Key Points
High expression of MAF protein in myeloma due to t(14;16) translocation confers innate resistance to PIs.
PIs prevent GSK3-mediated degradation of MAF protein, which further augments the resistance to PIs in t(14:16) myeloma.
Abstract
Multiple myeloma (MM) patients with the t(14;16) translocation have a poor prognosis, and unlike other molecular subgroups, their outcome has not improved with the introduction of bortezomib (Bzb). The mechanism underlying innate resistance of MM to Bzb is unknown. In the present study, we have investigated how MAF overexpression impacts resistance to proteasome inhibitor (PI) therapy (Bzb and carfilzomib). High levels of MAF protein were found in t(14;16) cell lines; cell lines from the t(4;14) subgroup had intermediate levels, whereas cell lines from the other subgroups had low levels. High expression of MAF protein in t(14;16) was associated with significantly higher PI half-maximum inhibitory concentration values compared with other molecular subgroups. PI exposure abrogated glycogen synthase kinase 3β (GSK3β)-mediated degradation of MAF protein, resulting in increased MAF protein stability and PI resistance. Subsequent studies using loss-of-function and gain-of-function models showed that silencing MAF led to increased sensitivity to PIs, enhanced apoptosis, and activation of caspase-3, -7, -8, -9, poly (ADP-ribose) polymerase, and lamin A/C. In contrast, overexpression of MAF resulted in increased resistance to PIs and reduced apoptosis. These results define the role of MAF and GSK3 in the resistance of t(14;16) MM to PIs and identifies a novel mechanism by which MAF protein levels are regulated by PIs, which in turn confers resistance to PIs.
Introduction
Multiple myeloma (MM) is a malignancy of terminally differentiated clonal plasma cells that display significant molecular heterogeneity. Approximately 5% of newly diagnosed cases express high levels of MAF, and are defined cytogenetically by either the MAF expressing t(14;16) or MAFb expressing t(14;20). Despite only constituting a small group, MAF is an important therapeutic target in MM as t(4;14)-overexpressing cases also express MAF at intermediate levels.1
Early studies revealed that MAF messenger RNA (mRNA) is overexpressed in myeloma cells harboring the translocation t(14;16)(q32;q23), which fuses the immunoglobulin heavy chain and MAF gene loci.2,3 The t(14;16) is associated with poor clinical outcomes with a high proportion of patients having high-risk disease, extramedullary disease4 and primary plasma cell leukemia.5-10 The outcome of these cases has not improved since the introduction of bortezomib (Bzb) which is in sharp contrast to t(4;14) patients. For example, historically, we have shown that t(4;14) and t(14;16) cases had inferior survival9,10 and that the addition of Bzb, to the total therapy regimen, provided a survival advantage for t(4;14) patients. In contrast, patients with a t(14;16) did not benefit from the addition of Bzb.11-13
High expression of MAF mRNA due to the t(14;16) translocation is seen in 25% of human myeloma cell lines (HMCLs)1,2,14 but only 2% to 7% of primary MM samples.3,15,16 High MAF mRNA is also detected in 25% of t(4;14) HMCLs.1,14 The high MAF mRNA in these 2 subgroups appears to be mediated via transmembrane activator and calcium modulator and cyclophilin ligand interactor.17 Despite the extended studies on MAF mRNA transcription in MM, the association of MAF mRNA expression with MAF protein has not been fully explored.
MAF is the cellular homolog of V-MAF, the transforming gene of the avian retrovirus AS42.18 It is a member of the basic leucine zipper transcriptional factor family that regulates gene transcription by cyclic adenosine monophosphate–response elements.19 It has been identified as an oncogene important in the development of human lymphoid malignancies,20,21 and aberrant MAF expression contributes to MM pathogenesis by deregulation of cyclin D2,14,22 CCR1,14 ARK5,23 ITGB7.14,17,24 The increase in ARK5 expression is associated with enhanced invasion23 and extramedullary disease.4 Upregulation of cyclin D2 promotes cell cycle progression14,25 and a high proliferative index.10,26 Upregulation of ITGB7 and E-cadherin enhances MM cell adhesion with stromal cells and promotes secretion of vascular endothelial growth factor.14,24,25
In addition to its role as an oncogene in MM, we hypothesize that high MAF protein confers innate resistance to Bzb. We demonstrate that resistance to proteasome inhibitors (PIs) is associated with the high MAF protein expression and identify a novel mechanism by which MAF protein stabilization is regulated by glycogen synthase kinase 3β (GSK3β) activity and inhibition of PIs.
Material and methods
For detailed methods, please see supplemental Methods, available on the Blood Web site.
Cell lines and primary CD138+ plasma cells
HMCLs are listed in supplemental Methods and described elsewhere.27-30 Cells were cultured in growth medium as described previously.31 Primary CD138-expressing MM cells were isolated as previously described.32 MAF gene expression was determined by gene expression profiling analysis of CD138 plasma cells from MM patients.33 Human bone marrow (BM) stromal cells (HBMSCs) from MM patients were generated as described previously.34
Lentiviral-mediated transduction to generate MAF stable clones
The retroviral production of pReceiver vector or the vector containing MAF complementary DNA from Genecopoeia was generated according to the manufacturer’s instructions. HMCLs were infected with lentiviral particles as previously described.35 Stable clonal cell lines expressing MAF were generated by puromycin selection as previously described.28
Silencing MAF expression by short hairpin RNA
Knockdown of MAF gene was performed in HMCLs expressing high levels of MAF using a lentiviral expressing system as described in supplemental Methods.
MTT assay for cell proliferation
Viability of MM cells was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide (MTT) assay as described in supplemental Methods. Bzb, carfilzomib (CFZ), MG132, cycloheximide (CHX), and SB216763 were as described previously.34,36,37
Detection of apoptotic cells
MM cells in growth media alone or cocultured with HBMSCs were treated with or without serial concentrations of PIs, and apoptotic cells were stained by the fluorescein isothiocyanate (FITC)-Annexin V Apoptosis Detection kit according to the manufacturer’s instructions and detected by flow cytometry using the FACSVerse System (BD) as described in supplemental Methods.
Coculture with BM stromal cells
Coculture of MM cells with HBMSC line HS27A31 or with primary HBMSCs was performed to determine the effect of MAF on MM apoptosis in the presence of the BM microenvironment.38
Immunoblotting
Proteins were isolated from cells treated with or without Bzb, CFZ, MG132, SB216763, or LiCl. Specific proteins were detected by immunoblotting analysis (IBA) using anti-c-maf, caspase-3, -6, -7, -8, and -9, poly (ADP-ribose) polymerase (PARP), lamin A/C antibodies described elsewhere.28
CHX chase assay
MM cells were cultured in growth media with CHX. Cell lysate was subject to IBA. The autoradiographs were scanned and densitometry performed.34
Immunoprecipitation and phosphatase treatment of MAF protein
MAF protein in whole-cell lysate was purified by immunoprecipitation assay and dephosphorylated by alkaline phosphatase (ALP) treatment as described previously.27
TCF/LEF luciferase report gene assay
T-cell factor/lymphoid enhancer factor (TCF/LEF) activity was used as a surrogate method for GSK3β activity as SB21673 induces transcriptional activity of TCF/LEF by inhibiting GSK3β activity39 as previously described.34
Immunofluorescence staining
Immunofluorescence staining was used to determine MAF protein expression in cell nuclei using anti-MAF antibody and 4′,6-diamidino-2-phenylindole (DAPI).36
Real-time qPCR
For quantitation of mRNA expression, total RNA was extracted from MM cell lines and complementary DNA was generated from 1 μg of RNA as described previously.34 Quantitative polymerase chain reaction (qPCR) was performed using QuantStudio 6 and 7 Flex Real-Time PCR Systems. The indicated genes were amplified using methods described in supplemental Methods.35,40
Statistical analysis
The statistical significance of differences between experimental groups was analyzed by the Student t test using the Microsoft Excel software statistical package. P values <.05 by 2-tailed test were considered significant. Progression-free survival (PFS) and overall survival (OS) of patients in the total therapy 3 (TT3) study were calculated by the Kaplan-Meier method.41 Differences between survival curves were analyzed by the log-rank test.42
Results
Clinical impact of t(14;16)
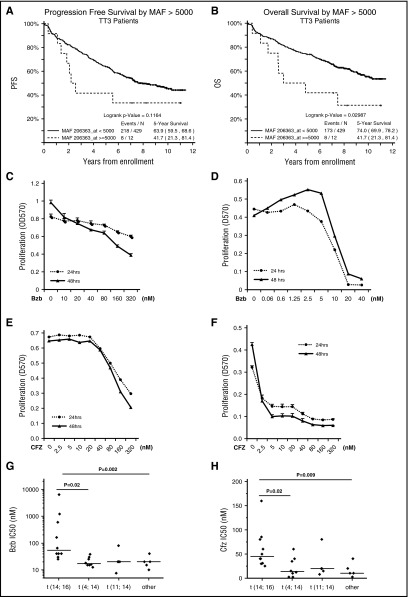
We analyzed the impact of MAF expression on PFS and OS for patients entered into the TT3 clinical study who were treated with Bzb induction and maintenance. We show that high MAF expression is associated with poor PFS and OS compared with other subgroups (Figure 1A-B).
Figure 1.
MM cells harboring t(14;16) are resistant to PIs in vitro and are associated with a poor prognosis in MM patients. PFS (A) and OS (B) according to MAF expression status in patients in the TT3 clinical study. APR1 (C,E) and H929 (D,F) and were seeded at 2 × 104 per well in 96-well plates and cultured in growth media in the presence of the indicated concentrations of Bzb (C-D) or CFZ (E-F) for 24 and 48 hours. Proliferation was measured by MTT assay. Results are presented as mean ± standard error (SE) (n = 4). Data are representative of 3 separate experiments. MM cells lines were treated with serial concentrations of Bzb (G) and CFZ (H) for 48 hours and proliferation was measured by MTT assay. The IC50 of Bzb (G) and CFZ (H) were classified based on molecular subgroups.
t(14;16) subgroup is insensitive to PIs
To investigate the association of the limited therapeutic effect of Bzb in t(14;16)s, we determined the half-maximum inhibitory concentration (IC50) of Bzb for 27 HMCLs harboring t(14;16), t(4;14), or other cytogenetic abnormalities. We selected ARP1, a t(14;16)-positive cell line, and H929, a t(4;14)-positive cell line, with high comparable levels of MAF mRNA, to treat with Bzb and measure proliferation (Figure 1C-D). The IC50 of Bzb in ARP1 was 30-fold higher than for H929. Similar results were seen with CFZ (Figure 1E-F). These results demonstrate that the sensitivity of APR1 to PIs was significantly less than H929.
To compare the differential sensitivity between t(14;16)s and other subgroups, the IC50 of Bzb in 27 HMCLs was measured using the MTT assay. The mean Bzb IC50 for 10 HMCLs with t(14;16) was significantly higher than those either with t(4;14), t(11;14), or subgroups lacking these translocations (Figure 1G). Similar significantly higher IC50 for CFZ was observed in t(14;16)s in comparison with other subgroups (Figure 1H). Taken together, these results suggest that the t(14;16) subgroup is more resistant to PIs than other subgroups.
MAF protein level is partially associated with mRNA level
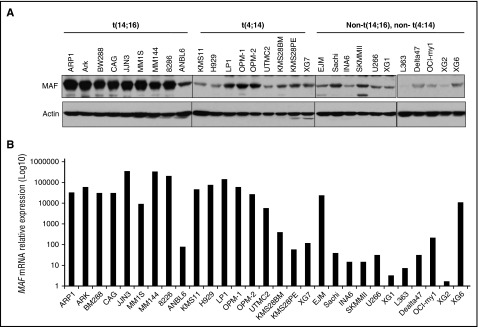
To correlate the MAF mRNA to the protein level in HMCLs based on molecular subgroups,24,43 the basal level of MAF protein was determined by immunoblotting analysis. The highest levels of MAF protein were detected in 8 of 10 t(14;16) cell lines (Figure 2A). The majority (6 of 9) of HMCLs harboring t(4;14) had lower levels of MAF protein than those with a t(14;16). HMCLs from other subgroups had low MAF. In contrast, MAF mRNA levels were similarly high in the majority of t(14;16) and t(4;14) HMCLs. These results are consistent with previous reports.14 Interestingly, MAF mRNA levels were comparable with protein level in 8 of 9 t(14;16) lines. Six of 9 t(4;14) lines which lacked correspondingly high levels of the protein had comparable mRNA levels to cases with a t(14;16) (Figure 2). These results indicate that MAF protein is only partially correlated with the mRNA and that protein synthesis and/or degradation may contribute to the differences seen between subgroups.
Figure 2.
MAF protein was heterogeneously expressed and only partially correlated with mRNA levels in MM cells. Cell lysates were isolated from HMCLs harboring t(14;16), t(4;14), or cell lines without these translocations and resolved on 8% SDS-polyacrylamide gels. Protein was transferred to membranes and blotted with anti-MAF antibody using immunoblotting analysis (A). The level of MAF mRNA in HMCLs was determined by RT-qPCR (B).
GSK3β-mediated phosphorylation induces degradation of MAF protein
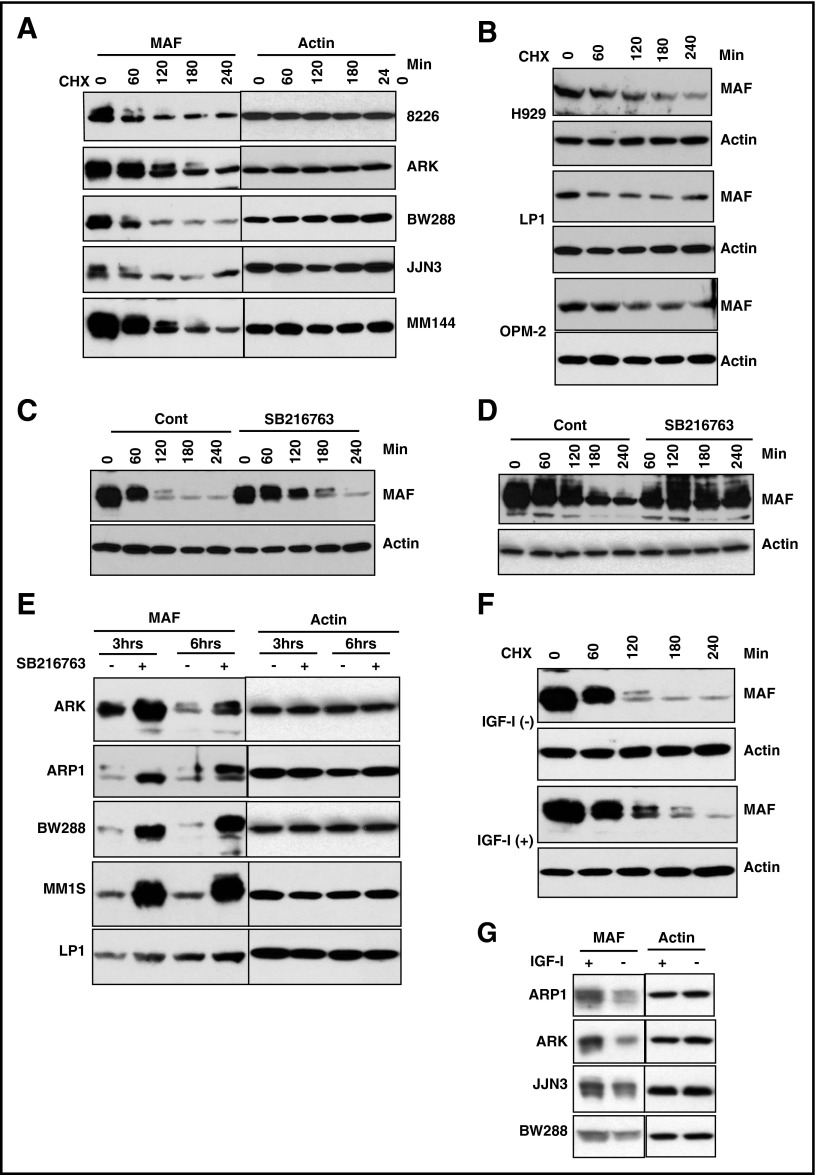
To determine whether protein degradation regulates MAF, we used CHX to inhibit new protein synthesis from mRNA and determined protein half-life.44 MAF protein decreased by 60 minutes in 3 of 5 t(14;16) HMCLs and by 80 minutes and 120 minutes in 2 further lines, whereas the actin protein level remained stable (Figure 3A; supplemental Figure 1A). Additionally, a slow migrating band shifted from the main band (lower band) was observed in all 5 t(14;16) lines (Figure 3A,C). Importantly, the migrated bands disappeared more quickly with a shorter half-life than the major band (Figure 3A,C; supplemental Figure 2A), indicating that slowly migrating MAF protein decays faster in t(14;16).
Figure 3.
The degradation of MAF protein was abrogated by inhibition of GSK3β activity. HMCL cells harboring t(14;16) (A) or t(4;14) (B) were treated with 5 μg/mL CHX for indicated time points. MM144 (C), 8226 (D), or other HMCLs (E) were treated with or without SB216763 in the presence of CHX for indicated times. M144 cells (F) were cultured in serum-free medium for 16 hours and treated with or without 100 ng/mL IGF-1 for indicated times in the presence of 5 μg/mL CHX. A panel of MM cell lines (G) cultured in serum-free medium for 16 hours was treated with or without 100 ng/mL IGF-1 for 6 hours in the presence of CHX. Cell lysates were isolated and MAF protein levels determined. The anti-MAF antibody recognizes 2 bands, of which the upper band represents the phosphorylated form. An anti-β-actin was used to indicate protein loading for each cell line.
Given that MAF protein levels are not comparable with mRNA levels in t(4;14) cells, we selected H929, OPM-2, and LP1 as representatives to examine the degradation status and half-life of MAF compared with t(14;16) cases. The basal levels of MAF protein in t(4;14) were lower than in t(14;16) (Figure 2B). The exposure times of the films for the t(4;14) lines were longer (H929, 10 times; OPM2 and LP1, 5 times) than those of t(14;16)s, indicating that the endogenous level of MAF protein in t(4;14) cells is lower than in t(14;16). Additionally, the slowly migrated isoform of MAF in t(14;16) was not seen in t(4;14) cells. The half-life of total MAF protein in t(4;14) is at an intermediate level compared with t(14;16) lines (supplemental Figure 1A-B). The lack of MAF upper band may explain why the half-life of total MAF protein in t(4;14) was not shorter than in (14;16) despite the low baseline level of the protein in t(4;14) cells.
Posttranslation modification of protein such as phosphorylation is the most likely cause of the slow migration seen in IBA.27,45 To determine whether the slowly migrating isoforms of MAF protein are phosphorylated, MAF protein immunoprecipitated from HMCLs was treated with ALP to dephosphorylate the protein and analyzed by IBA. Treatment of the cells with ALP led to the disappearance and reduction of the upper bands (supplemental Figure 3). These results indicate the slowly migrating protein represents a phosphorylated form of MAF protein.
Phosphorylation by GSK3 has been shown in other tissues to govern the degradation of MAF-A protein.46 Serial experiments were therefore conducted to assess how inhibition of GSK3 activity affected MAF in myeloma. Because GSK3β is constitutively active in myeloma,28 we evaluated the effect of SB216763, a well-known specific inhibitor of GSK3,39 on GSK3β activity. Treatment of 4 HMCLs with SB216763 led to significant inhibition of GSK3β activity (supplemental Figure 4A). The role of SB216763 in regulation of GSK3β was further confirmed by showing an increase in TCF/LEF transcriptional activity in response to SB216763 (supplemental Figure 4B), as previously induction of TCF/LEF has been shown to be a surrogate indicator of SB216763-mediated inhibition of GSK3β activity.39
Given that SB216763 significantly inhibited GSK3β activity, we next determined its effect on GSK3β-regulated stability of MAF. Treatment of MM144 with SB216763 led to an increase in MAF protein (Figure 3C). Similar increases in MAF were observed in 8226 cells (Figure 3D). The stabilization of MAF by SB16763 was also seen in an extended panel of t(14;16) lines (Figure 3E). These results suggest GSK3β activity is required for the degradation of MAF protein in t(14;16) cells.
IGF-1 inhibits GSK3β activity in many cell types as well as in MM cells28,47; therefore, to validate whether GSK3β activity is required for MAF degradation, we assessed the effect of IGF-1 on MAF protein. As expected, IGF-1 treatment induced phosphorylation of ser9 of GSK3 (supplemental Figure 5) which is an indication of GSK3β inhibition in response to IGF-1.28,48,49 Treatment of MM144 with IGF-1 showed an increase in MAF (Figure 3F). IGF-1–induced accumulation of MAF protein was also seen in other t(14;16) cell lines (Figure 3G). No impact in MAF mRNA was seen (data not shown). We conclude that IGF-1–induced stabilization of MAF protein is via inhibition of GSK3 activity and is independent of MAF mRNA. Finally, to explore whether MAPK is involved in regulating MAF protein stability, we used U0126, a well-known MEK1/2 inhibitor.28 Treatment of JJN3, MM144, and 8226 cells with U0126 had no effect on MAF protein (data not shown). Treatment of these cells with SB20350 and SP6000125, inhibitors to p38 and Jun N-terminal protein kinases, alone or in combination, also had no effect on MAF protein (data not shown). Taken together, these results suggest that GSK3 activity regulates degradation of MAF protein in t(14;16) cells independently of the MAPK pathway and p38 kinases.
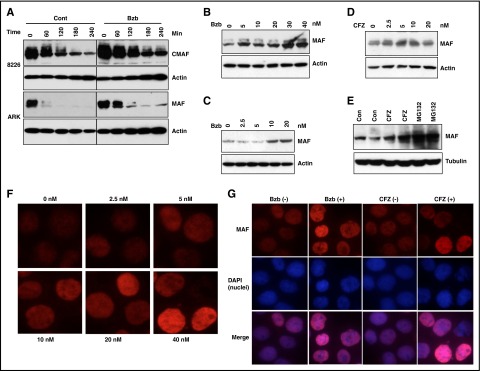
PI exposure prevents degradation of MAF protein
Having shown that MAF protein is regulated by inhibition of GSK3β activity, we determined whether PIs had an effect on MAF stability. Treatment of 8226 and ARK with Bzb led to an increase in MAF protein (Figure 4A). Moreover, Bzb treatment of ARP1 and MM144 (Figure 4C-B) resulted in a dose-dependent increase in MAF. To confirm whether this effect was PI specific, we confirmed our results using CFZ. Dose-dependent increases in MAF were seen in MM144 following treatment with CFZ, with similar increases observed in ARP1 with CFZ and MG132 (Figure 4D-E). It should be noted that PI treatment did not affect MAF protein in t(4;14) cells (data not shown). Because MAF is a transcription factor,19 we determined the effect of PIs on nuclear localization of MAF protein. Treatment of MM144 with Bzb led to an increase of MAF in the nucleus (Figure 4F). Similar increases were seen in the nucleus and cytoplasm of 8226 cells in response to Bzb and CFZ (Figure 4G and data not shown). Taken together, these results suggest that inhibition of proteasome activity by PIs prevents degradation of MAF protein in t(14;16), but not in other subgroups.
Figure 4.
PIs induced accumulation of MAF protein in MM cells. 8226 and ARK (A) cells were treated with 20 nM Bzb in the presence of 5 μg/mL CHX for indicated times. APR1 (B,E), MM144 (C-D) were treated with serial concentrations of Bzb (B-C), CFZ (D), or with 20 nM CFZ or 10 μM MG132 (E) for 6 hours in the presence of CHX. Cell lysates were isolated and MAF protein analyzed. Anti-β-actin or anti-tubulin was used to indicate protein loading for each cell line. MM144 cells were treated with serial concentrations of Bzb for 12 hours; MAF nuclei and cytoplasm expression was determined by immunofluorescence staining (F). 8226 cells were treated with indicated concentrations (0 or 20 nM) of Bzb or CFZ for 12 hours. MAF nuclei and cytoplasm expression was determined by immunofluorescence staining with DAPI counterstaining (G). Images were taken with a fluorescent microscope and digital camera.
Silencing MAF restores sensitivity of MM cells to PIs whereas its overexpression induces resistance to PIs
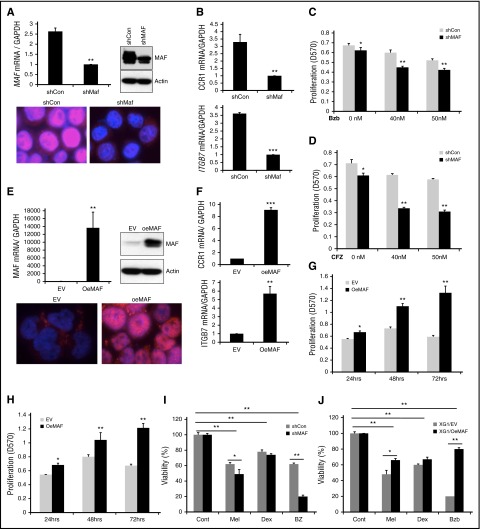
To further determine whether Bzb-induced stabilization of MAF protein confers resistance to Bzb, we generated a loss of function of MAF cell line by silencing its expression in 8226 cells2,17,24 using a short hairpin RNA (shRNA) lentiviral system. Specific MAF knockdown was confirmed at the mRNA and protein level (>75%) (Figure 5A). Additionally, the functionality of knockdown of MAF was confirmed by a decrease in target gene expression of ITGB7 and CCR1 in shMAF compared with shCon (Figure 5B). The proliferation of shMAF in response to Bzb was significantly lower than shCon (Figure 5C; supplemental Figure 6A). This effect of silencing MAF on sensitivity to PIs was also seen in 8226 treated with CFZ (Figures 5D and 6B). A similar increase in sensitivity to Bzb was seen in shMAF ANBL6 cells (data not shown).
Figure 5.
Manipulation of MAF expression led to alteration in sensitivity of MM cells to PIs. 8226 cells were infected with lentiviral containing shRNA specific to MAF gene (shMAF) or shRNA containing scramble sequences (shCon) for 48 hours. The level of MAF mRNA in shMAF and shCon cells was determined by RT-qPCR. Results are presented as mean ± SE (n = 3). **P < .01 for shMAF vs shCon. The level of MAF protein in whole-cell lysates was determined by immunoblotting analysis. MAF protein expression in both cytoplasm and nuclei of shMAF and shCon cells was analyzed by immunofluorescent staining (A). The mRNA levels of CCR1 and ITGB7 in shMAF and shCon cells were measured by RT-qPCR analysis (B). Results are presented as mean ± SE (n = 3). **P < .01 or ***P < .001 for shMaf vs shCon. shMAF and shCon cells were treated with the indicated concentrations of Bzb (C) or CFZ (D) for 48 hours, and proliferation measured by MTT assay. Results are presented as mean ± SE (n = 4). Data are representative of 3 separate experiments. *P < .01 vs control. ***P < .001 vs control. Total RNA was obtained from XG1EV and XG1OeMAF and the levels of MAF mRNA (E), ITGB7 and CCR1 (F) was determined by RT-qPCR analysis. Results are presented as mean ± SE (n=3). **P < 0 .01 or ***P < 0 .001 for XG1EV vs XG1OeMAF. The levels of MAF protein in whole-cell lysate or in the cytoplasm and nucleus were determined (E). XG1OeMAF and XG1EV cells were treated with 20 nM Bzb (G) or CFZ (H) for indicated time points, and proliferation measured by MTT assay. Results are presented as mean ± SE (n = 4). Data are representative of 3 separate experiments. *P < .01 vs control. **P < .001 vs control. 8226/shMAF and 8226/shCon (I) or XG1OeMAF and XG1EV (J) cells were treated with or without Mel (50 μM), Bzb (40 nM), or Dex (250 μM) for 24 hours. Cell viability was measured by MTT assay. Results are presented as the percentage of alive cells relative to untreated cells. Each experiment was repeated n = 6 and reported as the mean value ± standard deviation (SD). *P < 0 .01 shMAF vs shCon; **P < 0 .001 shMAF vs shCon; **P < 0 .001 untreated vs treated shCon or untreated vs treated shMAF (I) and *P < 0 .01 XG1OeMAF vs XG1EV; **P < 0 .001 XG1OeMAF vs XG1EV; **P < 0 .001 untreated vs treated XG1EV or untreated vs treated XG1OeMAF (J).
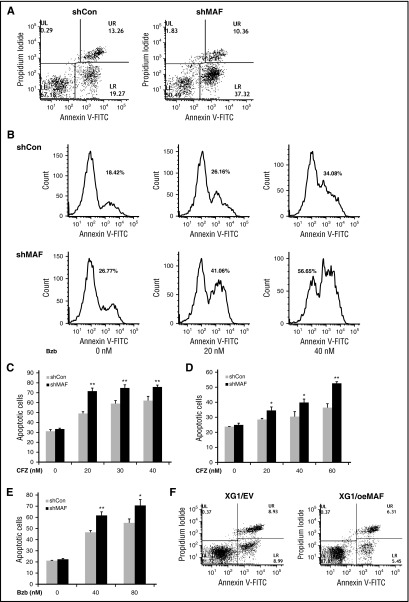
Figure 6.
MAF protein prevents MM cells from PI-induced apoptosis. 8226/shCon (left) or 8226/shMAF (right) cells were treated with 30 nM Bzb for 16 hours, and then stained using Annexin V–FITC and propidium iodide and apoptotic cell numbers determined by flow cytometry analysis. The lower right (LR) area represents early apoptotic cells (Annexin V–FITC positive), the upper right (UR) area represents late apoptotic and/or necrotic cells (Annexin V–FITC and propidium iodide positive); the lower left area represents viable cells (unstained) (A). Apoptotic cell numbers in 8226/shCon or 8226 shMAF treated with indicated serial concentrations of Bzb for 16 hours were determined by Annexin V–FITC and propidium iodide staining and flow cytometry analysis. The data are shown by histogram analysis (B). The percentages of apoptotic cells in the presence of increasing concentrations of CFZ were determined by the same methods. Data are representative of 3 separate experiments. **P < .001 shMaF vs shCon. (C). Cells were cocultured with BM stromal cells and treated with indicated concentrations of CFZ (D) or Bzb (E) and apoptotic cells were analyzed as described in the description of panel A. Data are representative of 3 separate experiments. *P < .01 or **P < .001 shMAF vs shCon. XG1EV and XG1OeMAF cells were treated with 20 nM Bzb for 16 hours and apoptotic cells measured by Annexin V–FITC and PI staining, and flow cytometry analysis as described in the description of panel A (F).
To validate the ability of MAF-mediated resistance to PIs, we developed a gain-of-function MAF stable clone from XG1 cells, lacking a t(14;16) using a lentiviral expressing system containing an empty vector (designated as XG1EV) or MAF gene (designated as XG1OeMAF). Expression of MAF mRNA and protein in XG1OeMAF cells was higher than in XG1EV (Figure 5E), as was the expression of ITGB7 and CCR1 target genes (Figure 5F). XG1OeMAF exhibited resistance to Bzb and CFZ compared with XG1EV (Figure 5G-H; supplemental Figure 6C-D). These results suggest that high MAF expression confers intrinsic MM resistance to PIs. We next explored whether the abundance of MAF protein also affected sensitivity to other therapeutic reagents such as dexamethasone (Dex) and melphalan (Mel). Knockdown MAF expression in 8226 cells significantly enhanced sensitivity of cells to Bzb, moderately increased sensitivity to Mel, but had no effect on Dex (Figure 5I). In contrast, overexpression of MAF in XG1 cells led to a decrease in sensitivity to Bzb and Mel, with no effect on Dex (Figure 5J). These results indicate that the effect of MAF protein is mainly related to PIs.
Silencing MAF enhances PI-induced apoptosis in myeloma
To elucidate the molecular mechanism underlying MAF-mediated resistance to PIs, we measured the effect of silencing MAF on PI-induced apoptosis. The numbers of apoptotic cells in shMAF were higher following treatment with Bzb compared with shCon (Figure 6A-B). Similar increases in apoptosis were seen in shMAF in response to CFZ (Figure 6C). These results indicate that silencing MAF enhances PI-induced apoptosis. To further validate the effect of MAF on PI-induced apoptosis, we examined the effect of PIs on apoptosis in XG1OeMAF and XG1EV. As expected, reduced apoptosis was seen in XG1OeMAF compared with XG1EV when treated with Bzb (Figure 6F), indicating that high MAF expression in MM overcomes PI-induced apoptosis.
To determine whether the BM microenvironment affects the role of MAF in PI-mediated apoptosis, shMAF and shCon cells were cocultured with HBMSCs derived from MM patients. Treatment of shMAF cells with CFZ or Bzb (Figure 6D/E, led to an increase in apoptotic cells, suggesting that MAF protein also prevents PI-induced apoptosis even in the presence of the BM microenvironment.
MAF prevents PI-induced apoptosis via activation of caspases
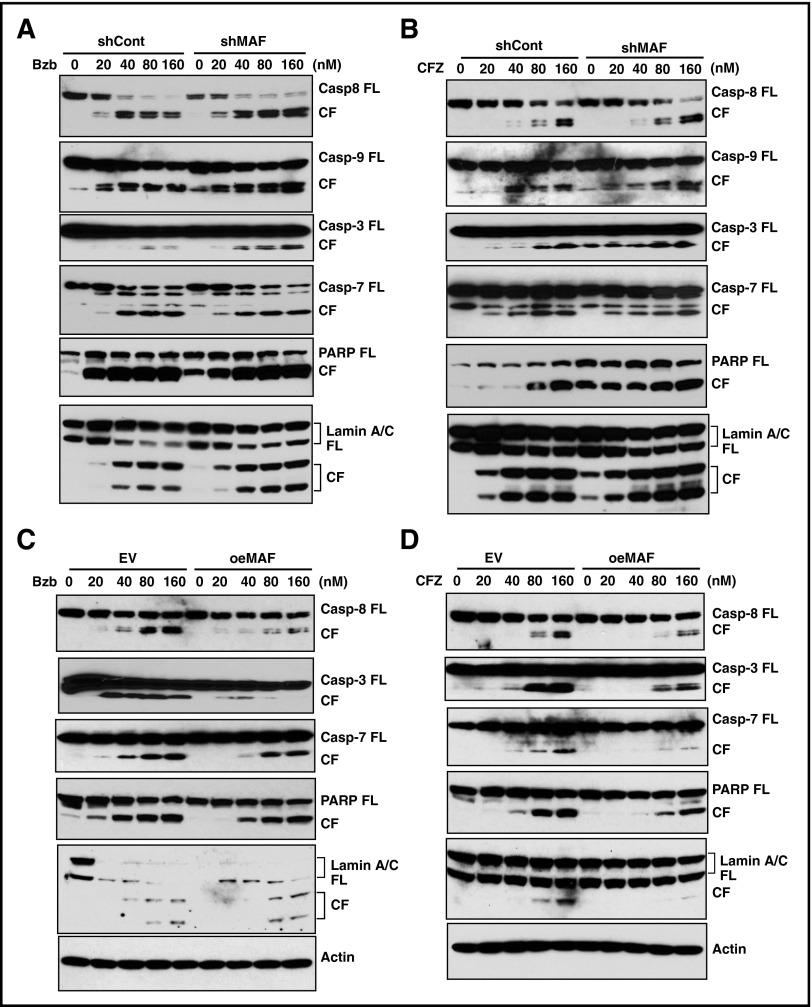
Because the caspases are important proteases of the apoptotic machinery,50 we next determined the role of MAF in PI-mediated apoptosis by evaluating the effect of Bzb on caspase activity in 8226/shMaf and 8226/shCon cells. The cleavage fragments (CFs) of caspase-8, an initiator caspase that functions as an active executioner of the caspase pathway,50 in shMAF were higher than those in shCon (Figure 7A). A similar increase in caspase-9 CFs, another initiator of caspase-induced apoptosis, was observed in shMAF. These results indicate that knockdown of MAF enhances Bzb-induced activation of caspase-8 and -9.
Figure 7.
MAF protein abrogated PI-induced activation of caspases, PARP, and lamin A/C. 8226/shMAF and shCon (A-B) or XG1OeMaf and XG1EV (C-D) cells were treated with serial concentrations of Bzb (A,C) or CFZ (B,D) for 16 hours. Cell lysates were resolved in 4% to 12% SDS-PAGE. Full-length (FL) and CFs of each indicated protein were determined by IBA using antibodies specifically recognizing FL and CFs of caspases-3, -6, -7, -8, and -9, as well as PARP and lamin A/C.
As caspases-3, -6, and -7 are known substrates of active caspase-8 and -9,50 we next assessed these cleavage forms in both shMAF and shCon cells. Increases in CFs of caspase-3 and -7 in shMAF were seen following Bzb treatment, compared with shCon, with no difference in caspase-6 (Figure 7A and data not shown). These results indicate MAF enhances Bzb-induced activation of caspase-3 and -7, but not caspase-6.
Next, we examined the effect of MAF on Bzb-induced activation of PARP, which helps to maintain cell viability and is one of major substrates of activated caspase-3.51,52 Increases in PARP CFs were seen in shMAF compared with shCon. Moreover, the intact 116-kDa PARP protein was degraded with an increase in PARP CFs in shMAF in response to Bzb. Thus, knockdown of MAF promotes degradation of PARP protein and enhances Bzb-induced activation of PARP.
Lamins are nuclear membrane structural components important for DNA replication and chromatin organization, and their cleavage results in nuclear deregulation and cell death.53 An increase in lamin A/C CFs in shMAF was seen following exposure to Bzb (Figure 7A). Similar increases in the CFs of caspases-3, -7, -8, -9, PARP, and lamin A/C were observed in shMAF treated by CFZ (Figure 7B). Therefore, knockdown of MAF enhances PI-induced activation of caspases-8 and -9, which in turn led to activation of caspases-3 and -7, finally resulting in the degradation of PARP and lamin A/C.
To further validate the mechanism underlying reduced PI-induced apoptosis in cells expressing MAF, we assessed the effect of MAF overexpression on caspase activation. As expected, Bzb-induced CF of caspases-8, -7, and -3, PARP, and lamin A/C were decreased in XG1OeMaf compared with XG1EV. Similar effects were seen in response to CFZ (Figure 7C-D and data not shown). These results provide further evidence that overexpression of MAF prevents activation of caspases, PARP, and lamin A/C during PI-induced apoptosis.
Discussion
In this work, we show that t(14;16)-positive cell lines tolerate exposure to Bzb and CFZ at doses up to 30-fold higher than the IC50 of HMCLs with t(4;14), t(11;14), or other molecular subgroups which are more sensitive. This laboratory observation is consistent with clinical studies in which patients in the t(14;16)/MAF subgroup do not benefit from the addition of the Bzb, in contrast to t(4;14) cases which have gained a major advantage.11-13 An assessment of the mechanisms by which t(14:16) patients with high MAF expression are resistant to PIs is important to both understand the biology of the disease as well as to provide a basis for future targeted therapeutic approaches.
It is recognized that dysregulation of MAF transcription in t(14;16) results from juxtaposing the MAF gene with the strong enhancer of the immunoglobulin H (IgH) locus.1,2,14,17,26,54 We demonstrate that MAF mRNA levels in t(14;16) are at a similar level to some cell lines with a t(4;14), but are lower in t(11;14) and other subgroups. These results are in agreement with previous studies1,14 using reverse transcription qPCR (RT-qPCR) analysis.34,40
High MAF transcription in t(4;14) is reported to be driven via MMSET and its activation of downstream MEK/ERK/Fos signaling.1 To validate the effect of MMSET on high expression of MAF transcription, we determined mRNA and protein levels in KMS11 and KMS11/TKO, in which the MMSET gene is disrupted by homologous recombination leading to the inactivation of the rearranged IGH-MMSET allele and KMS11/NKTO in which the wild-type MMSET or nontranslocated MMSET allele was inactivated.30 We did not find a difference in MAF mRNA or protein expression between KMS11, KMS11/TKO, and KMS11-NTKO using q-RT-PCR analysis and immunoblotting analysis (Y.-W.Q., S.Y., G.J.M., and F.E.D., unpublished data), suggesting that MMSET did not directly effect MAF transcription.
We have identified a new mechanism for the regulation of the stability of MAF protein via GSK3β activity and the proteasome. We found that levels of MAF protein are only partly correlated with the levels of its transcription. Eighty percent of HMCLs with t(14;16) have high levels of MAF protein: only ANBL6 had an intermediate level and KMS11, which has both a t(14;16) and t(4;14),55 had a low level. The baseline levels of MAF protein in t(4;14)s are much lower than in the t(14;16) subgroup despite comparable levels of mRNA, which is agreement with previous reports.1,14 Low levels of MAF protein in HMCLs expressing high mRNA suggest that posttranscriptional/translational regulation can govern the abundance of MAF protein. These results explain conflicting data in the literature in which high frequencies (30%) of MAF protein in the nucleus of primary MM plasma cells detected by immunohistochemistry were not in agreement with the frequency of the t(14;16) detected by fluorescence in situ hybridization analysis.56 High levels of MAF protein detected by immunoblotting analysis were seen in 80% of HMCLs. Besides MAF translocation, the mutation of other genes in cell lines might regulate MAF expression and protein stability.1
The use of inhibitors specific for GSK3β, MEK, or p38 kinase clearly demonstrated that inhibition of the MEK kinase and P38 kinase pathways had no effect on MAF protein degradation, whereas degradation was abrogated by GSK3β inhibition. We provide evidence that inhibition of GSK3β activity by IGF-1 prevents degradation of MAF independent of its mRNA transcriptional regulation. These data are consistent with blockade of GSK3β activity resulting in accumulation of MAF protein in MM cells.37 Previously, GSK3β has been shown to regulate MAFA protein stabilization, another member of the maf-family.46,57 In rat pancreatic β cells, GSK3 phosphorylates MAFA leading to MAFA degradation,46 and multiple MAFA serine and threonine residues have been identified as phosphorylated by GSK3β.57 The data presented in our study are consistent with these previous studies as the phosphorylation of MAF family proteins was independent of MEK inhibition.57 Sii-Felice et al demonstrated that the co-overexpression of the MAF family, with p38 in 293T fibroblast cells, led to gel-mobility shift of MAF-family members.58 These data are different to what we observed in myeloma where p38 did not seem to be involved. The discrepancy between our study and this previous report may be reconciled by differences in methods and cell types. For instance, although we used a p38 inhibitor to test whether p38 activity was required for regulating stability of MAF protein in MM cells, Sii-Felice et al used overexpression of MAF and p38 genes. Furthermore, we evaluated the stability of MAF protein in MM cells, whereas Sii-Felice et al used fibroblast 293T cells. It has been documented previously that the degradation of MAF-family proteins is differentially regulated in a cell-type–specific manner as proteasome-mediated stability of MAFA protein was observed in pancreatic β cells, but not in NIH3T3 fibroblasts.57 Taken together, GSK3β activity plays a central role in regulating the stability of MAF protein in MM and targeting GSK3β by increasing its activity might be a therapeutic option.
Many proteins including transcriptional factors are known to be substrates of GSK3β.59 Having observed that inhibition of GSK3β led to an accumulation of MAF protein, the question of whether PIs also regulate the stability of MAF was examined. Both Bzb and CFZ induce stabilization of MAF protein in a dose-dependent manner in t(14;16) cells. Thus, PIs play a key role in regulating the stability of MAF protein. It is important to note that PI-induced stabilization of MAF did not occur in cells lacking a t(14;16). The reason for this is unknown, although a number of possibilities can be considered. First, the level of the mRNA may be important. Second, it is likely that the activity of GSK3β is different between the subgroups as we observe that the GSK3β-mediated phosphorylated MAF protein only occurs in t(14;16) cells not in other subgroups. Finally, it is possible that there is a different amount of ubiquitination between the subgroups. In agreement with this proposal, a recent study has reported that glucocorticoid-induced degradation of MAF is associated with upregulation of ubiquitin-C mRNA.60 A further recent study has also suggested that ubiquitin ligase HERC4 interacts with MAF protein and promotes proteasome-mediated degradation, suggesting the level of HERC4 may be important.61
GSK3β induces phosphorylation of MAF and once phosphorylated, MAF undergoes proteasome-mediated degradation. In the presence of active GSK3β, phosphorylated MAF disappeared faster than unphosphorylated MAF as demonstrated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). In contrast, the loss of phosphorylation, by inhibition of GSK3β activity using a GSK3β inhibitor, resulted in loss of MAF degradation, leading to the slow disappearance of total MAF and the upper MAF protein band. Generally, phosphorylated proteins have a higher molecular weight and thus migrate more slowly in SDS-PAGE and appear as an upper band. The size of this upper band depends on the number of phosphorylation sites in the protein. Multiple phosphorylated sites are shared between MAF and MAFA in the N-terminal region,19 these are induced by GSK3β as well as unidentified kinase.57 For example, phosphorylation of Ser49, Thr53 and Thr57, and Ser61 of the MAFA is phosphorylated by GSK3β, whereas phosphorylation of Ser72 and Ser342 has yet to be identified.57 It is currently unknown which specific phosphorylated sites are responsible for the upper band of MAF in myeloma cells. Additionally, phosphorylation of MAFA by GSK3β requires priming phosphorylation of Ser65 by an unknown priming kinase, which is independent of GSK3β activity and thus is not affected by a GSK3β inhibitor. Taken together, these may account for the slow disappearance of the upper band of the MAF protein in the presence of the GSK3 inhibitor. Further studies are needed to answer these questions.
In conclusion, our findings suggest that high MAF protein in t(14;16) confers intrinsic resistance to PIs via abrogating PI-induced apoptosis and activation of caspases. The stabilization of MAF protein in t(14;16) is regulated by GSK3 and proteasome activity. PIs induce accumulation of MAF protein, which in turn enhances resistance to PIs. Identification of the important role of MAF protein in mediating resistance of t(14:16) myeloma to PIs and its underlying mechanism is not only critical to the better understanding of MM biology but also provides a basis for novel therapeutic approaches for targeting this subgroup.
Acknowledgments
The authors thank the faculty, staff, and patients of the Myeloma Institute. Thanks to Sarah Johnson for assistance with flow cytometry. The authors also thank the Laboratory of Microbiology, Core Facility of University of Arkansas for Medical Sciences for their help with image acquisition.
This work was supported in part by a grant from the National Institutes of Health, National Cancer Institute (grant number CA055819) and 2 Multiple Myeloma Research Foundation awards (Y.-W.Q.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: Y.-W.Q. conceptualized and designed the research, performed experiments, analyzed and interpreted the results, made figures, and wrote paper; S.Y. performed experiments and analyzed data; Y.C. performed experiments; A.F.B. and R.E. analyzed data; J.E. helped design the experimental studies and edited the manuscript; B.B. and F.v.R. provided patient materials; and G.J.M. and F.E.D. helped design experimental studies, provided patient materials, interpreted the data, and edited the manuscript.
Conflict-of-interest disclosure: B.B. is a coinventor on patents and patent applications related to use of gene expression profiling in cancer medicine that have been licensed to Myeloma Health, LLC, but has no financial interests in this company. The remaining authors declare no competing financial interests.
Correspondence: Ya-Wei Qiang, Myeloma Institute, Winthrop P. Rockefeller Cancer Institute, Room 914, University of Arkansas for Medical Sciences, 4301 W Markham St, Slot 776, Little Rock, AR 72205; e-mail: yqiang@uams.edu; and Faith E. Davies, Myeloma Institute, University of Arkansas for Medical Sciences, 4301 W Markham St, Slot 816, Little Rock, AR 72205; e-mail: fedavies@uams.edu.
References
- 1.Annunziata CM, Hernandez L, Davis RE, et al. A mechanistic rationale for MEK inhibitor therapy in myeloma based on blockade of MAF oncogene expression. Blood. 2011;117(8):2396-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chesi M, Bergsagel PL, Shonukan OO, et al. Frequent dysregulation of the c-maf proto-oncogene at 16q23 by translocation to an Ig locus in multiple myeloma. Blood. 1998;91(12):4457-4463. [PubMed] [Google Scholar]
- 3.Fonseca R, Barlogie B, Bataille R, et al. Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res. 2004;64(4):1546-1558. [DOI] [PubMed] [Google Scholar]
- 4.Usmani SZ, Heuck C, Mitchell A, et al. Extramedullary disease portends poor prognosis in multiple myeloma and is over-represented in high-risk disease even in the era of novel agents. Haematologica. 2012;97(11):1761-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avet-Loiseau H, Malard F, Campion L, et al. ; Intergroupe Francophone du Myélome. Translocation t(14;16) and multiple myeloma: is it really an independent prognostic factor? Blood. 2011;117(6):2009-2011. [DOI] [PubMed] [Google Scholar]
- 6.Munshi NC, Anderson KC, Bergsagel PL, et al. ; International Myeloma Workshop Consensus Panel 2. Consensus recommendations for risk stratification in multiple myeloma: report of the International Myeloma Workshop Consensus Panel 2. Blood. 2011;117(18):4696-4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergsagel PL, Mateos MV, Gutierrez NC, Rajkumar SV, San Miguel JF. Improving overall survival and overcoming adverse prognosis in the treatment of cytogenetically high-risk multiple myeloma. Blood. 2013;121(6):884-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd KD, Ross FM, Chiecchio L, et al. ; NCRI Haematology Oncology Studies Group. A novel prognostic model in myeloma based on co-segregating adverse FISH lesions and the ISS: analysis of patients treated in the MRC Myeloma IX trial. Leukemia. 2012;26(2):349-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12(5):335-348. [DOI] [PubMed] [Google Scholar]
- 10.Shaughnessy JD Jr, Zhan F, Burington BE, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109(6):2276-2284. [DOI] [PubMed] [Google Scholar]
- 11.Nair B, van Rhee F, Shaughnessy JD Jr, et al. Superior results of Total Therapy 3 (2003-33) in gene expression profiling-defined low-risk multiple myeloma confirmed in subsequent trial 2006-66 with VRD maintenance. Blood. 2010;115(21):4168-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pineda-Roman M, Zangari M, Haessler J, et al. Sustained complete remissions in multiple myeloma linked to bortezomib in total therapy 3: comparison with total therapy 2. Br J Haematol. 2008;140(6):625-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinhold N, Heuck CJ, Rosenthal A, et al. Clinical value of molecular subtyping multiple myeloma using gene expression profiling. Leukemia. 2016;30(2):423-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurt EM, Wiestner A, Rosenwald A, et al. Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer Cell. 2004;5(2):191-199. [DOI] [PubMed] [Google Scholar]
- 15.Fabris S, Agnelli L, Mattioli M, et al. Characterization of oncogene dysregulation in multiple myeloma by combined FISH and DNA microarray analyses. Genes Chromosomes Cancer. 2005;42(2):117-127. [DOI] [PubMed] [Google Scholar]
- 16.Rasmussen T, Knudsen LM, Dahl IM, Johnsen HE. C-MAF oncogene dysregulation in multiple myeloma: frequency and biological relevance. Leuk Lymphoma. 2003;44(10):1761-1766. [DOI] [PubMed] [Google Scholar]
- 17.Moreaux J, Hose D, Jourdan M, et al. TACI expression is associated with a mature bone marrow plasma cell signature and C-MAF overexpression in human myeloma cell lines. Haematologica. 2007;92(6):803-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishizawa M, Kataoka K, Goto N, Fujiwara KT, Kawai S. v-maf, a viral oncogene that encodes a “leucine zipper” motif. Proc Natl Acad Sci USA. 1989;86(20):7711-7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eychène A, Rocques N, Pouponnot C. A new MAFia in cancer. Nat Rev Cancer. 2008;8(9):683-693. [DOI] [PubMed] [Google Scholar]
- 20.Morito N, Yoh K, Fujioka Y, et al. Overexpression of c-Maf contributes to T-cell lymphoma in both mice and human. Cancer Res. 2006;66(2):812-819. [DOI] [PubMed] [Google Scholar]
- 21.Murakami YI, Yatabe Y, Sakaguchi T, et al. c-Maf expression in angioimmunoblastic T-cell lymphoma. Am J Surg Pathol. 2007;31(11):1695-1702. [DOI] [PubMed] [Google Scholar]
- 22.Mattioli M, Agnelli L, Fabris S, et al. Gene expression profiling of plasma cell dyscrasias reveals molecular patterns associated with distinct IGH translocations in multiple myeloma. Oncogene. 2005;24(15):2461-2473. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki A, Iida S, Kato-Uranishi M, et al. ARK5 is transcriptionally regulated by the large-MAF family and mediates IGF-1-induced cell invasion in multiple myeloma: ARK5 as a new molecular determinant of malignant multiple myeloma. Oncogene. 2005;24(46):6936-6944. [DOI] [PubMed] [Google Scholar]
- 24.Neri P, Ren L, Azab AK, et al. Integrin β7-mediated regulation of multiple myeloma cell adhesion, migration, and invasion. Blood. 2011;117(23):6202-6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Natkunam Y, Tedoldi S, Paterson JC, et al. Characterization of c-Maf transcription factor in normal and neoplastic hematolymphoid tissue and its relevance in plasma cell neoplasia. Am J Clin Pathol. 2009;132(3):361-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergsagel PL, Kuehl WM, Zhan F, Sawyer J, Barlogie B, Shaughnessy J Jr. Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood. 2005;106(1):296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiang YW, Endo Y, Rubin JS, Rudikoff S. Wnt signaling in B-cell neoplasia. Oncogene. 2003;22(10):1536-1545. [DOI] [PubMed] [Google Scholar]
- 28.Qiang YW, Kopantzev E, Rudikoff S. Insulinlike growth factor-I signaling in multiple myeloma: downstream elements, functional correlates, and pathway cross-talk. Blood. 2002;99(11):4138-4146. [DOI] [PubMed] [Google Scholar]
- 29.Zhan F, Hardin J, Kordsmeier B, et al. Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood. 2002;99(5):1745-1757. [DOI] [PubMed] [Google Scholar]
- 30.Lauring J, Abukhdeir AM, Konishi H, et al. The multiple myeloma associated MMSET gene contributes to cellular adhesion, clonogenic growth, and tumorigenicity. Blood. 2008;111(2):856-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiang YW, Walsh K, Yao L, et al. Wnts induce migration and invasion of myeloma plasma cells. Blood. 2005;106(5):1786-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiang YW, Shaughnessy JD Jr, Yaccoby S. Wnt3a signaling within bone inhibits multiple myeloma bone disease and tumor growth. Blood. 2008;112(2):374-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhan F, Huang Y, Colla S, et al. The molecular classification of multiple myeloma. Blood. 2006;108(6):2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiang YW, Hu B, Chen Y, et al. Bortezomib induces osteoblast differentiation via Wnt-independent activation of beta-catenin/TCF signaling. Blood. 2009;113(18):4319-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiang YW, Chen Y, Stephens O, et al. Myeloma-derived Dickkopf-1 disrupts Wnt-regulated osteoprotegerin and RANKL production by osteoblasts: a potential mechanism underlying osteolytic bone lesions in multiple myeloma. Blood. 2008;112(1):196-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu B, Chen Y, Usmani SZ, et al. Characterization of the molecular mechanism of the bone-anabolic activity of carfilzomib in multiple myeloma. PLoS One. 2013;8(9):e74191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herath NI, Rocques N, Garancher A, Eychène A, Pouponnot C. GSK3-mediated MAF phosphorylation in multiple myeloma as a potential therapeutic target. Blood Cancer J. 2014;4:e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiang YW, Kitagawa M, Higashi M, Ishii G, Morimoto C, Harigaya K. Activation of mitogen-activated protein kinase through alpha5/beta1 integrin is required for cell cycle progression of B progenitor cell line, Reh, on human marrow stromal cells. Exp Hematol. 2000;28(10):1147-1157. [DOI] [PubMed] [Google Scholar]
- 39.Coghlan MP, Culbert AA, Cross DA, et al. Selective small molecule inhibitors of glycogen synthase kinase-3 modulate glycogen metabolism and gene transcription. Chem Biol. 2000;7(10):793-803. [DOI] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402-408. [DOI] [PubMed] [Google Scholar]
- 41.Kaplan EL, Maier P. Nonparametric estimation from incomplete observation. J Am Stat Assoc. 1958;53:457-481. [Google Scholar]
- 42.Wilcoxon F. Individual comparisions by ranking methods. Biometrics. 1945;1:80-83. [Google Scholar]
- 43.Moreaux J, Klein B, Bataille R, et al. A high-risk signature for patients with multiple myeloma established from the molecular classification of human myeloma cell lines. Haematologica. 2011;96(4):574-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider-Poetsch T, Ju J, Eyler DE, et al. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol. 2010;6(3):209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Semënov MV, Snyder M. Human dishevelled genes constitute a DHR-containing multigene family. Genomics. 1997;42(2):302-310. [DOI] [PubMed] [Google Scholar]
- 46.Rocques N, Abou Zeid N, Sii-Felice K, et al. GSK-3-mediated phosphorylation enhances Maf-transforming activity. Mol Cell. 2007;28(4):584-597. [DOI] [PubMed] [Google Scholar]
- 47.Shaw M, Cohen P, Alessi DR. Further evidence that the inhibition of glycogen synthase kinase-3beta by IGF-1 is mediated by PDK1/PKB-induced phosphorylation of Ser-9 and not by dephosphorylation of Tyr-216. FEBS Lett. 1997;416(3):307-311. [DOI] [PubMed] [Google Scholar]
- 48.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378(6559):785-789. [DOI] [PubMed] [Google Scholar]
- 49.Cross DA, Alessi DR, Vandenheede JR, McDowell HE, Hundal HS, Cohen P. The inhibition of glycogen synthase kinase-3 by insulin or insulin-like growth factor 1 in the rat skeletal muscle cell line L6 is blocked by wortmannin, but not by rapamycin: evidence that wortmannin blocks activation of the mitogen-activated protein kinase pathway in L6 cells between Ras and Raf. Biochem J. 1994;303(Pt 1):21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McIlwain DR, Berger T, Mak TW. Caspase functions in cell death and disease. Cold Spring Harb Perspect Biol. 2013;5(4):a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tewari M, Quan LT, O’Rourke K, et al. Yama/CPP32 beta, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell. 1995;81(5):801-809. [DOI] [PubMed] [Google Scholar]
- 52.Oliver FJ, de la Rubia G, Rolli V, Ruiz-Ruiz MC, de Murcia G, Murcia JM. Importance of poly(ADP-ribose) polymerase and its cleavage in apoptosis. Lesson from an uncleavable mutant. J Biol Chem. 1998;273(50):33533-33539. [DOI] [PubMed] [Google Scholar]
- 53.Gruenbaum Y, Wilson KL, Harel A, Goldberg M, Cohen M. Review: nuclear lamins--structural proteins with fundamental functions. J Struct Biol. 2000;129(2-3):313-323. [DOI] [PubMed] [Google Scholar]
- 54.Fonseca R, Bergsagel PL, Drach J, et al. ; International Myeloma Working Group. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23(12):2210-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stengel C, Cheung CW, Quinn J, Yong K, Khwaja A. Optimal induction of myeloma cell death requires dual blockade of phosphoinositide 3-kinase and mTOR signalling and is determined by translocation subtype. Leukemia. 2012;26(8):1761-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang H, Qi Q, Xu W, Patterson B. c-Maf nuclear oncoprotein is frequently expressed in multiple myeloma. Leukemia. 2007;21(7):1572-1574. [DOI] [PubMed] [Google Scholar]
- 57.Han SI, Aramata S, Yasuda K, Kataoka K. MafA stability in pancreatic beta cells is regulated by glucose and is dependent on its constitutive phosphorylation at multiple sites by glycogen synthase kinase 3. Mol Cell Biol. 2007;27(19):6593-6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sii-Felice K, Pouponnot C, Gillet S, et al. MafA transcription factor is phosphorylated by p38 MAP kinase. FEBS Lett. 2005;579(17):3547-3554. [DOI] [PubMed] [Google Scholar]
- 59.Jope RS, Johnson GV. The glamour and gloom of glycogen synthase kinase-3. Trends Biochem Sci. 2004;29(2):95-102. [DOI] [PubMed] [Google Scholar]
- 60.Mao X, Stewart AK, Hurren R, et al. A chemical biology screen identifies glucocorticoids that regulate c-maf expression by increasing its proteasomal degradation through up-regulation of ubiquitin. Blood. 2007;110(12):4047-4054. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Z, Tong J, Tang X, et al. The ubiquitin ligase HERC4 mediates c-Maf ubiquitination and delays the growth of multiple myeloma xenografts in nude mice. Blood. 2016;127(13):1676-1686. [DOI] [PubMed] [Google Scholar]