Lung cancer is the leading cause of cancer-related death in the United States and worldwide. While the 5-year survival for lung cancer is a dismal 15%, there have been significant advances in treating patients with non-small cell lung cancer (NSCLC) in the past decade. These recent advances stem from the shift to classifying patients with NSCLC into distinct subtypes based upon the molecular driver, which include KRAS mutations (~25%), EGFR mutations (~15%), ALK translocations (~8%), and MET amplification/mutations (~6-7%) (1,2). A subset of patients, such as those patients with EGFR- and ALK-driven NSCLC, have benefitted significantly from the use of tyrosine-kinase inhibitors (TKIs) that specifically inhibit these oncogenes. While those patients with EGFR-mutant NSCLC have benefitted from the use of EGFR TKIs, therapeutic resistance to these targeted therapies is inevitable (3). Studies characterizing the resistance mechanisms to EGFR inhibitors (EGFRi) have identified multiple mechanisms, which include gatekeeper mutations (T790M) within EGFR (49%), MET amplification (5%), and transformation to small-cell lung cancer (14%) (3). Interestingly, an epithelial-mesenchymal transition (EMT) phenotype has been identified as a driver of resistance to EGFRi in potentially as many as 20% of EGFR-mutant lung cancer patients (4). Initial studies investigating the role of EMT in EGFRi resistance identified the AXL kinase and its ligand GAS6 as drivers of EMT-mediated EGFRi resistance (4). However, little is known about other drivers of EMT-mediated resistance. Additional studies are needed to identify both novel drivers of EMT and therapeutic strategies to overcome EMT-mediated EGFRi resistance.
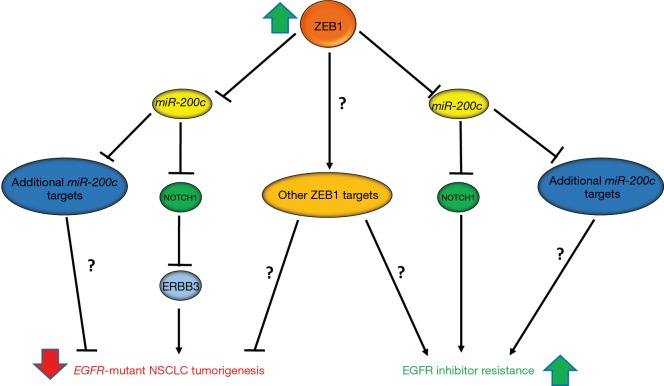
Interestingly, Zhang et al. recently demonstrated that the EMT transcription factor, ZEB1 is a potential driver of EMT-mediated resistance to EGFRi in EGFR-mutant NSCLC (5). In this report, the authors demonstrated that ZEB1 has opposing functions in EGFR-mutant NSCLC, both inhibiting EGFR-driven tumorigenesis and promoting resistance to EGFRi (Figure 1). The function of ZEB1 as a tumor suppressor appears to be oncogenic driver specific as ZEB1 expression inhibited both soft agar and xenograph growth in EGFR-mutant NSCLC cell lines, while it promoted soft agar and xenograph growth in KRAS-mutant NSCLC. However, this tumor suppressive role of ZEB1 seems to be independent of its ability to induce EMT, given that ZEB1 induced EMT in both KRAS- and EGFR-mutant cell lines. Further investigation of the mechanisms by which ZEB1 suppressed EGFR-driven NSCLC revealed that ZEB1 suppression of miR-200c expression was required for this tumor suppressive phenotype. ZEB1 has been previously established to inhibit miR-200c expression, and suppression of miR-200c is required for ZEB1-mediated EMT and invasion (5,6). Interestingly, the authors were able to demonstrate that a target of miR-200c, NOTCH1, directly suppresses ERRB3 expression, a member of the EGFR-family of receptors that has been shown to be required for EGFR-driven lung cancer (5). While the authors clearly demonstrated that the ZEB1-miR-200c-NOTCH1 signaling axis is important for suppression of ERBB3 (Figure 1), given the lack of phenotypic data presented, it remains an unanswered question if ZEB1 suppression of ERBB3 is required for ZEB1-suppression of EGFR-driven NSCLC. Additionally, the data presented suggests that there are other miR-200c targets and potentially other ZEB1 target genes or signaling pathways that play a role in the tumor suppressive function of ZEB1 (Figure 1).
Figure 1.
ZEB1 suppresses EGFR-mutant non-small cell lung cancer (NSCLC) tumorigenesis and mediates EGFR inhibitor resistance. In the discussed report ZEB1 was shown to inhibit EGFR-mutant NSCLC by repressing miR-200c expression, which subsequently allows for a target gene of miR-200c, NOTCH1, to repress ERBB3 expression. ZEB1 suppression of ERBB3 expression appears to be an important mechanism through which ZEB1 suppresses EGFR-mutant tumorigenesis. However, there may be additional miR-200c targets and potentially other ZEB1 targets that are important for the tumor suppressive role of ZEB1. Conversely, ZEB1 suppression of miR-200c expression and subsequent derepression of NOTCH1 was also shown to be required for ZEB1-mediated EGFR tyrosine kinase inhibitor resistance in NSCLC cell lines. Similar to the tumor suppressive function of ZEB1, ZEB1-mediated resistance most likely involves miR-200c targets, beyond NOTCH1, and may require additional ZEB1 target genes. Future studies are required to further delineate the mechanisms of ZEB1-mediated suppression of EGFR-mutant NSCLC and EGFR inhibitor resistance.
While a previous report demonstrated that ZEB1 is required for EGFRi resistance in a subset of EGFR-driven NSCLC, Zhang et al. provided additional evidence for ZEB1 as a mediator of therapeutic resistance to EGFRi (5,7). The authors demonstrated that ZEB1 overexpression is sufficient to cause gefitinib resistance in a gefitinib sensitive EGFR-mutant cell line. Additionally, this resistance is dependent upon the ability of ZEB1 to suppress miR-200c and partially dependent upon NOTCH1 expression (Figure 1). The authors demonstrated γ-secretase inhibitors (BMS-708163 and DAPT) are able to partially overcome gefitinib resistance in a gefitinib resistant cell line. However, the in vitro evidence provided with γ-secretase inhibitors suggests a relatively minor role for NOTCH1 in the setting of ZEB1-mediated resistance. The data presented suggests that there are other miR-200c targets or ZEB1 target genes that are important for ZEB1-mediated resistance (Figure 1). Additionally, given the non-selectivity and dose-limiting systemic toxicities of γ-secretase inhibitors, there may be a limited role for such therapies in the setting of EMT-mediated resistance (8). However, monoclonal antibody therapies that specifically target NOTCH1 and one of its ligands, Delta-like 4 (DLL4), have been developed and some of which are in early phase trials, may provide a more advantageous therapeutic strategy given their selectivity for the NOTCH1 signaling pathway (8). Further studies, specifically in vivo, are needed to explore efficacy and potential toxicities of therapies that combine EGFR and NOTCH1 inhibition. Additionally, in order to determine the clinical relevance of ZEB1-mediated therapeutic resistance, studies are needed to determine how frequently ZEB1 is overexpressed in patient tumors in the EGFRi resistance setting.
While other EMT transcription factors (EMT-TF), such as ZEB2, the TWIST and SNAIL proteins, have been implicated in therapeutic resistance, it remains unknown if other EMT-TFs play a role in resistance to EGFRi. Interestingly, ZEB2 has been shown to inhibit radiation-induced apoptosis by suppressing the DNA-damage response. The ability of ZEB2 to suppress DNA-damage induced apoptosis seems to be largely independent of its ability to induce EMT (9,10). Additionally, TWIST1 has been shown to lead to chemoresistance in both breast and lung cancer, through direct induction of AKT and modulation of BCL-2 family members, respectively (11,12). SNAIL1 and SNAIL2 have both been linked to chemotherapy and radiation resistance through their ability to inhibit p53-mediated apoptosis, by repressing expression of multiple genes within the p53 pathway (10,13). In addition to the evidence that multiple EMT-TFs mediate therapeutic resistance, there is extensive data that EMT-TFs are often co-expressed in tumors and in many model systems, EMT-TFs can increase expression, often directly, of other EMT-TFs (6,14,15). Given the interconnectedness of the EMT-TFs signaling networks and potential redundancy in their functions, additional studies are needed to determine if other EMT-TFs mediate resistance to EGFRi and if ZEB1-driven resistance occurs independently of other EMT-TFs.
An aspect of EMT-TFs that remains largely unexplored is whether the EMT-TF mediated therapeutic resistance is EMT-dependent or -independent. A previous report on AXL-driven erlotinib-resistance in EGFR-driven NSCLC revealed that AXL-mediated resistance required EMT (4). More specifically, both AXL-mediated EGFRi resistance and AXL-mediated migration and adhesion in this model system required vimentin expression (4). While Zhang et al. and others have implicated ZEB1 in EGFRi resistance, it is largely unclear if EMT is simply a consequence of ZEB1 overexpression or if EMT is required for ZEB1-mediated resistance (5,7). Interestingly, TWIST1 has been shown to inhibit oncogene-induce senescence (OIS) and apoptosis in lung cancer, largely independently of EMT (16,17). A previous study demonstrated that ZEB1 can suppress OIS following EGFR overexpression in esophageal cells (18). The ability of ZEB1 to suppress OIS in this model system occurred in the context of EMT. However, ZEB1 was recently shown to mediate radioresistance via increasing CHK1 stabilization and subsequent homologous recombination, a mechanism that seems to be independent of its EMT-function (10,19).
Future studies elucidating the role of EMT in ZEB-1 mediated EGFRi resistance are needed and may reveal new therapeutic targets and strategies to overcome EGFRi resistance. Given that AXL kinase has been established as an important driver of EMT in lung cancer (4), if ZEB1-mediated resistance proves to require EMT, the relationship between AXL and ZEB1 should be explored. A previous study in hematologic cells demonstrated that activation of AXL can induce expression of TWIST1 (20). A similar relationship may exist for ZEB1. If ZEB1 and AXL signaling pathways prove to be connected, use of small molecule AXL kinase inhibitors may be a therapeutic option to overcome ZEB1-mediated EGFRi resistance. Alternatively, if ZEB1 proves to mediate EGFRi resistance independently of EMT, ZEB1 suppression of apoptosis and modulation of members of the BCL-2 family of proteins should be explored. Given the recent FDA approval of the BCL-2 inhibitor, ABT-199, and additional BCL-2, BCL-XL, and MCL-1 inhibitors currently under investigation, there are potential novel strategies to use these BCL-2 inhibitors to overcome ZEB1-suppression of apoptosis in response to EGFRi. Ultimately, the identification and development of direct inhibitors of ZEB1 and other EMT-TFs may have significant clinical impact; however, this has remained an elusive goal.
In summary, Zhang et al. demonstrate unique divergent functions of ZEB1 as both a tumor suppressor and driver of EGFRi resistance in EGFR-driven NSCLC. The authors show that ZEB1 suppression of ERBB3 expression through the ZEB1-miR-200c-NOTCH1 signaling axis may be a mechanism by which ZEB1 suppresses EGFR-driven NSCLC tumorigenesis. Contrasting this tumor suppressive function of ZEB1, the authors present evidence that ZEB1 mediates EGFRi resistance in EGFR-mutant NSCLC cell lines. ZEB1-mediated resistance requires ZEB1-suppresion of miR-200c, partially requires NOTCH1 expression, and is associated with EMT. Furthermore, the authors demonstrate that gefitinib resistance can be partially overcome with γ-secretase inhibitors, providing evidence for use of inhibitors of the NOTCH1 signaling pathway in the setting of EGFRi resistance. Future studies are needed to further elucidate the mechanisms through which ZEB1-mediates therapeutic resistance which may lead to novel combination therapies to combat EGFRi resistance.
Acknowledgements
None.
Footnotes
Provenance: This is an invited Commentary commissioned by the Section Editor Long Jiang (Second Affiliated Hospital, Institute of Respiratory Diseases, Zhejiang University School of Medicine, Hangzhou, China).
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Cancer Genome Atlas Research Network . Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014;311:1998-2006. 10.1001/jama.2014.3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. 10.1126/scitranslmed.3002003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Z, Lee JC, Lin L, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet 2012;44:852-60. 10.1038/ng.2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang T, Guo L, Creighton CJ, et al. A genetic cell context-dependent role for ZEB1 in lung cancer. Nat Commun 2016;7:12231. 10.1038/ncomms12231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2014;15:178-96. 10.1038/nrm3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshida T, Song L, Bai Y, et al. ZEB1 Mediates Acquired Resistance to the Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors in Non-Small Cell Lung Cancer. PLoS One 2016;11:e0147344. 10.1371/journal.pone.0147344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takebe N, Miele L, Harris PJ, et al. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol 2015;12:445-64. 10.1038/nrclinonc.2015.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sayan AE, Griffiths TR, Pal R, et al. SIP1 protein protects cells from DNA damage-induced apoptosis and has independent prognostic value in bladder cancer. Proc Natl Acad Sci U S A 2009;106:14884-9. 10.1073/pnas.0902042106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ansieau S, Collin G, Hill L. EMT or EMT-Promoting Transcription Factors, Where to Focus the Light? Front Oncol 2014;4:353. 10.3389/fonc.2014.00353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin HO, Hong SE, Woo SH, et al. Silencing of Twist1 sensitizes NSCLC cells to cisplatin via AMPK-activated mTOR inhibition. Cell Death Dis 2012;3:e319. 10.1038/cddis.2012.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ansieau S, Morel AP, Hinkal G, et al. TWISTing an embryonic transcription factor into an oncoprotein. Oncogene 2010;29:3173-84. 10.1038/onc.2010.92 [DOI] [PubMed] [Google Scholar]
- 13.Kurrey NK, Jalgaonkar SP, Joglekar AV, et al. Snail and slug mediate radioresistance and chemoresistance by antagonizing p53-mediated apoptosis and acquiring a stem-like phenotype in ovarian cancer cells. Stem Cells 2009;27:2059-68. 10.1002/stem.154 [DOI] [PubMed] [Google Scholar]
- 14.Casas E, Kim J, Bendesky A, et al. Snail2 is an essential mediator of Twist1-induced epithelial mesenchymal transition and metastasis. Cancer Res 2011;71:245-54. 10.1158/0008-5472.CAN-10-2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol 2014;16:488-94. 10.1038/ncb2976 [DOI] [PubMed] [Google Scholar]
- 16.Tran PT, Shroff EH, Burns TF, et al. Twist1 suppresses senescence programs and thereby accelerates and maintains mutant Kras-induced lung tumorigenesis. PLoS Genet 2012;8:e1002650. 10.1371/journal.pgen.1002650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burns TF, Dobromilskaya I, Murphy SC, et al. Inhibition of TWIST1 leads to activation of oncogene-induced senescence in oncogene-driven non-small cell lung cancer. Mol Cancer Res 2013;11:329-38. 10.1158/1541-7786.MCR-12-0456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohashi S, Natsuizaka M, Wong GS, et al. Epidermal growth factor receptor and mutant p53 expand an esophageal cellular subpopulation capable of epithelial-to-mesenchymal transition through ZEB transcription factors. Cancer Res 2010;70:4174-84. 10.1158/0008-5472.CAN-09-4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang P, Wei Y, Wang L, et al. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat Cell Biol 2014;16:864-75. 10.1038/ncb3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merindol N, Riquet A, Szablewski V, et al. The emerging role of Twist proteins in hematopoietic cells and hematological malignancies. Blood Cancer J 2014;4:e206. 10.1038/bcj.2014.22 [DOI] [PMC free article] [PubMed] [Google Scholar]