Abstract
Imputation is commonly used in genome-wide association studies to expand the set of genetic variants available for analysis. Larger and more diverse reference panels, such as the final Phase 3 of the 1000 Genomes Project, hold promise for improving imputation accuracy in genetically diverse populations such as Hispanics/Latinos in the USA. Here, we sought to empirically evaluate imputation accuracy when imputing to a 1000 Genomes Phase 3 versus a Phase 1 reference, using participants from the Hispanic Community Health Study/Study of Latinos. Our assessments included calculating the correlation between imputed and observed allelic dosage in a subset of samples genotyped on a supplemental array. We observed that the Phase 3 reference yielded higher accuracy at rare variants, but that the two reference panels were comparable at common variants. At a sample level, the Phase 3 reference improved imputation accuracy in Hispanic/Latino samples from the Caribbean more than for Mainland samples, which we attribute primarily to the additional reference panel samples available in Phase 3. We conclude that a 1000 Genomes Project Phase 3 reference panel can yield improved imputation accuracy compared with Phase 1, particularly for rare variants and for samples of certain genetic ancestry compositions. Our findings can inform imputation design for other genome-wide association studies of participants with diverse ancestries, especially as larger and more diverse reference panels continue to become available.
Introduction
Imputation is commonly used in genome-wide association studies (GWAS) to expand the set of genetic variants available for analysis (1). The approach uses a set of densely genotyped reference samples to infer genotypes at unmeasured variants in study samples, using the set of overlapping variants as an imputation basis. While early imputations were done with reference panels from either phase 2 (2) or phase 3 (3) of the HapMap Project, more recently the 1000 Genomes Project has become a common source of reference panels (4). Even larger reference panels are now available, such as the UK10K Cohorts Project (5) and the Haplotype Reference Consortium (6), which combines multiple sequencing efforts. The evolving landscape of imputation reference panels has expanded: (a) the number and types of variants available for imputation, (b) the number of available reference samples and (c) the diversity of genetic ancestries represented by reference sample populations. The latter two points are particularly important for imputing into samples from ancestrally diverse populations. Previous studies have demonstrated the association between imputation accuracy and several reference panel features, including panel size and diversity (4,6–11), quality of genotyping and sequencing (7) and quality of phasing (6,11). Here, we compare 1000 Genomes reference panels when imputing into a genetically diverse set of US Hispanic/Latino participants.
Expanding genetic studies into non-Europeans
The importance of extending genetic studies beyond predominately European sample sets and developing new approaches for the genetic analysis of samples with diverse ancestry is well-recognized within the genetics community (12). Rosenberg and colleagues enumerated challenges and opportunities associated with such an expansion. One challenge is obtaining reference samples that will yield accurate imputation results. Larger and more diverse reference panels have been shown to improve imputation accuracy in a variety of scenarios, including for rare variants in European ancestry samples (8), for worldwide populations in the Human Genome Diversity Project (9) and for admixed samples such as African Americans (13,14) and Hispanic Americans.
Hispanic/Latino genetic admixture
Due to geographical, historical and sociopolitical factors, Hispanic/Latino individuals in the USA are predominantly genetically admixed among three ancestry groups: Europeans, West Africans and Native Americans; however, different Hispanic/Latino groups show substantial heterogeneity in the proportions of these three ancestries (15–19). For reliable imputation in Hispanic/Latino individuals, special attention is required because of genetic heterogeneity both within and among different Hispanic/Latino groups. The reference panel needs to include samples from contemporary proxies of the three main source populations. Additionally, the imputation algorithm should allow for changes in genetic ancestry along the chromosome within each Hispanic/Latino study sample—the local ancestry (20). Many approaches have been developed to impute in the context of local ancestry, including MaCH-admix (14) and IMPUTE2 (4).
The 1000 Genomes Project
The 1000 Genomes Project recently released the final Phase 3 of the Project, comprising 2504 samples from 26 populations worldwide (7). Compared with Phase 1, Phase 3 includes 12 additional populations, including samples from the Caribbean, West Africa and South Asia. A combination of low-coverage whole genome sequencing, deep-coverage exome sequencing and array genotyping yielded a data set of over ∼81 M variants in these Phase 3 samples, including SNPs, copy number variations and more complex structural variants.
The 1000 Genomes Project has previously demonstrated improvements in imputation accuracy with a Phase 3 reference as compared with Phase 1, attributing these improvements to both increased sample size and higher genotyping and sequencing quality in Phase 3 (7). Our aim is to assess whether and to what extent the greater sample size and diversity of the Phase 3 reference panel as compared with Phase 1 will yield higher imputation accuracy in admixed population samples with recent ancestry from multiple continents, and particularly study samples from US Hispanic/Latino populations. Furthermore we aim to understand how variant-level imputation accuracy is affected across the minor allele frequency (MAF) spectrum and how sample-level imputation accuracy relates to genetic ancestry across different Hispanic/Latino groups from the Hispanic Community Health Study/Study of Latinos (HCHS/SOL) study (see Table 1).
Table 1.
Participant characteristics
| Genetic analysis group | Recruitment center |
Totals | |||
|---|---|---|---|---|---|
| Bronx | Chicago | Miami | San Diego | ||
| Cuban | 157 | 59 | 2012 | 29 | 2257 |
| Dominican | 1103 | 18 | 57 | 2 | 1180 |
| Puerto Rican | 1532 | 600 | 83 | 27 | 2242 |
| Central American | 96 | 385 | 840 | 76 | 1397 |
| Mexican | 171 | 1723 | 35 | 2821 | 4750 |
| South American | 175 | 293 | 418 | 35 | 921 |
| Not assigned | 37 | 1 | 11 | 7 | 56 |
| Totals | 3271 | 3079 | 3456 | 2997 | 12 803 |
Counts of HCHS/SOL samples by recruitment center and genetic analysis group, for 12 803 unique genotyped samples.
Results
Three primary imputation settings were tested: 1000 Genomes Phase 1 reference with maximum khap and Phase 3 reference with both half the maximum and maximum khap. Additional tests are reported in the Supplementary Material: Phase 3 “downsampled” to the size of Phase 1 and Phase 3 excluding East and South Asian reference samples.
Empirical masking of supplemental array genotypes
We compared imputed results with observed genotypes for a subset of 1424 HCHS/SOL samples genotyped on a supplemental array in addition to the primary array used for imputation (see Supplementary Material, Fig. S1). These results are for 21 394 chromosome 22 variants that passed quality control/quality assurance (QA/QC) in the supplemental array and were imputed with both the 1000 Genomes Phase 1 and Phase 3 reference panels. We present dosage r2: the squared Pearson correlation between observed {0, 1, 2} and imputed [0, 2] allelic dosage, both by variant, over all samples and by sample, over all variants.
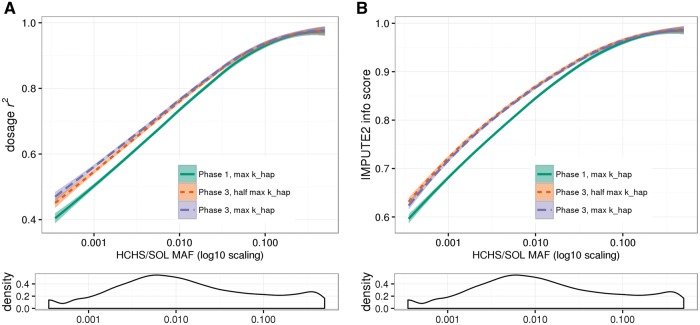
Figure 1A shows dosage r2 by variant across the MAF spectrum, where MAF is calculated in the HCHS/SOL supplemental array. As expected, imputation accuracy increases with MAF across all three settings. There was, however, improved accuracy with a Phase 3 reference panel as compared with Phase 1 across most of the MAF spectrum, particularly for less common and rare variants. Maximizing khap with the Phase 3 reference (purple line) yields a minor increase in imputation accuracy compared with half the maximum khap (orange line), albeit less than the improvement seen when moving from a Phase 1 to a Phase 3 reference. These by-variant dosage r2 metrics are summarized in Table 2, divided into three MAF bins: MAF < 1%, MAF 1–5% and MAF > 5%.
Figure 1.
Empirical masking results in supplemental array variants. Panel (A): By-variant dosage r2 between observed and imputed allelic dosages for supplemental array variants (Illumina HumanOmni2.5S), from empirical masking on chromosome 22. Panel (B): IMPUTE2 info metric at supplemental array variants. The x-axis for both plots is log10 scaling of MAF calculated from supplemental array genotypes. For plotting purposes, data were smoothed with the local regression smoothing method “loess,” which uses a t-based approximation to compute confidence bounds (shaded regions). Below the main plots are density curves illustrating the distribution of variants across the MAF spectrum. Note that 3819 monomorphic variants have been omitted from these plots, as they have missing (NA) dosage r2 values.
Table 2.
Imputation accuracy comparison by MAF category from empirical masking
| MAF category | Number of variants | Imputation setting 1: Phase 1, khap=max mean; median (SD) | Imputation setting 2: Phase 3, khap=half max mean; median (SD) | Imputation setting 3: Phase 3, khap=max mean; median (SD) |
|---|---|---|---|---|
| MAF < 1% | 11 161 | 0.606; 0.655 (0.311) | 0.643; 0.711 (0.303) | 0.653; 0.725 (0.303) |
| 1%≤MAF < 5% | 5262 | 0.803; 0.885 (0.218) | 0.822; 0.903 (0.203) | 0.826; 0.907 (0.203) |
| MAF ≥ 5% | 4971 | 0.948; 0.989 (0.112) | 0.952; 0.991 (0.106) | 0.953; 0.991 (0.106) |
| All variants | 21 394 | 0.742; 0.862 (0.289) | 0.766; 0.884 (0.275) | 0.772; 0.892 (0.273) |
Summaries of by-variant dosage r2 metrics from the empirical masking of supplemental array variants on chromosome 22: mean, median and SD. MAF is the observed MAF from the HCHS/SOL supplemental array genotyping (n = 1424 samples). Variants are limited to those passing quality filters in the supplemental array data set and imputed with both a Phase 1 and Phase 3 1000 Genomes Project panel. Variants either observed or imputed to be monomorphic have missing (NA) dosage r2 and are therefore omitted from these summaries.
Figure 1B shows IMPUTE2 info score for supplemental array variants. In general, info scores follow the same trend as by-variant dosage r2: values increase with increasing MAF, and a Phase 3 reference yields higher values compared with Phase 1 across the MAF spectrum. Unlike dosage r2, however, the info scores are slightly lower when maximizing khap with the Phase 3 reference (purple line) compared with half the maximum khap (orange line). This info score pattern is also found in the full set of imputed chromosome 22 variants and is consistent with previous observations (4) (discussed further below).
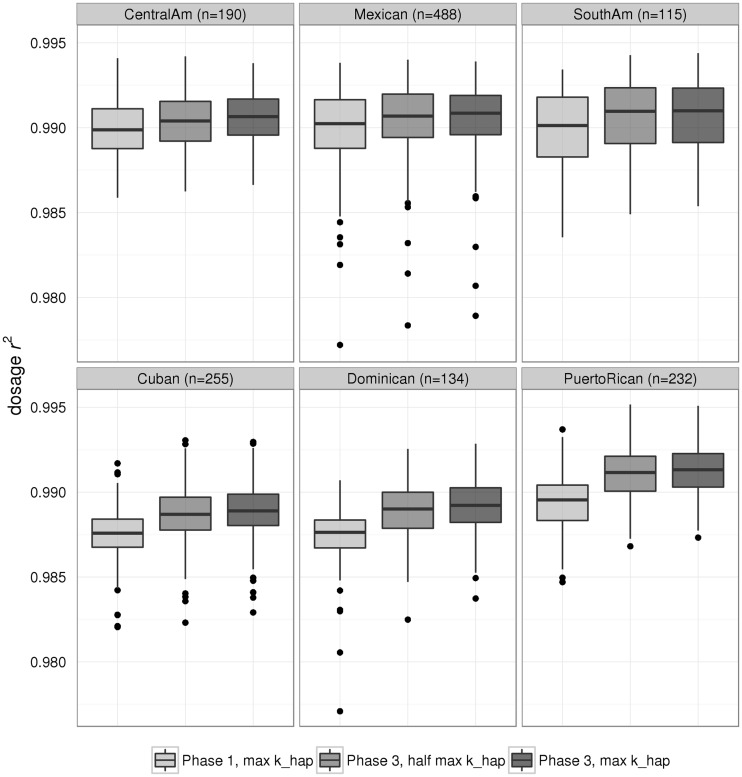
The by-sample empirical dosage r2 is presented in Figure 2, with HCSC/SOL samples divided by genetic analysis group. The three Mainland groups are in the top row of boxplots; the three Caribbean groups are in the bottom row. While imputation accuracy was quite high across all samples in all three settings (i.e. dosage r2 > 0.977), the average accuracy was slightly higher in Mainland samples compared with Caribbean (P-value < 2×10 − 16, ANOVA F-test of settings 1 and 2, see Table 3 for mean values). This finding was consistent across all three imputation settings. In general, imputation accuracy was higher when imputing to a Phase 3 reference compared with Phase 1 (P-value < 2×10 − 16, ANOVA F-test of settings 1 and 2, see Table 3 for mean values). Notably, the improvement was greater for Caribbean than for Mainland samples: comparing setting 2 with setting 1, mean dosage r2 increased by 0.0015 for Caribbean groups and by 0.0005 for Mainland groups. When comparing imputation settings 1 and 2, the interaction of region and reference panel was highly significant (P-value = 1.5×10 − 10, ANOVA F-test). As seen with by-variant dosage r2, by-sample metrics from the Phase 3 imputations increased slightly when using all available reference haplotypes (maximum khap); however, the relative improvement was less than moving from Phase 1 to Phase 3.
Figure 2.
Distributions of by-sample dosage r2 between observed and imputed allelic dosages. These distributions are over chromosome 22 variants from the supplemental Omni2.5SS array, presented as boxplots separately by genetic analysis group. The three Mainland groups are in the top row, and the three Caribbean groups are in the bottom row. Sample counts within each group are included in the panel headers. Excluded from these plots are 10 HCHS/SOL samples that have missing genetic group (i.e. are genetic outliers), thus there are 1414 total samples plotted here.
Table 3.
Imputation accuracy comparison by genetic analysis group and region from empirical masking.
| Region/ group | Number of samples | Imputation setting 1: Phase 1, khap=max mean (SD) | Imputation setting 2: Phase 3, khap=half max mean (SD) | Imputation setting 3: Phase 3, khap=max mean (SD) |
|---|---|---|---|---|
| Mainland | 772 | 0.99005 (0.00197) | 0.99056 (0.00184) | 0.99066 (0.00174) |
| Central American | 184 | 0.98995 (0.00173) | 0.99047 (0.00165) | 0.99063 (0.00154) |
| Mexican | 475 | 0.99011 (0.00203) | 0.99057 (0.00186) | 0.99067 (0.00176) |
| South American | 113 | 0.98994 (0.00207) | 0.99066 (0.00205) | 0.9907 (0.00192) |
| Caribbean | 616 | 0.98822 (0.00182) | 0.98967 (0.00196) | 0.98986 (0.00192) |
| Cuban | 253 | 0.98754 (0.00144) | 0.98875 (0.00171) | 0.98892 (0.00166) |
| Dominican | 132 | 0.98736 (0.00171) | 0.98889 (0.00159) | 0.98913 (0.00151) |
| Puerto Rican | 231 | 0.98946 (0.00156) | 0.99111 (0.00150) | 0.99131 (0.00147) |
Summaries of by-sample dosage r2 metrics from the empirical masking of supplemental array variants on chromosome 22. Mean dosage r2 values and SD are given for samples grouped both by region (Caribbean and Mainland) and by genetic analysis group. These summaries are restricted to the set of mutually unrelated samples from supplemental array genotyping with non-missing genetic analysis group (n = 1388).
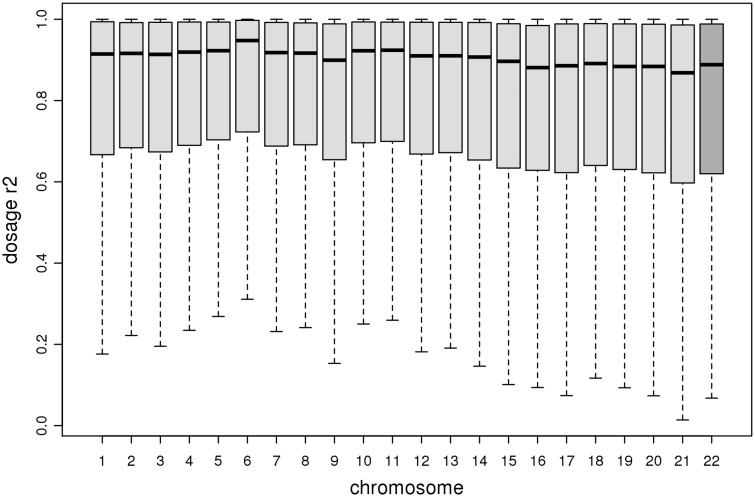
Above, we present empirical masking results for chromosome 22 only. Ultimately, we implemented imputation setting 2 (1000 Genomes Phase 3 reference, half the maximum khap) genome-wide and calculated dosage r2 across the full set of imputed variants also genotyped on the supplemental array (n = 1 654 398 variants). Boxplots of by-variant dosage r2 across all autosomes are shown in Figure 3, illustrating that imputation accuracy is consistently high across chromosomes, and that chromosome 22 is similar to other chromosomes not evaluated for the other imputation settings.
Figure 3.
Empirical masking results genome-wide. Boxplots of by-variant dosage r2 from imputation setting 2 (1000 Genomes Phase 3, half maximum khap) implemented genome-wide. Autosomes are ordered along the x-axis; chromosome 22 (used for testing) is a darkened color compared with the other autosomes. Outliers are excluded from these boxplots.
Overall imputation quality
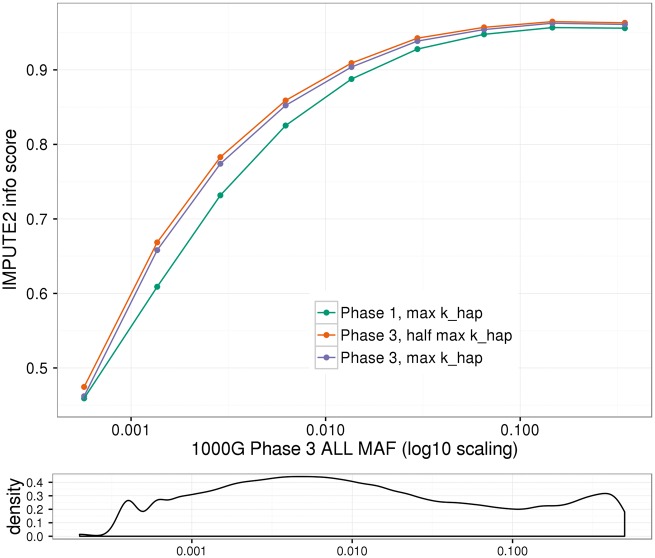
Next we present overall imputation quality for variants imputed in the full set of 12 803 HCHS/SOL samples genotyped on the primary array. IMPUTE2 info scores across the three imputation settings are summarized in Figures 4 and 5, and Table 4. Note that we have restricted these comparisons to chromosome 22 variants imputed across all three imputation settings, i.e. imputed with both a Phase 1 and a Phase 3 reference (n = 308 017 variants). In general, imputation quality improved with increasing MAF and with the Phase 3 reference panel as compared with Phase 1. Within the Phase 3 imputations, the mean info score decreased slightly when maximizing khap as compared with half the maximum khap (see Table 4), a previously observed phenomenon (4) that we discuss further below.
Figure 4.
Info score across all imputed variants. Info scores are plotted separately by imputation setting, for chromosome 22 variants imputed across all three settings (i.e. imputed with both 1000 Genomes Phase 1 and Phase 3 reference panels; n = 308 017 variants). For plotting, variants were binned by MAF in the worldwide (ALL) 1000 Genomes Project Phase 3 panel. The mean MAF per bin is plotted on the x-axis with log10 scaling; the mean info score over variants in the bin is plotted on the y-axis. The density plot in the sub-panel illustrates the density of variants across the (log10 scaled) MAF spectrum.
Figure 5.
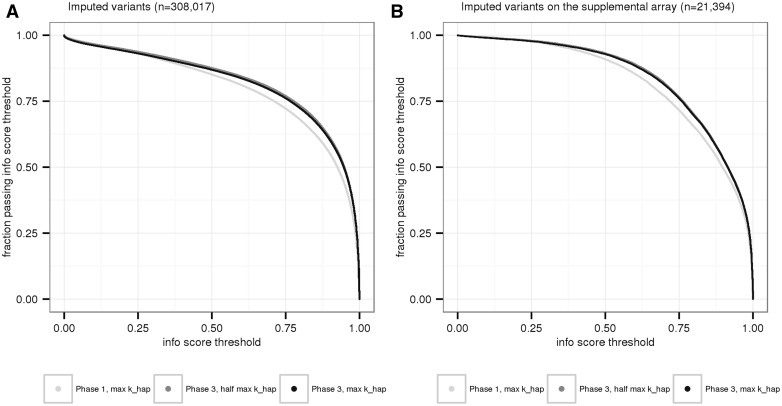
Fraction of imputed variants passing a range of info score thresholds. Each point along the x-axis represents an info score cutoff value. The corresponding y-axis values are the fraction of imputed variants passing the info score cutoff, in each of the three imputation settings. The points plotted in panel (A) are for the 308 017 chromosome 22 variants imputed across all three imputation settings (i.e. imputed with both 1000 Genomes Phase 1 and Phase 3 reference panels). The points plotted in panel (B) are for the subset of imputed variants that are also on the supplemental genotyping array and thus were used in the empirical masking experiments. In both plots, the dark gray series representing a 1000 Genomes Phase 3 reference at half maximum khap is shifted just slightly right of Phase 3 maximum khap in black.
Table 4.
Imputation quality comparison by MAF category, all imputed variants
| MAF category | Number of variants | Imputation setting 1: Phase 1, khap=max mean; median (SD) | Imputation setting 2: Phase 3, khap=half max mean; median (SD) | Imputation setting 3: Phase 3, khap=max mean; median (SD) |
|---|---|---|---|---|
| MAF<1% | 163 825 | 0.681; 0.785 (0.292) | 0.721; 0.845 (0.293) | 0.711; 0.837 (0.300) |
| 1%≤MAF<5% | 67 138 | 0.911; 0.969 (0.145) | 0.928; 0.977 (0.126) | 0.923; 0.977 (0.136) |
| MAF≥5% | 77 054 | 0.954; 0.993 (0.111) | 0.962; 0.995 (0.097) | 0.960; 0.995 (0.104) |
| All variants | 308 017 | 0.799; 0.925 (0.263) | 0.826; 0.948 (0.254) | 0.820; 0.947 (0.261) |
The imputation quality “info” score output by IMPUTE2 for chromosome 22, calculated across all samples in the primary array data set (n = 12 803), summarized with mean, median and SD. Variants were binned according the worldwide (ALL) 1000 Genomes Project Phase 3 MAF. Variants are limited to those imputed across all three imputation settings (i.e. imputed with both 1000 Genomes Phase 1 and Phase 3 reference panels).
One way to interpret info scores is to define the fraction of imputed variants passing a given threshold as the “imputation yield,” or fraction of imputed variants one would carry forward into downstream analysis. We illustrate this concept in Figure 5, showing the fraction of imputed variants passing an increasingly stringent info threshold across each imputation setting. For example, 67.8% of Phase 1 imputed variants and 73.1% of Phase 3 (half maximum khap) imputed variants pass an info score cutoff of 0.8. Note that Figure 5A includes all imputed chromosome 22 variants (n = 308 017), whereas Figure 5B is the subset of empirically masked variants from the supplemental array (n = 21 394).
Discussion
Within the HCHS/SOL study cohort of 12 803 Hispanic/Latino individuals, we have assessed the accuracy of imputation using a 1000 Genome Project Phase 1 compared with a Phase 3 reference panel. Furthermore, with a Phase 3 reference, we have tested the effects of varying the IMPUTE2 khap parameter, or the number of reference haplotypes used to impute each study sample (4). By comparing imputed genotypes with observed genotypes in a supplemental array experiment for a subset of samples, we have empirically evaluated imputation accuracy, in addition to examining overall imputation quality as measured by the IMPUTE2 info score. Ultimately we have identified characteristics that affect imputation quality at both the variant and sample level, which we discuss further below.
Phase 3 reference improves imputation accuracy for rare variants more than for common variants
The by-variant metrics from both empirical masking (dosage r2) and overall imputation quality (info score) demonstrate the challenge of accurately imputing rare variants (MAF < 1%). We show that using a Phase 3 reference panel improves imputation accuracy for rare variants compared with a Phase 1 reference, even when using similar khap values (i.e. khap = 2184 in setting 1 and khap = 2504 in setting 2). We attribute this primarily to the increased diversity of Phase 3, which presumably allows for a more appropriately tailored selection of reference haplotypes for each study sample as compared with Phase 1. As noted by the 1000 Genomes Project, increased sequencing depth in Phase 3 may also be a contributing factor (7). The improvement in imputation accuracy afforded by using Phase 3 over Phase 1 attenuates with increasing MAF, likely because of the sufficient representation of more common variants in Phase 1.
Within the Phase 3 reference, we also tested two khap values: half of the maximum (2504 haplotypes) and the maximum (5108 haplotypes). While maximizing khap yielded some improvement in the empirical masking metrics for rare variants (Fig. 1A), the effect on info score was in the opposite direction (Fig. 1B), although the effects in both cases are small. Previously, developers of the IMPUTE2 software made a similar observation and explained that using the full set of reference haplotypes may actually decrease model fit because of inclusion of divergent haplotypes (4). This is especially likely when the reference panel is very diverse (i.e. has a high degree of population structure) and therefore contains samples that are probably uninformative for imputing a given study haplotype. In such a scenario, restricting khap can slightly improve accuracy in addition to the practical benefit of reducing computation time. Supplementary tests support these observations: Phase 3 “downsampled” to the size of Phase 1 with maximized khap yielded lower quality compared with Phase 1 with maximized khap; the quality decrease was attenuated by lowering khap (see Supplementary Material, Fig. S2). (Note that we initially chose to maximize khap in setting 1 because of our findings from previous experiments in HCHS/SOL where khap of 2184 yielded higher accuracy, as measured by IMPUTE2 internal masking metrics, compared with khap values of 500 and 1000; data not shown.)
Another possible explanation for the apparently different effects of khap, using either empirical masking or info score, is the different samples underlying the two metrics: info was calculated based on the full set of 12 803 samples, whereas empirical dosage r2 was calculated in the subset of 1424 supplemental array samples. The 1424 samples have approximately the same proportions of genetic analysis groups as the full set of 12 803 samples, though there may be subtler differences that underlie the different patterns of the two metrics. However, the difference between the two khap conditions is minor for both metrics: info and dosage r2. Ultimately, choosing a khap involves considering both expected imputation accuracy and computational resources, as run times increase linearly with increasing khap values. We and others (4) have shown that intermediate khap values can yield high quality imputation results and that in some circumstances maximizing khap may in fact slightly decrease quality, as measured by IMPUTE2 info scores.
Phase 3 reference improves imputation accuracy for Caribbean groups more than for mainland groups
The by-sample metrics from empirical masking show that larger and more diverse reference samples benefit some types of samples more than others. As seen in Figure 2, imputation accuracy improves in all genetic analysis groups when using a Phase 3 reference over Phase 1. However, the average increase in accuracy is roughly 3-fold higher for Caribbean as compared with Mainland groups (mean dosage r2 increase of 0.0015 and 0.0005, respectively, when comparing setting 2 with setting 1; i.e. Phase 3 versus 1).
We attribute the increased improvement for Caribbean over Mainland samples to the additional samples available in 1000 Genomes Phase 3. Specifically, Phase 3 includes new samples from existing Phase 1 populations and populations new to Phase 3. Of particular relevance to HCHS/SOL Caribbean samples is the addition of the African Caribbean in Barbados population (ACB, 96 samples) and the increase in sample size for the Puerto Rican in Puerto Rico population (PUR, 55 Phase 1 samples and 104 Phase 3 samples). Furthermore, previous studies of Hispanic/Latino admixture have shown that groups from the Caribbean have higher proportions of West African ancestry compared with non-Caribbean groups (15–17). New West African populations in Phase 3 are the Gambian in Western Division, The Gambia (GWD, n = 113), Mende in Sierra Leone (MSL, n = 85) and Esan in Nigeria (ESN, n = 99). Therefore we expect the addition of West African samples in Phase 3 is also contributing to the improved accuracy in HCHS/SOL Caribbean groups.
Imputation accuracy also improved across Mainland groups, albeit to a lesser degree than for Caribbean. We attribute this primarily to the enhanced number and diversity of reference haplotypes available in Phase 3 compared with Phase 1, though again sequencing depth may also play a role. Phase 3 includes a new South American population, Peruvian in Lima, Peru (PEL, n = 85), which we expect to be relevant for imputing Mainland samples. In addition, although Caribbean samples may have higher proportions of West African ancestry compared with Mainland, the addition of West African samples in Phase 3 may still benefit Mainland samples.
Implementing genome-wide imputation in HCHS/SOL
We conducted these imputation analyses primarily to assess the impact of switching from a Phase 1 to a Phase 3 reference panel in HCHS/SOL. A secondary aim was to test different khap values within Phase 3 to determine whether maximizing khap was worth the additional computation time. Given the parallelization enabled by our compute cluster, we estimated that genome-wide imputation for half of the maximum khap would take ∼17 calendar days, whereas maximizing khap would require 34 calendar days. Ultimately we chose to impute genome-wide using half the maximum khap, given the tradeoff between marginal improvements in imputation accuracy and increased computational burden. Furthermore, there was evidence that maximizing khap in setting 3 led to slightly decreased mean info scores, a previously observed phenomenon (4).
Power to detect genotype–phenotype associations
The sample size required to detect an association using imputed genotypes is inversely proportional to r2, the squared correlation between imputed allelic dosage and true allelic dosage (see Appendix in 21). Therefore, differences in imputation accuracy result in differences in power to detect genotype–phenotype associations, as shown previously for imputation-based genomic coverage of SNP arrays (21).
Limitations, strengths and future research
We have demonstrated that a 1000 Genomes Project Phase 3 reference panel can yield improved imputation accuracy compared with Phase 1, particularly for rare variants and for samples of certain genetic ancestry compositions. These findings can inform imputation analysis design for other GWAS, in particular those with admixed samples. However, given the substantial genetic variability within and between admixed populations, including Hispanic/Latino, the extent to which our findings extend to other cohorts will need to be evaluated empirically.
Here, we have only evaluated chromosome 22, as it is customary to use one autosome for testing and evaluation. While we did not test the three settings on all chromosomes, we ultimately implemented setting 2 genome-wide and have compared empirical masking r2 metrics on chromosome 22 with the remaining autosomes and found no substantial differences (see Fig. 3). In addition, we have not assessed the effect of imputation panel choice on the X chromosome, which has a unique genetic ancestry profile (16). While we would expect the larger and more diverse set of reference haplotypes available in Phase 3 to similarly improve X chromosome imputation, this again would require further research.
There are additional aspects of using a 1000 Genomes Phase 3 reference panel that our experiments were not designed to address. First, we have limited our analysis to Phase 3 variants that are also available in Phase 1 to compare a consistent set of variants across imputation settings. However, Phase 3 contains over twice the number of variants in Phase 1 and thus offers many more variants for imputation and, by extension, downstream association testing. Thus investigators considering which Phase to use as a reference should consider the number and frequency distribution of variants available for imputation.
Additionally, in our empirical masking experiments we were limited to variants genotyped on the Illumina HumanOmni2.5S supplemental array. Variants selected to be on genotyping arrays may not be representative of genetic variation more broadly; e.g. they may have specific MAF distributions and/or linkage disequilibrium (i.e. “tagging”) properties. Therefore the absolute value of our empirical masking metrics may not be representative of all variants. However, we have compared the same set of supplemental array variants across the three imputation settings and thus have an unbiased estimate of relative changes in imputation accuracy.
Strengths of our study include the large and diverse set of Hispanic/Latino samples available in the HCHS/SOL study. Often empirical masking experiments involve small numbers of samples, sometimes obtained from the same source populations as the reference panel samples. Here, we imputed into a large cohort of US Hispanic/Latino samples unconnected to the reference sample source populations, thus providing a more realistic assessment. Furthermore, defining genetic analysis groups enabled us to observe the effects of reference panel diversity in these specific groups.
In addition, the supplementary array data set enabled us to empirically (rather than only theoretically) evaluate imputation accuracy, by comparing observed genotypes with imputed results. This is important as info scores may not accurately reflect imputation accuracy, particularly at rare variants (11). Comparison of empirical r2 to info score indicated that the results are largely consistent, although we see a small difference in evaluation of effects of the khap parameter.
In conclusion, the final Phase of the 1000 Genomes Project appears to be an improved imputation reference panel compared with earlier phases, in particular for imputation of rare variants in GWAS that include diverse, admixed individuals. Ongoing efforts to assemble even larger reference panels by pooling resources across sequencing projects will continue to improve imputation quality, particularly at rare variants, in addition to offering even larger collections of variants to impute. Indeed, initial reports on the HRC reference demonstrate more accurate imputation over 1000 Genomes Phase 3 alone in European-ancestry populations (6). We anticipate the addition of more diverse populations in Phase 2 of the HRC (http://www.haplotype-reference-consortium.org/participating-cohorts) last accessed June 10, 2016 will further aid imputation into diverse, admixed individuals such as Hispanics/Latinos.
Materials and Methods
Samples and genotyping
The HCHS/SOL is a community-based cohort study of over 16 000 self-identified Hispanic/Latino adults from four US urban areas: the Bronx, San Diego, Chicago and Miami (22). The HCHS/SOL cohort includes a range of self-reported Hispanic/Latino backgrounds, with the primary groups being Central American, Cuban, Dominican, Mexican, Puerto Rican and South American. The overarching goals of HCHS/SOL are to describe the prevalence of risk and protective factors for a range of chronic diseases, as well as to describe all-cause mortality from cardiovascular and pulmonary disease. Ethical oversight of HCHS/SOL activities is conducted by Institutional Review Boards at each of the field centers and the study coordinating center, as described by Sorlie et al. (22). All participants included in this article provided informed consent to genetic studies at their first visit.
Two sets of array genotyping were used in these imputation assessments: one primary and one supplemental (see Supplementary Material, Fig. S1). Both genotyping data sets underwent standard QA/QC measures (23) to create a cleaned set of genotypes comprising only high quality variants and samples. The primary genotyping array was a custom version of the Illumina HumanOmni2.5-8v1-1array, consisting of ∼150 000 custom variants added to the standard Omni2.5M array (G.J.P., K.D.T. and J.I.R., unpublished data). After QA/QC procedures, this primary array data set comprised 2 395 790 variants and 12 803 unique study samples. Supplemental genotyping on the Illumina HumanOmni2.5S array was done in a subset of HCHS/SOL samples. After QA/QC procedures, this supplemental array data set comprised 1 924 262 variants and 1424 unique study samples. Here, the supplemental array genotypes were used only for assessing accuracy of imputation based on the primary array genotypes. Genotyping and initial genotype data cleaning were done by Illumina Microarray Services and Los Angeles Biomedical Research Institute. Subsequent genotype data cleaning and imputation analyses were performed at the HCHS/SOL Genetic Analysis Center (GAC) at the University of Washington.
In addition to self-identified background group, the GAC has previously defined genetic analysis groups based on both self-identified background group and principal component analysis of the primary array genotypes (18). There are six such genetic analysis groups: Cuban, Dominican, Puerto Rican, Mexican, Central American and South American (see Table 1); the concordance with self-identified background groups is very high (range 92–98%, mean 96%). The six genetic analysis groups are further classified into two regions: Mainland (Mexican, Central American and South American) and Caribbean (Cuban, Dominican and Puerto Rican).
Imputation protocol
The GAC has developed an imputation pipeline consisting of formatting and preparing the study data set (including plus strand alignment), pre-phasing, imputation and post-imputation analysis of quality metrics. Initially HCHS/SOL study data were in a genome-wide, binary PLINK (24) data set with genotypes expressed in TOP alleles. First we subset the data into chromosome-specific PLINK files, converting TOP alleles to the plus strand of the human genome reference based on strand information in the Illumina array manifest (25). At this stage we also filtered variants to those passing quality filters and representing unique genomic positions. Next we pre-phased the study data using SHAPEIT2 software (26) (version 2.r778). The SHAPEIT2 haplotypes were used as the basis for all subsequent imputations.
To impute the HCHS/SOL primary array data set, we used IMPUTE2 software (4,27) (version 2.3.0) and a reference panel from either Phase 1 or Phase 3 of the 1000 Genomes Project (7,28). Phase 1 contains 1092 samples from 14 populations worldwide, which are grouped into four superpopulations: AFR (African), AMR (Americas), ASN (Asian) and EUR (European). Phase 3 contains 2504 samples from 26 populations, which are grouped into superpopulations similarly to Phase 1 except with the further division of ASN into EAS (East Asian) and South Asian (SAS). Phase 3 has ∼81 M variants genome-wide compared with ∼38 M total variants in Phase 1. We limited imputation target variants to those with at least two copies of the minor allele observed in any one of the superpopulations.
Imputation settings
We considered three imputation settings to assess the accuracy of imputing into HCHS/SOL. Each setting was evaluated using chromosome 22 from the primary HCHS/SOL genotyping data set: 12 803 study samples genotyped on the custom HumanOmni2.5-8v1-1array, filtered and pre-phased as described above. We varied two aspects of imputation for these comparisons: (a) 1000 Genomes Project Phase and (b) the effective number of reference haplotypes used to impute each sample, the IMPUTE2 parameter khap (4). Setting 1 was imputing to Phase 1 with a khap of 2184, the maximum value for a panel with 1092 individuals. Setting 2 was imputing to Phase 3 with a khap of 2504, half the maximum value for a panel with 2504 individuals. Setting 3 was imputing to Phase 3 with a khap of 5008, the maximum value. In Supplementary Material, we evaluate three additional settings: Phase 3 “downsampled” to the size of Phase 1, with both maximum and half the maximum khap; and Phase 3 explicitly excluding Asian ancestry superpopulations (EAS and SAS).
The IMPUTE2 manual suggests setting khap to the number of reference haplotypes expected to be useful for the given set of study samples (see IMPUTE2 manual, https://mathgen.stats.ox.ac.uk/impute/impute_v2.html#using_multipop_panels) Last accessed June 10, 2016. In practice it may be difficult to determine the appropriate khap value a priori, and often empirically testing various khap values is the best option to find the value that maximizes imputation accuracy. The default value for khap is 500, which for 1000 Genomes Phase 1 would be utilizing ∼23% of the 2184 available reference haplotypes. An important practical consideration is that computation time increases approximately linearly with increasing khap.
Assessments of imputation accuracy
We evaluated imputation accuracy within and across the three imputation settings in two ways: empirical masking and imputed quality metrics. In the empirical masking experiments, we compared genotypes observed only in the HCHS/SOL supplemental array data set with genotypes imputed using the primary array data set as the imputation basis. This comparison was carried out in all 1424 HCHS/SOL samples included in both the primary and supplemental array data sets. We calculated the squared correlation between observed and imputed allelic dosages, or dosage r2, both by-variant (over all samples) and by-sample (over all variants).
When evaluating by-sample dosage r2, we grouped HCHS/SOL samples both by genetic analysis group and by region (Mainland or Caribbean) to assess differences in imputation accuracy both within and across imputation settings. Specifically, we performed analysis of variance (ANOVA) F tests with region as a main effect, 1000 Genomes reference panel as a main effect (Phase 1 versus Phase 3), and the interaction of region and reference panel. We repeated this analysis using the six genetic analysis groups rather than region. A subset consisting of 1388 mutually unrelated samples was used for these tests.
In addition to empirical masking, we examined the distribution of imputation accuracy metrics (IMPUTE “info” score) across the full set of imputed variants. The info score is a statistical measure of imputation quality, which correlates with the r2 metrics reported by other imputation software (see 1 and Supplementary Material). Info scores range between 0 and 1, with scores near 1 indicating reliable, high quality imputation. When comparing info scores, we only included variants imputed across all three imputation settings, i.e. imputed with both Phase 1 and Phase 3 reference panels (n = 308 017 variants). Overlapping variants were determined by matching on chromosome, base pair position (hg19/GRCh37) and plus strand alleles. An additional ∼300K chromosome 22 variants imputed with Phase 3 but not with Phase 1 were excluded from these comparisons. In addition, we examined info score in the empirically masked supplemental array variants to investigate whether the distribution differed from the full set of imputed variants. Metrics were visualized with R statistical and graphing software packages ggplot2 1.0.1 (29) and RColorBrewer 1.1-2 (30).
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
We thank the participants and staff of the HCHS/SOL study for their contributions to this study. This article has been reviewed by the HCHS/SOL Publications Committee for scientific content and consistency of data interpretation with previous HCHS/SOL publications. The views expressed in this article are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services. We additionally thank Jonathan Marchini and Bryan Howie for consultation provided during manuscript preparation.
Conflict of Interest statement. None declared.
Funding
The baseline examination of HCHS/SOL was carried out as a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (N01-HC65233), University of Miami (N01-HC65234), Albert Einstein College of Medicine (N01-HC65235), Northwestern University (N01-HC65236), and San Diego State University (N01-HC65237). The following Institutes/Centers/Offices contribute to the HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research (NIDCR), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Neurological Disorders and Stroke, NIH Institution-Office of Dietary Supplements. The Genetic Analysis Center at University of Washington was supported by NHLBI and NIDCR contracts (HHSN268201300005C AM03 and MOD03). Genotyping efforts were supported by NHLBI HSN 26220/20054C, National Center for Advancing Translational Science Clinical Translational Science Institute grant UL1TR000124, and NIDDK Diabetes Research Center grant DK063491.
References
- 1.Marchini J., Howie B. (2010) Genotype imputation for genome-wide association studies. Nat. Rev. Genet., 11, 499–511. [DOI] [PubMed] [Google Scholar]
- 2.Frazer K., Ballinger D., Cox D., Hinds D., Stuve L., Gibbs R., Belmont J., Boudreau A., Hardenbol P., Leal S., et al. (2007) A second generation human haplotype map of over 3.1 million SNPs. Nature, 449, 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altshuler D., Gibbs R., Peltonen L., Dermitzakis E., Schaffner S., Yu F., Bonnen P., de Bakker P., Deloukas P., Gabriel S., et al. (2010) Integrating common and rare genetic variation in diverse human populations. Nature, 467, 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howie B., Marchini J., Stephens M. (2011) Genotype Imputation with Thousands of Genomes. G3 Genes Genomes Genet., 1, 457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walter K., Min J.L., Huang J., Crooks L., Memari Y., McCarthy S., Perry J.R., Xu C., Futema M., Lawson D., et al. (2015) The UK10K project identifies rare variants in health and disease. Nature, 526, 82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Haplotype Reference Consortium (2015) A reference panel of 64,976 haplotypes for genotype imputation. bioRxiv. doi: http://dx.doi.org/10.1101/035170. [Google Scholar]
- 7.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. (2015) A global reference for human genetic variation. Nature, 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jostins L., Morley K.I., Barrett J.C. (2011) Imputation of low-frequency variants using the HapMap3 benefits from large, diverse reference sets. Eur. J. Hum. Genet., 19, 662–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang L., Li Y., Singleton A.B., Hardy J.A., Abecasis G., Rosenberg N.A., Scheet P. (2009) Genotype-imputation accuracy across worldwide human populations. Am. J. Hum. Genet., 84, 235–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Browning B., Browning S. (2009) A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet., 84, 210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J., Howie B., McCarthy S., Memari Y., Walter K., Min J.L., Danecek P., Malerba G., Trabetti E., Zheng H.F., et al. (2015) Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat. Commun., 6, 8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenberg N.A., Huang L., Jewett E.M., Szpiech Z.A., Jankovic I., Boehnke M. (2010) Genome-wide association studies in diverse populations. Nat. Rev. Genet., 11, 356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shriner D., Adeyemo A., Chen G., Rotimi C.N. (2010) Practical considerations for imputation of untyped markers in admixed populations. Genet. Epidemiol., 34, 258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu E.Y., Li M., Wang W., Li Y. (2013) MaCH-admix: genotype imputation for admixed populations. Genet. Epidemiol., 37, 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bryc K., Durand E.Y., Macpherson J.M., Reich D., Mountain J.L. (2015) The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am. J. Hum. Genet., 96, 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryc K., Velez C., Karafet T., Moreno-Estrada A., Reynolds A., Auton A., Hammer M., Bustamante C.D., Ostrer H. (2010) Colloquium paper: genome-wide patterns of population structure and admixture among Hispanic/Latino populations. Proc. Natl. Acad. Sci. U. S. A, 107 (Suppl. 2), 8954–8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burchard E.G., Borrell L.N., Choudhry S., Naqvi M., Tsai H.J., Rodriguez-Santana J.R., Chapela R., Rogers S.D., Mei R., Rodriguez-Cintron W, et al. (2005) Latino populations: a unique opportunity for the study of race, genetics, and social environment in epidemiological research. Am. J. Public Health, 95, 2161–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conomos M.P., Laurie C.A., Stilp A.M., Gogarten S.M., McHugh C.P., Nelson S.C., Sofer T., Fernandez-Rhodes L., Justice A.E., Graff M., et al. (2016) Genetic diversity and association studies in US Hispanic/Latino populations: applications in the Hispanic Community Health Study/Study of Latinos. Am. J. Hum. Genet., 98, 165–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manichaikul A., Palmas W., Rodriguez C.J., Peralta C.A., Divers J., Guo X., Chen W.M., Wong Q., Williams K., Kerr K.F., et al. (2012) Population structure of Hispanics in the United States: the multi-ethnic study of atherosclerosis. PLoS Genet., 8, e1002640.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sankararaman S., Sridhar S., Kimmel G., Halperin E. (2008) Estimating local ancestry in admixed populations. Am. J. Hum. Genet., 82, 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson S.C., Doheny K.F., Pugh E.W., Romm J.M., Ling H., Laurie C.A., Browning S.R., Weir B.S., Laurie C.C. (2013) Imputation-Based Genomic Coverage Assessments of Current Human Genotyping Arrays. G3 Genes Genomes Genet., 3, 1795–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sorlie P.D., Aviles-Santa L.M., Wassertheil-Smoller S., Kaplan R.C., Daviglus M.L., Giachello A.L., Schneiderman N., Raij L., Talavera G., Allison M., et al. (2010) Design and implementation of the Hispanic Community Health Study/Study of Latinos. Ann. Epidemiol., 20, 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laurie C.C., Doheny K.F., Mirel D.B., Pugh E.W., Bierut L.J., Bhangale T., Boehm F., Caporaso N.E., Cornelis M.C., Edenberg H.J., et al. (2010) Quality control and quality assurance in genotypic data for genome-wide association studies. Genet. Epidemiol., 34, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M., Bender D., Maller J., Sklar P., de Bakker P., Daly M., et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson S.C., Laurie C.C., Doheny K.F., Mirel D.B. (2012) Is ‘forward’ the same as ‘plus’?… and other adventures in SNP allele nomenclature. Trends Genet., 28, 361–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delaneau O., Zagury J.F., Marchini J. (2013) Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods, 10, 5–6. [DOI] [PubMed] [Google Scholar]
- 27.Howie B., Donnelly P., Marchini J. (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet., 5, e1000529.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McVean G., Altshuler D.M., Durbin R.M., Abecasis G., Bentley D.R., Chakravarti A., Clark A.G., Donnelly P., Eichler E.E. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature, 491, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wickham H. (2009) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York. [Google Scholar]
- 30.Neuwirth E. (2014) RColorBrewer: ColorBrewer palettes. R package version 1.1-2 http://CRAN.R-project.org/package=RColorBrewer.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.