Abstract
Paracoccidioides brasiliensis and Paracoccidioides lutzii are dimorphic fungi and are the etiological agents of paracoccidioidomycosis (PCM). Adhesion is one of the most important steps in infections with Paracoccidioides and is responsible for the differences in the virulence of isolates of these fungi. Because of the importance of adhesion to the establishment of an infection, this study focused on the preliminary development of a new therapeutic strategy to inhibit adhesion by Paracoccidioides, thus inhibiting infection and preventing the disease. We used two phage display libraries to select peptides that strongly bind to the Paracoccidioides cell wall to inhibit adhesion to host cells and extracellular matrix (ECM) components (laminin, fibronectin, and type I and type IV collagen). This approach allowed us to identify four peptides that inhibited up to 64% of the adhesion of Paracoccidioides to pneumocytes in vitro and inhibited the adhesion to the ECM components by up to 57%. Encouraged by these results, we evaluated the ability of these peptides to protect Galleria mellonella from Paracoccidioides infection by treating G. mellonella larvae with the different peptides prior to infection with Paracoccidioides and observing larval survival. The results show that all of the peptides tested increased the survival of the larvae infected with P. brasiliensis by up to 64% and by up to 60% in those infected with P. lutzii. These data may open new horizons for therapeutic strategies to prevent PCM, and anti-adhesion therapy could be an important strategy.
Keywords: adhesion, anti-adhesive peptides, phage display, Paracoccidioides spp., paracoccidioidomycosis
Introduction
Paracoccidioidomycosis (PCM) is a systemic mycosis caused by the dimorphic fungi Paracoccidioides brasiliensis and Paracoccidioides lutzii that occurs in Latin America and in Brazil, which is the country with the largest number of endemic areas for this disease in the world. The incidence of the disease in endemic areas has been estimated to be approximately one to three clinical cases per 100,000 inhabitants per year (Coutinho et al., 2002), and it is an important public health issue.
An infection with Paracoccidioides spp. begins with the adhesion of fungal cells to host cells, which is mediated by a special class of proteins present on the cell wall known as adhesins (Silva, 2008). The pathogen’s ability to colonize and invade the host tissue is strictly dependent on these proteins and the adhesion capacity of the fungus. de Oliveira et al. (2015), using in vivo animal models, demonstrated that the ability to express adhesins and adhere to the host are decisive factors in the virulence of different Paracoccidioides spp. isolates.
The inhibition or blocking of fungal adhesion to host cells may be an innovative and efficient way to prevent infection. Such strategies would reduce fungal colonization of different host tissues and nutrient acquisition and, consequently, facilitate the actions of the host immune system to fight the infection. This type of therapy is known as anti-adhesion therapy and can be an effective way to improve the efficacy of PCM treatments. In support of this idea, de Oliveira et al. (2015) demonstrated that when blocking two important Paracoccidioides spp. adhesins, enolase and 14-3-3, using specifics antibodies, host organisms were able to resist fungal infection and showed increased survival.
Anti-adhesive compounds can competitively inhibit adhesion by mimicking either microbes or host cell ligands. Alternatively, antibodies that recognize the surface epitopes on the pathogen can be used to block adhesion and, in the process, can actively or passively immunize the host (Krachler and Orth, 2013).
Resistance to anti-adhesive agents may also be expected to emerge. Resistance to drugs arises spontaneously in a population through mutations. The persistent use of antibiotics will result in the death of all non-resistant bacteria. Therefore, only those with mutations conferring resistance can propagate, resulting in the quick spread of resistance in a population. However, unlike with the use of antibiotics, which kill or stop the growth of susceptible microorganisms, during anti-adhesion therapy, nonresistant strains can continue to propagate and be transmitted to new hosts. Any wild-type strains would continue to compete with resistant strains in untreated individuals. This would potentially allow sensitive and resistant organisms to propagate and be transmitted at equivalent rates, dramatically slowing the emergence of a predominantly resistant population (Ofek et al., 2003; Cozens and Read, 2012).
Phage display has been extremely important for identifying and characterizing new high-affinity ligands and their receptors in the context of specific diseases. It has also been helpful for the identification of molecules with different applications, from diagnostic biomarkers to potential targets for use in the treatment of different infections (Lionakis et al., 2005; Antonara et al., 2007; Mullen et al., 2007; Shkoporov et al., 2008; Posadas et al., 2012), since the selected peptides frequently have biological activity related to the nature of the molecule or cell in a study.
These characteristics of phage display allow for the identification of therapeutic targets relevant to many biological processes in an organism and simultaneously allow for the isolation and characterization of peptide antagonists or agonists for specific targets. Therefore, peptides isolated via phage display can be exploited for the development of novel therapeutic agents for use in rational drug design, targeted therapy, gene therapy, vaccine production, diagnostic testing, and other applications (Sergeeva et al., 2006; Giordano et al., 2009).
This study proposed the use of the phage display technique to select peptides that interacted with molecules and receptors involved in the interaction of Paracoccidioides spp. with host and in that way, evaluated these selected peptides as candidates for anti-adhesive molecules to be a new alternative therapy against PCM.
Materials and Methods
Paracoccidioides Isolates
Two Paracoccidioides isolates were used: Pb18 (P. brasiliensis) and Pl01 (P. lutzii), maintained in vitro in Fava Netto’s medium at 37°C (Fava Netto, 1961). The handle of Paracoccidioides spp. cultures was made in a Class II Biological Safety Cabinet according recommendations of American Type Culture Collection (ATCC) and Brazilian Ministry of Health, Secretariat of Science, Technology and Strategic Inputs (Ministério da Saúde and Estratégicos, 2006).
Phage Display Peptides Libraries X6 and CX8C
Phage libraries were produced using the vector fUSE55 and the methodology developed by George Smith (Smith and Scott, 1993), with modifications to improve the efficiency and the number of final clones (Koivunen et al., 1993; Koivunen et al., 1999). The libraries were produced and provided by Dr. Ricardo José Giordano from the Laboratory of Vascular Biology, University of Sao Paulo (USP) and were X6 (a linear library) and CX8C (a cyclic library; X represents any amino acid, and C = cysteine that allows for cyclical peptides) libraries (Michaloski et al., Submitted).
Selection of Peptides That Bind to P. brasiliensis
The BRASIL (Biopanning and Rapid Analysis of Selective Interactive Ligands) technique (Giordano et al., 2001) was used for the selection of peptides that bind to Paracoccidioides spp. This technique consists of the selection of phages that bind to live cells and the use of a two-phase system to separate the cells and phages in a single centrifugation step. First, 109 transduction units (TU) of each phage display library were separately incubated with Saccharomyces cerevisiae (S288C) (106 cells) in 100 μl of PBS. After 3 h on ice, the suspensions containing the phages and the cells were transferred to the top of a nonmiscible organic lower phase known as BRASIL oil [dibutyl phthalate:cyclohexene (9:1) v/v] and centrifuged (10,000 × g, 10 min, 25°C). After centrifugation, the supernatants containing phages that did not bind to S. cerevisiae were recovered and transferred to a suspension of 106 Pb18 cells. After 3 h on ice, the cell suspensions were again transferred on to BRASIL oil and centrifuged at 10,000 × g for 10 min. The pellets of Pb18 cells that formed at the bottom of the organic phase were collected, and the phages bound to the cells were recovered via infection with log-phase E. coli K91 in LB medium supplemented with kanamycin and tetracycline (100 μg/mL and 20 μg/mL, respectively) and incubated at 37°C, with rotation at 250 rpm, for 20 h. The amplified phages were recovered from the culture medium via precipitation with polyethylene-glycol/NaCl and were used for subsequent rounds of selection (second and third) to further enrich for phages from each library that bound to Pb18 cells.
Binding Assays
Paracoccidioides brasiliensis, P. lutzii, and S. cerevisiae cells were incubated with 109 TU of the phage to be tested, as described above. After 2 h of incubation on ice, they were transferred to the top of BRASIL oil as previously described in the item 2.4 and centrifuged (10,000 × g, 10 min, 25°C). The pellet was placed in a new tube, and the phages were recovered via infection with E. coli K91. Serial dilutions were seeded onto LB/Kan/Tet plates to quantify the number of phage bound to each receiver. As a control P. brasiliensis and P. lutzii cells were incubated with 109 of the insertless phage Fd-tet. Three independent experiments were performed.
Inhibition of Paracoccidioides spp. Adhesion to Extracellular Matrix (ECM) Components by the Selected Peptides Using Enzyme Linked Immunosorbent Assay (ELISA)
Ninety-six-well plates were separately pre-coated with 10 μg/mL of each tested ECM component (laminin, fibronectin, and type I and type IV collagen) in a 0.05 M carbonate buffer (pH 9.6) at room temperature for 1 h. The plates were then washed with PBS/Tween-20, 0.05% (PBS-T) and blocked with 1% bovine serum albumin (BSA) for 2 h at 37°C. The two Paracoccidioides isolates (Pb18 and Pl01, 106 cells/mL) were incubated with 200 μg/mL of each studied peptide, which were synthesized by the Chinese Peptide Company (China), at 37°C, with rotation at 250 rpm, for 1 h. These inocula were incubated with the ECM components for 15 h at 37°C. The plates were washed with PBS-T, and an anti-cell free Paracoccidioides antibody (1:100 in PBS-T and 0.5% BSA) (Blotta and Camargo, 1993) was added for 1 h at 37°C. Another wash cycle was performed, and a secondary antibody, anti-rabbit IgG-HRP (1:2000 in PBS-T and 0.5% BSA), was added for 1 h at 37°C. The colorimetric reaction was observed by adding a solution of 0.2 M sodium phosphate, 0.1 M citric acid, 0.4 mg/mL o-phenylenediamine, and 30% hydrogen peroxide for 10 min, followed by the addition of 3 M hydrochloric acid. Absorbance was measured at 490 nm and converted to percent inhibition. For this conversion, Paracoccidioides spp. cells without treatment were used as a control, and the absorbance of this control was set as 100% adhesion. The difference in the absorbance in the treated Paracoccidioides spp. cells relative to absorbance in the control was considered the percentage of adhesion inhibition. This experiment was performed in triplicate, with three independent experiments.
Inhibition of Paracoccidioides spp. Adhesion to A549 Pneumocytes Cell Line by the Selected Peptides Using ELISA
The continuous culture of A549 cells line took place in F-12 Nutrient Mixture (HAM-F12) medium supplemented with 10% fetal bovine serum (FBS) in sterile plastic bottles maintained at 36.5°C with 5% CO2. After 3–4 days, the bottles were trypsinized by washing the cell monolayer with 1 mL of a 0.2% trypsin ATV solution (Adolfo Lutz) supplemented with 0.02% EDTA. After 1–2 min, the cells were homogenized with variable volumes of HAM-F12 medium supplemented with 10% of FBS to neutralize the trypsin. The total volume of the cell suspension was determined to achieve a concentration of 106 cells/mL. For the assay, 105 A549 pneumocytes were added to each well of a 96-well plate and then infected over 15 h (37°C with 5% CO2) with the different Paracoccidioides isolates (Pb18 and Pl01, 106 cells/mL) that were previously incubated with 200 μg/mL of each studied peptide at 37°C for 1 h, with rotation at 250 rpm. After incubation, ELISAs were performed as described in the Section “Inhibition of Paracoccidioides spp. Adhesion to Extracellular Matrix (ECM) Components by the Selected Peptides Using Enzyme Linked Immunosorbent Assay (ELISA).” In this experiment, untreated Paracoccidioides spp. cells were also used as controls. This experiment was performed in triplicate, with three independent experiments.
Determination of the Antifungal Activity of the Peptides
The minimum inhibitory concentration (MIC) of each peptide was determined following the standardized method in the M27-A3 document CLSI (2008). Amphotericin B (Sigma) was used as a control, and the final concentrations of peptides used ranged from 400 to 0.008 μg/mL after the addition of the inocula. Both peptides and amphotericin B were prepared with RPMI 1640 medium (Gibco) supplemented with 2% glucose (RPMI 1640-2%). The fungal cells were suspended in PBS, and the number of viable cells was estimated by staining them with Trypan blue and counting them with a haemocytometer. A suspension with 106 cells/mL was diluted at 1:50 in PBS, and then was diluted at 1:20 in RPMI 1640-2% Glc. The plates were incubated at 35°C, with rotation at 150 rpm, for 48 h, and the MABA (Microplate AlamarBlue Assay) was employed. Samples were incubated for an additional 24 h, for a total of 72 h for the final determination of MIC values, according de Paula e Silva et al. (2013). Three independent experiments were performed.
Determination of the Cytotoxicity of the Peptides Using the MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide) Cell Viability Test
The cytotoxicity of the selected peptides was determined using the A549 pneumocyte linage. The peptides were evaluated at concentrations ranging from 400 to 25 μg/mL using the MTT cell viability test according Denizot and Lang (1986) and Ferrari et al. (1990). For this, plates were prepared with 106 cells/well in HAM-F12 medium. Before the experiments, the medium was removed, and 100 μL of each tested dilution of the peptides in HAM-F12 was placed in contact with the cells. The plates were incubated at 37°C under 5% CO2 in the dark for 24 h. The solutions were then removed, 10 μL of MTT solution (5 mg/L) was added, and the plates were incubated again at 37°C for 4 h. After incubation, the MTT solution was removed, 100 μl of isopropanol was added to dissolve the precipitate, and the plates were read spectrophotometrically at 595 nm DO. The dead control was 10% hydrogen peroxide. The live control was untreated cells, and the reading from this control was considered as 100% living cells. This experiment was performed in triplicate, with three independent experiments, and the percentage of viable cells was calculated using the following formula:
In vivo Toxicity of the Peptides in Galleria mellonella
The toxicity of the peptides was evaluated using 2, 4, 8, 50, 100, 200, and 400 μg/larva of each peptide. For this assay, groups of 16 larvae were treated with the various concentrations tested and incubated at 37°C. The survival of the larvae was evaluated every day for 7 days after treatment. As controls, we used a group of larvae treated with PBS and a group of untreated larvae. Three independent experiments were performed.
Capacity of Peptides to Prevent the Infection of G. mellonella with Paracoccidioides spp.
Larval groups (n = 16) were separately treated with 100 μg/larvae of each peptide for 3 h. The larvae were subsequently infected with 5 × 106 cells/larva of Paracoccidioides spp. isolates (Pb18 or Pb01) according to Scorzoni et al. (2015), and larval survival was then observed daily for 1 week. The peptide treatments and fungal inoculations were performed with 10 μL Hamilton syringes (Hamilton, USA). Larval death was evaluated based on the visual confirmation of a lack of movement after the larvae were touched with tweezers. As controls, groups of larvae treated with PBS and infected with Pb18 or Pb01 and a group of uninfected larvae only treated with PBS were used. Three independent experiments were performed.
Effect of Peptide Treatment on Haemocyte Density
Groups of 16 larvae were inoculated with 100 μg/larvae of each peptide and incubated at 37°C for 3 h. The peptide inoculations were performed with 10 μL Hamilton syringes (Hamilton, USA). After incubation, the haemolymph from each larva was collected separately and diluted 1:10 in PBS. The cells were counted in a haemocytometer under a bright field microscope. A group treated with PBS was used as the control. Three independent experiments were performed.
Statistical Analysis
Statistical analyses and graph design were performed using GraphPad Prism version 5.00 (GraphPad Software, San Diego, CA, USA), and the results were considered significant when p < 0.05. Binding assay, MTT test and the haemocyte density test results were analyzed using one-way ANOVAs and Tukey’s post hoc tests. Survival assays were analyzed using Mantel–Cox log-rank tests.
Results
Selection of Peptides That Bound to the Surface of P. brasiliensis Cells via Phage Display Using BRASIL Technique
We employed the BRASIL technique to select peptides that bound to P. brasiliensis using linear (X6) and cyclic (CX8C) peptide libraries of different sizes to increase the variety of peptides available for study. A pre-clearing step was added to our selection protocol to favor the selection of peptides that specifically bound to this species. Phage libraries were first incubated with the non-pathogenic yeast S. cerevisiae to remove ubiquitously binding peptides. The remaining unbound phages were transferred to the pathogenic P. brasiliensis strain, and bound phages were recovered for further rounds of selection. After three rounds of selection, we observed an enrichment of phages bound to P. brasiliensis compared to the second round (2.3 times for the X6 library and five times for CX8C library), indicating an increase in phages that could bind to the fungal cells with a high affinity.
At the end of the selection process, 96 individual phage clones from each library were randomly picked from the third round and sequenced, and 84 and 76 sequences were obtained from the X6 and CX8C libraries, respectively. Four peptides were selected because they were displayed by multiple phages (Table 1): three peptides from the X6 library and one from CX8C library. These peptides were selected for further study because they were likely to have higher affinities for P. brasiliensis cells.
Table 1.
Peptide sequences and frequency.
| Peptides sequences | N | Percentage (%) |
|---|---|---|
| LVGRVV | 5 | 5.9 |
| LDFVVG | 5 | 5.9 |
| CSVSALGGAC | 3 | 3.9 |
| VVAGSV | 3 | 3.5 |
Selected peptides amino acids sequences. The other sequences obtained from both libraries demonstrated a frequency of 1.3% or less and because of this were not selected for further studies.
Binding Assays
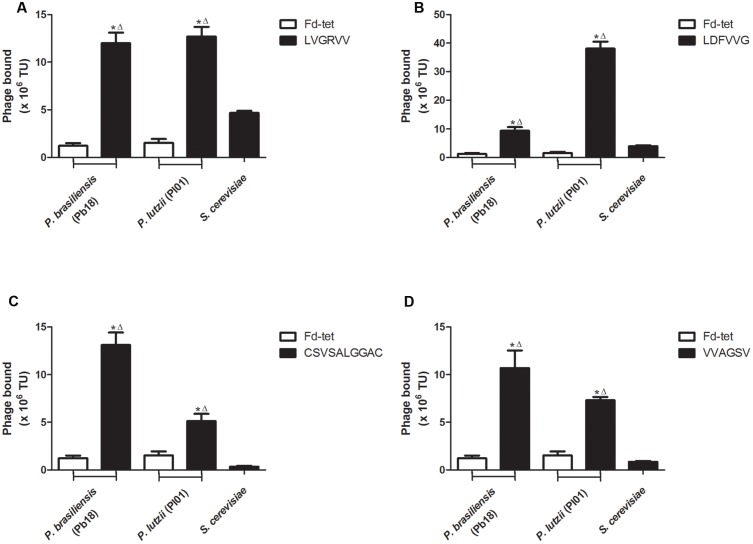
We evaluated the binding of each peptide to P. brasiliensis and P. lutzii and to the non-pathogenic yeast S. cerevisiae. As a control, we evaluated the binding of the insertless phage Fd-tet to P. brasiliensis and P. lutzii cells. We observed that all peptides showed greater binding to P. brasiliensis (phage LVGRVV, 9.7-fold; phage LDFVVG, 7.5-fold; phage CSVSALGGAC, 10.5-fold; and phage VVAGSV, 8.1-fold) and P. lutzii (phage LVGRVV, 9.7-fold; phage LDFVVG, 30.8-fold; phage CSVSALGGAC, 4.1-fold; and phage VVAGSV, 5.9-fold) compared with the binding of the Fd-tet control phage (Figure 1). None of the phages tested were found to significantly bind to S. cerevisiae, indicating that the pre-clearing step efficiently removed the peptides that bound to these cells. In summary, our validation assays confirmed that all selected peptides bound to specific cell surface ligands of the two species of the genus Paracoccidioides, P. brasiliensis (isolate Pb18) and P. lutzii (isolate Pl01).
FIGURE 1.
Selected peptides bind to Paracoccidioides cells. Binding assays of the four selected phages to Paracoccidioides brasiliensis (Pb18), Saccharomyces cerevisiae, and Paracoccidioides lutzii (Pl01) isolates. Binding of LVGRVV (A), LDFVVG (B), CSVSALGGAC (C), and VVAGSV (D) phages. Binding of the control phage Fd-tet to P. brasiliensis (Pb18) and P. lutzii is indicated. TU, transduction units. ∗p < 0.05 (compared with Fd control); Δp < 0.05 (compared with the S. cerevisiae control).
Inhibition of Paracoccidioides spp. Adhesion to ECM Components and A549 Pneumocytes by the Selected Peptides Using an Inhibition Enzyme Linked Immunosorbent Assay (inh-ELISA)
Assays were conducted using four different ECM components (laminin, fibronectin, and collagen types I and IV) (Figure 2) and the A549 pneumocyte line (Figure 3). The results indicate the strong potential of the peptides to inhibit the adhesion of P. brasiliensis and P. lutzii and indicate that these peptides might bind to some components of the fungal cell wall important for the Paracoccidioides-host interaction.
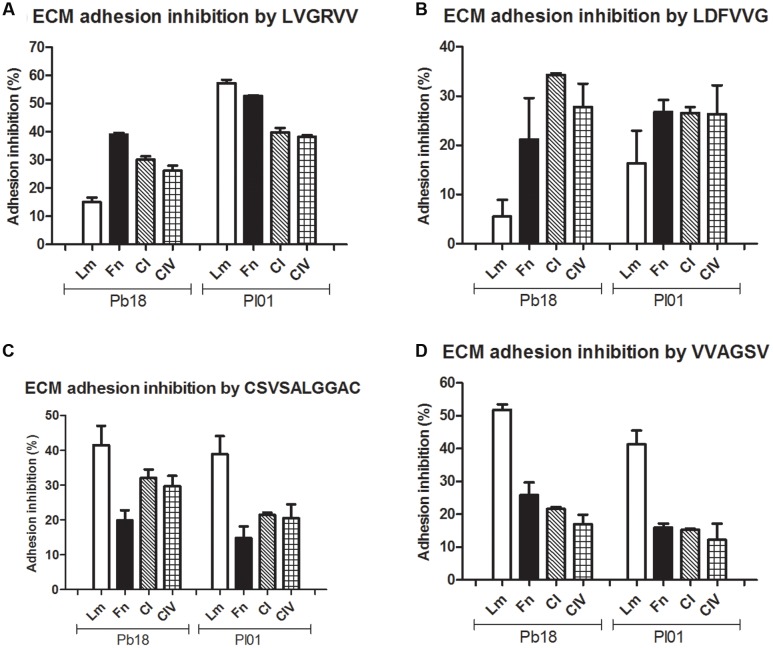
FIGURE 2.
Inhibition of P. brasiliensis and P. lutzii adhesion by the selected peptides to extracellular matrix (ECM) components laminin, fibronectin, type I and type IV collagen tested via inhibition ELISAs. (A) Inhibition by LVGRVV, (B) inhibition by LDFVVG, (C) inhibition by CSVSALGGAC, and (D) inhibition by VVAGSV.
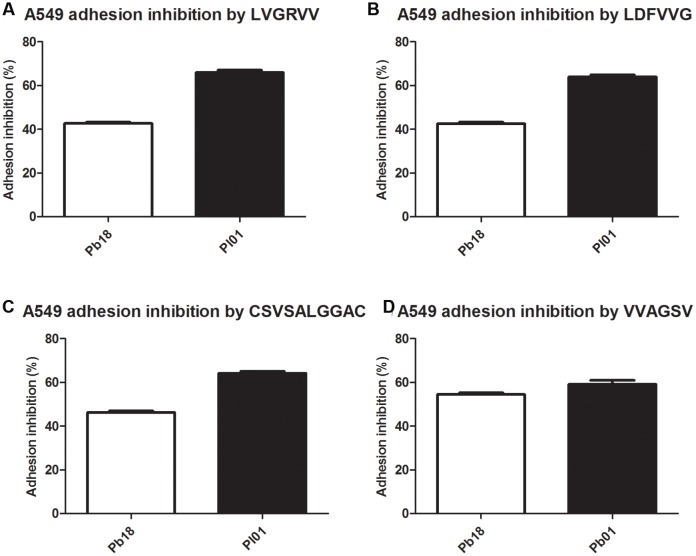
FIGURE 3.
Inhibition of P. brasiliensis and P. lutzii adhesion to A549 cells by the selected peptides tested via inhibition ELISAs. (A) Inhibition by LVGRVV, (B) inhibition by LDFVVG, (C) inhibition by CSVSALGGAC, and (D) inhibition by VVAGSV.
All the peptides tested inhibited the adhesion to all of the ECM components, with inhibition ranging from 10 to 60% depending on the strain and the ECM component (Figure 2). LVGRVV more efficiently inhibited the adhesion of P. brasiliensis to fibronectin (39%) and the adhesion of P. lutzii to laminin (57%). LDFVVG efficiently inhibited the adhesion of both fungi to fibronectin and type I and type IV collagen (from 21 to 34% inhibition). CSVSALGGAC and VVAGSV efficiently inhibited the adhesion of both fungi to laminin, with an inhibition rate ranging from 38 to 51%.
All tested peptides were able to inhibit the adhesion of P. brasiliensis and P. lutzii to A549 pneumocytes in a similar manner. The inhibition of the adhesion of the Pb18 and Pl01 strains to A549 pneumocytes ranged from 40 to 60%, respectively, for all the studied peptides (Figure 3).
Determination of the Antifungal Activities of the Peptides
The peptides did not kill or inhibit the growth of P. brasiliensis and P. lutzii at any tested concentration, which ranged from 400 to 0.008 μg/mL (data not shown).
Determination of the Cytotoxicity of the Peptides Using the MTT Cell Viability Test
The peptides did not show cytotoxicity. The percentage of viable cells was over 92% for LVGRVV and LDFVVG and was 100% for CSVSALGGAC and VVAGSV at a concentration of 200 μg/mL, which was the concentration used in the inh-ELISA. All concentrations lower than 100 μg/mL showed no cytotoxicity (data not shown).
Toxicity of Peptides in G. mellonella
No larvae died at any concentration tested, demonstrating that the peptides are nontoxic to the larvae. These results complemented the in vitro tests in which the peptides showed no toxicity to A549 pneumocytes.
Protection of G. mellonella from Infection with P. brasiliensis and P. lutzii by the Peptides
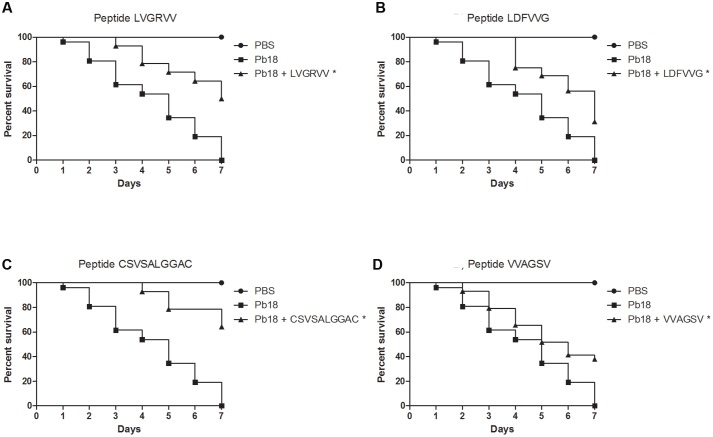
All peptides significantly (p < 0.05) protected the larvae against P. brasiliensis infections (Figure 4). Survival rates of 50%, 31%, 64%, and 37% were observed for larvae treated with the LVGRVV, LDFVVG, CSVSALGGAC, and VVAGSV peptides, respectively. The survival time increased when the larvae were treated with each of the peptides. Untreated larvae began to die after 1 day, but the treated larvae began to die after 3 days for LVGRVV, 4 days for LDFVVG and CSVSALGGAC and 2 days for VVAGSV, and in all cases, 7 days was not sufficient to kill all treated larvae, whereas 7 days were sufficient to kill all untreated larvae.
FIGURE 4.
Capacity of selected peptides to protect G. mellonella larvae against P. brasiliensis infection. The graphs show the percentage of G. mellonella larvae treated with the selected peptides and challenged with P. brasiliensis. (A) Treatment with LVGRVV, (B) treatment with LDFVVG, (C) treatment with CSVSALGGAC, and (D) treatment with VVAGSV. ∗Statistical significance (p < 0.05) is relative to Pb18 (infected and untreated larvae).
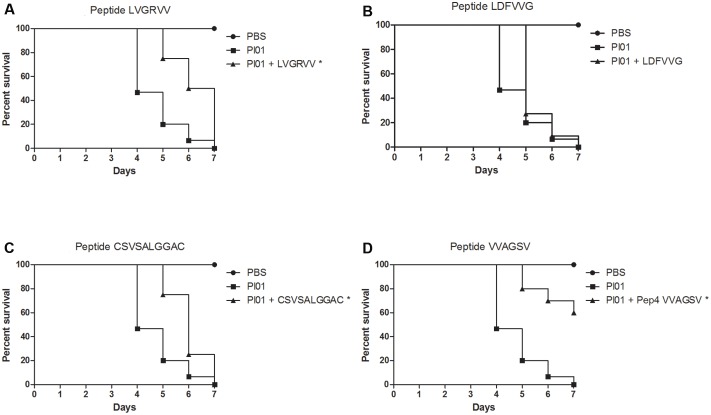
Figure 5 shows the results obtained for peptide-treated larvae infected with P. lutzii. Only the VVAGSV peptide was able to significantly (p < 0.05) protect the larvae against P. lutzii infections, resulting in a survival rate of 60% at the end of the seventh day of infection. However, the LVGRVV and CSVSALGGAC peptides were able to significantly (p < 0.05) increase the survival time. Treatment with LDFVVG did not affect larval survival after P. lutzii infection. The survival time increased when the larvae were treated with each peptide. Untreated larvae began to die after 4 days, whereas larvae treated with the different peptides began to die after 5 days. After treatment with the VVAGSV peptide, 7 days were not sufficient to kill all of the larvae (Figure 5).
FIGURE 5.
Capacity of the selected peptides to protect G. mellonella larvae against P. lutzii infection. The graphs show the percentage of G. mellonella larvae treated with the selected peptides and challenged with P. lutzii. (A) Treatment with LVGRVV, (B) treatment with LDFVVG, (C) treatment with CSVSALGGAC, and (D) treatment with VVAGSV. ∗Statistical significance (p < 0.05) is relative to Pl01 (infected and untreated larvae).
Determination of Haemocyte Density after Treatment with the Selected Peptides
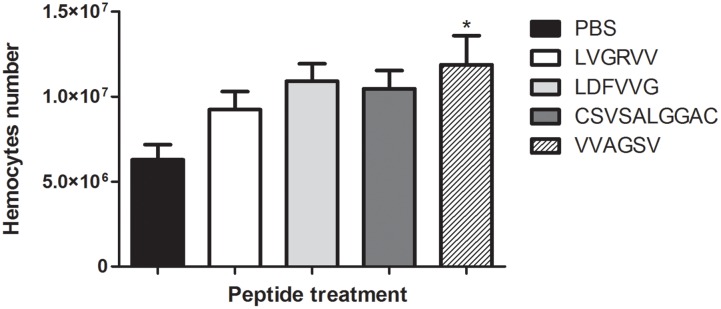
Larvae were treated with the selected peptides for 3 h, and we observed that peptide VVAGSV stimulated significantly the haemocyte production (p < 0.05) compared with production in the PBS control (Figure 6).
FIGURE 6.
Haemocyte density. The graph shows alterations in haemocyte density in the different G. mellonella larvae groups after 3 h of treatment with LVGRVV, LDFVVG, CSVSALGGC, and VVAGSV peptides. ∗p < 0.05.
Discussion
Paracoccidioides spp. express various adhesins, and the adhesion mediated by these molecules is the most important step in a successful infection. Therefore, inhibiting adhesion to host cells may be an interesting way to protect the host from Paracoccidioides infections. The use of molecules that interfere with the adhesion of pathogens, known as anti-adhesion therapy, appears to be an efficient way to prevent infection because if the fungus cannot adhere, it is incapable of colonizing the host tissue, and its acquisition of nutrients is impeded, which facilitates resistance by the host’s immune system.
In this study, four peptides selected by phage display promoted the in vitro inhibition (up to 57%) of P. brasiliensis and P. lutzii adhesion to different ECM components and to A549 pneumocytes (up to 64%). These results indicate that the anti-adhesive properties of these peptides could prevent the adhesion of Paracoccidioides spp. We demonstrated that these peptides did not have antifungal activity, which means that the reduction in the adhesion of both fungi was not caused by a loss of fungal viability during the experiments. In addition, the cytotoxicity of these peptides was tested using the MTT test, and no cytotoxicity was observed in A549 pneumocytes or in vivo in G. mellonella. These results indicate the possible safe therapeutic use of these substances.
We preliminarily tested the efficiency of these peptides to protect the host against Paracoccidioides infections in G. mellonella, an alternative animal model for pathogenicity and virulence studies on fungal pathogens (Brennan et al., 2002; Slater et al., 2011) that has recently been used to study the pathogenicity and virulence of Paracoccidioides spp. (de Oliveira et al., 2015; Marcos et al., 2015; Scorzoni et al., 2015).
The treatment of G. mellonella prior to P. brasiliensis infection demonstrated that the peptides were able to prevent larval death for up to 7 days after infection, with survival rates of 50% (LVGRVV), 31% (LDFVVG), 64% (CSVSALGGAC), and 37% (VVAGSV), and treatments increased the survival time of the larvae. For P. lutzii, only the VVAGSV peptide increased the survival rate of larvae after infection, to 60%; however, the LVGRVV and CSVSALGGAC peptides were able to significantly increase larval survival times. These results revealed that all peptides have the potential to prevent Paracoccidioides spp. infection. We also observed that treatment with the peptides led to an increase in larval survival times when infected with both fungi. We believe that the peptides bind to molecules on the fungal cells important for adhesion, obstructing infection by the fungus. Furthermore, these peptides might compete with the fungus for binding sites on the host cell, also inhibiting fungal adhesion and facilitating the ability of the host to resolve the infection or making the infectious process difficult for the fungus. These characteristics show us that all the selected peptides are potential anti-adhesive therapeutic agents.
The stimulation of the immune response to counter the undesirable effects caused by the disease and reduce fungal burden are objectives to be explored for the effective treatment of severe PCM cases (Travassos and Taborda, 2012). However, since PCM primarily affects rural workers, the disease is associated with significant social and economic factors that mostly affect less affluent populations (Shikanai-Yasuda et al., 2006; de Amorim et al., 2013), so the development of new molecules that could work as a vaccine directed at high-risk populations could be an important step in effective prophylaxis for this disease.
The body cavity of G. mellonella contains haemolymph, which has functions similar to those of blood in mammals, carrying nutrients and signal molecules and participating in gas exchange (Bergin et al., 2003). Additionally, haemolymph contains cells known as haemocytes and antimicrobial peptides that can immobilize and kill invading microbes (Cotter et al., 2000).
We observed that when larvae were treated with the VVAGS peptide, haemocyte density increased (p < 0.05). These results suggest that this peptide can modulate the immune system of the larvae, increasing haemocyte production. This appeared to trigger a protective effect in the host, resulting in the protection of the larvae against Paracoccidioides spp. infections, which is important in anti-fungal therapy.
A peptide known as p10, derived from the P. brasiliensis adhesin gp43, has been shown to be recognized by the T lymphocytes of mice and humans and has a protective effect, and isogenic mice developed a 200-fold less intense infection when immunized with this peptide (Taborda et al., 1998). Furthermore, a combination of p10 immunization with various antifungal drugs showed an additive protective effect, with a significant reduction in CFU counts after 60 and 120 days of infection. Histological analyses revealed little or no P. brasiliensis cells in mouse tissue, and the use of p10 combined with drugs improved the standard therapy with antifungal drugs and reduced the duration of treatment (Marques et al., 2006). This type of combined therapy was also effective in anergic mice, and anergy is a common in patients suffering from acute and sub-acute PCM (Marques et al., 2008). Currently, studies using a DNA vaccine coding for p10 have shown satisfactory results; the administration of the vaccine 1 month before or after infection induced a significant reduction in the fungal burden in the lungs of mice, indicating that immunization with this vaccine is a powerful tool for the prevention and treatment of PCM (Rittner et al., 2012).
The selected peptides demonstrated anti-adhesive activity, inhibiting the adhesion of P. brasiliensis and P. lutzii to pneumocytes and ECM components. The treatment of infected G. mellonella with the peptides increased larval survival and increased haemocyte density, demonstrating a possible immunomodulatory effect in the larvae that can affect their survival. More studies are necessary to understand the mechanisms involved during the use of these peptides, including studies with murine models and studies focused on the combined use of the peptides and traditional antifungal therapies. However, our study shows that these molecules might provide new insights on the treatment and prevention of PCM.
Author Contributions
HdO, AF-A, RG, and MM-G conceived and designed the experiments. HdO, JM, JdS, LS, AS, CM, PA, and DY performed the experiments. HdO, JM, JdS, LS, AS, CM, PA, DY, AA, RG, and MM-G analyzed the data. HdO, RG, and MM-G drafted the manuscript. All the authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This study was supported by Fundação de Apoio à Pesquisa do Estado de São Paulo (FAPESP) 2015/14023-8 (HdO), 2013/10917-9 (LS), 2016/17048-4 (CM) 2015/03700-9 (MM-G) and 2008/54806-8 (RG), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Programa de Apoio ao Desenvolvimento Científico da Faculdade de Ciências Farmacêuticas da UNESP (PADC/FCF).
References
- Antonara S., Chafel R. M., LaFrance M., Coburn J. (2007). Borrelia burgdorferi adhesins identified using in vivo phage display. Mol. Microbiol. 66 262–276. 10.1111/j.1365-2958.2007.05924.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergin D., Brennan M., Kavanagh K. (2003). Fluctuations in haemocyte density and microbial load may be used as indicators of fungal pathogenicity in larvae of Galleria mellonella. Microbes Infect. 5 1389–1395. 10.1016/j.micinf.2003.09.019 [DOI] [PubMed] [Google Scholar]
- Blotta M. H., Camargo Z. P. (1993). Immunological response to cell-free antigens of Paracoccidioides brasiliensis: relationship with clinical forms of paracoccidioidomycosis. J. Clin. Microbiol. 31 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan M., Thomas D. Y., Whiteway M., Kavanagh K. (2002). Correlation between virulence of Candida albicans mutants in mice and Galleria mellonella larvae. FEMS Immunol. Med. Microbiol. 34 153–157. 10.1111/j.1574-695X.2002.tb00617.x [DOI] [PubMed] [Google Scholar]
- CLSI (2008). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeast. Wayne, PA: Clinical and Laboratory Standards Intitute. [Google Scholar]
- Cotter G., Doyle S., Kavanagh K. (2000). Development of an insect model for the in vivo pathogenicity testing of yeasts. FEMS Immunol. Med. Microbiol. 27 163–169. 10.1111/j.1574-695X.2000.tb01427.x [DOI] [PubMed] [Google Scholar]
- Coutinho Z. F., Silva D., Lazera M., Petri V., Oliveira R. M., Sabroza P. C., et al. (2002). Paracoccidioidomycosis mortality in Brazil (1980-1995). Cad. Saude Publica 18 1441–1454. 10.1590/S0102-311X2002000500037 [DOI] [PubMed] [Google Scholar]
- Cozens D., Read R. C. (2012). Anti-adhesion methods as novel therapeutics for bacterial infections. Exp. Rev. Anti Infect. Ther 10 1457–1468. 10.1586/eri.12.145 [DOI] [PubMed] [Google Scholar]
- de Amorim J., Magalhães A., Muñoz J. E., Rittner G. M., Nosanchuk J. D., Travassos L. R., et al. (2013). DNA vaccine encoding peptide P10 against experimental paracoccidioidomycosis induces long-term protection in presence of regulatory T cells. Microbes Infect. 15 181–191. 10.1016/j.micinf.2012.11.007 [DOI] [PubMed] [Google Scholar]
- de Oliveira H. C., da Silva J. F., Scorzoni L., Marcos C. M., Rossi S. A., de Paula e Silva A. C. A., et al. (2015). Importance of adhesins in virulence of Paracoccidioides spp. Front. Microbiol. 6:303 10.3389/fmicb.2015.00303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paula e Silva A. C., Oliveira H. C., Silva J. F., Sangalli-Leite F., Scorzoni L., Fusco-Almeida A. M., et al. (2013). Microplate alamarBlue assay for Paracoccidioides susceptibility testing. J. Clin. Microbiol. 51 1250–1252. 10.1128/JCM.02914-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denizot F., Lang R. (1986). Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J. Immunol. Methods 89 271–277. 10.1016/0022-1759(86)90368-6 [DOI] [PubMed] [Google Scholar]
- Fava Netto C. (1961). Contribuição para o estudo imunológico da blastomicose de Lutz (Blastomicose sul americana). Rev. Inst. Adolfo Lutz 21 99–194. [Google Scholar]
- Ferrari M., Fornasiero M. C., Isetta A. M. (1990). MTT colorimetric assay for testing macrophage cytotoxic activity in vitro. J. Immunol. Methods 131 165–172. 10.1016/0022-1759(90)90187-Z [DOI] [PubMed] [Google Scholar]
- Giordano R. J., Cardó-Vila M., Lahdenranta J., Pasqualini R., Arap W. (2001). Biopanning and rapid analysis of selective interactive ligands. Nat. Med. 7 1249–1253. 10.1038/nm1101-1249 [DOI] [PubMed] [Google Scholar]
- Giordano R. J., Edwards J. K., Tuder R. M., Arap W., Pasqualini R. (2009). Combinatorial ligand-directed lung targeting. Proc. Am. Thorac. Soc. 6 411–415. 10.1513/pats.200903-014AW [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen E., Arap W., Rajotte D., Lahdenranta J., Pasqualini R. (1999). Identification of receptor ligands with phage display peptide libraries. J. Nucl. Med. 40 883–888. [PubMed] [Google Scholar]
- Koivunen E., Gay D. A., Ruoslahti E. (1993). Selection of peptides binding to the alpha 5 beta 1 integrin from phage display library. J. Biol. Chem. 268 20205–20210. [PubMed] [Google Scholar]
- Krachler A. M., Orth K. (2013). Targeting the bacteria-host interface: strategies in anti-adhesion therapy. Virulence 4 284–294. 10.4161/viru.24606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionakis M. S., Lahdenranta J., Sun J., Liu W., Lewis R. E., Albert N. D., et al. (2005). Development of a ligand-directed approach to study the pathogenesis of invasive aspergillosis. Infect. Immun. 73 7747–7758. 10.1128/IAI.73.11.7747-7758.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos C. M., da Silva J. F., de Oliveira H. C., Assato P. A., Singulani J. L., Lopez A. M., et al. (2015). Decreased expression of 14-3-3 in Paracoccidioides brasiliensis confirms its involvement in fungal pathogenesis. Virulence 7 72–84. 10.1080/21505594.2015.1122166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques A. F., da Silva M. B., Juliano M. A., Munhõz J. E., Travassos L. R., Taborda C. P. (2008). Additive effect of P10 immunization and chemotherapy in anergic mice challenged intratracheally with virulent yeasts of Paracoccidioides brasiliensis. Microbes Infect. 10 1251–1258. 10.1016/j.micinf.2008.07.027 [DOI] [PubMed] [Google Scholar]
- Marques A. F., da Silva M. B., Juliano M. A., Travassos L. R., Taborda C. P. (2006). Peptide immunization as an adjuvant to chemotherapy in mice challenged intratracheally with virulent yeast cells of Paracoccidioides brasiliensis. Antimicrob. Agents Chemother. 50 2814–2819. 10.1128/AAC.00220-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ministério da Saúde S. d. C., Estratégicos T. e., I (2006). “Classificação de risco dos agentes biológicos.,” in Secretaria de Ciência, Tecnologia e Insumos Estratégicos Departamento do Complexo Industrial e Inovação em Saúde, ed. Tecnologia D. D. C. E. (Brasília: Editora do Ministério da Saúde; ). [Google Scholar]
- Mullen L. M., Nair S. P., Ward J. M., Rycroft A. N., Williams R. J., Henderson B. (2007). Comparative functional genomic analysis of Pasteurellaceae adhesins using phage display. Vet. Microbiol. 122 123–134. 10.1016/j.vetmic.2006.12.022 [DOI] [PubMed] [Google Scholar]
- Ofek I., Hasty D. L., Sharon N. (2003). Anti-adhesion therapy of bacterial diseases: prospects and problems. FEMS Immunol. Med. Microbiol. 38 181–191. 10.1016/S0928-8244(03)00228-1 [DOI] [PubMed] [Google Scholar]
- Posadas D. M., Ruiz-Ranwez V., Bonomi H. R., Martín F. A., Zorreguieta A. (2012). BmaC, a novel autotransporter of Brucella suis, is involved in bacterial adhesion to host cells. Cell Microbiol. 14 965–982. 10.1111/j.1462-5822.2012.01771.x [DOI] [PubMed] [Google Scholar]
- Rittner G. M., Muñoz J. E., Marques A. F., Nosanchuk J. D., Taborda C. P., Travassos L. R. (2012). Therapeutic DNA vaccine encoding peptide P10 against experimental paracoccidioidomycosis. PLoS Negl. Trop Dis. 6:e1519 10.1371/journal.pntd.0001519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorzoni L., de Paula e Silva A. C., Singulani J. E. L, Leite F. S., de Oliveira H. C., Moraes da Silva R. A., et al. (2015). Comparison of virulence between Paracoccidioides brasiliensis and Paracoccidioides lutzii using Galleria mellonella as a host model. Virulence 6 766–776. 10.1080/21505594.2015.1085277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeeva A., Kolonin M. G., Molldrem J. J., Pasqualini R., Arap W. (2006). Display technologies: application for the discovery of drug and gene delivery agents. Adv. Drug Deliv. Rev. 58 1622–1654. 10.1016/j.addr.2006.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai-Yasuda M. A., Telles Filho F. E. Q., Mendes R. P., Colombo A. L., Moretti M. L. (2006). Guidelines in paracoccidioidomycosis. Rev. Soc. Bras. Med. Trop. 39 297–310. [DOI] [PubMed] [Google Scholar]
- Shkoporov A. N., Khokhlova E. V., Kafarskaia L. I., Pavlov K. A., Smeianov V. V., Steele J. L., et al. (2008). Search for protein adhesin gene in Bifidobacterium longum genome using surface phage display technology. Bull. Exp. Biol. Med. 146 782–785. 10.1007/s10517-009-0423-4 [DOI] [PubMed] [Google Scholar]
- Silva J. D. F. D. (2008). Estudo de Diferentes Isolados de Paracoccidioides brasiliensis Quanto ao Padrão de Adesão e Expressão de Ligantes da Matriz Extracelular. Master thesis, Faculdade de Ciências Farmacêuticas de Araraquara FCFAr - UNESP, Araraquara. [Google Scholar]
- Slater J. L., Gregson L., Denning D. W., Warn P. A. (2011). Pathogenicity of Aspergillus fumigatus mutants assessed in Galleria mellonella matches that in mice. Med. Mycol. 49(Suppl. 1), S107–S113. 10.3109/13693786.2010.523852 [DOI] [PubMed] [Google Scholar]
- Smith G. P., Scott J. K. (1993). Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 217 228–257. 10.1016/0076-6879(93)17065-D [DOI] [PubMed] [Google Scholar]
- Taborda C. P., Juliano M. A., Puccia R., Franco M., Travassos L. R. (1998). Mapping of the T-cell epitope in the major 43-kilodalton glycoprotein of Paracoccidioides brasiliensis which induces a Th-1 response protective against fungal infection in BALB/c mice. Infect. Immun. 66 786–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travassos L. R., Taborda C. P. (2012). New advances in the development of a vaccine against paracoccidioidomycosis. Front. Microbiol. 3:212 10.3389/fmicb.2012.00212 [DOI] [PMC free article] [PubMed] [Google Scholar]