Abstract
Acute pharmacological activation of adenosine monophosphate (AMP)-kinase using 5-aminoimidazole-4-carboxamide-1-b-D-ribofuranoside (AICAR) has been shown to improve muscle mitochondrial function by increasing mitochondrial biogenesis. We asked whether prolonged AICAR treatment is beneficial in a mouse model of slowly progressing mitochondrial myopathy (Cox10-Mef2c-Cre), and whether the compensatory mechanism is indeed an increase in mitochondrial biogenesis. We treated the animals for 3 months and found that sustained AMP-dependent kinase activation improved cytochrome c oxidase activity, rescued the motor phenotype and delayed the onset of the myopathy. This improvement was observed whether treatment started before or after the onset of the disease. We found that AICAR increased skeletal muscle regeneration thereby decreasing the levels of deleted Cox10-floxed alleles. We conclude that although increase in mitochondrial biogenesis and other pathways may contribute, the main mechanism by which AICAR improves the myopathy phenotype is by promoting muscle regeneration.
Introduction
Mitochondrial myopathies are caused by deficits in the oxidative phosphorylation system (OXPHOS) in skeletal muscle. The main symptoms of mitochondrial myopathies are muscle weakness, atrophy and exercise intolerance (1). Mitochondrial myopathies are often accompanied by neurodegeneration as the CNS is also very sensitive to OXPHOS defects. Mitochondrial myopathies can be caused by mutations in genes encoded by the mitochondrial DNA (mtDNA) or the nuclear DNA (nDNA) (2,3), and the incidence in the population is higher than one in 5000 children (4).
With a few exceptions (5) there are no effective treatments mitochondrial myopathies. However, in the pursuit of effective treatments several experimental therapeutics approaches have shown promise. Some of the approaches are disease-specific, such as gene replacement (6,7) and mtDNA heteroplasmy shifts (8–10). Others are general therapeutic strategies that can be used for the treatment of diverse disease-associated OXPHOS defects such as the induction of mitochondrial biogenesis, the use of vitamins and antioxidants, nutritional interventions, exercise and others (reviewed in 11).
We and others have previously shown that activation of mitochondrial biogenesis in cultured cells (12,13) with respiratory chain defects can boost residual OXPHOS capacity, preventing the OXPHOS defect. The OXPHOS defect in mouse models of mitochondrial myopathy caused by complex IV (CIV) deficiency was improved by increasing mitochondrial biogenesis by overexpression of the peroxisome proliferator-activated receptor γ (PPARγ) coactivator-1α, PGC-1α (14–16) PGC-1α interacts and activates several transcription factors such as nuclear respiratory factors (NRF1 and 2) that increase the transcription of the OXPHOS genes, therefore increasing mitochondrial capacity (17).
Several strategies have been tried in mouse models to increase the activity of PGC-1α and mitochondrial biogenesis for the correction of the OXPHOS defects. Bezafibrate, a PPAR pan-agonist that induces PGC-1α expression, showed some beneficial effects in mouse models, but also liver toxicity (15,18,19). PGC-1α activity can be increased upon deacetylation by NAD+ dependent Sirtuins, so strategies to increases the NAD+ pool in vivo and therefore the activity of Sirtuins have been tried. In agreement, supplementation of the diet with the vitamin B3-analogue nicotinamide riboside (NR) a NAD+ precursor, increased NAD+ levels and Sirtuins activity in vivo (20) and increased mitochondrial function in mouse models for mitochondrial diseases (21,22)
PGC-1α activity is also stimulated by phosphorylation, which depends on the activities of several kinases including the adenosine monophosphate-dependent kinase (AMPK) (23). Thus, one of the pathways to induce PGC-1α-dependent mitochondrial biogenesis has been reported to be through the activation of AMPK. AMPK regulates energy homeostasis and is activated by cellular stress, exercise, hormones, and also by the widely prescribed type 2 diabetes drug Metformin and 5-aminoimidazole-4-carboxamide-1-b-D-ribofuranoside (AICAR), an AMP analog (24). Interestingly, administration of AICAR activated AMPK in the skeletal muscle of wild-type mice, promoting metabolic changes similar to exercise (25). It was also shown that PGC-1α is required for AICAR-induced expression of mitochondrial proteins in mouse skeletal muscle (26). Indeed, pharmacological activation of AMPK using AICAR increased the levels of PGC-1α and the respiratory complex activities in three mouse models of cytochrome c oxidase (COX) deficiency (15). In addition, AICAR treatment increased mitochondrial mass and improved muscle function in a mouse model of Duchenne Muscular Dystrophy (27). Therefore, these results showed that pharmacological activation of AMPK using AICAR improved muscle mitochondrial function in vivo.
However, all the aforementioned studies were performed in acute settings and nothing is known about the effects of prolonged treatments or the initiation of treatment after the disease onset, which are the most likely scenarios in a clinical setting. Therefore, we decided to treat a mouse model with a mild myopathy (Cox10 floxed allele deleted by the muscle-specific Mef2c-cre, heretofore referred to as ‘Cox10-Mef2c’), with AICAR five times a week during 3 months and in two different paradigms: pre-symptomatic and post-symptomatic. Cox10 encodes for a heme farnesyl transferase that catalyzes the first step of the biosynthesis of heme a, an essential prosthetic group for the maturation of COXI, and therefore indispensable for the function of COX (28). Several missense mutations in the Cox10 gene have been found in patients with different clinical presentations: leukodystrophy and tubulopathy (29,30); anemia and Leigh syndrome; anemia, sensorineural deafness and fatal infantile hypertrophic cardiomyopathy (31); Leigh-like syndrome (32). Also, mutations in Cox10 have been associated with a relatively mild clinical phenotype comprising myopathy, demyelinating neuropathy, premature ovarian failure, short stature, hearing loss, pigmentary maculopathy and renal tubular dysfunction (33). Here, we report that prolonged AICAR treatment ameliorated the motor phenotype in Cox10-Mef2c mice and restored COX activity mostly by increasing fiber regeneration, likely with the contribution of activation of unfolded mitochondrial protein response and autophagy in skeletal muscle. Surprisingly, we did not observe a robust induction in mitochondrial biogenesis. In addition, we showed that sustained AICAR treatment was also effective after the disease ensues.
Results
Pre-symptomatic AICAR treatment restored and maintained running endurance and COX activity in a myopathy model
We previously generated a myopathy mouse model, the Cox10-Mef2c, by specific ablation of the Cox10 gene in skeletal muscle using the Cre-loxP system (34). To obtain Cox10-Mef2c animals, mice homozygous for floxed Cox10 (Cox10f/f) were bred with transgenic mice (Mef2c-cre) expressing cre recombinase under the control of the myocyte enhancer factor 2 promoter (35,36). This promoter expresses Cre in all muscle fiber sub-types starting in the embryo (E10). In the absence of COX10, COXI cannot mature, Complex IV (CIV) cannot assemble and consequently Cox10-Mef2c mice showed reduced CIV activity in skeletal muscle, resulting in a progressive myopathy (Supplementary Material, Fig. S1A and B). The onset of the myopathy occurs at 3 months of age, when the animals presented impaired endurance (Supplementary Material, Fig. S1C). In order to analyze if AICAR improves the phenotype of the myopathy animals we tested their performance on a treadmill every 2 weeks.
First, we examined the effects of 500 mg/kg of AICAR (5 days a week) during 3 months by intraperitoneal injection (IP) in pre-symptomatic Cox10-Mef2c animals (treatment started at 1.5 months, Fig. 1A). This prolonged treatment was able to activate AMPK in vivo in skeletal muscle as indicated by the increase in the phosphorylation of the threonine 172 of the kinase (Fig. 1B), and similar to what has been reported for the short treatments (15). The prolonged AMPK activation induced a significant increase in the running endurance in both control and knockout (KO) animals (Fig. 1C). In the KO mice, increased endurance was already observed 2 weeks after the beginning of the AICAR treatment, and by the end of the treatment (3 months later) these mice ran longer (41.4% more) than the vehicle-treated Cox10-Mef2c (Fig. 1C, age 4.5 months). Remarkably, the recovery of the endurance phenotype was maintained for an additional 3 months after the end of the treatment of the Cox10-Mef2c mice (Fig. 1C, age 7.5 months), suggesting that the changes induced by AICAR were stable and maintained over time. Similar results were obtained in the control mice treated with AICAR (Fig. 1C).
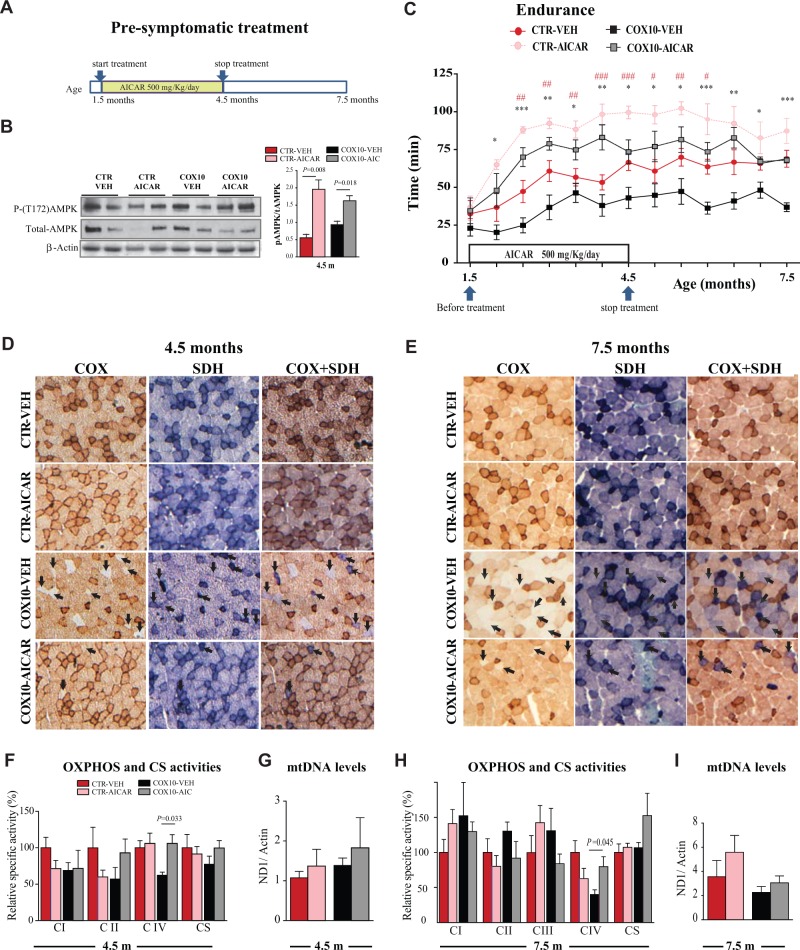
Figure 1.
Pre-symptomatic AICAR treatment improved a mitochondrial myopathy and the effect was still observed 3 months after the end of the treatment. (A) Scheme of the Pre-symptomatic AICAR treatment. (B) Representative western blots showing levels of phospho-AMPK and total-AMPK in quadriceps at the age of 4.5 months. (C) Running endurance measured as a function of time. Control littermate mice were treated with vehicle or AICAR (500 mg/kg/day, 12 weeks). Cox10-Mef2c mice were treated with vehicle or AICAR (500 mg/kg/day, 12 weeks). (D and E) COX, SDH and COX/SDH histochemical double staining in cross sections of quadriceps. Arrows show COX-deficient fibers that stain strong for the SDH activity (blue) and weakly for the COX activity (light brown/white). (F and H) OXPHOS complex activities and CS activity expressed as percentage of the control-vehicle group in homogenates from skeletal muscle (quadriceps). (G and I) mtDNA levels in homogenates from quadriceps. Data in (C) (n = 10), (B, D, E, G and I) (n = 5), and (F and H)) (n= 6) are presented as mean ± SEM, and unpaired Student’s two-tailed t-test was done for statistical significance. * P< 0.05, ** P< 0.001 and *** P< 0.0001 represents the difference between Cox10-vehicle and Cox10-AICAR; #P< 0.05 ##P< 0.001 and ###P< 0.0001 represents the difference between control (CTR)-vehicle and CTR-AICAR group.
Histological analysis of the quadriceps showed a reduction in COX-deficient fibers at both ages in the AICAR-treated Cox10-Mef2c (Fig. 1D and E, arrows). COX-deficient fibers stain for succinate dehydrogenase (SDH) activity (blue) but only weakly for COX activity (light brown/white). The wild-type littermates showed no COX-negative fibers. To analyze the CIV activity in the muscle, we performed an enzymatic assay in homogenates from quadriceps. At the age of 4.5 months, Cox10-Mef2c animals showed decreased CIV activity (62% ± 4 of control levels), that was fully recovered after AICAR treatment (106% ± 12 of control levels) (Fig. 1F). Although there was a decline of the CIV activity from 106% at 4.5 months to 80% of control values 3 months post treatment (7.5 months of age) in the KO mice treated with AICAR, the CIV activity on the treated mice was double than the one observed in untreated KO animals (40% ± 7 of control values, Fig. 1H). These data indicate that the recovery in the running endurance observed 3 months after the end of the treatment can be explained by the improved CIV activity. To analyze if prolonged AICAR treatment increased other OXPHOS complexes in the Cox10-Mef2c model, we also measured CI, CII and CIII enzymatic activities in homogenates from quadriceps. After 3 months of AICAR treatment we did not observe significant changes in the activities of these OXPHOS complexes, although complex II and citrate synthase (CS) showed a trend toward an increase (Fig. 1F and H).
To determine if AICAR treatment induced an increase in mitochondrial biogenesis, we measured CS activity (Fig. 1F and H) and mtDNA levels (Fig. 1G and I) in homogenates from quadriceps. Although results showed a trend toward an increase in the AICAR-treated groups, no statistical significance was reached, suggesting that AICAR-treated groups do not have a major increase in mitochondrial content. We measured the steady-state level of several mitochondrial proteins, including: NDUFB8 (CI subunit), SDHA (CII subunit), UQCRC2 (CIII subunit), COXI (CIV subunit) and ATPase-α (CV subunit) in quadriceps homogenates normalized to actin (Supplementary Material, Fig. S2). With the exception of the CIV subunit COXI, that was increased, there were no other changes, suggesting that there was no more global mitochondrial mass in the animals after prolonged AICAR treatment. In addition, we measured the levels of PGC-1α in the same homogenates but we detected only a trend toward an increase (Supplementary Material, Fig. S2). However, we did not determine the levels of PGC-1α potentially activated by phosphorylation. These data indicate that pre-symptomatic AICAR treatment delayed the progression of the myopathy in the Cox10-Mef2c model and the mechanism responsible was not a simple increase in mitochondrial biogenesis.
At 1.5 months of age, when treatment started, there were no differences in body weight between the animals assigned to the different groups (Supplementary Material, Fig. S3C). To determine if AICAR treatment had an effect in body weight, mice were weighted every 2 weeks during the 3 months treatment. No differences were found between vehicle and AICAR-treated mice, indicating that pre-symptomatic AICAR treatment had no effect in body weight (Supplementary Material, Fig. S2C, 4.5 m). At the age 7.5 m, the myopathy Cox10-Mef2c mice weight significantly less than the control littermates (Supplementary Material, Fig. S3C) and body weight was restored in the AICAR-treated Cox10-Mef2c group (Supplementary Material, Fig. S3C, 7.5 m).
Prolonged AICAR-induced AMPK activation in rats remodels adipocyte metabolism by up-regulating pathways that favor energy dissipation versus lipid storage, causing a decrease in fat storage (37,38). In order to determine if AICAR promoted a reduction in white adipose tissue (WAT) content, we measured the gonadal fat deposits in our animals after the 3 months of treatment. We found that pre-symptomatic AICAR treatment decreased gonadal fat mass to body weight ratio in both control and KO mice (Supplementary Material, Fig. S3A and B), without altering body weight (Supplementary Material, Fig. S3C), confirming effective activation of AMPK signaling in WAT in addition to skeletal muscle. We also analyzed the adipocyte morphology in sections from WAT and found decreased adipocyte area in AICAR-treated mice (Supplementary Material, Fig. S3D and E). We measured the same parameters in the animals at 7.5 months, 3 months after the end of the treatment (Supplementary Material, Fig. S3E). The WAT/Body weight was restored to untreated values (Supplementary Material, Fig. S3B), but the adipocyte area was smaller in the AICAR-treated groups (Supplementary Material, Fig. S3E). These results suggest that in addition to the skeletal muscle tissue, AICAR induced metabolic changes that were maintained over time in WAT.
To determine if prolonged AICAR treatment had side effects in mice, we collected blood and analyzed the levels of markers of liver, kidney, pancreas and muscle damage (Supplementary Material, Fig. S4). No differences were found between groups suggesting that these organs were not damaged.
Post-symptomatic AICAR treatment restored and maintained running endurance and COX activity in a myopathy model
To determine if prolonged AICAR treatment is also effective after the disease onset, we injected the drug in the Cox10-Mef2c mice at the age of 4.5 months, when endurance capacity was already reduced to 68% of the control levels (Fig. 2B and Supplementary Material, Fig. S1). This protocol is referred to as post-symptomatic AICAR treatment, and the duration, dose, and route of administration of the drug was the same as the pre-symptomatic treatment (Fig. 2A). At the end of the treatment Cox10-Mef2c mice treated with AICAR ran 27% longer than the vehicle-treated Cox10-Mef2c. On average, AICAR-treated Cox10-Mef2c ran 68 min versus 49 min vehicle-treated Cox10-Mef2c (Fig. 2B, age 7.5 m). We followed the animals up to 4 months after the end of the treatment, age 11.5 months, and observed that the recovery in the endurance phenotype was maintained in the AICAR-Cox10-Mef2c group (Fig. 2B). AICAR-KO-treated mice ran on average 70 min, whereas at the same age the running endurance time in the vehicle-treated Cox10-Mef2c was reduced to 37 min, as expected from the progression of the myopathy (Fig. 2B). We also analyzed the enzymatic activity of the single OXPHOS complexes by spectrophotometric assays in muscle homogenates. Similarly to what we observed in the pre-symptomatic treatment, the activity of CIV relative to control was significantly increased in the post-symptomatic AICAR-treated Cox10-Mef2c (92%) versus vehicle-treated Cox10-Mef2c group (58%) (Fig. 2E). These results were substantiated by histochemical COX staining, which showed reduced COX-negative fibers in the AICAR-treated Cox10-Mef2c group (Fig. 2C, arrows). Complex I, II and III activities were not affected after AICAR treatment, and CS activity was significantly increased only in the AICAR-treated control group (Fig. 2E), but not in the AICAR-treated Cox10-Mef2c mice.
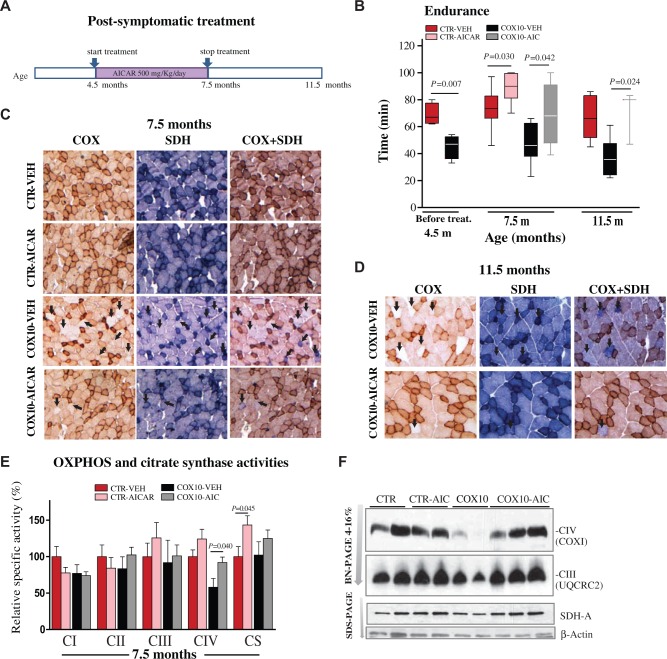
Figure 2.
Post-symptomatic AICAR improved running endurance and restored Complex IV activity and assembly in COX10-Mef2c mice. (A) Scheme of the post-symptomatic AICAR treatment. (B) Endurance measured as a function of time and represented in whisker plots. Cox10-Mef2c mice showed reduced endurance before the treatment started, age 4.5 m. AICAR-treatment during 3 months restored running endurance to control levels in the Cox10-Mef2c mice (age 7.5 m). The recovery was maintained for additional 4 months after the end of the treatment. Unpaired Student’s two-tailed t-test was performed and P values are shown. Significance was set at P < 0.05, (n ≥ 5 in all groups, with the exception of AICAR-treated Cox10-Mef2c mice of 11.5 months old were n = 3) (C-D) COX, SDH and COX/SDH histochemical double staining in cross sections of quadriceps. Arrows show COX-deficient fibers, which stain strongly for the SDH activity (blue) and weakly for the COX activity (light brown/white) (n = 5). (E) OXPHOS complex activities and citrate synthase activity expressed as percentage of the Control-vehicle group in homogenates from skeletal muscle (quadriceps). Data (n = 5) are presented as mean ± SEM, and unpaired Student’s two-tailed t-test was done for comparison of the two myopathy groups. (F) Steady-state levels of respiratory complexes in muscle homogenates from 7.5 month-old control and Cox10-Mef2c mice. Respiratory complexes in quadriceps (20 µg) from vehicle and AICAR-treated control and Cox10-Mef2c mice were separated by BN-PAGE (4–16% gels). (G) Quantification of BN-PAGE in (F) showing complexes levels normalized to SDH-A (CII) and to control animals.
To further characterize the molecular mechanism of the recovery, we analyzed the assembly of individual OXPHOS complexes in Blue Native Gels. We observed that the steady-state levels of the CIV holocomplex were markedly reduced in the Cox10-Mef2c mice at 7.5 months (Fig. 2F). However, CIV holocomplex levels were restored after AICAR-treatment in the Cox10-Mef2c mice (Fig. 2F and G). Therefore, we concluded that post-symptomatic AICAR treatment also restored both the endurance phenotype and the CIV activity in skeletal muscle. To investigate the molecular basis of the improved of the phenotype, which was still observed 4 months after the end of the treatment, we performed histochemical COX staining in skeletal muscle. At the age of 11.5 months, AICAR-treated Cox10-Mef2c showed a lower number of COX-negative fibers compared with the vehicle-treated Cox10-Mef2c (Fig. 2D, arrows).
Mitochondrial biogenesis was not significantly increased in post-symptomatic AICAR-treated Cox10-Mef2c mice
In order to study the mechanisms responsible for the correction of CIV activity in the Cox10-Mef2c mice treated with AICAR, we analyzed the steady-state levels of representative subunits for each OXPHOS complex by western blot analysis of skeletal muscle homogenates. COX1, an mtDNA-encoded CIV subunit, was reduced in Cox10-Mef2c mice compared with controls, and fully restored to control levels in AICAR-treated Cox10-Mef2c (Fig. 3A and B). In both control and Cox10-Mef2c homogenates, the steady-state levels of representative subunits of other OXPHOS complexes (I–V) were not altered after AICAR treatment (Fig. 3A and B). Then, we analyzed mtDNA levels in two different muscles (quadriceps femoralis and gastrocnemius) and found no significant differences in AICAR-treated groups compared with the respective vehicle-treated groups (Fig. 3C). Because PGC-1α is a master regulator of mitochondrial biogenesis and was reported to be increased in vivo by AICAR (39), we analyzed the PGC-1α levels in skeletal muscle of the animals with the post-symptomatic treatment. We found no changes in the protein levels of PGC-1α between the Cox10-Mef2c groups (Fig. 3A and B), but did not determine its phosphorylation status. Taken together, these data indicate that post-symptomatic AICAR-treatment restored COX levels and activity in the Cox10-Mef2c model. However, global increased mitochondrial biogenesis is unlike to explain the recovery observed, suggesting a different mechanism.
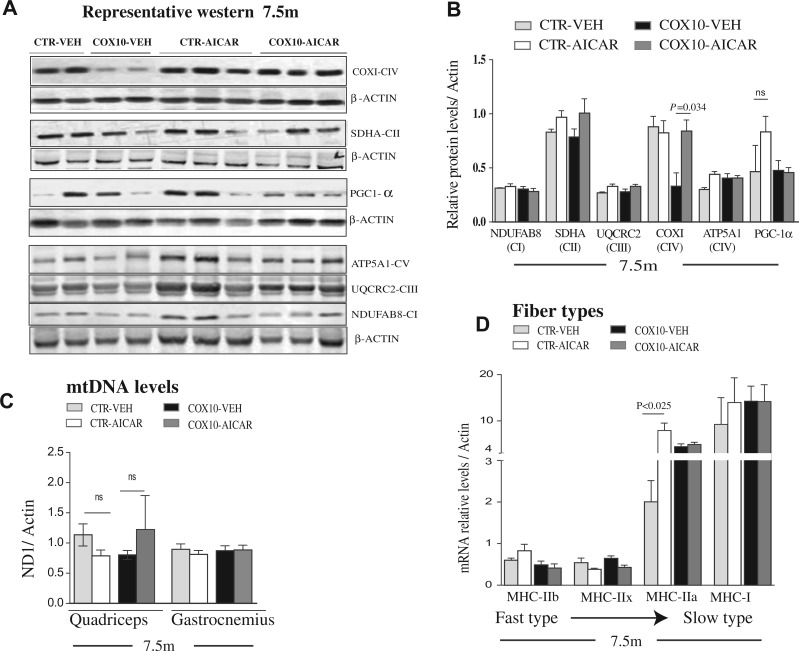
Figure 3.
Post-symptomatic AICAR treatment does not increased mitochondrial biogenesis in skeletal muscle. (A) Representative western blot showing steady-state levels of COXI (CIV), SDHA (CII), PGC-1α, ATPase-α (CV), UQCRC2 (CIII) and NDUFB8 (CI) in quadriceps homogenates at 7.5 months old, after post-symptomatic AICAR treatment. (B) Quantification of the protein levels showed in A, relative to β-Actin and to the vehicle-treated control group. (C) mtDNA levels in homogenates from quadriceps femoralis and gastrocnemius determined by qPCR. (D) Relative expression of transcripts coding for the contractile proteinsMHC: IIb, IIx, IIa and I in the quadriceps femoralis muscle Data (n = 5/group) are presented as mean ± SEM, and unpaired Student’s two-tailed t-test was done for pair wise comparisons (untreated versus treated).
AICAR treatment changes fiber type composition
Skeletal muscle is an adaptive tissue composed of multiple fibers types that differ in their metabolic and contractile properties including glycolytic fast-twitch (type IIb and IIx) fibers, mixed oxidative/glycolytic fast twitch (type IIa) fibers and oxidative-slow-twitch (type I) fibers (40,41). The switching of muscle fibers from fast to slow twitch is associated with increased endurance (42–44). Therefore, we tested whether the improvement in running endurance in the AICAR-treated Cox10-Mef2c mice, was associated with a switch from fast to slow-twitch (more oxidative) fiber types. For this aim, we analyzed the expression of transcripts coding for the myosin heavy chain (MHC) subtypes: IIb, IIx, IIa and I. Quantitative RT- polymerase chain reaction (RT-PCR) in RNA from the quadriceps femoralis muscle showed no significant change in the transcripts levels of MHC-IIb, MHC-IIx and MHC-I after AICAR treatment (Fig. 3D). However, MHC-IIa transcripts levels were increased 3.5-fold in skeletal muscle of AICAR-treated control group compared with the vehicle-treated one (Fig. 3D). These results were substantiated by immunohistochemistry (Supplementary Material, Fig. S5), and suggest an increase of this more oxidative fiber type after AICAR treatment in wild-type mice. Interestingly, MHC-IIa transcript levels were also increased in untreated Cox10-Mef2c animals (by 2-fold) compared with the untreated wild-type animals, suggesting a switch to slow fibers in the myopathy model (Fig. 3D and Supplementary Material, Fig S5). However, treatment with AICAR did not enhance these changes (as observed in wild-type mice, Fig. 3D).
Skeletal muscle transcriptome induced by AICAR in Cox10-Mef2c mice
To dissect the transcriptional alterations associated with the restored CIV activity and endurance phenotype, we studied the muscle transcriptome induced by AICAR in quadriceps muscle in the myopathy model. Microarray analysis identified 39 genes differentially expressed after 12 weeks of treatment in the skeletal muscle of the AICAR-treated Cox10-Mef2c compared with the vehicle-treated Cox10-Mef2c (Supplementary Material, Table S1). Enrichment pathway analysis showed that the differentially expressed genes included some of the already known downstream effectors of AMPK, like PPAR-δ, p70S6 kinase and the histone acetylase PCAF (p300/CBP associated transcription factor) (24). Other transcripts activated by AICAR included the thyroid hormone receptor (TR-alpha), retinoid acid receptor alpha and beta, AKT pathway and Inositol 3-Phosphate receptor (IP3 receptor) among others (Supplementary Material, Table S2). We detected changes in the MicroRNA133a-1 (Mir133a-1), which was down-regulated (2.63-fold) and in the cysteine and glycine-rich protein 3 (Csrp3), which was up-regulated (2.84-fold, Supplementary Material, Table S1). Mir133a-1 is a negative regulator of myogenesis (45), while Csrp3 is a transcription coactivator that facilitates myogenic differentiation (46,47).
We also analyzed the transcriptome data by filtering genes that had their expression changed by AICAR in the myopathy model, but not in the controls. This normalized analyses showed that 48 genes were differentially regulated by AICAR only in the myopathy model (Supplementary Material, Table S3). These genes belong to several functional classes, and 20% are transcription factors or co-activators/co-repressors of transcription factors (Supplementary Material, Table S3). Other functional classes included: proteasome, chaperones, autophagy, apoptosis, inflammation, angiogenesis and myogenesis. The analysis of the transcriptome showed several candidates that could mediate a beneficial response upon AMPK activation in the myopathic tissue. The levels of another micro RNA (MicroRNA-1) were decreased 1.82-fold in the quadriceps of AICAR-treated Cox10-Mef2c when compared with AICAR-treated control mice (Supplementary Material, Table S3). MicroRNA-1 targets Histone Deacetylase 4, and is a well-established inhibitor of muscle differentiation (45). Therefore, its down-regulation could also contribute to the regeneration of the skeletal muscle.
In summary, the transcriptome analysis suggested that prolonged AICAR treatment induced several regulatory pathways that include: transcription regulation, protein folding, regulation of inflammatory processes, and cell growth, proliferation and differentiation.
AICAR induces muscle fiber regeneration reducing the percentage of recombined floxed-Cox10 allele in skeletal muscles of Cox10-Mef2c
Based on the transcriptome studies, we next tested whether the recovered CIV activity and motor performance were associated with AICAR-induced skeletal muscle regeneration. This was suggested by the down-regulation of Mir133a-1 (Supplementary Material, Table S1) and Mir1 (Supplementary Material, Table S2), negative regulators of muscle proliferation and differentiation, respectively (45), and by the up-regulation of Csrp3, a positive regulator of myogenesis. In newly formed myofibers, the nuclei are centrally located (arrows in Fig. 4D) and then migrate to the periphery as the muscle fiber ages. Histological analysis of the quadriceps showed increased newly formed myofibers in the AICAR-treated Cox10-Mef2c compared with the vehicle-treated Cox10-Mef2c mice (by 2-fold, Fig. 4A–E). Similarly, we detected 1.8-fold increase in the number of fibers presenting central nuclei in the gastrocnemius muscle of AICAR-treated Cox10-Mef2c compared with the vehicle-treated KO (Supplementary Material, Fig. S6F), confirming that AICAR enhanced the myofiber regeneration in the mitochondrial myopathy model. In contrast, no differences in the number of newly formed fibers were found in the AICAR-treated wild-type animals compared with the vehicle-treated ones.
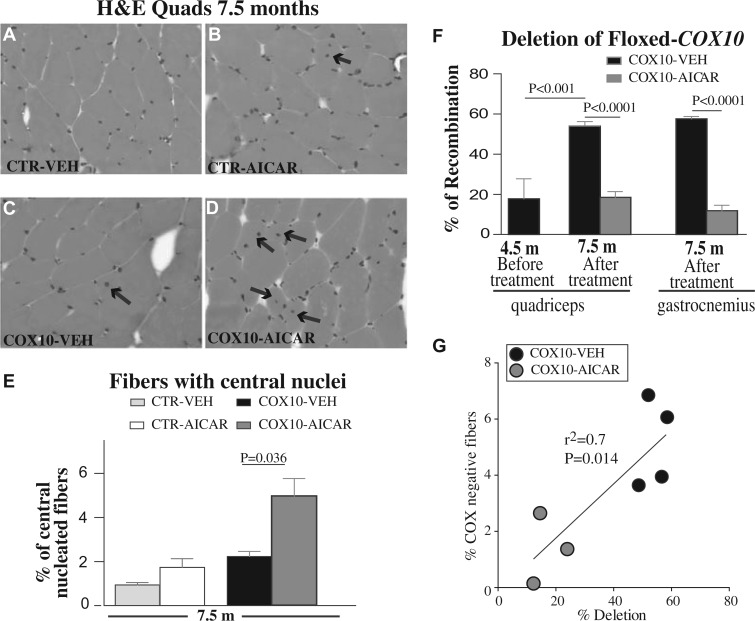
Figure 4.
Post-symptomatic AICAR increased the number of fiber with central nuclei in skeletal muscle of Cox10-Mef2c mice and reduced the deletion of floxed Cox10 allele. (A-D) H&E staining of quadriceps from control and Cox10-Mef2c mice after 3 months treatment with AICAR or vehicle. Arrows indicate the centralized nuclei. (E): Quantification of the number of centralized nuclei in the different groups (n = 5). Data are presented as mean ± SEM (800 myofibers/sample were analyzed (n ≥ 5/group and treatment). Unpaired Student’s two-tailed t-test was used for pairwise comparisons. (F) % of recombination of floxed-Cox10 allele was reduced after AICAR treatment in quadriceps and gastrocnemius. Data are presented as mean ± SEM (n = 5). One-way analysis of variance was done for multiple comparisons, followed by Bonferroni’s test; P< 0.001 represents the difference between Cox10-Mef2c before the treatment (age 4.5 months) and vehicle-treated Cox10-Mef2c after 3 months (age 7.5 months). (G) Correlation between COX-negative fibers and % deletion of floxed-Cox10 allele in quadriceps from Cox10-Mef2c mice after 3 months treatment with AICAR or vehicle (n ≥ 3/group and treatment and age 7.5 months).
Because the skeletal muscle of AICAR-treated Cox10-Mef2c mice showed increased number of newly formed myofibers, we suspected that the restored CIV activity could be related to reduced deletion of Cox10 gene in these newly formed fibers. To validate this hypothesis we analyzed the % of deletion of the floxed Cox10 allele in skeletal muscle of Cox10-Mef2c in the vehicle and AICAR-treated groups (expressed as % of recombination Fig. 4F). Quantitative PCR (qPCR) of genomic DNA from quadriceps and gastrocnemius of vehicle-treated Cox10-Mef2c confirmed the loss of exon 6 of the Cox10 gene (Fig. 4F). For quadriceps, at the age of 4.5 months (right before post-symptomatic treatment starts) we determined that ∼18% of all alleles were deleted (Fig. 4F). Three months later, and in agreement with the progression of the disease, recombination in the vehicle-Cox10-Mef2c group increased to ∼54%. However, the % of recombination did not increase in the AICAR-treated Cox10-Mef2c group (Fig. 4F, 7.5m). A similar finding was detected in the gastrocnemius muscle of the AICAR-treated Cox10-Mef2c. Accordingly, the number of COX-negative fibers positively correlated with the % of floxed allele deletion (Fig. 4G). Likewise, we detected a similar reduction in floxed allele deletion in the pre-symptomatic AICAR treatment (Supplementary Material, Fig. S7). Because the beneficial effects of the AICAR treatment were still observed 3 months after the end of the treatment, we calculated the % of recombination of floxed-Cox10 at that time point (Supplementary Material, Fig. S7, 7.5 m). Three months after stopping AICAR treatment, the % of deletion increased in the AICAR-treated Cox10-Mef2c mice (compared to 4.5 m of age, Supplementary Material, Fig. S7). However, it was still lower than the % of recombination in the vehicle-treated Cox10-Mef2c group at the same age (Supplementary Material, Fig. S7, 7.5 m). These data indicate that AICAR-treatment increased the number of newly formed fibers and reduced the percentage of deletion of floxed-Cox10 gene in skeletal muscle of Cox10-Mef2c animals, therefore increasing the levels of a functional Cox10 gene and ameliorating the myopathy phenotype.
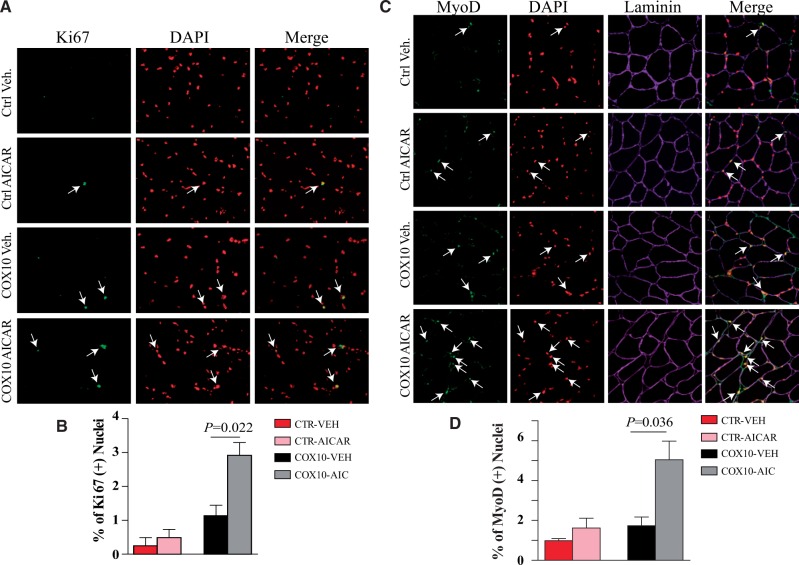
To confirm that there was an increase in muscle regeneration we stained muscle sections with MyoD and Ki67, markers of immature muscle (48) and cell proliferation (49), respectively. Accordingly, we observed an increase in both markers after treating the Cox10-Mef2c mice with AICAR (Fig. 5).
Figure 5.
Post-symptomatic AICAR increased the number of Ki67 and myoD positive cells in skeletal muscle of Cox10-Mef2c mice. Muscle sections from CTR and Cox10 mice (Veh. Or AICAR treated) for 3 months were immunostained for Ki67 (A) and MyoD (C). Sections were also stained for DAPI and in the case of C, also laminin. White arrows show nuclei that were positive for Ki67 (A) or positive for MyoD (C). The number of positive nuclei for Ki67 (proliferating cells) or MyoD (immature muscle) were quantified (B and D, respectively). n = 4 mice (≥ 600 fibers per quadriceps/mouse). Data are presented as mean ± SEM. Groups compared by the Student’s t-test.
The role of autophagy and mitochondrial unfolded protein response in the AICAR treatment of a mitochondrial myopathy model
Although muscle regeneration appears to play a major role in the improved phenotype, we further explored other mechanisms that could contribute to the improved muscle function.
To test if autophagy was increased in our model, we measured the levels of the autophagy marker LC3B in homogenates from quadriceps by western blot. Microtubule-associated protein 1 light chain 3 (LC3B) was increased in the AICAR-treated Cox10-Mef2c group. LC3B-II, the lipidated form that is more active in autophagy was not detectable in most samples, but the high levels of LC3B-I suggest an increase in the autophagy machinery (Supplementary Material, Fig. S8A). WIPI-2 (WD repeat domain phosphoinositide-interacting protein-2) is the mammalian homolog of Atg18 protein (Autophagy-related protein 18) and participates in the formation of the autophagosome by recognition of the phosphatidylinositol 3-phosphate (PtdIns3P) (50,51). Western blot results showed that WIPI-2 was increased in the AICAR-treated Cox10-Mef2c group compared with the vehicle-treated Cox10-Mef2c (Supplementary Material, Fig. S8A). WIPI-2 RNA levels were also increased in the transcriptome studies (Supplementary Material, Table S3). However, the levels of other proteins related to the autophagy process showed no differences (Supplementary Material, Fig. S8B).
Both mitochondrial proteins, HSP60 and HSP70 were increased in AICAR-treated Cox10-Mef2c mice compared with vehicle-treated Cox10-Mef2c (Supplementary Material, Fig. S8C). This indicates that the mitochondrial UPR may also contribute to the amelioration of the phenotype.
Discussion
In this work, we showed that prolonged treatment with the AMP-mimetic AICAR improved the motor phenotype and the COX activity in the skeletal muscle of the Cox10-Mef2c-cre mice. These beneficial effects were associated with the presence of new muscle fibers with reduced number of deleted Cox10 alleles. Our results agree with previous studies showing that AICAR treatment restored COX deficiency in mouse models with isolated COX deficiency (15), corroborating the potential of AICAR to treat mitochondrial myopathies. Besides the molecular mechanisms (discussed below), we made three novel observations that are relevant to its potential therapeutic use: 1) We showed that AICAR-activated AMPK improved the COX deficiency after the onset of the disease, therefore demonstrating the efficacy of this compound once the disease has manifested. This result has great significance as it better reflects clinical situations. 2) We showed that the improvement was maintained months after the drug was discontinued, which also provided clues to its mechanisms (discussed in more details below). 3) We showed the safety of the prolonged administration of AICAR, at least in rodents, since we did not observe overt toxicity.
It has been shown that AICAR can enhance endurance in wild-type mice by re-programing skeletal muscle gene expression (25). This study showed that AMPK simultaneously target multiple transcriptional programs that increased oxidative metabolism, angiogenesis and glucose sparing, pathways that are directly relevant to muscle performance. Because one of the pathways activated by AMPK is mitochondrial biogenesis and function (52) we expected to induce strong mitochondrial biogenesis by AICAR treatment. However, our results showed that prolonged AICAR treatment increased some but not all mitochondrial markers, and these increases were modest. We did not detect significant increases in mtDNA levels, neither in CS activity in the skeletal muscle of the Cox10-Mef2c after AICAR exposure, although we did detect an increase in CS in treated controls. Our results differ from previous studies showing that AICAR robustly enhances mitochondrial biogenesis in three different mouse models with CIV deficiency (15). However, there are several differences between these studies that could explain this discrepancy. We treated the animals chronically for 12 weeks with AICAR, while the length of the AICAR treatment in the Viscomi et al. (15) study was shorter, 4 weeks. It is possible that short AICAR-mediated AMPK activation increased PGC-1α protein levels, but this effect cannot be maintained after prolonged administration. Also, the fact that the dose of AICAR used in our study (500 mg/kg/day) was twice as high as the one used in Viscomi et al. (250 mg/kg/day) could be responsible of the activation of different target-pathways.
Therefore, although increased autophagy and mitochondrial unfolded protein response (mtUPR) may create a favorable environment that helps improve mitochondrial dysfunction, we believe that the main mechanism responsible for the phenotypic improvements relates to muscle regeneration. Cox10-Mef2c mice showed a trend toward an increase in newly formed muscle fibers, which suggests an adaptive response to the presence of CIV deficiency. AICAR treatment led to a further increase in the presence of newly formed fibers. Several studies showed that AMPK is required for skeletal muscle regeneration after cardiotoxin injury (53). Skeletal muscle repair was delayed in the AMPKα1 full body knock-out (KO) mice from day 2 post injury compared with wild-type mice (54). AMPKα1 KO muscle presented a significant higher number of necrotic myofibers, and consequently the number of newly formed myofibers (centrally nucleated) was delayed (54). Specific deletion of AMPKα1 in macrophages also impaired skeletal muscle regeneration after injury. AMPKα1 in the macrophages has been shown to be necessary for the activation of an anti-inflammatory state, which is required for muscle regeneration.
In addition, administration of Metformin, which directly activates AMPK, protects skeletal muscle from cardiotoxin-induced damage (55). In metformin treated muscle, 5 days after injury, the degenerating fibers were replaced by regenerating tissue, as shown by the presence of centro-nucleated fibers and by smaller fiber size. In contrast, muscle from untreated mice showed fewer centro-nucleated regenerating fibers and massive presence of inflammatory cells filling up the degenerating fibers (55). Recently, it has been shown that a periodic diet that mimics fasting, and therefore thought to triggers the stress nutrient deprivation signals such as AMPK, promotes regeneration of several tissues in mice, including skeletal muscle (56). In our study, the histological and immunohistological analyses (H&E, MyoD and Ki67) confirmed that AICAR-induced muscle regeneration in the myopathy model and restored CIV activity and endurance by increasing the number of new skeletal muscle fibers that were not COX deficient yet. The reduced levels of deletion of the floxed-Cox10 gene in the skeletal muscle of the AICAR-treated mutant, which correlates with the COX deficiency, provides strong evidence supporting this model. The microarray data indicated that AICAR induced the down-regulation of inhibitors of muscle proliferation and differentiation (Micro RNA133a-1 and Micro-RNA-1), and the up-regulation of the positive regulator of myogenesis transcription factor Csrp3. Therefore, our combined data support a model in which AICAR-induced muscle regeneration is the main mechanism responsible for the improved muscle CIV activity and recovered motor phenotype (Fig. 6). We did not see AICAR-induced muscle regeneration in control animals. This can be explained by the absence of inflammation in the skeletal muscle of control since AMPKα-1 is crucial for the resolution of inflammation during skeletal muscle regeneration (54).
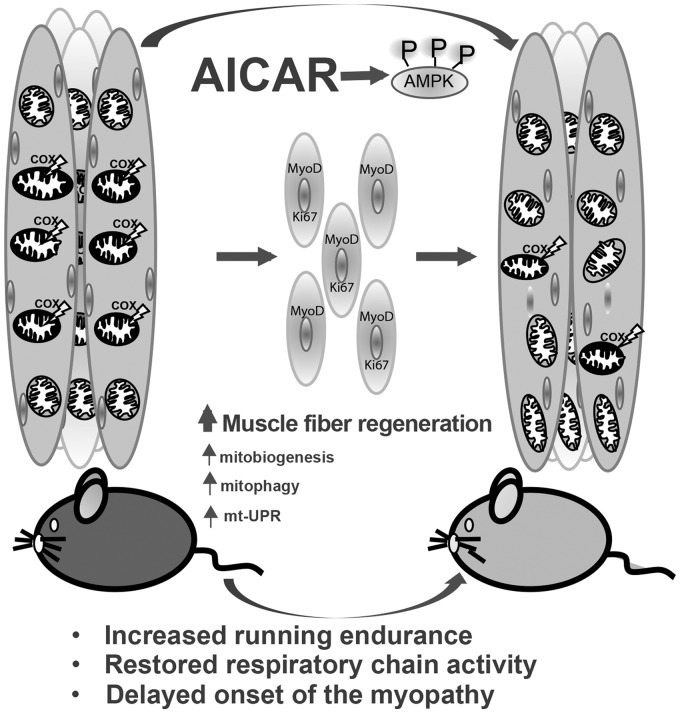
Figure 6.
General mechanisms associated with prolonged AICAR treatment in a mitochondrial myopathy model. We found that AICAR treatment led to an increase in muscle regeneration, reflected by increased central nucleated fibers and increased MyoD and Ki67-positive nuclei. This resulted in a concomitant marked decrease in deleted Cox10 alleles. Milder differences were also found in some mitochondrial, autophagy and mt-UPR markers.
Our results clearly showed that the beneficial effects of AICAR were maintained 3 months after the end of treatment. This long lasting effect can be explained indeed by the presence of new skeletal-muscle fibers, which in our model would take some time to become completely deficient for COX activity. Accordingly, we detected decreased levels of deletion of floxed-Cox10 gene in skeletal muscle of Cox10-Mef2c animals after AICAR treatment compared with the Cox10 group treated with saline. The lower % of deletion in the Cox10 gene would restore CIV activity and running endurance. However, once the treatment ended, the % of Cox10 deletion started to increase in both groups (AICAR and vehicle). Because of its lower starting point, the proportion of Cox10 deletion in muscle was still lower even 3 months after the end of the treatment in the groups that were treated with AICAR.
In addition, AMPK activation by AICAR has the potential to stimulates autophagy in skeletal muscle (57), which could also have beneficial effects on muscle function by selective elimination of the defective mitochondria (24). Acute AMPK stimulation by AICAR has been shown to initiate autophagy through different mechanisms involving activation of ULK1 (mammalian homolog of Atg1), inhibition of mTORC1, phosphorylation of raptor and activation the tuberous sclerosis complex (58,59). In AICAR-treated Cox10-Mef2c mice, there was significant up-regulation of the transcripts and protein levels of the mitophagy-associated protein WIPI-2, which participates in the phagosome formation and is required for the formation of LC3B (a microtubule-associated protein implicated in autophagy) positive phagosomes (50). We also observed increased expression of LC3B. Interestingly, WIPI-2 protein sequence has the consensus recognition motif of AMPK (60), suggesting it is a putative direct target of AMPK and of AICAR by extension. In addition, our data showed that AICAR-treatment increased the mtUPR in the KO mice, as indicated by the presence of increased levels of the mitochondrial chaperones HSP60 and HSP70. However, all these changes in the autophagy and UPR machinery may be secondary and not be the major drivers of the phenotypic improvement.
If this model (Fig. 6) is correct, AICAR treatment would be beneficial mostly to myopathies that affect older, mature muscle fibers more severely than younger, newly regenerated fibers. This probably include a large group of conditions where aggregates and oxidative products accumulate. It could also benefit myopathies caused by heteroplasmic mtDNA mutations, as regenerating fibers were found to have lower percentages of mutant genomes (61).
In summary, we propose that pharmacological activation of the AMPK, using AICAR, have the capacity to restore CIV activity and ameliorate the phenotype in the myopathy mouse model Cox10-Mef2c mostly by increasing muscle fiber regeneration, and to a lesser extend or secondarily, by increasing the levels of some mitochondrial proteins, increasing mitophagy and mitochondrial unfolded protein response (Fig. 6). Clinically approved pharmacological AMPK activators exist (e.g. Metformin) and AICAR has been used in clinical trials successfully for diabetes and ischemic reperfusion injury following coronary artery bypass surgery (62–64). Thus, this approach has the potential of being beneficial to some patients with mitochondrial myopathies.
Materials and Methods
Mice procedures
All animals used in this work were females in a C57Bl/6J background, backcrossed for at least 10 generations. All experiments and animal husbandry were performed according to a protocol approved by the University of Miami Institutional Animal Care and Use Committee. Mice were housed in a virus-antigen-free facility of the University of Miami, Division of Veterinary Resources in a 12-h light/dark cycle at room temperature and fed ad libitum. Mice homozygous for floxed Cox10 were generated previously in this lab (34) and the transgenic Mef2c-cre animals were obtained from Dr Brian L. Black at UCSF. In order to knock out Cox10 in skeletal muscle, mice homozygous for floxed Cox10 (Cox10f/f) were bred with transgenic mice (Mef2c-cre) expressing cre recombinase under the control of the myocyte enhancer factor 2 promoter (36).
AICAR treatments
AICAR was purchased from Toronto Research Chemicals, Cat#A611700. Two different treatment protocols were tested. Pre-symptomatic treatment start before the onset of the myopathy (mice were 1.5 month at initiation, 4.5 months of age at termination) and the post-symptomatic treatment start after onset of the disease (mice were 4.5 months at initiation, 7.5 m at termination). In both cases, AICAR was administered continuously, during 3 months (5 days a week), by IP at dose of 500 mg/kg dissolved in vehicle (saline).
Endurance test
A treadmill apparatus (Columbus Instruments) was used to determine exercise capacity under an endurance paradigm, before treatment started and every 2 weeks once started. A gradually accelerating program, with speed initially at 6.5 m/min and increasing by 0.5 m/min every 3 min, was used. Animals ran until exhaustion, which is defined by >10 falls/min into the motivational grid.
Blood work
Blood was withdrawn from deeply anesthetized animals by cardiac puncture. Serum was obtained, and the levels of liver, kidney, pancreas enzymes and creatine phosphokinase were determined by the Comparative Pathology Laboratory at the University of Miami, Miller School of Medicine
Histochemistry
Muscle tissue was frozen in isopentane liquid nitrogen. Cross-sections (10 µm) were stained for COX, SDH and combined activities as described in (65). Briefly, sections were incubated in 100 mm sodium phosphate buffer pH 7.4 containing 0.5 mg/ml diaminobenzidine, 0.2 mg/ml cytochrome c and 40 mg/ml sucrose at 37°C for 40 min. Succinate dehydrogenase or SDH activity stain was performed as described in Ref. (34) by incubating sections in 10 mm sodium phosphate buffer pH 7.5 containing 1.6 mg/ml EDTA, 0.65 mg/ml KCN, 0.06 mg/ml phenazine methosulfate, 1.3 mg/ml succinic acid and 1.2 mg/ml nitroblue tetrazolium at 37°C for 20 min. For double COX/SDH stain, sections were first stained with COX reagents, washed three times in PBS and then incubated with SDH reagents.
Determination of enzyme activities
Homogenates of quadriceps were prepared in PBS containing a protease inhibitor mixture (Roche Diagnostics) in a volume of ∼10× the weight. Tissue was minced first with scissors and then disrupted by 10–15 strokes using a motor driven homogenizer. Homogenates were centrifuged at 100g for 5 min and supernatants used for enzymatic assays. Complex I–IV and CS activities were measured spectrophotometrically as described previously (66,67). Assay results were normalized to protein concentration obtained by the Bradford method (68) Specific activity was determined and values represented as percentage of control values performed simultaneously.
Western blots
Protein extracts were prepared from homogenates of quadriceps as mentioned above. Before loading, sodium dodecyl sulfate (SDS) was added to the homogenate at the final concentration of 4%. Homogenates were then centrifuged at 14 000g at 4 °C, and the supernatant was collected for analysis. Protein concentration was determined by Lowry assay using the BCA kit (BioRad). Approximately 20–40 µg of protein were separated by SDS–polyacrylamide gel electrophoresis (PAGE) in 4–20% acrylamide gels and transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were blocked with 5% non-fat milk in 0.1% Tween 20 in PBS and subsequently incubated with specific antibodies. Primary antibodies, which were incubated overnight at 4 °C. Antibodies against Phospho-AMPK(T172), Total-AMPK, LC3B, WIPI2, ATG7 and BECLIN (1:1000) were obtained from Cell Signaling; COX1, NDUFA9, SDHA, UQCRC2, OXPHOS cocktail rodent mixture and PGC-1α (1:1000) were obtained from Abcam; Actin (1:5000) from Sigma. Horseradish peroxidase-conjugated secondary antibodies were used at 1:2000 dilution (Cell Signaling) and signal was developed by chemiluminescence using the SuperSignal West Pico reagent (Thermo Scientific).
Blue native–PAGE
To identify and estimate the levels of individual respiratory complexes, quadriceps homogenates were treated with 1% lauryl maltoside (Sigma) and mitochondrial complexes separated by blue native–PAGE (BN–PAGE) in 4–16% acrylamide gradient gels (Invitrogen) (69,70). Twenty micrograms of proteins were separated by PAGE, transferred to a PVDF membrane (Bio-Rad), and incubated sequentially with antibodies against several subunits of the different mitochondrial respiratory complexes.
mtDNA levels
Genomic DNA was extracted from quadriceps and gastrocnemius by fast pulverization of the tissue in liquid nitrogen and DNA extraction using standard proteinase K, phenol, chloroform extraction and isopropyl alcohol precipitation. The ratio of mitochondrial to nDNA was determined by quantitative real-time PCR using 2 ng of genomic DNA in a 20 µl reaction mixture using Sso Advanced Universal SYBR Green Supermix (Biorad) following PCR conditions stipulated by the manufacturer in a CFX96 Real Time PCR system (Biorad). Primers for the mtDNA were ND1-F: 5′-CAG CCT GAC CCA TAG CCA TA-3′ and ND1-B: 5′-ATT CTC CTT CTG TCA GGT CGA A-3′ and for the genomic DNA β-actin-F: 5′-GCG CAA GTA CTC TGT GTG GA-3′ and β-actin-B: 5′-CAT CGT ACT CCT GCT TGC TG-3′. To determine relative quantity of mtDNA in each sample, we used the comparative Ct method (71) and expressed as a ratio of ND1/Actin.
qPCR of genomic DNA
To quantify the deletion of the floxed Cox10 allele qPCR was performed using 20 ng of genomic DNA in a 20 µl assay using Sso Advanced Universal SYBR Green Supermix (Biorad) on a CFX96 Real Time PCR system (Biorad) in triplicates. A standard curve was generated using genomic DNA from Cox10flox/flox and Cox10flox/+ animals. The values were standardized to an independent gene (Epidermal Growth Factor) as described previously (72). A 167-base-pair Cox10flox-specific fragment was obtained with primers 5′-CGGGGATCAATTCGAGCTCGCC-3′ and 5′-CACTGACGCA GCGCC AGCATCTT-3′. The percentage of deletion was calculated by assuming that recombination occurs in both floxed alleles in Cre-expressing cells (73).
Microarray analysis
Genome-wide analysis was performed in quadriceps from vehicle-treated control, AICAR-treated control, vehicle-treated Cox10-Mef2c and AICAR-treated Cox10-Mef2c mice. RNA was extracted using QIAGEN kit, following manufacture indications. Preparations of transcription products, oligonucleotide array hybridization and scanning were performed by using Affymetrix high-density oligonucleotide array mouse genome chip 1.0 according to Affymetrix protocols. Microarray was performed in Microarray and Gene Expression Core, Miami Institute for Human Genomics and University of Miami Miller School of Medicine. First Expression Console software was used for data import, normalization and quality control. And for the differential expression the Transcriptome Analysis Console Software was used.
Immunohistology
Paraffin muscle sections (10 µm) were processed for hematoxylin and eosin staining for visualization of muscle fibers. Central nucleation (distinguishing feature of regenerating muscle fibers) were determined using Image J software.
Frozen muscle sections (10 µm) were blocked with 5% bovine serum albumin for 1 h at RT, and permeabilized with 1% Triton X-100. Sections were incubated with anti-Ki67 antibody (Abcam, 1:400), anti-Laminin (Sigma, 1:100) and anti MyoD (D7F2, Developmental Studies Hybridoma Bank,1:400) and anti- MHCIIa antibody (SC-71, Developmental Studies Hybridoma Bank, 1:400), over night at 4 °C. Slides were then incubated with Alexa-fluor secondary Ab for 2 h at RT, (Invitrogen) and mounted with Vectashield mounting medium for fluorescence plus D.A.P.I. Images were captured with an Olympus BX51 confocal microscope.
Statistical analysis
Two-tailed, unpaired Student t-test was used to determine the statistical significance between two groups (GraphPad Software). Multiple groups were compared using a one-way analysis of variance followed by a Bonferroni post hoc comparison (GraphPad Software). Error bars represent SEM. Significance when P < 0.05.
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
We are grateful to Ms. Aline Hida for technical assistance and to Mr. Isaac Biju for the microarray analysis.
Conflict of Interest Statement. None declared.
Funding
This work was supported by the US National Institutes of Health Grants 1R01NS079965, 5R01EY010804 and 1R01AG036871; the Muscular Dystrophy Association and the United Mitochondrial Disease Foundation. We acknowledge support from the NEI center grant P30-EY014801 from the National Institutes of Health (NIH).
References
- 1.DiMauro S., Schon E.A. (2008) Mitochondrial disorders in the nervous system. Ann. Rev. Neurosci., 31, 91–123. [DOI] [PubMed] [Google Scholar]
- 2.Wallace D.C., Fan W., Procaccio V. (2010) Mitochondrial energetics and therapeutics. Annu. Rev. Pathol., 5, 297–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schon E.A., DiMauro S., Hirano M. (2012) Human mitochondrial DNA: roles of inherited and somatic mutations. Nat. Rev., Genet., 13, 878–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorman G.S., Schaefer A.M., Ng Y., Gomez N., Blakely E.L., Alston C.L., Feeney C., Horvath R., Yu-Wai-Man P., Chinnery P.F. et al. (2015) Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann. Neurol., 77, 753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirano M., Garone C., Quinzii C.M. (2012) CoQ(10) deficiencies and MNGIE: two treatable mitochondrial disorders. Biochim. Biophys. Acta, 1820, 625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flierl A., Chen Y., Coskun P.E., Samulski R.J., Wallace D.C. (2005) Adeno-associated virus-mediated gene transfer of the heart/muscle adenine nucleotide translocator (ANT) in mouse. Gene Ther., 12, 570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torres-Torronteras J., Viscomi C., Cabrera-Perez R., Camara Y., Di Meo I., Barquinero J., Auricchio A., Pizzorno G., Hirano M., Zeviani M. et al. (2014) Gene therapy using a liver-targeted AAV vector restores nucleoside and nucleotide homeostasis in a murine model of MNGIE. Mol. Ther., 22, 901–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bacman S.R., Williams S.L., Pinto M., Peralta S., Moraes C.T. (2013) Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nat. Med., 19, 1111–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gammage P.A., Rorbach J., Vincent A.I., Rebar E.J., Minczuk M. (2014) Mitochondrially targeted ZFNs for selective degradation of pathogenic mitochondrial genomes bearing large-scale deletions or point mutations. EMBO Mol. Med., 6, 458–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hashimoto M., Bacman S.R., Peralta S., Falk M.J., Chomyn A., Chan D.C., Williams S.L., Moraes C.T. (2015) MitoTALEN: a general approach to reduce mutant mtDNA loads and restore oxidative phosphorylation function in mitochondrial diseases. Mol. Ther., 23, 1592–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peralta S., Torraco A., Iommarini L., Diaz F. (2015) Mitochondrial diseases part III: therapeutic interventions in mouse models of OXPHOS deficiencies. Mitochondrion, 23, 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bastin J., Aubey F., Rötig A., Munnich A., Djouadi F. (2008) Activation of peroxisome proliferator-activated receptor pathway stimulates the mitochondrial respiratory chain and can correct deficiencies in patients' cells lacking its components. J. Clin. Endocrinol. Metab., 93, 1433–1441. [DOI] [PubMed] [Google Scholar]
- 13.Srivastava S., Diaz F., Iommarini L., Aure K., Lombes A., Moraes C.T. (2009) PGC-1alpha/beta induced expression partially compensates for respiratory chain defects in cells from patients with mitochondrial disorders. Hum. Mol. Genet., 18, 1805–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wenz T., Rossi S.G., Rotundo R.L., Spiegelman B.M., Moraes C.T. (2009) Increased muscle PGC-1alpha expression protects from sarcopenia and metabolic disease during aging. Proc. Natl. Acad. Sci. U S A., 106, 20405–20410. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Viscomi C., Bottani E., Civiletto G., Cerutti R., Moggio M., Fagiolari G., Schon E.A., Lamperti C., Zeviani M. (2011) In vivo correction of COX deficiency by activation of the AMPK/PGC-1α axis. Cell Metab., 14, 80–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dillon L.M., Williams S.L., Hida A., Peacock J.D., Prolla T.A., Lincoln J., Moraes C.T. (2012) Increased mitochondrial biogenesis in muscle improves aging phenotypes in the mtDNA mutator mouse. Hum. Mol. Genet., 21, 2288–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scarpulla R.C. (2008) Nuclear control of respiratory chain expression by nuclear respiratory factors and PGC-1-related coactivator. Ann. N. Y. Acad. Sci., 1147, 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dillon L.M., Hida A., Garcia S., Prolla T.A., Moraes C.T. (2012) Long-term bezafibrate treatment improves skin and spleen phenotypes of the mtDNA mutator mouse. PLoS One, 7, e44335.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yatsuga S., Suomalainen A. (2012) Effect of bezafibrate treatment on late-onset mitochondrial myopathy in mice. Hum. Mol. Genet., 21, 526–535. [DOI] [PubMed] [Google Scholar]
- 20.Canto C., Houtkooper R.H., Pirinen E., Youn D.Y., Oosterveer M.H., Cen Y., Fernandez-Marcos P.J., Yamamoto H., Andreux P.A., Cettour-Rose P. et al. (2012) The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab., 15, 838–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerutti R., Pirinen E., Lamperti C., Marchet S., Sauve A.A., Li W., Leoni V., Schon E.A., Dantzer F., Auwerx J. et al. (2014) NAD(+)-dependent activation of Sirt1 corrects the phenotype in a mouse model of mitochondrial disease. Cell Metab., 19, 1042–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan N.A., Auranen M., Paetau I., Pirinen E., Euro L., Forsstrom S., Pasila L., Velagapudi V., Carroll C.J., Auwerx J. et al. (2014) Effective treatment of mitochondrial myopathy by nicotinamide riboside, a vitamin B3. EMBO Mol. Med., 6, 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez-Marcos P.J., Auwerx J. (2011) Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis. Am. J. Clin. Nutr., 93, 884S–8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mihaylova M.M., Shaw R.J. (2011) The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol., 13, 1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narkar V.A., Downes M., Yu R.T., Embler E., Wang Y.X., Banayo E., Mihaylova M.M., Nelson M.C., Zou Y., Juguilon H. et al. (2008) AMPK and PPARdelta agonists are exercise mimetics. Cell, 134, 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leick L., Fentz J., Bienso R.S., Knudsen J.G., Jeppesen J., Kiens B., Wojtaszewski J.F., Pilegaard H. (2010) PGC-1{alpha} is required for AICAR-induced expression of GLUT4 and mitochondrial proteins in mouse skeletal muscle. Am. J. Physiol. Endocrinol. Metab., 299, E456–E465. [DOI] [PubMed] [Google Scholar]
- 27.Jahnke V.E., Van Der Meulen J.H., Johnston H.K., Ghimbovschi S., Partridge T., Hoffman E.P., Nagaraju K. (2012) Metabolic remodeling agents show beneficial effects in the dystrophin-deficient mdx mouse model. Skelet. Muscle, 2, 16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tzagoloff A., Nobrega M., Gorman N., Sinclair P. (1993) On the functions of the yeast COX10 and COX11 gene products. Biochem. Mol. Biol. Int., 31, 593–598. [PubMed] [Google Scholar]
- 29.Valnot I., von Kleist-Retzow J.C., Barrientos A., Gorbatyuk M., Taanman J.W., Mehaye B., Rustin P., Tzagoloff A., Munnich A., Rotig A. (2000) A mutation in the human heme A: farnesyltransferase gene (COX10) causes cytochrome c oxidase deficiency. Hum. Mol. Genet., 9, 1245–1249. [DOI] [PubMed] [Google Scholar]
- 30.Williams S.L., Valnot I., Rustin P., Taanman J.W. (2004) Cytochrome c oxidase subassemblies in fibroblast cultures from patients carrying mutations in COX10, SCO1, or SURF1. J. Biol. Chem., 279, 7462–7469. [DOI] [PubMed] [Google Scholar]
- 31.Antonicka H., Leary S.C., Guercin G.H., Agar J.N., Horvath R., Kennaway N.G., Harding C.O., Jaksch M., Shoubridge E.A. (2003) Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum. Mol. Genet., 12, 2693–2702. [DOI] [PubMed] [Google Scholar]
- 32.Coenen M.J., van den Heuvel L.P., Ugalde C., Ten Brinke M., Nijtmans L.G., Trijbels F.J., Beblo S., Maier E.M., Muntau A.C., Smeitink J.A. (2004) Cytochrome c oxidase biogenesis in a patient with a mutation in COX10 gene. Ann. Neurol., 56, 560–564. [DOI] [PubMed] [Google Scholar]
- 33.Pitceathly R.D., Taanman J.W., Rahman S., Meunier B., Sadowski M., Cirak S., Hargreaves I., Land J.M., Nanji T., Polke J.M. et al. (2013) COX10 mutations resulting in complex multisystem mitochondrial disease that remains stable into adulthood. JAMA Neurol., 70, 1556–1561. [DOI] [PubMed] [Google Scholar]
- 34.Diaz F., Thomas C.K., Garcia S., Hernandez D., Moraes C.T. (2005) Mice lacking COX10 in skeletal muscle recapitulate the phenotype of progressive mitochondrial myopathies associated with cytochrome c oxidase deficiency. Hum. Mol. Genet., 14, 2737–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dodou E., Xu S.M., Black B.L. (2003) mef2c is activated directly by myogenic basic helix-loop-helix proteins during skeletal muscle development in vivo. Mech. Devel., 120, 1021–1032. [DOI] [PubMed] [Google Scholar]
- 36.Heidt A.B., Black B.L. (2005) Transgenic mice that express Cre recombinase under control of a skeletal muscle-specific promoter from mef2c. Genesis, 42, 28–32. [DOI] [PubMed] [Google Scholar]
- 37.Gaidhu M.P., Fediuc S., Anthony N.M., So M., Mirpourian M., Perry R.L., Ceddia R.B. (2009) Prolonged AICAR-induced AMP-kinase activation promotes energy dissipation in white adipocytes: novel mechanisms integrating HSL and ATGL. J. Lipid Res., 50, 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaidhu M.P., Bikopoulos G., Ceddia R.B. (2012) Chronic AICAR-induced AMP-kinase activation regulates adipocyte lipolysis in a time-dependent and fat depot-specific manner in rats. Am. J. Physiol., 303, C1192–C1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jager S., Handschin C., St-Pierre J., Spiegelman B.M. (2007) AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc. Natl. Acad. Sci. U S A., 104, 12017–12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pette D., Staron R.S. (2000) Myosin isoforms, muscle fiber types, and transitions. Microsc. Res. Tech., 50, 500–509. [DOI] [PubMed] [Google Scholar]
- 41.Fluck M., Hoppeler H. (2003) Molecular basis of skeletal muscle plasticity–from gene to form and function. Rev. Physiol. Biochem. Pharmacol., 146, 159–216. [DOI] [PubMed] [Google Scholar]
- 42.Booth F.W., Thomason D.B. (1991) Molecular and cellular adaptation of muscle in response to exercise: perspectives of various models. Physiol. Rev., 71, 541–585. [DOI] [PubMed] [Google Scholar]
- 43.Yoshioka M., Tanaka H., Shono N., Snyder E.E., Shindo M., St-Amand J. (2003) Serial analysis of gene expression in the skeletal muscle of endurance athletes compared to sedentary men. FASEB J., 17, 1812–1819. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y.X., Zhang C.L., Yu R.T., Cho H.K., Nelson M.C., Bayuga-Ocampo C.R., Ham J., Kang H., Evans R.M. (2004) Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol., 2, e294.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J.F., Mandel E.M., Thomson J.M., Wu Q., Callis T.E., Hammond S.M., Conlon F.L., Wang D.Z. (2006) The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet., 38, 228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arber S., Halder G., Caroni P. (1994) Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell, 79, 221–231. [DOI] [PubMed] [Google Scholar]
- 47.Kong Y., Flick M.J., Kudla A.J., Konieczny S.F. (1997) Muscle LIM protein promotes myogenesis by enhancing the activity of MyoD. Mol. Cell. Biol., 17, 4750–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mokalled M.H., Johnson A.N., Creemers E.E., Olson E.N. (2012) MASTR directs MyoD-dependent satellite cell differentiation during skeletal muscle regeneration. Genes Devel., 26, 190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scholzen T., Gerdes J. (2000) The Ki-67 protein: from the known and the unknown. J. Cell. Physiol., 182, 311–322. [DOI] [PubMed] [Google Scholar]
- 50.Polson H.E., de Lartigue J., Rigden D.J., Reedijk M., Urbe S., Clague M.J., Tooze S.A. (2010) Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy, 6, 506–522. [DOI] [PubMed] [Google Scholar]
- 51.Proikas-Cezanne T., Takacs Z., Donnes P., Kohlbacher O. (2015) WIPI proteins: essential PtdIns3P effectors at the nascent autophagosome. J. Cell Sci., 128, 207–217. [DOI] [PubMed] [Google Scholar]
- 52.Jorgensen S.B., Wojtaszewski J.F., Viollet B., Andreelli F., Birk J.B., Hellsten Y., Schjerling P., Vaulont S., Neufer P.D., Richter E.A. et al. (2005) Effects of alpha-AMPK knockout on exercise-induced gene activation in mouse skeletal muscle. FASEB J., 19, 1146–1148. [DOI] [PubMed] [Google Scholar]
- 53.Mounier R., Theret M., Lantier L., Foretz M., Viollet B. (2015) Expanding roles for AMPK in skeletal muscle plasticity. Trends Endocrinol. Metab., 26, 275–286. [DOI] [PubMed] [Google Scholar]
- 54.Mounier R., Theret M., Arnold L., Cuvellier S., Bultot L., Goransson O., Sanz N., Ferry A., Sakamoto K., Foretz M. et al. (2013) AMPKalpha1 regulates macrophage skewing at the time of resolution of inflammation during skeletal muscle regeneration. Cell Metab., 18, 251–264. [DOI] [PubMed] [Google Scholar]
- 55.Langone F., Cannata S., Fuoco C., Lettieri Barbato D., Testa S., Nardozza A.P., Ciriolo M.R., Castagnoli L., Gargioli C., Cesareni G. (2014) Metformin protects skeletal muscle from cardiotoxin induced degeneration. PLoS One, 9, e114018.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brandhorst S., Choi I.Y., Wei M., Cheng C.W., Sedrakyan S., Navarrete G., Dubeau L., Yap L.P., Park R., Vinciguerra M. et al. (2015) A periodic diet that mimics fasting promotes multi-system regeneration, enhanced cognitive performance, and healthspan. Cell Metab., 22, 86–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanchez A.M., Csibi A., Raibon A., Cornille K., Gay S., Bernardi H., Candau R. (2012) AMPK promotes skeletal muscle autophagy through activation of forkhead FoxO3a and interaction with Ulk1. J. Cell. Biochem., 113, 695–710. [DOI] [PubMed] [Google Scholar]
- 58.Lee J.W., Park S., Takahashi Y., Wang H.G. (2010) The association of AMPK with ULK1 regulates autophagy. PLoS One, 5, e15394.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Egan D.F., Shackelford D.B., Mihaylova M.M., Gelino S., Kohnz R.A., Mair W., Vasquez D.S., Joshi A., Gwinn D.M., Taylor R. et al. (2011) Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science, 331, 456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hardie D.G. (2010) Transcription. Targeting the core of transcription. Science, 329, 1158–1159. [DOI] [PubMed] [Google Scholar]
- 61.Taivassalo T., Fu K., Johns T., Arnold D., Karpati G., Shoubridge E.A. (1999) Gene shifting: a novel therapy for mitochondrial myopathy. Hum. Mol. Genet., 8, 1047–1052. [DOI] [PubMed] [Google Scholar]
- 62.Boon H., Bosselaar M., Praet S.F., Blaak E.E., Saris W.H., Wagenmakers A.J., McGee S.L., Tack C.J., Smits P., Hargreaves M. et al. (2008) Intravenous AICAR administration reduces hepatic glucose output and inhibits whole body lipolysis in type 2 diabetic patients. Diabetologia, 51, 1893–1900. [DOI] [PubMed] [Google Scholar]
- 63.Cuthbertson D.J., Babraj J.A., Mustard K.J., Towler M.C., Green K.A., Wackerhage H., Leese G.P., Baar K., Thomason-Hughes M., Sutherland C. et al. (2007) 5-aminoimidazole-4-carboxamide 1-beta-D-ribofuranoside acutely stimulates skeletal muscle 2-deoxyglucose uptake in healthy men. Diabetes, 56, 2078–2084. [DOI] [PubMed] [Google Scholar]
- 64.Drew B.G., Kingwell B.A. (2008) Acadesine, an adenosine-regulating agent with the potential for widespread indications. Exp. Opin. Pharmacother., 9, 2137–2144. [DOI] [PubMed] [Google Scholar]
- 65.Sciacco M., Bonilla E. (1996) Cytochemistry and immunocytochemistry of mitochondria in tissue sections. Methods Enzymol., 264, 509–521. [DOI] [PubMed] [Google Scholar]
- 66.Barrientos A., Fontanesi F., Diaz F. et al. (2009) Evaluation of the mitochondrial respiratory chain and oxidative phosphorylation system using polarography and spectrophotometric enzyme assays. Curr. Protoc. Hum. Genet, 63, 19.3:19.3.1–19.3.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martinez B., del Hoyo P., Martin M.A., Arenas J., Perez-Castillo A., Santos A. (2001) Thyroid hormone regulates oxidative phosphorylation in the cerebral cortex and striatum of neonatal rats. J. Neurochem., 78, 1054–1063. [DOI] [PubMed] [Google Scholar]
- 68.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 69.Calvaruso M.A., Smeitink J., Nijtmans L. (2008) Electrophoresis techniques to investigate defects in oxidative phosphorylation. Methods, 46, 281–287. [DOI] [PubMed] [Google Scholar]
- 70.Diaz F., Barrientos A., Fontanesi F. et al. (2009) Evaluation of the mitochondrial respiratory chain and oxidative phosphorylation system using blue native gel electrophoresis. Curr. Protoc. Hum. Genet, 63, 19.4:19.4.1–19.4.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schmittgen T.D., Livak K.J. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc., 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- 72.Funfschilling U., Supplie L.M., Mahad D., Boretius S., Saab A.S., Edgar J., Brinkmann B.G., Kassmann C.M., Tzvetanova I.D., Mobius W. et al. (2012) Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature, 485, 517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Diaz F., Garcia S., Padgett K.R., Moraes C.T. (2012) A defect in the mitochondrial complex III, but not complex IV, triggers early ROS-dependent damage in defined brain regions. Hum. Mol. Genet., 21, 5066–5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.