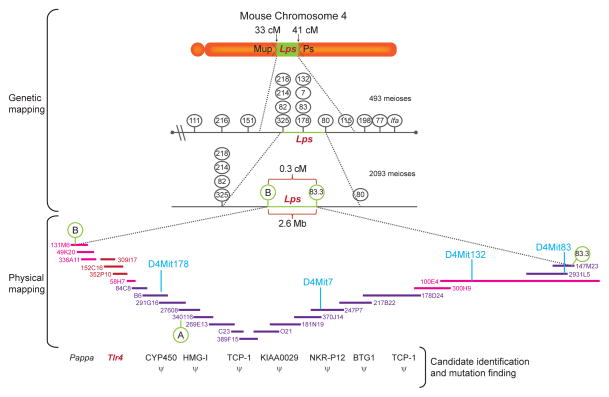
Figure 1.
Positional cloning, exemplified here by the stages used in the cloning of the Lps locus, consisted of genetic mapping, physical mapping, and candidate and mutation identification steps. In genetic mapping, the phenotype was proved to emanate from a point between a proximal and distal marker on the chromosome, based on phenotypic documentation of crossover(s) between each of these markers and the causative mutation. In physical mapping, all of the genomic DNA was captured in a set of overlapping BAC and/or YAC clones spanning the critical region. In the candidate identification step, these clones were examined for gene content. Ultimately, the causative mutation was identified because it was present only in the mutant strain, and not in control strains.
A great number of disease phenotypes were solved in both humans and mice. In humans, most of the classical monogenic diseases that had long been etiologically elusive were solved. Among these were cystic fibrosis (13–15), Huntington’s disease (16;17), Rett’s syndrome (18), and Duchenne’s muscular dystrophy (19;20), to cite but a few examples. In mice, where invasive phenotypic analysis was more a possibility than it was in humans, classical diseases were solved as well. But so too were more “academic” phenotypes that led to enhanced understanding of specific biological questions. For example, mutations causing massive obesity (21), failure of LPS sensing (22), and disorders of circadian rhythms (23) were identified. Each told a story that applied in humans as well as mice.
Back to forward genetics: deliberately creating phenotypes in order to explain them
While there was something magical about finding the cause of a phenotype, whether that phenotype was quantitative (e.g., body weight, or the magnitude of the TNF response) or qualitative (e.g. coat color, or the presence of a tremor), the process remained slow. Confident that the situation would improve with the promised sequence of the human and mouse genomes, some investigators persevered. There was a shortage of phenotypic variation within common inbred strains of mice and their closely related substrains. Therefore, we and others created new phenotypes at random using the chemical mutagen N-ethyl-N-nitrosourea (ENU) (24–26). Many of these ENU-induced phenotypes were solved (23;27–35). It became apparent that ENU-induced phenotypes almost always emanated from changes in coding sense (missense, makesense, nonsense, start loss, or splicing errors).
But despite much success, there were limitations. One could not know how much damage was done to the genome using ENU, and therefore, one could not be confident of the saturation achieved with a given amount of mutagenesis. And more importantly, one could create far more phenotype than could be solved.
The golden age begins
The speed of positional cloning improved as a result of two breakthroughs. First, the annotated mouse genome sequence was published as a draft in 2002 (36). This eliminated the second step in positional cloning: physical mapping. Second, it became possible to sequence whole mammalian genomes, and eventually whole mammalian exomes (37;38). In mice, at least, this allowed one to see all the candidate mutations that might be responsible for a phenotype in a given pedigree.
But the genetic mapping step remained necessary and arduous. In general, mapping required the creation of a homozygous stock, outcross of the stock to a marker strain and backcross to the homozygous stock. Then, in the F2 generation, phenotypic analysis combined with genotyping at a panel of markers in linkage with all points in the genome could establish a critical region. Certain refinements made mapping a bit quicker over time. In order to prevent the disappearance of phenotype as a result of modifying mutations present in distant strains, the closely related C57BL/10J strain was used to map mutations that had been induced on a C57BL/6J background. Using microsatellite markers and later SNPs identified by high throughput sequencing, an adequate panel was devised for this purpose (39). Mapping by bulk segregation analysis, pioneered by plant geneticists (40), could also be used to map mouse mutations. It permitted mixtures of DNA from many F2 mice to be analyzed at each locus to confine the location of a mutation responsible for a phenotype (39;41–44). Despite all these innovations, mapping remained the bottleneck in the forward genetic process.
During the latter half of 2013, our laboratory developed a technology for highly automated genetic mapping (Figure 2). This technology depends on the identification of all ENU-induced mutations in every pedigree by whole exome sequencing of each G1 mouse; then upon genotyping to determine the zygosity of each mutation in all G2 and G3 mice prior to phenotypic screening. G2 mice may be either REF (homozygous reference allele) or HET (heterozygous), while G3 mice may be either REF, HET, or VAR (homozygous variant allele) at each of the ancestral mutation sites. With this information in hand, data gathered from the phenotypic analysis of G3 mice could be computationally analyzed to identify statistically significant associations between phenotype and genotype using recessive, additive, and dominant models of inheritance. The process would only declare phenotypes that were heritable, and would simultaneously display the causative mutation along with information about the affected protein. This “instant positional cloning” method was a tremendous breakthrough, as it allowed the resolution of phenotype in real time (45;46). Phenotypes could be solved as quickly as they were detected. Moreover, the amount of saturation of the genome was, for the first time, calculable within definable limits. As of this writing, about 28% saturation of the genome has been achieved. Saturation is defined as the outright destruction of genes or damage of genes to the point of phenotypic detection, with observation of mutated alleles three times or more in the homozygous state.
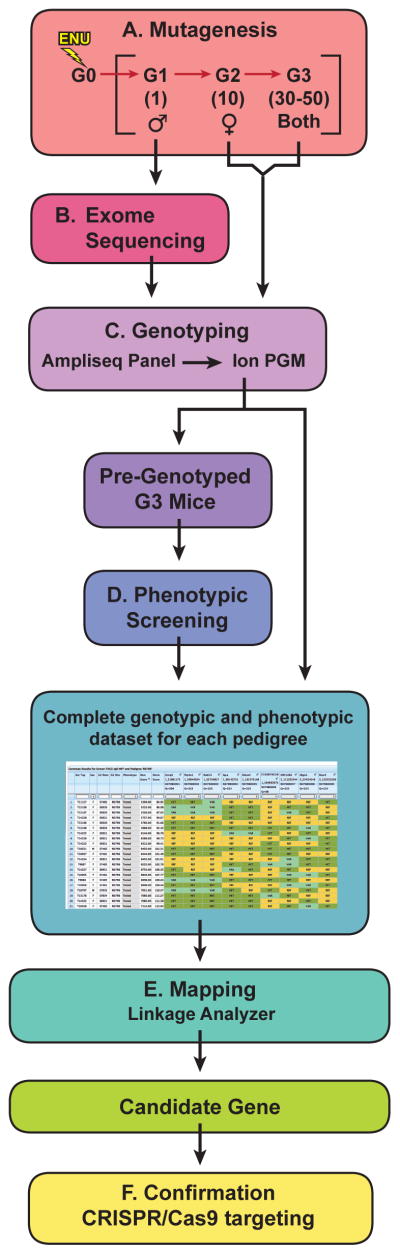
Figure 2.
Pipeline for mutagenesis, screening, and mapping to identify causative mutations in real time. (A) Male C57BL/6J mice mutagenized with ENU are bred to produce 30–50 third generation (G3) mice carrying mutations in homozygous and heterozygous state. (B) The G1 male founder of each pedigree is subjected to exome sequencing, and (C) data are used to generate Ampliseq panel primers for amplification of mutated loci from G2 and G3 mouse DNA, followed by Ion PGM 200-bp sequencing. Genotyping data are uploaded to the database prior to (D) phenotypic screening. Quantitative phenotype data are also entered into the database and used with genotype data for (E) mapping by Linkage Analyzer, a program developed in the lab. (F) Confirmation of candidate genes depends on duplication of the mutant phenotype by a second allele, which may be generated by CRISPR/Cas9 targeting.
The identification of new components of the innate immune system
When mapping is essentially “free of charge,” it is logical to perform all the informative screens one possibly can. We developed automated mapping technology primarily to examine aspects of innate and adaptive immune function in the mouse, but did not ignore other phenotypes. Metabolic, neurobehavioral, developmental, and metabolic phenotypes were pursued as well. At present, a total of 851 phenotypic variants have putative mutational solutions, with mutations falling into 617 genes. Not all of these mutational solutions will ultimately prove to be correct. Most are examined by CRISPR/Cas9 targeting for verification. But the great majority have already been found to be correct or will ultimately be found to be correct.
The positional cloning such a large number of induced phenovariants, including quantitative rather than qualitative traits, would have seemed laughable three years ago. Now it is a practical reality. So too is the mutating to approach saturation, screening for complex traits, and solving models of rather common human phenotypes as diverse as allergy, inflammatory colitis, and drug addiction. One may also begin to consider deep saturation screens aimed at disease suppression. This approach will likely yield actionable drug targets.
An example can be seen in the steady analysis of TLR signaling defects identified over the last several years (Figure 3). By keeping six of the TLRs under surveillance and screening for TNF and type I IFN production in response to TLR ligands, we have identified a total of 70 mutations in 34 genes that interfere with TLR signaling. Nine of these mutations were considered highly novel and are described briefly below.
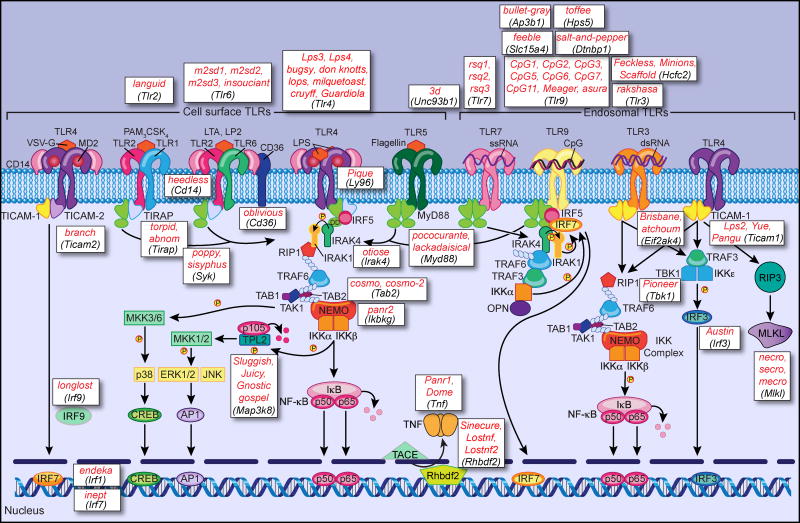
Figure 3.
The TLR signaling pathway and ENU-induced mutations that impair TLR-dependent TNF or type I IFN secretion. Following receptor activation, the TLRs recruit combinations of the adapter proteins MyD88, TICAM-1, TICAM-2, and TIRAP. In the MyD88-dependent pathway utilized by all TLRs except TLR3, MyD88 recruits a complex containing the IRAK serine kinases, which then recruits the E3 ubiquitin ligase TRAF6. TRAF6 adds K63-linked polyubiquitin (small white ovals) to itself and to NEMO. The TRAF6 complex recruits TAK1, TAB1, and TAB2, and activation of TAK1 leads to activation of MAP kinase cascades and the IKK complex. The IKK complex phosphorylates IκB, p105, and TPL2, resulting in IκB and p105 ubiquitination and degradation (small pink circles), and thereby releasing NF-κB into the nucleus and permitting TPL2 to become activated, respectively. Activation of p38, ERK, and JNK kinases leads to CREB and AP1 activation, and their subsequent activation of transcription. In plasmacytoid dendritic cells, TLR7 and TLR9 activate a pathway resulting in IRF7-dependent production of type I IFN. The TICAM-1-dependent pathway is utilized only by TLR3 and TLR4, and activates both NF-κB and IRF3 to produce proinflammatory cytokines and type I IFN, respectively. In addition, necroptosis may be induced via RIP3 and MLKL. ENU-induced mutations (red italics) and the genes they affect (black italics) were identified in our lab by forward genetic analysis, first through positional cloning and recently through real-time automated genetic mapping.
Signal transduction from TLRs ultimately induces the expression of numerous genes required for the inflammatory response, including inflammatory cytokines, chemokines, and costimulatory molecules important for adaptive immune activation. At the level of the receptors, we identified CD36 as a co-receptor for TLR2/TLR6 (47). We also discovered several proteins necessary for the proper localization of nucleic acid sensing TLRs in the subcellular compartments from which they signal: By analysis of the 3d phenotype, we showed that trafficking of TLR3, TLR7, and TLR9 from the endoplasmic reticulum to endosomes requires UNC93B1 (48). The feeble phenotype, specific to plasmacytoid dendritic cells, revealed that the amino acid transporter Slc15a4 is necessary for TLR7 and TLR9 signaling (49); Slc15a4 (49) and TLR9 itself (50) must be trafficked to lysosome-related organelles, a process dependent on the AP-3 complex, dysbindin, and Hps5. Recently we found that HCFC2, a protein of previously unknown function, is required to permit the expression of Tlr3. In the absence of HCFC2, TLR3 and several other proteins related to the antiviral response are poorly expressed, leading to susceptibility to influenza virus, mouse cytomegalovirus (MCMV), and perhaps other pathogens (51).
Directly downstream from receptor activation, four adapter proteins, MyD88, TIRAP (also called MAL), TICAM-1 (also called TRIF), and TICAM-2 (also called TRAM), bind to activated TLRs and recruit necessary signaling components. The function of TICAM-1 as an adapter for TLR3 and TLR4 was first revealed through our analysis of the Lps2 mutation (52). Further downstream, following NF-κB activation and cytokine gene expression, the protein iRhom2 is necessary for TNF secretion, as revealed by the sinecure phenotype (53); iRhom2 chaperones TACE, the TNF processing enzyme, to the ER to release bioactive TNF.
The advantages of ENU saturation mutagenesis for discovery of gene function
It is sometimes assumed that all gene function will be deciphered by genetic analysis of humans, coupled with human phenotyping. Why study mice when one can infer the causes of inherited diseases directly from humans?
In part, the answer is that one can’t always do so. Humans are genetically diverse, whereas mouse mutagenesis is performed on a defined (minimally variable) genetic background. Therefore one may study the effects of small parcels of mutations at a time, pedigree by pedigree, and solve Mendelian phenotypes (and even digenic phenotypes, rare though they may be) with high confidence. In humans, the situation is far different. Large pedigrees cannot be constructed at will, millions of genetic differences distinguish the two parents that contribute genetic information to a proband, and many overtly damaged genes exist in each of us in either a homozygous or complex heterozygous state. It is not at all simple to find new causes of clinical phenotypes, even if one may safely identify known causes.
GWAS studies, in which attempts are made to link common polymorphisms to clinical phenotypes, have been spectacularly unsuccessful in achieving that goal. This reflects many complicating facts. Identical clinical phenotypes sometimes result from mutations at many loci. Much of the pathogenic mutation load is of recent rather than ancient mutational origin. Therefore, even if only mutations at a single locus are capable of causing a given clinical phenotype, many different mutations on different backgrounds may do so. GWAS, which does not interrogate every nucleotide pair as sequencing does, often cannot assign culpability to a particular gene even if linkage is strong (many genes fall under suspicion). Even when sequencing is the tool that is used rather than GWAS, the challenge of determining the cause of phenotype is far greater in humans than it is in mice.
Beyond this, humans cannot be subjected to invasive phenotyping as mice can be. They cannot be inoculated with lethal infectious agents, given dangerous toxins, subjected to severe physical stresses, nor isolated for long periods of time. Some phenotypes simply cannot be approached in humans. But it is as well to acknowledge that some phenotypes, perhaps fewer, cannot be approached in mice.
Summary
Innate immunity has been partly demystified although full mechanistic understanding remains beyond our grasp. Innate immunity has had a cancer connection, best exemplified by the TNF story (54). Adaptive immunity also has its link to cancer, increasingly prominent with the still evolving use of checkpoint inhibitors. All cancers are genetic diseases, with somatic mutations occurring within a particular germline genomic context. We may still learn much about cancers using the tools of classical genetics. How do malignant cells interact with host cells and tissues? When a particular cancer is introduced into a syngeneic host with a defined genome, do specific host mutations influence its course? Are there mutations that prevent metastasis or augment it? What pathways mediate paraneoplastic effects? These questions are more accessible than ever before, and may perhaps become even more accessible in the near future.
Footnotes
Conflict of Interest Statement
The author declares no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Morgan TH. The Genetics of Drosophila. Garland Publishing, Inc; New York: 1925. [Google Scholar]
- 2.Muller HJ. Artificial Transmutation of the Gene. Science. 1927;66:84–87. doi: 10.1126/science.66.1699.84. [DOI] [PubMed] [Google Scholar]
- 3.Heppner G, Weiss DW. High Susceptibility of Strain A Mice to Endotoxin and Endotoxin-Red Blood Cell Mixtures. J Bacteriol. 1965;90:696–703. doi: 10.1128/jb.90.3.696-703.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coutinho A, Forni L, Melchers F, Watanabe T. Genetic Defect in Responsiveness to the B Cell Mitogen Lipopolysaccharide. Eur J Immunol. 1977;7:325–328. doi: 10.1002/eji.1830070517. [DOI] [PubMed] [Google Scholar]
- 5.Coutinho A, Meo T. Genetic Basis for Unresponsiveness to Lipopolysaccharide in C57BL/10Cr Mice. Immunogenetics. 1978;7:17–24. doi: 10.1007/BF01843983. [DOI] [PubMed] [Google Scholar]
- 6.Watson J, Riblet R. Genetic Control of Responses to Bacterial Lipopolysaccharides in Mice. I. Evidence for a Single Gene that Influences Mitogenic and Immunogenic Respones to Lipopolysaccharides. J Exp Med. 1974;140:1147–1161. doi: 10.1084/jem.140.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kabir S, Rosenstreich DL. Binding of Bacterial Endotoxin to Murine Spleen Lymphocytes. Infec Immun. 1977;15:156–164. doi: 10.1128/iai.15.1.156-164.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watson J, Riblet R. Genetic Control of Responses to Bacterial Lipopolysaccharides in Mice. II. A Gene that Influences a Membrane Component Involved in the Activation of Bone Marrow-Derived Lymphocytes by Lipolysaccharides. J Immunol. 1975;114:1462–1468. [PubMed] [Google Scholar]
- 9.Beutler E. The Hemolytic Effect of Primaquine and Related Compounds: A Review. Blood. 1959;14:103–139. [PubMed] [Google Scholar]
- 10.La Du BN, Zannoni VG, Laster L, Seegmiller JE. The Nature of the Defect in Tyrosine Metabolism in Alcaptonuria. J Biol Chem. 1958;230:251–260. [PubMed] [Google Scholar]
- 11.Pauling L, Itano HA. Sickle Cell Anemia a Molecular Disease. Science. 1949;110:543–548. doi: 10.1126/science.110.2865.543. [DOI] [PubMed] [Google Scholar]
- 12.Ingram VM. Abnormal Human Haemoglobins. I. the Comparison of Normal Human and Sickle-Cell Haemoglobins by Fingerprinting. Biochim Biophys Acta. 1958;28:539–545. doi: 10.1016/0006-3002(58)90516-x. [DOI] [PubMed] [Google Scholar]
- 13.Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, Buchwald M, Tsui LC. Identification of the Cystic Fibrosis Gene: Genetic Analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 14.Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL. Identification of the Cystic Fibrosis Gene: Cloning and Characterization of Complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 15.Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M, Rozmahel R, Cole JL, Kennedy D, Hidaka N. Identification of the Cystic Fibrosis Gene: Chromosome Walking and Jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 16.A Novel Gene Containing a Trinucleotide Repeat that is Expanded and Unstable on Huntington’s Disease Chromosomes. the Huntington’s Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 17.Bates GP, MacDonald ME, Baxendale S, Youngman S, Lin C, Whaley WL, Wasmuth JJ, Gusella JF, Lehrach H. Defined Physical Limits of the Huntington Disease Gene Candidate Region. Am J Hum Genet. 1991;49:7–16. [PMC free article] [PubMed] [Google Scholar]
- 18.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett Syndrome is Caused by Mutations in X-Linked MECP2, Encoding Methyl-CpG-Binding Protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 19.Lindenbaum RH, Clarke G, Patel C, Moncrieff M, Hughes JT. Muscular Dystrophy in an X; 1 Translocation Female Suggests that Duchenne Locus is on X Chromosome Short Arm. J Med Genet. 1979;16:389–392. doi: 10.1136/jmg.16.5.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray JM, Davies KE, Harper PS, Meredith L, Mueller CR, Williamson R. Linkage Relationship of a Cloned DNA Sequence on the Short Arm of the X Chromosome to Duchenne Muscular Dystrophy. Nature. 1982;300:69–71. doi: 10.1038/300069a0. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM. Positional Cloning of the Mouse Obese Gene and its Human Homologue. Nature. 1994;372:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- 22.Poltorak A, He X, Smirnova I, Liu M, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg MA, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS Signaling in C3H/HeJ and C57BL/10ScCr Mice: Mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 23.Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and Mapping of a Mouse Gene, Clock, Essential for Circadian Behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shedlovsky A, McDonald JD, Symula D, Dove WF. Mouse Models of Human Phenylketonuria. Genetics. 1993;134:1205–1210. doi: 10.1093/genetics/134.4.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herron BJ, Lu W, Rao C, Liu S, Peters H, Bronson RT, Justice MJ, McDonald JD, Beier DR. Efficient Generation and Mapping of Recessive Developmental Mutations using ENU Mutagenesis. Nat Genet. 2002;30:185–189. doi: 10.1038/ng812. [DOI] [PubMed] [Google Scholar]
- 26.Georgel P, Du X, Hoebe K, Beutler B. ENU Mutagenesis in Mice. Methods Mol Biol. 2008;415:1–16. doi: 10.1007/978-1-59745-570-1_1. [DOI] [PubMed] [Google Scholar]
- 27.Kibar Z, Underhill DA, Canonne-Hergaux F, Gauthier S, Justice MJ, Gros P. Identification of a New Chemically Induced Allele (Lp(m1Jus)) at the Loop-Tail Locus: Morphology, Histology, and Genetic Mapping. Genomics. 2001;72:331–337. doi: 10.1006/geno.2000.6493. [DOI] [PubMed] [Google Scholar]
- 28.Meehan TP, Tabeta K, Du X, Woodward LS, Firozi K, Beutler B, Justice MJ. Point Mutations in the Melanocortin-4 Receptor Cause Variable Obesity in Mice. Mamm Genome. 2006;17:1162–1171. doi: 10.1007/s00335-006-0073-z. [DOI] [PubMed] [Google Scholar]
- 29.Smyth I, Du X, Taylor MS, Justice MJ, Beutler B, Jackson IJ. The Extracellular Matrix Gene Frem1 is Essential for the Normal Adhesion of the Embryonic Epidermis. Proc Natl Acad Sci U S A. 2004;101:13560–13565. doi: 10.1073/pnas.0402760101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barnes MJ, Aksoylar H, Krebs P, Bourdeau T, Arnold CN, Xia Y, Khovananth K, Engel I, Sovath S, Lampe K, Laws E, Saunders A, Butcher GW, Kronenberg M, Steinbrecher K, Hildeman D, Grimes HL, Beutler B, Hoebe K. Loss of T Cell and B Cell Quiescence Precedes the Onset of Microbial Flora-Dependent Wasting Disease and Intestinal Inflammation in Gimap5-Deficient Mice. J Immunol. 2010;184:3743–3754. doi: 10.4049/jimmunol.0903164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berger M, Krebs P, Crozat K, Li X, Croker BA, Siggs OM, Popkin D, Du X, Lawson BR, Theofilopoulos AN, Xia Y, Khovananth K, Moresco EM, Satoh T, Takeuchi O, Akira S, Beutler B. An Slfn2 Mutation Causes Lymphoid and Myeloid Immunodeficiency due to Loss of Immune Cell Quiescence. Nat Immunol. 2010;11:335–343. doi: 10.1038/ni.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brandl K, Tomisato W, Li X, Neppl C, Pirie E, Falk W, Xia Y, Moresco EM, Baccala R, Theofilopoulos AN, Schnabl B, Beutler B. Yip1 Domain Family, Member 6 (Yipf6) Mutation Induces Spontaneous Intestinal Inflammation in Mice. Proc Natl Acad Sci U S A. 2012;109:12650–12655. doi: 10.1073/pnas.1210366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du X, She E, Gelbart T, Truksa J, Lee P, Xia Y, Khovananth K, Mudd S, Mann N, Moresco EM, Beutler E, Beutler B. The Serine Protease TMPRSS6 is Required to Sense Iron Deficiency. Science. 2008;320:1088–1092. doi: 10.1126/science.1157121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Croker B, Crozat K, Berger M, Xia Y, Sovath S, Schaffer L, Eleftherianos I, Imler JL, Beutler B. ATP-Sensitive Potassium Channels Mediate Survival during Infection in Mammals and Insects. Nat Genet. 2007;39:1453–1460. doi: 10.1038/ng.2007.25. [DOI] [PubMed] [Google Scholar]
- 35.Shedlovsky A, King TR, Dove WF. Saturation Germ Line Mutagenesis of the Murine t Region Including a Lethal Allele at the Quaking Locus. Proc Natl Acad Sci U S A. 1988;85:180–184. doi: 10.1073/pnas.85.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mouse Genome Sequencing Consortium. Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O’Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES. Initial Sequencing and Comparative Analysis of the Mouse Genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 37.Pareek CS, Smoczynski R, Tretyn A. Sequencing Technologies and Genome Sequencing. J Appl Genet. 2011;52:413–435. doi: 10.1007/s13353-011-0057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metzker ML. Sequencing Technologies - the Next Generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 39.Xia Y, Won S, Du X, Lin P, Ross C, La Vine D, Wiltshire S, Leiva G, Vidal SM, Whittle B, Goodnow CC, Koziol J, Moresco EM, Beutler B. Bulk Segregation Mapping of Mutations in Closely Related Strains of Mice. Genetics. 2010;186:1139–1146. doi: 10.1534/genetics.110.121160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michelmore RW, Paran I, Kesseli RV. Identification of Markers Linked to Disease-Resistance Genes by Bulked Segregant Analysis: A Rapid Method to Detect Markers in Specific Genomic Regions by using Segregating Populations. Proc Natl Acad Sci U S A. 1991;88:9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arnold CN, Xia Y, Lin P, Ross C, Schwander M, Smart NG, Muller U, Beutler B. Rapid Identification of a Disease Allele in Mouse through Whole Genome Sequencing and Bulk Segregation Analysis. Genetics. 2011;187:633–641. doi: 10.1534/genetics.110.124586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arnold CN, Pirie E, Dosenovic P, McInerney GM, Xia Y, Wang N, Li X, Siggs OM, Karlsson Hedestam GB, Beutler B. A Forward Genetic Screen Reveals Roles for Nfkbid, Zeb1, and Ruvbl2 in Humoral Immunity. Proc Natl Acad Sci U S A. 2012;109:12286–12293. doi: 10.1073/pnas.1209134109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blasius AL, Arnold CN, Georgel P, Rutschmann S, Xia Y, Lin P, Ross C, Li X, Smart NG, Beutler B. Slc15a4, AP-3, and Hermansky-Pudlak Syndrome Proteins are Required for Toll-Like Receptor Signaling in Plasmacytoid Dendritic Cells. Proc Natl Acad Sci U S A. 2010;107:19973–19978. doi: 10.1073/pnas.1014051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krieg L, Milstein O, Krebs P, Xia Y, Beutler B, Du X. Mutation of the Gastric Hydrogen-Potassium ATPase Alpha Subunit Causes Iron-Deficiency Anemia in Mice. Blood. 2011;118:6418–6425. doi: 10.1182/blood-2011-04-350082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang T, Zhan X, Bu CH, Lyon S, Pratt D, Hildebrand S, Choi JH, Zhang Z, Zeng M, Wang KW, Turer E, Chen Z, Zhang D, Yue T, Wang Y, Shi H, Wang J, Sun L, SoRelle J, McAlpine W, Hutchins N, Zhan X, Fina M, Gobert R, Quan J, Kreutzer M, Arnett S, Hawkins K, Leach A, Tate C, Daniel C, Reyna C, Prince L, Davis S, Purrington J, Bearden R, Weatherly J, White D, Russell J, Sun Q, Tang M, Li X, Scott L, Moresco EM, McInerney GM, Karlsson Hedestam GB, Xie Y, Beutler B. Real-Time Resolution of Point Mutations that Cause Phenovariance in Mice. Proc Natl Acad Sci U S A. 2015;112:E440–9. doi: 10.1073/pnas.1423216112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simon MM, Moresco EM, Bull KR, Kumar S, Mallon AM, Beutler B, Potter PK. Current Strategies for Mutation Detection in Phenotype-Driven Screens Utilising Next Generation Sequencing. Mamm Genome. 2015;26:486–500. doi: 10.1007/s00335-015-9603-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoebe K, Georgel P, Rutschmann S, Du X, Mudd S, Crozat K, Sovath S, Shamel L, Hartung T, Zahringer U, Beutler B. CD36 is a Sensor of Diacylglycerides. Nature. 2005;433:523–527. doi: 10.1038/nature03253. [DOI] [PubMed] [Google Scholar]
- 48.Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, Mudd S, Mann N, Sovath S, Goode J, Shamel L, Herskovits AA, Portnoy DA, Cooke M, Tarantino LM, Wiltshire T, Steinberg BE, Grinstein S, Beutler B. The Unc93b1 Mutation 3d Disrupts Exogenous Antigen Presentation and Signaling Via Toll-Like Receptors 3, 7 and 9. Nat Immunol. 2006;7:156–164. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- 49.Blasius AL, Arnold CN, Georgel P, Rutschmann S, Xia Y, Lin P, Ross C, Li X, Smart NG, Beutler B. Slc15a4, AP-3, and Hermansky-Pudlak Syndrome Proteins are Required for Toll-Like Receptor Signaling in Plasmacytoid Dendritic Cells. Proc Natl Acad Sci U S A. 2010;107:19973–19978. doi: 10.1073/pnas.1014051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sasai M, Linehan MM, Iwasaki A. Bifurcation of Toll-Like Receptor 9 Signaling by Adaptor Protein 3. Science. 2010;329:1530–1534. doi: 10.1126/science.1187029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun L, Jiang Z, Berger M, Du X, Choi JH, Wang J, Wang K, Acosta-Rodriguez VA, Mohawk JA, Quan J, Scott L, Hildebrand S, Li X, Tang M, Zhan X, Murray AR, LaVine D, Moresco EM, Takahashi JS, Beutler B. HCFC2 Needed for IRF1- and IRF2-Dependent Tlr3Transcription, and for Survival during Viral Infections. doi: 10.1084/jem.20161630. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoebe K, Du X, Georgel P, Janssen E, Tabeta K, Kim SO, Goode J, Lin P, Mann N, Mudd S, Crozat K, Sovath S, Han J, Beutler B. Identification of Lps2 as a Key Transducer of MyD88-Independent TIR Signaling. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 53.Siggs OM, Xiao N, Wang Y, Shi H, Tomisato W, Li X, Xia Y, Beutler B. IRhom2 is Required for the Secretion of Mouse TNFalpha. Blood. 2012;119:5769–5771. doi: 10.1182/blood-2012-03-417949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Old LJ. Tumor Necrosis Factor. Sci Am. 1988;258:59–60. 69–75. doi: 10.1038/scientificamerican0588-59. [DOI] [PubMed] [Google Scholar]