Abstract
Breast cancer is one of the most common malignancies among women worldwide. Genetic factors have been shown to play an important role in breast cancer aetiology. We conducted a two-stage genome-wide association study (GWAS) including 14 224 cases and 14 829 controls of East Asian women to search for novel genetic susceptibility loci for breast cancer. Single nucleotide polymorphisms (SNPs) in two loci were found to be associated with breast cancer risk at the genome-wide significance level. The first locus, represented by rs12118297 at 1p22.3 (near the LMO4 gene), was associated with breast cancer risk with odds ratio (OR) and (95% confidence interval (CI)) of 0.91 (0.88–0.94) and a P-value of 4.48 × 10− 8. This association was replicated in another study, DRIVE GAME-ON Consortium, including 16 003 cases and 41 335 controls of European ancestry (OR = 0.95, 95% CI = 0.91–0.99, P-value = 0.019). The second locus, rs16992204 at 21q22.12 (near the LINC00160 gene), was associated with breast cancer risk with OR (95% CI) of 1.13 (1.07–1.18) and a P-value of 4.63 × 10 − 8. The risk allele frequency for this SNP is zero in European-ancestry populations in 1000 Genomes Project and thus its association with breast cancer risk cannot be assessed in DRIVE GAME-ON Consortium. Functional annotation using the ENCODE data indicates that rs12118297 might be located in a repressed element and locus 21q22.12 may affect breast cancer risk through regulating LINC00160 expressions and interaction with oestrogen receptor signalling. Our findings provide additional insights into the genetics of breast cancer.
Introduction
Breast cancer is the most common malignancy among women in the United States and many other countries around the world (1). Genetic factors have been shown to play an important role in breast cancer aetiology (2,3). Since 2007, genome-wide association studies (GWAS) have identified approximately 100 common genetic susceptibility loci for breast cancer risk (3–32). To date, most GWAS have been conducted primarily among women of European ancestry, and genetic risk variants identified in these studies explain approximately 16% of familial breast cancer risk in European descendants (5). Many variants discovered in European ancestry populations showed a weak or no association with breast cancer risk in other ethnic groups (14,33–36). Therefore, it is necessary to conduct GWAS in non-European populations to discover additional genetic risk variants for breast cancer. In 2008, we initiated the Asia Breast Cancer Consortium (ABCC), a GWAS in East Asians to search for novel genetic susceptibility loci for breast cancer risk. Over the years, this consortium has grown into a large collaboration involving cases and controls recruited in studies conducted in multiple Asian countries (13). We have identified 10 novel susceptibility loci for breast cancer risk (13,15,19–22,29,37), and many of these loci were subsequently replicated in studies of European descendants (13,15,20,38). Studies from African and Latino-ancestry populations also have identified novel susceptibility variants associated with breast cancer risk (30,32). In this paper, we report novel findings from an expanded ABCC that included additional samples in the discovery stage and imputed the genome-wide scan data using data from the 1000 Genomes Project as reference (39).
Results
Association analyses among East Asian women
The current study included data from 29 053 women (14 224 cases and 14 829 controls) as part of the ABCC. All study participants were of East Asian ancestry and recruited from eight studies conducted in multiple countries (Table 1, Supplementary Material, Text S1). Our discovery stage (stage I) included three studies with genome-wide scan data comprising a total of 7619 cases and 6286 controls, including 4866 Chinese women (SBCGS) (13,29), 4298 Korean women (SeBCS1)(40), and 4741 Japanese women (BBJ1) (41,42). Imputation was performed within each study using Minimac2 (43). SBCGS and BBJ1 were imputed with the 1000 Genomes Project Phase 3 as reference and SeBCS1 was imputed with the 1000 Genomes Project Phase 1 as reference. Only single nucleotide polymorphisms (SNPs) imputed with high imputation quality (RSQR ≥ 0.5) and minor allele frequency (MAF) ≥ 0.01 were included in the discovery stage analyses. A meta-analysis of imputed data from SBCGS, SeBCS1 and BBJ1 was conducted using fixed-effects, inverse variance meta-analysis using the METAL software (44). In the discovery stage, we have evaluated the association of risk variants in 106 loci identified previously for breast cancer risk via GWAS. Among those 106 SNPs, 80 SNPs were available among all three breast cancer datasets in our current GWAS. We found that the 35 SNPs were associated with breast cancer risk at P-value < 0.05 with the same direction as observed in previous reports (Supplementary Material, Table S1).
Table 1.
Selected characteristics of studies included in the current analysis from the Asia Breast Cancer Consortium
| Study | Cases | Controls | Population | Study designa | Age (years)b | ER(+) (%)c | Postmenopausal (%)d |
|---|---|---|---|---|---|---|---|
| Stage I | 7619 | 6286 | |||||
| SBCGS | 2731 | 2135 | Chinese | Population-based | 51/50 | 55 | 41/41 |
| SeBCS1 | 2246 | 2052 | Korean | Hospital-based | 48/51 | 63 | 36/56 |
| BBJ1 | 2642 | 2099 | Japanese | Hospital-based | 57/56 | 63 | 79/72 |
| Stage II | 6605 | 8543 | |||||
| KOHBRA/KoGES | 1397 | 3209 | Korean | Hospital-based | 40/50 | 63 | 23/NA |
| HCES-Br | 3387 | 3186 | Korean | Population-based | 50/57 | 64 | 45/81 |
| SeBCS2 | 776 | 1,103 | Korean | Hospital-based | 48/48 | 63 | 36/37 |
| Nagoya | 644 | 644 | Japanese | Hospital-based | 51/51 | 73 | 49/49 |
| Nagano | 401 | 401 | Japanese | Hospital-based | 54/54 | 75 | 55/65 |
| Total | 14 224 | 14 829 |
Abbreviations: ER, Estrogen receptor; NA, Not available.
aCase-control study design was used.
bMean age of cases/controls with available data.
cProportion of ER-positive women among cases.
dProportion of postmenopausal status of cases/controls with available data.
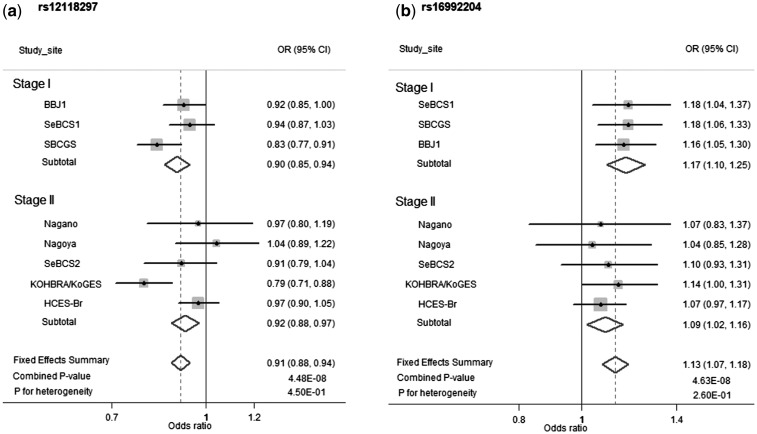
In order to select SNPs for fast-track replication (stage II), we used the following criteria: (i) an imputation score of RSQR > 0.8 in at least two studies with genome-wide scan data; (ii) an MAF of > 0.05 in all stage I studies with available data; (iii) P < 1.0 x 10 − 4 in the stage I meta-analysis; (iv) the same direction of association in all stage I studies; and (v) no strong linkage disequilibrium (LD) (r2 < 0.3 in Asians) with any of the known breast cancer susceptibility loci (5,13–15,19,20,29). The top 32 SNPs were selected for replication in an independent set of 6605 cases and 8543 controls from five studies participating in the ABCC. After filtering out SNPs with low quality among designable SNPs, 28 SNPs were evaluated in stage II, in which 5 SNPs were associated with breast cancer risk at P < 0.05 (Supplementary Material, Table S2). In the combined data from both stages, the association for two SNPs reached the genome-wide significance level (P < 5.0 x 10 − 8): rs12118297 at 1p22.3 with OR (95% CI) of 0.91 (0.88–0.94) and P = 4.48 × 10 − 8, and rs16992204 at 21q22.12 with OR (95% CI) of 1.13 (1.07–1.18) and P = 4.63 × 10 − 8 (Table 2). With the exception of the Nagoya study for rs12118297, the direction of the association between breast cancer risk and each of the two SNPs was consistent across all studies included in the present analysis (P for heterogeneity > 0.05) (Fig. 1).
Table 2.
Results for the association of two newly identified genetic loci with breast cancer risk
| SNP (allelesa) | Locus | EAFc | Stage | Per-allele association |
NearestGenes | |
|---|---|---|---|---|---|---|
| (Positionb) | OR (95% CI)d | P e | ||||
| rs12118297 | 1p22.3 | 0.38 | Stage I | 0.90 (0.85–0.94) | 1.54 x 10−5 | |
| (T/G) | 87,779,217 | Stage II | 0.92 (0.88–0.97) | 6.00 x 10−4 | ||
| Combined | 0.91 (0.88–0.94) | 4.48 x 10−8 | LMO4 | |||
| rs16992204 | 21q22.12 | 0.12 | Stage I | 1.17 (1.10–1.25) | 8.79 x 10−7 | |
| (C/T) | 36,111,201 | Stage II | 1.09 (1.02– 1.16) | 4.40 x 10−3 | ||
| Combined | 1.13 (1.07–1.18) | 4.63 x 10−8 | LINC00160 | |||
aEffect/reference alleles.
bChromosome position (bp) based on NCBI Human Genome Build 37.
cEffect allele frequency based on controls from the current study.
dPer-allele OR (95% CI) was adjusted for age and the principal components in each study in stage I, and age and study sites in stage II; combined OR (95% CI) was obtained using fixed-effect meta-analysis in each stage.
eObtained from a weighted z statistic-based meta-analysis.
Figure 1.
Forest plots for risk variants in the two newly identified breast cancer risk loci by study site and stage. Per-allele OR estimates and fixed-effect summary OR estimates are presented. The size of the square box is proportional to the number of cases and controls in each study site.
In analyses stratified by study population, although the associations of both SNPs rs12118297 and rs16992204 were stronger for Chinese than for Korean and Japanese participants, heterogeneity tests were not statistically significant (Supplementary Material, Table S3). Both SNPs showed a stronger association for ER-positive breast cancer than ER-negative breast cancer, and the difference was statistically significant for rs16992204 (P = 0.05) (Supplementary Material, Table S4).
Evaluation of the two SNPs in European-ancestry women
To investigate the association of these two SNPs with breast cancer risk in women of European ancestry, we accessed data from the DRIVE GAME-ON Consortium (45), consisting of 16 003 cases and 41 335 controls. SNP rs12118297 showed a significant association with breast cancer risk in women of European ancestry at P =0.019. The OR for the association was 0.95 (95% CI = 0.91–0.99), consistent with the association observed in the East Asian population. The MAF of this SNP was much lower in European descendants (0.18) than in East Asians (0.38), and the strength of the association was weaker in European than in East Asian women (P for heterogeneity = 0.03). We were not able to evaluate rs16992204 in the DRIVE GAME-ON Consortium since this SNP showed an allele frequency of 0 in European ancestry in the 1000 Genomes Project. Therefore, it is likely that this SNP cannot be imputed to 1000 Genomes project for GWAS of European ancestry, like the DRIVE GAME-ON Consortium.
Expression quantitative trait loci (eQTL) analyses and functional annotation
To explore potentially regulated target genes for the newly identified loci, we conducted eQTL analysis to evaluate the association of rs12118297 and rs16992204 with the expression levels of genes within 1 Mb region in breast tumour tissue using data from The Cancer Genome Atlas (TCGA) (46) and the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) (47) (see Methods). Additionally, two publicly available eQTL databases, GTEx database (48) and HaploReg V4 (49) were also examined. To investigate whether nearby genes may be involved in breast carcinogenesis, we performed differential gene expression analysis between breast tumour tissue and adjacent normal tissue using data from 87 patients included in TCGA. The functional significance of both newly identified loci was evaluated using the Encyclopedia of DNA Elements (ENCODE) Project (50), HaploReg V4 (49), and RegulomeDB (51).
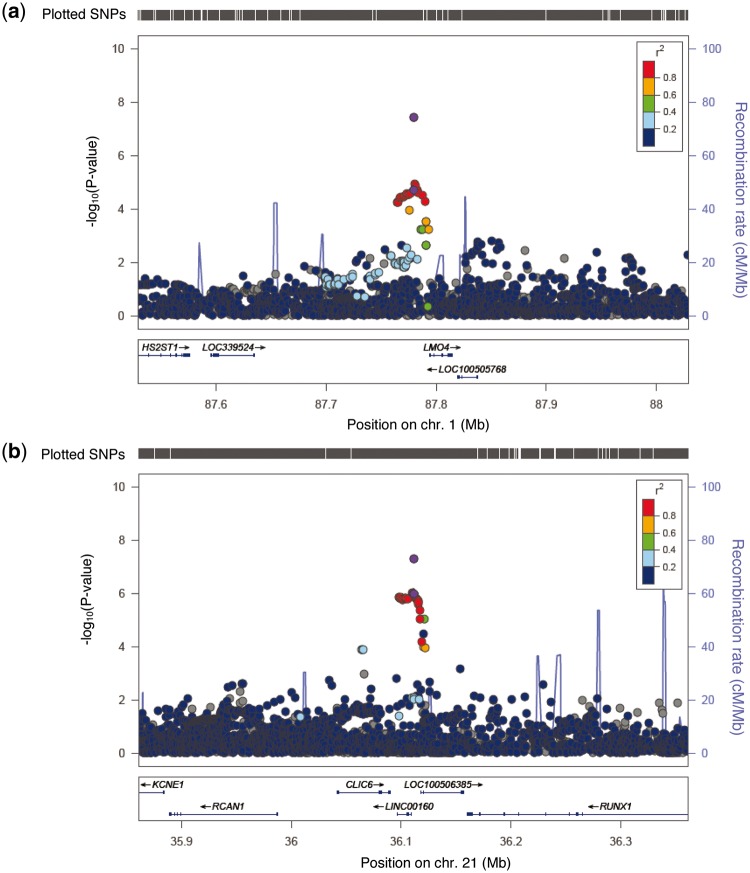
For locus 1p22.3, SNP rs12118297 is located 14 934 bp upstream of the LMO4 gene (LIM-only protein 4) (Fig. 2A). No significant associations were found between this SNP and expression of genes within 1 Mb region based on the eQTL analysis in GTEx, TCGA, or METABRIC data. However, a search of eQTL results from both HaploReg V4 and RegulomDB showed that this SNP was correlated with LMO4 gene expression in human monocytes (52) and brain tissue (cerebellum and temporal cortex) (53). The expression level of the LMO4 gene was significantly lower in tumour tissue than in adjacent normal tissue (P = 5.71 × 10 − 5) among breast cancer cases included in TCGA (Supplementary Material, Table S5). We also found some evidence of eQTLs for nearby genes. They were associated with expression levels of CLCA2 (chloride channel accessory 2) gene and SH3GLB1 (SH3-Domain GRB2-Like Endophilin B1) gene (P < 0.05), located ∼857 kb and ∼565 kb upstream of rs12118297, respectively (Supplementary Material, Table S7). The expression levels of both genes were significantly lower in tumour tissues than in adjacent normal tissues (CLCA2 gene, P = 1.10 × 10 − 4 and SH3GLB1 gene, P = 1.25 × 10 − 23) (Supplementary Material, Table S5). These findings support possible roles of CLCA2 and SH3GLB1 as potential tumour suppressors in breast carcinogenesis (54,55). ChromHMM annotation using ENCODE data suggests that rs12118297 might be located in a polycomb-repressed element. In RegulomeDB, this SNP has been annotated as a potentially functional SNP with a score of 1f, indicating that it may lie within a region containing a transcription factor (TF) binding site, matched TF motif and DNase I hypersensitive site. Consistently, the annotation using HaploReg indicated that it might be located in a predicted LRF motif (Supplementary Material, Table S6).
Figure 2.
Regional plots of association results for the two newly identified risk loci for breast cancer. (A) rs12118297. (B) rs16992204. Each plot shows the -log10 P-values (y-axis) for each SNP in a given genomic region on the x-axis based on NCBI Build 37. The marker SNPs are shown in purple circles and Refseq genes are shown beneath each plot. The top SNPs (rs12118297 and rs16992204) with purple circles are from the meta-analyses of all studies conducted among East Asians, and data shown for all other SNPs are from Stage I only. Pairwise LD with adjacent SNPs as measured by r2 values (according to the 1000 Genomes Project Phase 3 Asian data) is indicated by the color of each circle. (a) rs12118297. (b) rs16992204. Combined P-values for SNPs rs12118297 and rs16992204 were 4.48 x 10− 8 and 4.63 x 10− 8, respectively.
For locus 21q22.12, SNP rs16992204 is located 1722 bp upstream of the LINC00160 gene (Long Intergenic Non-Protein Coding RNA 160) (Fig. 2A). We could not evaluate whether this SNP was an eQTL in GTEx, TCGA, and METABRIC data because the MAF of this SNP is less than 0.01 in European populations. The expression level of the LINC00160 gene was significantly higher in tumour tissue than in adjacent normal tissue (P = 1.13 × 10 − 4) among breast cancer cases included in TCGA (Supplementary Material, Table S5). At the 21q22.12, we examined nearby genes and found some evidence of eQTLs with MAF ≥ 0.01 (Supplementary Material, Table S7). SNP rs16992204 was associated with expression of a nearby gene, KCNE1 (P = 0.03), which is located ∼283 kb downstream of this SNP at 21q22.12 (Supplementary Material, Table S7). The expression level of the KCNE1 gene was significantly lower in tumour tissues than in adjacent normal tissues (P = 4.97 × 10 − 24) (Supplementary Material, Table S5). Our analysis showed some evidence of eQTLs for nearby genes, including RUNX1 (Runt-Related Transcription Factor 1) gene and RCAN1 (Regulator Of Calcineurin 1) gene (P < 0.05), located ∼49 kb upstream and ∼124 kb downstream of rs16992204, respectively (Supplementary Material, Table S7). Expression levels of RUNX1 gene were significantly higher in tumour tissues than in adjacent normal tissues (P = 1.92 × 10 − 8) (Supplementary Material, Table S5). This result is consistent with recent studies showing that RUNX1 expression was correlated with breast cancer progression and metastasis (56,57). Furthermore, we found that the expression level of the RCAN1 gene was significantly lower in tumour tissues than in adjacent normal tissues (P = 5.18 × 10 − 22) which supports the role of RCAN1 as a potential breast cancer suppressor reported from previous studies (58,59). The function of the SNP rs16992204 is still not known.
Discussion
In this two-stage GWAS based on 14 224 cases and 14 829 controls of East Asian women, we identified two new breast cancer susceptibility loci at 1p22.3 (rs12118297) and 21q22.12 (rs16992204). The association of SNP rs12118297 with breast cancer risk was replicated in 16 003 cases and 41 335 controls of European ancestry from the DRIVE GAME-ON Consortium. We also found that SNP rs16992204 showed stronger association for ER-positive breast cancer than ER-negative breast cancer with statistically significant difference. These SNPs were not in LD with any of the previously reported GWAS loci for breast cancer.
Our first SNP, rs12118297 in the 1p22.3 region, is located 14 934 bp upstream of the LMO4 gene. The LMO4 gene belongs to a family of LIM-only transcriptional regulators that function as molecular adaptors for protein-protein interactions. The mechanism of LMO4 function is not yet fully known. Several studies indicated a role for the LMO4 gene as an oncogene (60,61) which is inconsistent with our differential gene expression results using TCGA data: decreased expression among breast tumour tissues with Log2 fold changes = -0.53 (Supplementary Material, Table S5). However, the expression level of LMO4 is largely affected by the change in the stoichiometry of LMO4-containing complexes, such as those comprising CtIP, BRCA1, DEAF1, and/or Ldb1 (62,63). Thus, given the primary function of LMO4, which is an adaptor for protein-protein interactions, it remains to be established whether LMO4 is amplified or deregulated by other means in breast cancer. Our finding about SNP rs12118297 identified in the present study for breast cancer risk was not associated with LMO4 expression level in breast tissues using data from GTEx, TCGA or METABRIC, however, it has been shown to be an eQTL for LMO4 in human monocytes (52) and brain tissue (53). Therefore, even though the underlying biology is still not known, our study suggests that it is possible that SNP rs12118297 affects breast cancer risk through genetic mechanisms associated with the LMO4 gene.
Our second SNP, rs16992204 in the 21q22.12 region, is located 1722 bp upstream of the LINC00160 gene. The LINC00160 gene has numerous classes of non-protein coding transcripts longer than 200 nucleotides. Recently, Jonsson et al. (64) reported that LINC00160 is a direct target of ER-α, and 17β-estradiol treatment up-regulated LINC00160 expressions in breast cancer MCF-7 and T47D cells. The ChIP-qPCR experiments confirmed that ER-α binds to LINC00160 in both MCF-7 and T47D cells (64). The LINC00160 was expressed at higher levels in ERα-positive tumours, compared with both ERα-negative tumours and normal tissue among TCGA samples (64). Furthermore, silencing of LINC00160 using siRNA reduces MCF-7 cell proliferation (64). We also found that the association of SNP rs16992204 with breast cancer risk was mainly observed in ER-positive breast cancer. Taken together, genetic variation at the locus 21q22.12 may affect breast cancer risk through regulating LINC00160 expressions and interaction with ER signalling. Our analysis showed no evidence for rs16992204 as cis-eQTL for this gene due to the very low MAF in European populations. However, we found some evidence of eQTLs for nearby genes with MAF ≥ 0.01 at 21q22.12 (Supplementary Material, Table S7). In particular, many studies have demonstrated a possible link of the RUNX1 gene with breast cancer development (46,65), and we found evidence of eQTLs for nearby genes.
In summary, we report common variants at two genomic loci as new genetic risk factors for breast cancer in East Asian populations, providing additional insights into the genetics and biology of breast cancer. We have explored possible biological mechanisms for the observed associations. In particular, in silico analyses support a functional significance of one of these common SNPs at 1p22.3/LMO4. However, the other biological mechanism may also be involved. Future studies, including fine-mapping and functional experimental investigations, are needed to gain additional insights into the biological basis for the genetic associations with breast cancer risk in these two loci identified in our study.
Materials and Methods
Study populations
All study participants provided written informed consent, and the protocols for all participating studies were approved by the relevant institutional review boards. Detailed descriptions of participating studies are included in the S1 File. Briefly, as part of the ABCC, this study includes 14 224 cases and 14 829 controls from eight studies (Table 1), including 4866 Chinese, 17 356 Korean, and 6831 Japanese women. Data for Chinese women came from four studies based in Shanghai (n = 4866; the Shanghai Breast Cancer Study (SBCS), the Shanghai Breast Cancer Survival Study (SBCSS), the Shanghai Endometrial Cancer Study (SECS; controls only) and the Shanghai Women’s Health Study (SWHS)) (13,66–68). Data for Korean women came from four studies: the Seoul Breast Cancer Study (SeBCS; n = 6177) (40), the Korea Genome Epidemiology Study (KoGES; n = 3209) (69), the Korean Hereditary Breast Cancer study (KOHBRA; n = 1397) (70), and the Hwasun Cancer Epidemiology Study-Breast (HCES-Br; n = 6573) (71–73). Data for Japanese women came from three studies: the Biobank Japan Project (BBJ1; n = 4741) (41), the Nagoya Study (n = 1288) (74), and the Nagano Breast Cancer Study (n = 802) (75) (Table 1).
Genotyping and quality control
Three GWAS were included in stage I, in which 4866 Chinese women, 4298 Korean women, and 4741 Japanese women were genotyped. Genotyping protocols for stage I have been described elsewhere (13,15,19,20,29,40–42). In the Chinese GWAS (SBCGS), samples were scanned primarily using Affymetrix Genome-Wide Human SNP Array 6.0, and the initial 300 samples were scanned using the Affymetric GeneChip Mapping 500K Array Set. In the present study, only data from Affymetrix SNP Array 6.0 were used to perform imputation. After quality control exclusions, the final data set included 2731 cases and 2135 controls for 668 499 markers. For the Korean GWAS (SeBCS1), Affymetrix Genome-Wide Human SNP Array 6.0 was used. After quality control exclusions, the final data set included 2246 cases and 2052 controls for 555 117 markers. For the Japanese GWAS (BBJ1), Illumina OmniExpress BeadChip was used. A total of 550 026 SNPs from 2642 cases and 2099 controls were included after quality control exclusions.
Genotyping in stage II was completed at the Vanderbilt Molecular Epidemiology Laboratory using the iPLEX Sequenom MassArray platform for 15 148 samples from the KOHBRA/KoGES, HCES-Br, SeBCS2, Nagoya, and Nagano studies. QC samples were used in the Sequenom assay, including one negative control (water), two blinded duplicates and two samples from the HapMap project in each 96-well plate. We excluded samples or SNPs that had a genotyping call rate of < 95%. We also excluded SNPs that had a concordance with the QC samples of < 95% or an unclear genotype call.
Statistical analysis
Imputation and haplotype estimation (phasing) were carried out for autosomal SNPs using Minimac2 and SHAPEIT(76) with the 1000 Genomes Project Phase 3 as the reference data for the Chinese and Japanese GWAS. The Korean GWAS was imputed using the 1000 Genomes Project Phase 1 as the reference. We only included SNPs with an MAF ≥ 0.01 and high imputation quality (RSQR ≥ 0.5) in three GWAS in the analyses. Association analyses of dosage data for imputed SNPs in each stage I study were analysed using the Mach2dat for SeBCS1 and Rvtests for SBCGS and BBJ1 (see URLs). The first five principal components estimated through EIGENSTRAT software (see URLs) (77) were included in the logistic regression models for adjustment of population structures. ORs associated with each SNP and 95% CIs were estimated under a log-additive model. To analyze genotype data, we used SAS version 9.3, which provides results identical to those generated with dosage data using Mach2dat and Rvtests. Summary ORs and 95% CIs for SNPs were obtained using fixed-effect inverse variance meta-analysis using METAL software (see URLs). Stratified analyses by ancestry and ER status were carried out. Heterogeneity across studies, among ancestry groups, and according to ER status was assessed with a Cochran’s Q test. In the combined analysis, a significant threshold P-value of < 5 × 10 − 8 was used to determine GWAS SNPs.
In collaboration with the DRIVE GAME-ON Consortium, data from 16 003 cases and 41 335 controls were assessed to conduct in silico replication of the SNPs included in stage II analyses (Supplementary Material, Text S1). Forest plots were generated using STATA version 23 and regional association plots were generated using LocusZoom (see URLs). To identify proxy SNPs, pairwise LD r2 was calculated based on the 1000 Genomes Project Phase 3 Asian populations. All genomic references are based on NCBI Human Genome Build 37, and P-values presented are based on 2-sided tests.
Imputation accuracy
In the current study, 714 individuals from stage I SBCGS data were genotyped for 28 SNPs using the iPLEX Sequenom MassArray platform. These genotypes were used to evaluate the imputation accuracy by examining the correlation between array genotypes and imputed dosages. Accuracy was calculated using Pearson correlation coefficient. The imputed data were highly consistent with the genotype data from Sequenom for the two SNPs identified in this study (Supplementary Material, Table S8, squared correlation coefficient (r2) = 0.99 for both loci). Similarly, imputated data and genotype data were consistent for the remaining 26 SNPs (squared correlation coefficient (r2) > 0.8).
eQTL analysis
We extracted the RNA-Seq V2 data (level 3) of 1006 breast cancer tumour tissues and 94 adjacent normal tissues from the TCGA data portal (see URLs). We also downloaded DNA methylation data which were measured by the Illumina HumanMethylation450 BeadChip from TCGA level 3 data. SNP data genotyped using the Affymetrix SNP 6.0 array were also retrieved. Genotype data within the 1 Mb regions flanking the two loci were extracted and then imputed using Minimac2 and SHAPEIT with the 1000 Genomes Project Phase 3 as the reference data. Copy number variation (CNV) data for genes within a 1Mb region of the two loci for TCGA samples were collected from the cBioPortal (see URLs) for tumour tissues. We analyzed a total of 621 breast tumour tissues in the European population and 55 breast tumour tissues for the Asian population separately, including matched CNV, genotype, methylation and expression data. The eQTL analysis was performed in tumour tissue as previously described (78,79). Briefly, we transformed the RNA-Seq by the Expectation Maximization (RSEM) value of each gene, and performed principal component correction in gene expression data to remove potential batch effects. Residual linear regression analysis was then used to detect eQTLs while adjusting for methylation and CNV, according to the approach proposed by Li et al. (78,79).
In addition to TCGA, we conducted eQTL analyses using the GTEx database (see URLs), and data from the METABRIC project (47). We extracted matched genotypes and gene expression levels in a total of 1981 breast cancer tumour tissues from the METABRIC project. Gene expression profiling was generated on the Illumina HT12 arrays and downloaded from the Synapse (syn1757063, see URLs). A total of 49 576 transcripts are included in gene expression profiling and have been normalized as described previously (47). Genotype data using the Affymetrix SNP 6.0 array were downloaded from EBI (EGAD00010000164, see URLs). We used R package CRLMM (see URLs) to process genotype calls from the original image array-based data for METABRIC (80,81). Only probes of high qualities with intensities more than 3000 at 95% calling rate were included. Imputation was performed on the genotypes for the 1Mb regions flanking the two loci using Minimac2 and SHAPEIT with the 1000 Genomes Project Phase 3 as the reference data. The eQTL analysis was performed using Matrix eQTL (82) to evaluate the association between genotypes and gene expression levels. In the current study, we focused only on the SNPs imputed with high imputation quality (RSQR ≥ 0.5) and an MAF ≥ 0.01 within the 1Mb regions flanking the two newly identified risk loci to identify cis-eQTLs.
Differential gene expression analysis
To perform differential expression analysis on genes near the newly identified loci, we extracted their expression values from a total of 87 patients, consisting of tumour tissue sample and the corresponding adjacent normal tissue sample from TCGA. We first performed surrogate variable analysis on gene expression between tumour and normal tissues to reduce potential batch effects and other artefacts (83). The full model includes the tumour-normal comparison of interest adjusted for the paired design, and a null model was adjusted only for the paired design. The total number of latent factors and the values of the surrogate variables were identified and estimated using the two models. After adjusting for the surrogate variables, limma software package from Bioconductor was used to extract differential expression of genes (84). False discovery rate-adjusted (Benjamini and Hochberg method) P-values are presented (85).
Assessment of regulatory functions
We investigated the potential function of the two newly identified loci using epigenomic data from ENCODE (see URLs). First, we investigated whether they are located in regulatory elements (i.e. promoter and enhancer) using ChromHMM annotation tracks in ENCODE from the UCSC Genome Browser (see URLs) including nine cell lines: HMEC (breast normal cell line), GM12878, H1-hESC, K562, HepG2, HSMM, HUVEC, NHEK, and NHLF (86). We also evaluated DNase I hypersensitive and TF binding sites in all cell lines analyzed by ENCODE, including breast normal cell line, HMEC, and breast cancer cell lines, T-47D and MCF-7. We assessed the histone modification markers H3K4Me1, H3K4Me3, and H3K27Ac in all cell lines analyzed by ENCODE using the layered histone tracks from the UCSC Genome Browser. Two publicly-available tools, RegulomeDB (see URLs) (51) and HaploReg V4 (see URLs) (49), were also used to evaluate candidate functional variants.
URLs.
1000 Genomes Project, http://www.1000genomes.org/ last accessed on March, 2016
UCSC Genome Browser, http://genome.ucsc.edu/ last accessed on March, 2016
DRIVE GAME-ON Consortium, http://gameon.dfci.harvard.edu last accessed on March, 2016.
Minimac2 & SHAPEIT, https://imputationserver.sph.umich.edu/ last accessed on March, 2016.
The Cancer Genome Atlas (TCGA), http://cancergenome.nih.gov/ last accessed on March, 2016
cBioPortal, http://www.cbioportal.org/public-portal/ last accessed on March, 2016.
Genotype-Tissue Expression (GTEx), http://www.ncbi.nlm.nih.gov/gtex/GTEX2/gtex.cgi last accessed on March, 2016.
ENCODE Project, http://genome.ucsc.edu/ last accessed on March, 2016.
EIGENSTRAT, http://genepath.med.harvard.edu/∼reich/EIGENSTRAT.htm last accessed on March, 2016.
LocusZoom, v1.1, http://csg.sph.umich.edu/locuszoom/ last accessed on March, 2016.
HaploReg V4, http://www.broadinstitute.org/mammals/haploreg/haploreg.php last accessed on March, 2016.
HapMap Project, http://hapmap.ncbi.nlm.nih.gov/ last accessed on March, 2016.
Rvtests, http://genome.sph.umich.edu/wiki/RvTests last accessed on March, 2016.
Mach2dat, http://genome.sph.umich.edu/wiki/Mach2dat:_Association_with_MACH_output last accessed on March, 2016.
METAL, http://www.sph.umich.edu/csg/abecasis/metal last accessed on March, 2016.
Synapse, https://www.synapse.org/ last accessed on March, 2016.
EBI, https://www.ebi.ac.uk/ last accessed on March, 2016.
R version 3.2.0, http://www.r-project.org/ last accessed on March, 2016.
Genotype Calling (CRLMM) R package, http://bioconductor.org/packages/crlmm/ last accessed on March, 2016.
SAS version 9.3, http://www.sas.com/ last accessed on March, 2016.
STATA version 23, http://www.stata.com/ last accessed on March, 2016.
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
The authors wish to thank the study participants and research staff for their contributions and commitment to this project. We thank (R.C, J.W., J. H., H. C., and K. K.) at Vanderbilt for their help with sample preparation, genotyping and statistical analyses for the project, and editing and preparing the manuscript.
Conflict of Interest statement. None declared.
Funding
This research was supported in part by the US National Institutes of Health grants R01CA124558, R01CA148667, R01CA064277, R37CA070867, UM1CA182910 (to W. Z.); R01CA118229, R01CA092585 (to X.-O. S.); R01CA122756 (to Q. C.); and R01CA137013 (to J. L.), Department of Defense Idea Awards BC011118 (to X.-O. S.) and BC050791 (to Q. C.), and Ingram Professorship and Research Reward funds (to W. Z.). Sample preparation and genotyping assays at Vanderbilt were conducted at the Survey and Biospecimen Shared Resources and Vanderbilt Microarray Shared Resource, which are supported in part by the Vanderbilt-Ingram Cancer Center (P30CA068485). Data analyses were conducted using the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University. The SeBCS was supported by the BRL (Basic Research Laboratory) program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2011-0001564). KOHBRA/KOGES was supported by a grant from the National R&D Program for Cancer Control, Ministry for Health, Welfare and Family Affairs, Republic of Korea (#1020350).
Studies participating in the ABCC include (Principal Investigator, grant support): the Shanghai Breast Cancer Study (W.Z. and X.-O. S., R01CA064277), the Shanghai Women’s Health Study (W. Zheng, R37CA070867), the Shanghai Breast Cancer Survival Study (X.-O. S., R01CA118229), the Shanghai Endometrial Cancer Study (X.-O. S., R01CA092585, controls only), the Seoul Breast Cancer Study [D.K., BRL (Basic Research Laboratory) program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2012-0000347)], the BioBank Japan Project (S.-K.L., the Ministry of Education, Culture, Sports, Sciences and Technology from the Japanese Government); the Hwasun Cancer Epidemiology Study-Breast (S.-S. K., the National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea, # 1020010), the Nagano Breast Cancer Study (S.T., Grants-in-Aid for the Third Term Comprehensive Ten-Year Strategy for Cancer Control from the Ministry of Health, Labor and Welfare of Japan, and for Scientific Research on Priority Areas, 17015049 and for Scientific Research on Innovative Areas, 221S0001, from the Ministry of Education, Culture, Sports, Science, and Technology of Japan), the Hospital-based Epidemiologic Research Program at Aichi Cancer Center [Grant-in-Aid for Scientific Research on Priority Areas of Cancer (No. 17015018) from the Japanese Ministry of Education, Culture, Sports, Science and Technology (K.T.) and the “Practical Research for Innovative Cancer Control (15ck0106177h0001)” from the Japan Agency for Medical Research and development, AMED (K. Matsuo), and Cancer Bio Bank Aichi. The DRIVE GAME-ON consortium is funded by NIH grant U19CA148065 (D.H.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agents. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kamangar F. (2006) Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J. Clin. Oncol., 24, 2137–2150. [DOI] [PubMed] [Google Scholar]
- 2.Mavaddat N., Antoniou A.C., Easton D.F., Garcia-Closas M. (2010) Genetic susceptibility to breast cancer. Mol. Oncol., 4, 174–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang B., Beeghly-Fadiel A., Long J., Zheng W. (2011) Genetic variants associated with breast-cancer risk: comprehensive research synopsis, meta-analysis, and epidemiological evidence. Lancet Oncol., 12, 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Easton D.F., Pooley K.A., Dunning A.M., Pharoah P.D.P., Thompson D., Ballinger D.G., Struewing J.P., Morrison J., Field H., Luben R. et al. (2007) Genome-wide association study identifies novel breast cancer susceptibility loci. Nature, 447, 1087–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michailidou K., Beesley J., Lindstrom S., Canisius S., Dennis J., Lush M.J., Maranian M.J., Bolla M.K., Wang Q., Shah M. et al. (2015) Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat. Genet., 47, 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michailidou K., Hall P., Gonzalez-Neira A., Ghoussaini M., Dennis J., Milne R.L., Schmidt M.K., Chang-Claude J., Bojesen S.E., Bolla M.K. et al. (2013) Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet., 45, 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fletcher O., Johnson N., Orr N., Hosking F.J., Gibson L.J., Walker K., Zelenika D., Gut I., Heath S., Palles C. et al. (2011) Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J. Natl. Cancer Inst., 103, 425–435. [DOI] [PubMed] [Google Scholar]
- 8.Stacey S.N., Manolescu A., Sulem P., Thorlacius S., Gudjonsson S.A., Jonsson G.F., Jakobsdottir M., Bergthorsson J.T., Gudmundsson J., Aben K.K. et al. (2008) Common variants on chromosome 5p12 confer susceptibility to estrogen receptor–positive breast cancer. Nat. Genet., 40, 703–706. [DOI] [PubMed] [Google Scholar]
- 9.Stacey S.N., Manolescu A., Sulem P., Rafnar T., Gudmundsson J., Gudjonsson S.A., Masson G., Jakobsdottir M., Thorlacius S., Helgason A. et al. (2007) Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet., 39, 865–869. [DOI] [PubMed] [Google Scholar]
- 10.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A. et al. (2007) A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet., 39, 870–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turnbull C., Ahmed S., Morrison J., Pernet D., Renwick A., Maranian M., Seal S., Ghoussaini M., Hines S., Healey C.S. et al. (2010) Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet., 42, 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghoussaini M., Fletcher O., Michailidou K., Turnbull C., Schmidt M.K., Dicks E., Dennis J., Wang Q., Humphreys M.K., Luccarini C. et al. (2012) Genome-wide association analysis identifies three new breast cancer susceptibility loci. Nat. Genet., 44, 312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng W., Long J., Gao Y.T., Li C., Zheng Y., Xiang Y.B., Wen W., Levy S., Deming S.L., Haines J.L. et al. (2009) Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet., 41, 324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng W., Zhang B., Cai Q., Sung H., Michailidou K., Shi J., Choi J.Y., Long J., Dennis J., Humphreys M.K. et al. (2013) Common genetic determinants of breast-cancer risk in East Asian women: a collaborative study of 23 637 breast cancer cases and 25 579 controls. Hum. Mol. Genet., 22, 2539–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai Q., Zhang B., Sung H., Low S.K., Kweon S.S., Lu W., Shi J., Long J., Wen W., Choi J.Y. et al. (2014) Genome-wide association analysis in East Asians identifies breast cancer susceptibility loci at 1q32.1, 5q14.3 and 15q26.1. Nat. Genet., 46, 886–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purrington K.S., Slager S., Eccles D., Yannoukakos D., Fasching P.A., Miron P., Carpenter J., Chang-Claude J., Martin N.G., Montgomery G.W. et al. (2014) Genome-wide association study identifies 25 known breast cancer susceptibility loci as risk factors for triple-negative breast cancer. Carcinogenesis, 35, 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siddiq A., Couch F.J., Chen G.K., Lindstrom S., Eccles D., Millikan R.C., Michailidou K., Stram D.O., Beckmann L., Rhie S.K. et al. (2012) A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum. Mol. Genet., 21, 5373–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Closas M., Couch F.J., Lindstrom S., Michailidou K., Schmidt M.K., Brook M.N., Orr N., Rhie S.K., Riboli E., Feigelson H.S. et al. (2013) Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat. Genet., 45, 392–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai Q., Long J., Lu W., Qu S., Wen W., Kang D., Lee J.Y., Chen K., Shen H., Shen C.Y. et al. (2011) Genome-wide association study identifies breast cancer risk variant at 10q21.2: results from the Asia Breast Cancer Consortium. Hum. Mol. Genet., 20, 4991–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long J., Cai Q., Shu X.O., Qu S., Li C., Zheng Y., Gu K., Wang W., Xiang Y.B., Cheng J. et al. (2010) Identification of a functional genetic variant at 16q12.1 for breast cancer risk: results from the Asia Breast Cancer Consortium. PLoS Genet, 6, e1001002.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi J., Sung H., Zhang B., Lu W., Choi J.Y., Xiang Y.B., Kim M.K., Iwasaki M., Long J., Ji B.T. et al. (2013) New breast cancer risk variant discovered at 10q25 in East Asian women. Cancer Epidemiol. Biomarkers Prev., 22, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long J., Delahanty R.J., Li G., Gao Y.T., Lu W., Cai Q., Xiang Y.B., Li C., Ji B.T., Zheng Y. et al. (2013) A common deletion in the APOBEC3 genes and breast cancer risk. J. Natl. Cancer Inst., 105, 573–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed S., Thomas G., Ghoussaini M., Healey C.S., Humphreys M.K., Platte R., Morrison J., Maranian M., Pooley K.A., Luben R. et al. (2009) Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat. Genet., 41, 585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas G., Jacobs K.B., Kraft P., Yeager M., Wacholder S., Cox D.G., Hankinson S.E., Hutchinson A., Wang Z., Yu K. et al. (2009) A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat. Genet., 41, 579–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antoniou A.C., Wang X., Fredericksen Z.S., McGuffog L., Tarrell R., Sinilnikova O.M., Healey S., Morrison J., Kartsonaki C., Lesnick T. et al. (2010) A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat. Genet., 42, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cox A., Dunning A.M., Garcia-Closas M., Balasubramanian S., Reed M.W.R., Pooley K.A., Scollen S., Baynes C., Ponder B.A.J., Chanock S. et al. (2007) A common coding variant in CASP8 is associated with breast cancer risk. Nat. Genet., 39, 352–358. [DOI] [PubMed] [Google Scholar]
- 27.Milne R.L., Burwinkel B., Michailidou K., Arias-Perez J.I., Zamora M.P., Menéndez-Rodríguez P., Hardisson D., Mendiola M., González-Neira A., Pita G. et al. (2014) Common non-synonymous SNPs associated with breast cancer susceptibility: findings from the Breast Cancer Association Consortium. Hum. Mol. Genet., 23, 6096–6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Couch F.J., Wang X., McGuffog L., Lee A., Olswold C., Kuchenbaecker K.B., Soucy P., Fredericksen Z., Barrowdale D., Dennis J. et al. (2013) Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet., 9, e1003212.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long J., Cai Q., Sung H., Shi J., Zhang B., Choi J.Y., Wen W., Delahanty R.J., Lu W., Gao Y.T. et al. (2012) Genome-wide association study in east asians identifies novel susceptibility loci for breast cancer. PLoS Genet., 8, e1002532.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fejerman L., Ahmadiyeh N., Hu D., Huntsman S., Beckman K.B., Caswell J.L., Tsung K., John E.M., Torres-Mejia G., Carvajal-Carmona L. et al. (2014) Genome-wide association study of breast cancer in Latinas identifies novel protective variants on 6q25. Nat. Commun., 5, 5260.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen F., Chen G.K., Stram D.O., Millikan R.C., Ambrosone C.B., John E.M., Bernstein L., Zheng W., Palmer J.R., Hu J.J. et al. (2012) A genome-wide association study of breast cancer in women of African ancestry. Hum. Genet., 132, 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haiman C.A., Chen G.K., Vachon C.M., Canzian F., Dunning A., Millikan R.C., Wang X., Ademuyiwa F., Ahmed S., Ambrosone C.B. et al. (2011) A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor–negative breast cancer. Nat. Genet., 43, 1210–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long J., Shu X.O., Cai Q., Gao Y.T., Zheng Y., Li G., Li C., Gu K., Wen W., Xiang Y.B. et al. (2010) Evaluation of breast cancer susceptibility loci in Chinese women. Cancer Epidemiol. Biomarkers Prev., 19, 2357–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng W., Cai Q., Signorello L.B., Long J., Hargreaves M.K., Deming S.L., Li G., Li C., Cui Y., Blot W.J. (2009) Evaluation of 11 breast cancer susceptibility loci in African-American women. Cancer Epidemiol. Biomarkers Prev., 18, 2761–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng W., Wen W., Gao Y.T., Shyr Y., Zheng Y., Long J., Li G., Li C., Gu K., Cai Q. et al. (2010) Genetic and clinical predictors for breast cancer risk assessment and stratification among Chinese women. J. Natl. Cancer Inst., 102, 972–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Long J., Zhang B., Signorello L.B., Cai Q., Deming-Halverson S., Shrubsole M.J., Sanderson M., Dennis J., Michailiou K., Easton D.F. et al. (2013) Evaluating genome-wide association study-identified breast cancer risk variants in African-American women. PLoS One, 8, e58350.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma X., Beeghly-Fadiel A., Lu W., Shi J., Xiang Y.B., Cai Q., Shen H., Shen C.Y., Ren Z., Matsuo K. et al. (2012) Pathway analyses identify TGFBR2 as potential breast cancer susceptibility gene: results from a consortium study among Asians. Cancer Epidemiol. Biomarkers Prev., 21, 1176–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xuan D., Li G., Cai Q., Deming-Halverson S., Shrubsole M.J., Shu X.O., Kelley M.C., Zheng W., Long J. (2013) APOBEC3 deletion polymorphism is associated with breast cancer risk among women of European ancestry. Carcinogenesis, 34, 2240–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.1000 Genomes Project Consortium. (2015) A global reference for human genetic variation. Nature, 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim H., Lee J.Y., Sung H., Choi J.Y., Park S.K., Lee K.M., Kim Y.J., Go M.J., Li L., Cho Y.S. et al. (2012) A genome-wide association study identifies a breast cancer risk variant in ERBB4 at 2q34: results from the Seoul Breast Cancer Study. Breast Cancer Res., 14, R56.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elgazzar S., Zembutsu H., Takahashi A., Kubo M., Aki F., Hirata K., Takatsuka Y., Okazaki M., Ohsumi S., Yamakawa T. et al. (2012) A genome-wide association study identifies a genetic variant in the SIAH2 locus associated with hormonal receptor-positive breast cancer in Japanese. J. Hum. Genet., 57, 766–771. [DOI] [PubMed] [Google Scholar]
- 42.Low S.K., Takahashi A., Ashikawa K., Inazawa J., Miki Y., Kubo M., Nakamura Y., Katagiri T. (2013) Genome-wide association study of breast cancer in the Japanese population. PLoS One, 8, e76463.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuchsberger C., Abecasis G.R., Hinds D.A. (2015) minimac2: faster genotype imputation. Bioinformatics, 31, 782–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willer C.J., Li Y., Abecasis G.R. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaminski B.M., Amos C.I., DeRycke E., Gillanders E.M., Gruber S.B., Henderson B.E., Hunter D.J., Lepage P.K., Sellers T.A., Seminara D. (2012) Genetic Associations and Mechanisms in Oncology (GAME-ON): a network approach to post-GWAS research. Cancer Epidemiol. Biomarkers Prev., 21, 78–78. [Google Scholar]
- 46.Network,T.C.G.A. (2012) Comprehensive molecular portraits of human breast tumours. Nature, 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Curtis C., Shah S.P., Chin S.F., Turashvili G., Rueda O.M., Dunning M.J., Speed D., Lynch A.G., Samarajiwa S., Yuan Y. et al. (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature, 486, 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48., Ardlie K.G., Deluca D.S., Segrè A.V., Sullivan T.J., Young T.R., Gelfand E.T., Trowbridge C.A., Maller J.B., Tukiainen T. Consortium,T.Gte. et al. (2015) The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science, 348, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward L.D., Kellis M. (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res., 40, D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Consortium T.E.P. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S. et al. (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res., 22, 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeller T., Wild P., Szymczak S., Rotival M., Schillert A., Castagne R., Maouche S., Germain M., Lackner K., Rossmann H. et al. (2010) Genetics and beyond—the transcriptome of human monocytes and disease susceptibility. PLoS One, 5, e10693.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zou F., Chai H.S., Younkin C.S., Allen M., Crook J., Pankratz V.S., Carrasquillo M.M., Rowley C.N., Nair A.A., Middha S. et al. (2012) Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet., 8, e1002707.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li X., Cowell J.K., Sossey-Alaoui K. (2004) CLCA2 tumour suppressor gene in 1p31 is epigenetically regulated in breast cancer. Oncogene, 23, 1474–1480. [DOI] [PubMed] [Google Scholar]
- 55.Runkle K.B., Meyerkord C.L., Desai N.V., Takahashi Y., Wang H.G. (2012) Bif-1 suppresses breast cancer cell migration by promoting EGFR endocytic degradation. Cancer Biol. Ther., 13, 956–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Browne G., Taipaleenmäki H., Bishop N.M., Madasu S.C., Shaw L.M., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. (2015) Runx1 is associated with breast cancer progression in MMTV-PyMT transgenic mice and its depletion in vitro inhibits migration and invasion. J. Cell. Physiol., 230, 2522–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferrari N., Mohammed Z.M.A., Nixon C., Mason S.M., Mallon E., McMillan D.C., Morris J.S., Cameron E.R., Edwards J., Blyth K. (2014) Expression of RUNX1 correlates with poor patient prognosis in triple negative breast cancer. PLoS One, 9, e100759.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gozdecka M., Lyons S., Kondo S., Taylor J., Li Y., Walczynski J., Thiel G., Breitwieser W., Jones N. (2014) JNK suppresses tumor formation via a gene-expression program mediated by ATF2. Cell Rep., 9, 1361–1374. [DOI] [PubMed] [Google Scholar]
- 59.Xing L., Salas M., Zhang H., Gittler J., Ludwig T., Lin C.S., Murty V.V., Silverman W., Arancio O., Tycko B. (2013) Creation and characterization of BAC-transgenic mice with physiological over-expression of epitope-tagged RCAN1 (DSCR1). Mamm. Genome, 24, 30–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Montañez-Wiscovich M.E., Seachrist D.D., Landis M.D., Visvader J., Andersen B., Keri R.A. (2009) LMO4 is an essential mediator of ErbB2/HER2/Neu-induced breast cancer cell cycle progression. Oncogene, 28, 3608–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stokes P.H., Liew C.W., Kwan A.H., Foo P., Barker H.E., Djamirze A., O’Reilly V., Visvader J.E., Mackay J.P., Matthews J.M. (2013) Structural basis of the interaction of the breast cancer oncogene LMO4 with the Tumour suppressor CtIP/RBBP8. J. Mol. Biol., 425, 1101–1110. [DOI] [PubMed] [Google Scholar]
- 62.Cubeddu L., Joseph S., Richard D.J., Matthews J.M. (2012) Contribution of DEAF1 structural domains to the interaction with the breast cancer oncogene LMO4. PLoS One, 7, e39218.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sum E.Y.M., Peng B., Yu X., Chen J., Byrne J., Lindeman G.J., Visvader J.E. (2002) The LIM domain protein LMO4 interacts with the cofactor CtIP and the tumor suppressor BRCA1 and inhibits BRCA1 activity. J. Biol. Chem., 277, 7849–7856. [DOI] [PubMed] [Google Scholar]
- 64.Jonsson P., Coarfa C., Mesmar F., Raz T., Rajapakshe K., Thompson J.F., Gunaratne P.H., Williams C. (2015) Single-molecule sequencing reveals estrogen-regulated clinically relevant lncRNAs in breast cancer. Mol. Endocrinol., 29, 1634–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang L., Brugge J.S., Janes K.A. (2011) Intersection of FOXO- and RUNX1-mediated gene expression programs in single breast epithelial cells during morphogenesis and tumor progression. Proc. Natl. Acad. Sci. U S A., 108, E803–E812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng W., Chow W.H., Yang G., Jin F., Rothman N., Blair A., Li H.L., Wen W., Ji B.T., Li Q. et al. (2005) The Shanghai Women’s Health Study: rationale, study design, and baseline characteristics. Am. J. Epidemiol., 162, 1123–1131. [DOI] [PubMed] [Google Scholar]
- 67.Shu X.O., Zheng Y., Cai H., Gu K., Chen Z., Zheng W., Lu W. (2009) Soy food intake and breast cancer survival. JAMA, 302, 2437–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gao Y.T., Shu X.O., Dai Q., Potter J.D., Brinton L.A., Wen W., Sellers T.A., Kushi L.H., Ruan Z., Bostick R.M. et al. (2000) Association of menstrual and reproductive factors with breast cancer risk: results from the Shanghai breast cancer study. Int. J. Cancer, 87, 295–300. [DOI] [PubMed] [Google Scholar]
- 69.Cho Y.S., Go M.J., Kim Y.J., Heo J.Y., Oh J.H., Ban H.J., Yoon D., Lee M.H., Kim D.J., Park M. et al. (2009) A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat. Genet., 41, 527–534. [DOI] [PubMed] [Google Scholar]
- 70.Han S.A., Park S.K., Ahn S.H., Lee M.H., Noh D.Y., Kim L.S., Noh W.C., Jung Y., Kim K.S., Kim S.W. et al. (2011) The Korean Hereditary Breast Cancer (KOHBRA) study: protocols and interim report. Clin. Oncol., 23, 434–441. [DOI] [PubMed] [Google Scholar]
- 71.Song H.R., Shin M.H., Kim H.N., Piao J.M., Choi J.S., Hwang J.E., Park Y.K., Ryang D.W., Cho D., Kweon S.S. (2013) Sex-specific differences in the association between ABO genotype and gastric cancer risk in a Korean population. Gastric Cancer, 16, 254–260. [DOI] [PubMed] [Google Scholar]
- 72.Cui L.H., Shin M.H., Kweon S.S., Kim H.N., Song H.R., Piao J.M., Choi J.S., Shim H.J., Hwang J.E., Kim H.R. et al. (2010) Methylenetetrahydrofolate reductase C677T polymorphism in patients with gastric and colorectal cancer in a Korean population. BMC Cancer, 10, 236.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kweon S.S., Shin M.H., Jeong S.K., Nam H.S., Lee Y.H., Park K.S., Ryu S.Y., Choi S.W., Kim B.H., Rhee J.A. et al. (2014) Cohort profile: the Namwon Study and the Dong-gu Study. Int. J. Epidemiol., 43, 558–567. [DOI] [PubMed] [Google Scholar]
- 74.Hamajima N., Matsuo K., Saito T., Hirose K., Inoue M., Takezaki T., Kuroishi T., Tajima K. (2001) Gene-environment interactions and polymorphism studies of cancer risk in the hospital-based epidemiologic research program at Aichi Cancer Center II (HERPACC-II). Asian Pac. J. Cancer Prev., 2, 99–107. [PubMed] [Google Scholar]
- 75.Itoh H., Iwasaki M., Hanaoka T., Kasuga Y., Yokoyama S., Onuma H., Nishimura H., Kusama R., Tsugane S. (2009) Serum organochlorines and breast cancer risk in Japanese women: a case-control study. Canc. Causes Contr., 20, 567–580. [DOI] [PubMed] [Google Scholar]
- 76.Delaneau O., Marchini J., Zagury J.F. (2012) A linear complexity phasing method for thousands of genomes. Nat. Methods, 9, 179–181. [DOI] [PubMed] [Google Scholar]
- 77.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet., 38, 904–909. [DOI] [PubMed] [Google Scholar]
- 78.Pickrell J.K., Marioni J.C., Pai A.A., Degner J.F., Engelhardt B.E., Nkadori E., Veyrieras J.B., Stephens M., Gilad Y., Pritchard J.K. (2010) Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature, 464, 768–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Q., Seo J.H., Stranger B., McKenna A., Pe’er I., Laframboise T., Brown M., Tyekucheva S., Freedman M.L. (2013) Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell, 152, 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carvalho B.S., Louis T.A., Irizarry R.A. (2010) Quantifying uncertainty in genotype calls. Bioinformatics, 26, 242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Scharpf R.B., Irizarry R.A., Ritchie M.E., Carvalho B., Ruczinski I. (2011) Using the R package crlmm for genotyping and copy number estimation. J. Stat. Softw., 40, 1–32. [PMC free article] [PubMed] [Google Scholar]
- 82.Shabalin A.A. (2012) Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics, 28, 1353–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. (2012) The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics, 28, 882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smyth G.K., Michaud J., Scott H.S. (2005) Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics, 21, 2067–2075. [DOI] [PubMed] [Google Scholar]
- 85.Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B., 57, 289–300. [Google Scholar]
- 86.Ernst J., Kellis M. (2012) ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods, 9, 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.