Abstract
Predicting response to endocrine therapy and survival in oestrogen receptor positive breast cancer is a significant clinical challenge and novel prognostic biomarkers are needed. Long-range regulators of gene expression are emerging as promising biomarkers and therapeutic targets for human diseases, so we have explored the potential of distal enhancer elements of non-coding RNAs in the prognostication of breast cancer survival. HOTAIR is a long non-coding RNA that is overexpressed, promotes metastasis and is predictive of decreased survival. Here, we describe a long-range transcriptional enhancer of the HOTAIR gene that binds several hormone receptors and associated transcription factors, interacts with the HOTAIR promoter and augments transcription. This enhancer is dependent on Forkhead-Box transcription factors and functionally interacts with a novel alternate HOTAIR promoter. HOTAIR expression is negatively regulated by oestrogen, positively regulated by FOXA1 and FOXM1, and is inversely correlated with oestrogen receptor and directly correlated with FOXM1 in breast tumours. The combination of HOTAIR and FOXM1 enables greater discrimination of endocrine therapy responders and non-responders in patients with oestrogen receptor positive breast cancer. Consistent with this, HOTAIR expression is increased in cell-line models of endocrine resistance. Analysis of breast cancer gene expression data indicates that HOTAIR is co-expressed with FOXA1 and FOXM1 in HER2-enriched tumours, and these factors enhance the prognostic power of HOTAIR in aggressive HER2+ breast tumours. Our study elucidates the transcriptional regulation of HOTAIR, identifies HOTAIR and its regulators as novel biomarkers of patient response to endocrine therapy and corroborates the importance of transcriptional enhancers in cancer.
Introduction
Breast cancer diagnosis and prognosis is currently based on histological grade, disease stage (including lymph node status) and expression of hormone receptors (oestrogen receptor (ER), progesterone receptor (PgR) and ERBB2 (HER2)) (1). As breast cancer is a very heterogeneous disease and clinical outcomes can be highly variable, the predictive power of these markers is sometimes limited. The majority of ER+ patients receive tamoxifen or other forms of endocrine therapies, although many display or develop resistance, with a 33% recurrence rate after 5-years of tamoxifen treatment (2). Such endocrine resistance is common but difficult to predict (reviewed in Osborne and Schiff, 2011 (3)). Patients with HER2 positive tumours may be treated with Trastuzumab (Herceptin), however relapse in the form of metastasis is a regular occurrence (4). Thus, there is a clear and on-going need to identify additional prognostic and predictive molecular biomarkers that will augment traditional indicators and ultimately improve patient management and better predict response to therapies in these two types of breast cancer (5).
Gene transcription is regulated by a complex interplay of proximal and distal cis-acting regulatory elements, including promoters and enhancers, and trans-acting factors such as protein transcription factors (TFs) and non-coding RNAs. Whilst promoter elements lie proximal to the transcriptional start site (TSS) and are responsible for basal gene transcription, enhancers can be kilobases or megabases from the gene they regulate and are associated with cell- and tissue-specific expression (6,7). Disruption of enhancers through epigenetic or genomic aberrations can therefore impact cell identity, a key feature of tumourigenesis (reviewed in Kron et al., 2014 (8) and Herz et al., 2014 (9)) and so it is no surprise that these defects have been associated with cancer initiation and progression. For example, several breast cancer-associated SNPs map to enhancer elements bound by oncogenic transcription factors which regulate the expression of breast cancer susceptibility genes (10,11). Defects in the factors that mediate enhancer function have also been associated with cancer, including CTCF in breast and bladder cancer, and MED12 in prostate cancer. Enhancer elements and the factors controlling them therefore have significant potential to predict breast cancer progression (8,12,13).
Non-coding RNA genes are regulated at both the transcriptional and posttranscriptional level. Whole genome chromatin immunoprecipitation (ChIP) sequence analysis has shown that most miRNA promoters are over 1kb in length and map to distances up to 10kb from the miRNA sequence (14–17). The regulation of long non-coding RNA (lncRNA) genes is less understood, however it is now known that histone modification marks are associated with the regulatory elements controlling lncRNA gene expression (18). In addition, this analysis has revealed a distinctive methylation pattern around the transcription start site (reviewed in Venkatesh and Workman 2012 (19)), strong evolutionary conservation and binding sites for transcription factors that play a key role in cell proliferation and differentiation (20). Enhancers have emerged as crucial regulators of lncRNAs and their status is associated with the regulation of critical processes including the determination of cell fate and identity (7,21). These enhancer elements can bind a plethora of transcription factors and physically interact with gene promoters through chromatin looping to alter transcription (reviewed in Fraser 2006 (22)).
Hox antisense intergenic RNA (HOTAIR) is a lncRNA that has been implicated in chromatin remodeling and transcription. HOTAIR RNA associates with the polycomb repressive protein complex 2 (PRC2) (23), which is part of a vital poly-protein structure used to condense chromatin and effectively control transcription (24). HOTAIR regulates a wide range of genes, including HOXD genes, genes associated with epithelial-to-mesenchymal transition (EMT) and cell cycle regulation (25–27). HOTAIR is a marker of metastasis and poor prognosis in a range of cancers, including breast cancer (27), where it is overexpressed in a third of metastatic breast tumours (26). The ability of HOTAIR or its regulators to predict therapeutic response and outcome in ER and HER2 positive breast cancer has yet to be determined.
Several elements and factors controlling the transcriptional and post-transcriptional regulation of HOTAIR have been described and implicated in cancer (reviewed in Hajjari and Salavaty, 2015 (28)). At the transcriptional level, a proximal HOTAIR promoter has been identified approximately 1kb upstream of the HOTAIR TSS and several transcription factors have been shown to bind this region and regulate HOTAIR expression, including c-Myc (29), IRF1 (30), ERα (31) and MLL proteins. A susceptibility single nucleotide polymorphism (SNP) for esophageal squamous cell carcinoma (ESCC) and breast cancer has been found located within an enhancer in intron 2 of HOTAIR (32,33). In ESCC, the TT allele is associated with increased expression through higher enhancer activity. Methylation of proximal CpG islands mapping approximately 1kb downstream of HOTAIR have been associated with clinical and pathological features of breast cancer (34). The role of long-range regulation in the control of HOTAIR has not been reported and is the subject of this manuscript.
In this paper, we have elucidated the role of long-range regulation in the control of HOTAIR expression and explored the potential of long-range elements and factors to be valuable biomarkers for therapeutic response and outcome in breast cancer. Here, we report that FOX proteins can regulate HOTAIR, and can enhance its ability to stratify both HER2-enriched and ER+ breast cancer.
Results
Identification of a distal enhancer downstream of the HOTAIR gene
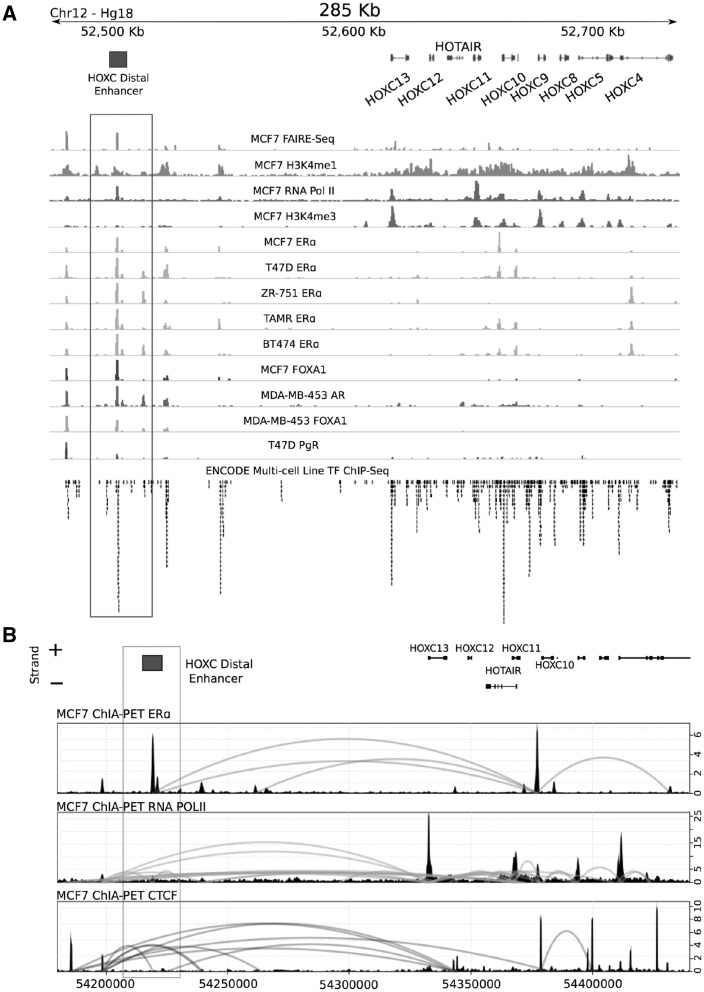
To identify transcription control elements and factors that regulate HOTAIR expression in breast cancer cells, a chromatin state map for the HOXC locus housing the HOTAIR gene was generated using publically available data on evolutionary conservation, histone modification, ChIP-Seq and RNA-Seq. This analysis revealed a series of highly conserved regions that exhibit histone modifications and TF-binding densities suggestive of cis-regulatory activity (Figure 1A, higher resolution of the HOXC locus: Supplementary Material, Fig. S1).
Figure 1.
Identification of a HOXC putative cis-regulatory element by investigating chromatin modifications and long-range interactions. (A) Histone modification and transcription factor binding across the HOXC locus, sourced from publically available ChIP-Seq data (Hg18), suggesting a putative enhancer ∼150 kb downstream of the HOTAIR gene. (B) ChIA-PET interactions for ESR1, RNA Polymerase II and CTCF with data sourced from GEO, GSE39495. Plots representing each interaction library include a wiggle track showing the binding of each transcription factor and a curved line that connects interacting genomic fragments indicating relative interaction frequency between fragments (right Y axis).
Of particular interest was a region located in a gene desert approximately 150 kb downstream of the HOTAIR TSS, marked by open chromatin (FAIRE-Seq), lysine mono-methylation (H3K4me1), RNA Polymerase II binding, as well as ER and PR binding, in multiple different breast cancer cell lines (Figure 1A). These marks are consistent with enhancer activity, and we therefore denoted this region as a putative distal enhancer, which we have termed HOXC Distal Enhancer (HDE). The enrichment of ERα and PgR binding at the HDE was associated with binding of FOXA1, a critical mediator of ER DNA binding in breast cancer cells. By interrogating ChIA-PET (Chromatin Interaction Analysis by Paired-End Tag Sequencing) data, we also found that the HDE engages in long-range interactions with the HOXC locus involving ERα, RNA Polymerase II and CTCF TFs (Figure 1B).
Mapping of enhancer-promoter interactions reveals an alternative promoter for HOTAIR
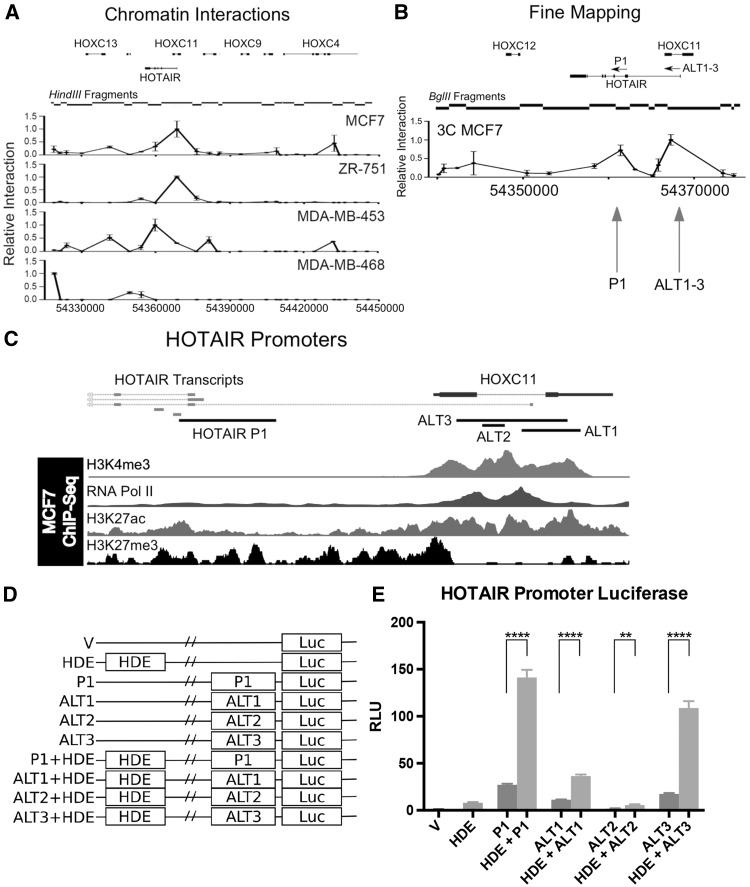
To investigate the interaction between the HDE and the HOTAIR promoter, chromosome conformation capture (3C) was performed. 3C libraries were generated for both ER+ (MCF7, ZR751) and ER- (MDA-MB-453 and MDA-MB-468) cell lines. Using the HindIII fragment containing the HDE as bait, 3C-qPCR relative interaction graphs were constructed for all fragments across the HOXC locus. In MCF7, ZR-751 and MDA-MB-453 cells, the HDE showed significant interactions with the HOTAIR start site (Figure 2A). Digestion with BglII effectively increased the resolution and revealed that the HDE interacts with two distinct regions within and upstream of the HOTAIR gene (Figure 2B). The first region corresponds to the promoter (P1) and the first intron of the canonical HOTAIR gene, and the second region, approximately 5kb upstream, corresponds to an alternate HOTAIR TSS, within intron-1 of HOXC11 (Figure 2C, alternative promoter cloned regions 1, 2 and 3, (ALT1, ALT2 and ALT3)). The alternate TSS is enriched for the histone mark H3K4me3 and RNA Polymerase II, suggesting bona fide promoter activity. The canonical HOTAIR promoter exhibits low levels of histone modifications that are suggestive of an active regulatory element (H3K4me3 and H3K27ac), perhaps suggesting a poised or inactive promoter in MCF7 cells. RT-PCR analysis demonstrated that an alternate longer form of the HOTAIR transcript is expressed in MCF7 cells originating from the alternative TSS (promoters ALT1-3) (Supplementary Material, Fig. S2).
Figure 2.
Mapping of enhancer-promoter interaction reveals alternative promoter for HOTAIR. (A) 3C analysis using libraries generated by HindIII in MCF7, ZR-75-1, MDA-MB-453 and MDA-MB-468. HindIII fragment length and position are shown in black corresponds to the center of each fragment within the graphs. (B) 3C fine mapping analysis using BglII in MCF7 breast cancer cells, position and fragment length relative to HOXC genes shown in black. X-axis is the relative genomic position corresponding to the start of BglII fragments. 3C Y-axis represents the relative interaction frequency between the HindIII/BglII fragments and the HDE. Error bars represent standard deviation. (C) HOTAIR promoter (P1) and alternative promoters (ALT1-3). Histone marks and RNA Pol II binding are characteristic of gene promoters. Black bars highlight the regions used for each pGL3 construct. (D) Schematic for luciferase reporter constructs used in E. (E) Luciferase reporter results for activity of HOTAIR alternative promoters in MCF7 cells. X-axis represents the construct used while the Y-axis is the relative light units (luciferase normalized to pRL-TK Renilla and pGL3-Basic (V)). Statistical significance was found via multiple two-tailed t-Tests between indicated columns, P-values are <0.05 (*), <0.01 (**), <0.001 (***) and <0.0001 (****).
The putative enhancer augments HOTAIR promoter activity
To determine whether the HDE can enhance the transcription of genes at the HOXC locus, we generated luciferase reporter constructs by sub-cloning the HDE upstream of the HOXC10, HOXC11 or HOTAIR promoters (Supplementary Material, Figures S3 and S4A). Luciferase assays were then performed in several breast cancer cell lines. We were unable to detect transcriptional activity from the HOXC11 promoter in these breast cancer cells and therefore excluded this construct from further analysis. Of the promoters tested, the HDE had the greatest effect on the canonical HOTAIR promoter, inducing a five-fold increase in luciferase expression compared to the HOTAIR promoter alone (Supplementary Material, Fig. S4B). To investigate the promiscuity of the HDE in affecting HOXC promoter activity we cloned it upstream of the SV40 promoter (Supplementary Material, Fig. S4C). Use of the SV40 promoter demonstrates a 12-fold increase in transcriptional activity when in combination with the HDE, suggesting that the HDE is non-specific in its activity on the HOTAIR and HOXC10 promoters (Supplementary Material, Fig. S4D). Three regions were cloned to test promoter activity at the alternative TSS, upstream of the TSS (ALT1), the centre of the histone markers and RNA Pol II binding (ALT2) and a larger fragment encompassing the alternative TSS and peaks of RNA Pol II binding. Luciferase reporter assays demonstrated promoter activities for P1, ALT1 and ALT3 that were significantly augmented by the HDE (Figures 2D and E). These luciferase data support the RNA Pol II ChIA-PET and 3C interactions in that the HDE specifically interacts with and augments HOTAIR promoters (Figure 1B). In support of these findings, analysis of MCF7 RNA-Seq data clearly demonstrates RNA produced from the longer alternative HOTAIR transcripts (Supplementary Material, Fig. S5).
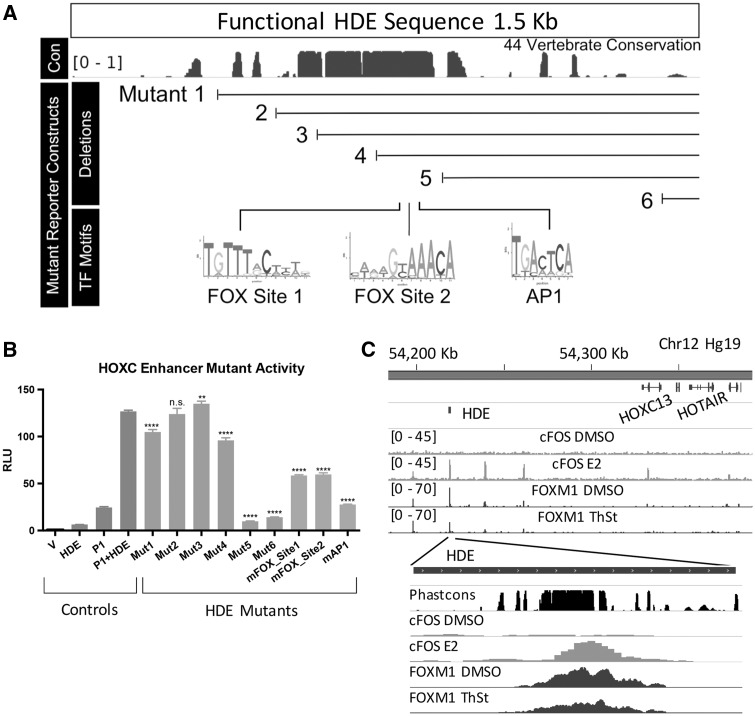
To identify the active sequences of the HDE, we mutagenized the HOTAIR-HDE canonical promoter (P1) reporter construct (Figure 3A), and localized HOTAIR enhancer activity to a 114 base-pair region in MCF7 cells (between mutants 4 and 5; Figure 3B). Interestingly, this region contains several highly conserved, predicted transcription factor binding sites, including consensus binding sequences for forkhead box proteins (FOX Site 1 and 2) and AP1, which are associated with oestrogen-mediated transcriptional activity (Figure 3B). Consistent with this, analysis of ChIP-Seq data demonstrated that both c-Fos (part of the AP1 complex) and FOXM1 bind to the HDE (Figure 3C). Interestingly, in MCF7 cells c-Fos binding appears to be dependent on the presence of oestrogen. Binding of FOXM1, also in MCF7 cells, is decreased following the addition of the antibiotic thiostrepton (ThSt), which is known to prevent binding of FOXM1 to regulatory elements and chromatin. Furthermore, mutagenizing the sites in the context of the P1-HDE construct significantly reduced the enhancer activity of the HDE (Figures 2D and 3B).
Figure 3.
The HOTAIR distal enhancer is dependent on FOX and AP1 binding. (A) Functional HDE sequence top and bottom the HDE mutant constructs for deletion and transcription factor motifs. Deletion mutant positions were chosen based on conservation with lines indicating remaining DNA. Up to three base pairs were edited within the TF motif sites for three separate constructs, one per motif listed. (B) Luciferase reporter assay measuring the transcriptional activity of the HOTAIR enhancer and the separate mutants, Y-axis as above. All mutants were generated using the P1 + HDE reporter vector as of Figure 2D. (C) Re-analyzed ChIP-Seq data ([GSE26831] (80), [GSE40767] (44)) demonstrating binding of both cFOS and FOXM1 to the HDE in MCF7 cells with the indicated treatment. All statistical tests performed were one-way ANOVAs with Tukey corrected multiple comparisons between (B) P1 + +oE, P-values are <0.05 (*), <0.01 (**), <0.001 (***) and <0.0001 (****).
HOTAIR is transcriptionally upregulated by FOX proteins
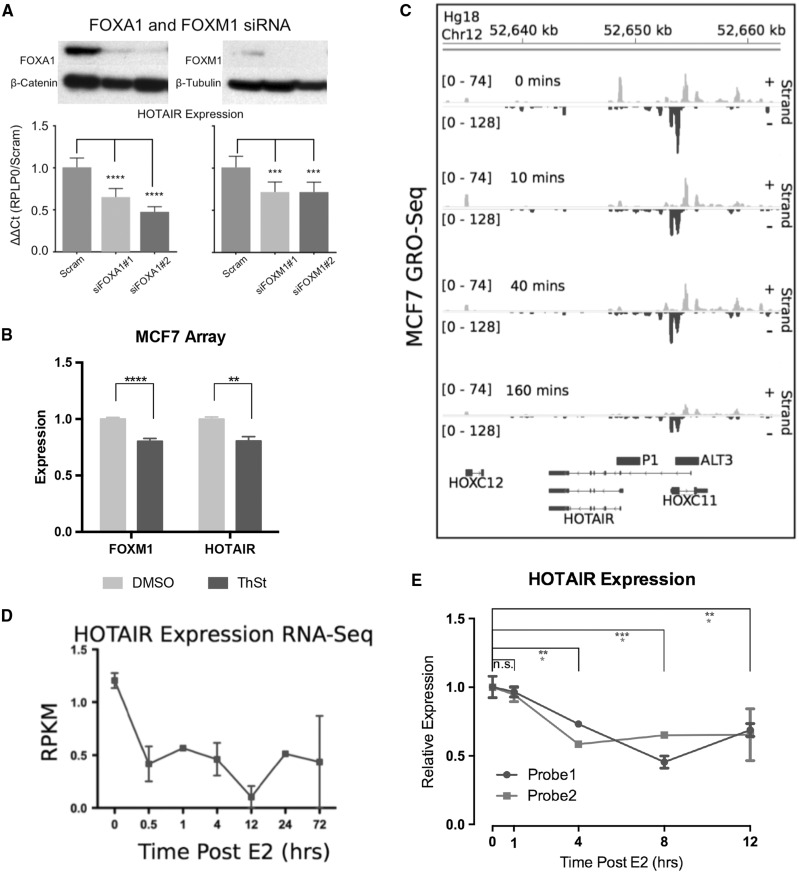
Given the role of FOX proteins in mediating enhancer activity of the HDE, we assessed the effect of FOXA1 and FOXM1 siRNA mediated knockdown on HOTAIR expression. FOXA1 and FOXM1 are key components of the ER signalling pathway in breast cancer and play important roles in disease progression, hence were prioritized for this study. This analysis showed a significant reduction in HOTAIR expression following knockdown of the FOX proteins (Figure 4A), suggesting that FOXM1 and FOXA1 positively regulate HOTAIR. In further support of these data, MCF7 cells treated with ThSt display significantly reduced the expression of HOTAIR and a known transcriptionally target of ThSt, FOXM1 (Figure 4B).
Figure 4.
HOTAIR is transcriptionally regulated by FOX proteins and repressed by oestrogen. (A) Top, western blot for indicated proteins, demonstrated siRNA knockdown in MCF7 cells and below qRT-PCR results for HOTAIR expression in each knockdown, normalized to RPLP0 and the scrambled control. (B) Expression of HOTAIR and FOXM1 following treatment of MCF7 cells by DMSO (vehicle) or thiostreptin (ThSt) (C) Re-analysis of MCF7 GRO-Seq data following E2 treatment [GSE27463] (81). The yellow histograms represent the positive strand and blue, the negative. The X-axis corresponds to the genomic position from Hg18 and the Y-axis the relative reads, HOTAIR promoters are shown above gene track. (D) RNA-Seq results for HOTAIR from E2 treated MCF7 cells over 72 hours (Reads per kilobase per million = RPKM). (E) qRT-PCR results of HOTAIR expression from a 10 nM E2 time course in MCF7 cells, representative of three separate experiments, data not shown. Y-axis represents relative expression (ΔΔCt of RPLP0/DMSO). Statistical analysis was performed via a one-way ANOVAs with Tukey’s corrected multiple comparisons between indicated columns or data points for A and E and via a two-tailed t-Test for B, P-values are <0.05 (*), <0.01 (**), <0.001 (***) and <0.0001 (****).
FOXA1 binds extensively throughout the HOXC locus (Supplementary Material Figure S1) and given the multiple 3C interactions between this locus and the HDE (Figure 2A), we assessed gene expression changes of the locus following depletion of FOXA1 in MDA-MB-453 cells. All HOXC genes and the miRNAs MIR196A and MIR615 responded with reduction in expression following FOXA1 depletion except for HOXC4 (Supplementary Material, Figs. S6A and B).
HOTAIR is transcriptionally repressed by oestrogen
AP1, FOXA1 and FOXM1 facilitate the activity of oestrogen-mediated transcription in breast cancer.
Given the role of AP1, FOXA1 and FOXM1 in augmenting the activity of the HDE and that c-Fos binding to the HDE is oestrogen-dependent (Figure 3C), we investigated the effects of oestrogen on HOTAIR expression. We re-analyzed GRO-Seq data from MCF7 E2 treated cells; a sensitive, genome-wide run-on sequencing assay that maps the position, amount, and orientation of transcriptionally engaged polymerases. This analysis revealed that the amount of polymerase engaged with the HOTAIR alternative promoter (negative strand) decreased over time, with minimal engagement by 160 min post-E2 treatment (Figure 4C). Importantly, we found that HOTAIR (canonical transcript) levels in MCF7 cells decreased in response to E2 treatment, as determined by RNA-Seq and qRT-PCR analysis using two different TaqMan® Probes, that detect both the canonical and alternative isoforms (Figures 4D and E). These data suggest that oestrogen negatively regulates HOTAIR transcription.
HOTAIR expression is inversely correlated with oestrogen receptor signalling and directly correlated with FOXM1 in breast cancer
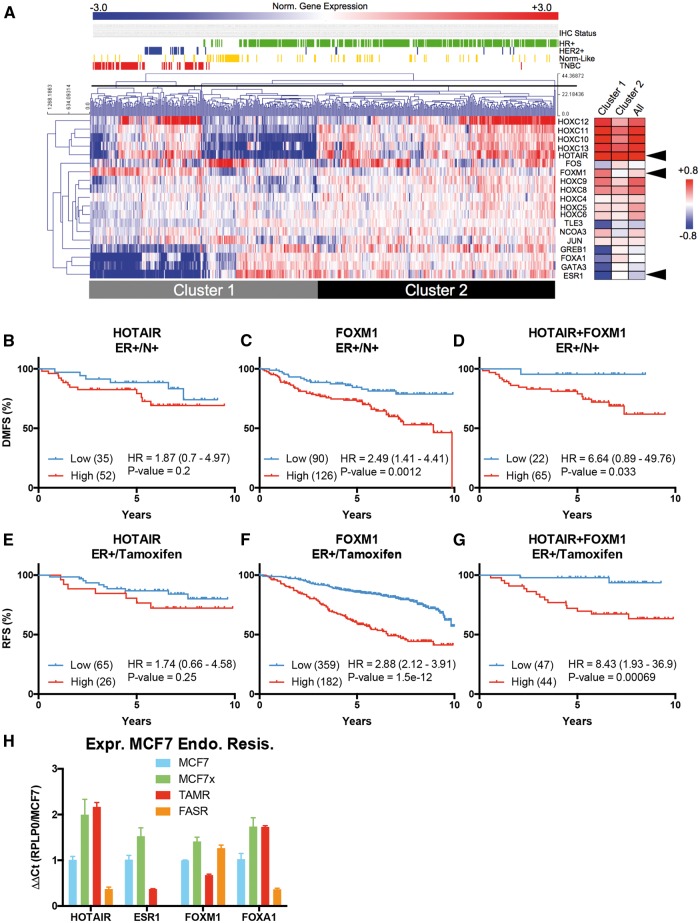
To investigate the relationship between HOTAIR and oestrogen signalling in breast cancer, we performed unsupervised hierarchical clustering of publically available (TCGA) breast cancer RNA-Seq data (Figure 5A). Given that numerous HOXC genes appear to be regulated by FOXA1 and possibly oestrogen, we clustered the expression of HOXC protein-coding genes, HOTAIR and known ERα cofactors. This analysis revealed a large cohort (Cluster 1 comprising 264 tumours and representing 49% of all cases) in which HOTAIR was inversely correlated with ESR1 and ERα cofactors: FOXA1, GATA3, GREB1, TLE3 and FOS (Figure 5A and Supplementary Material, Table S1). The proportion of breast cancer subtypes in Cluster 1 is heterogeneous, but is significantly enriched with triple-negative tumours (ER/PgR/HER2-negative by IHC), as well as those classified as basal-like, normal-like and HER2 by gene expression profiling (Figure 5A, Supplementary Material, Figures S7A and B).
Figure 5.
HOTAIR associates with FOXM1 to significantly stratify ER positive breast tumours. (A) Manhattan based hierarchical clustering of HOTAIR, HOXC genes, ERα and ERα cofactors in TCGA breast tumour data (75,78) with hormone receptor IHC status for each tumour displayed above. Pearson correlation coefficients for each cluster on the right. For full table and P-values see Supplementary Material, Table S1. Arrows highlight rows for HOTAIR, FOXM1 and ESR1 (ERα) (B-D) Kaplan-Meier curves for the stratification of distant metastasis free survival (DMFS) for patients with ER+ node positive (N+) tumours by HOTAIR or FOXM1 alone or their combination. Logrank hazards ratios with their 95% confidence intervals (CI) and P-values are shown and tumour number in the low or high expression groups indicated on right. (E-G) As above, except for relapse free survival (RFS) of ER+ tumours in patients that received tamoxifen (Tam). (H) HOTAIR, ESR1, FOXM1 and FOXA1 expression across MCF7 and MCF7 derived cell lines (ΔΔCt of RPLP0/MCF7, n = 2). All qRT-PCR error bars represent standard deviation.
HOTAIR and FOXM1 in combination predict response to therapy for ER+ tumours
Given the association between HOTAIR and FOXM1 expression and regulation of the HDE by FOX proteins, we sought to determine if these factors in combination may have prognostic potential in ER+ breast cancer. HOTAIR expression significantly stratifies the survival of patients with ER+ tumours and the combination of HOTAIR and FOXM1 expression increases this stratification (Supplementary Material, Table S2). The more aggressive ER+/N+ tumours were stratified by HOTAIR expression based on relapse free survival (RFS), however this was not observed for distant metastasis free survival (DMFS) (Supplementary Material, Table S3 and Figure 5B). FOXM1 alone is able to significantly stratify these tumours, however, the combination of HOTAIR and FOXM1 increased this stratification through an increase of the hazard ratio (Figures 5C and D).
HOTAIR and FOXM1 were also assessed for their potential to stratify patients on the basis of response to endocrine treatment and chemotherapy. Utilizing these biomarkers the RFS of patients treated with tamoxifen only (Tam), any endocrine therapy (ET) or the combination of endocrine and chemotherapy (CT) was stratified (Supplementary Material, Table S4, Figure 5E). HOTAIR alone could only stratify survival of patients who received CT in combination with ET, where high expression associated with poor survival. FOXM1 alone is a significant biomarker for response to tamoxifen in these ER+ tumours, however HOTAIR + FOXM1 display the largest hazard ratio where patients with low expression have a 95% chance of survival beyond 10 years (Figures 5F and G). Taken together these data indicate that HOTAIR in combination with FOXM1 enhances the value of these genes as biomarkers to predict response to any of the therapeutic options, especially patients with aggressive ER+ tumours.
Given the intersection of HOTAIR transcriptional regulation and the oestrogen-signalling pathway, we hypothesized that HOTAIR plays a role in endocrine resistance. To explore this, HOTAIR expression was determined in MCF7-derived in-vitro models of oestrogen deprivation or anti-oestrogen resistance: MCF7X, TAMR and FASR. These cell line models were derived following prolonged in vitro oestrogen deprivation (MCF7X), tamoxifen treatment (TAMR) or fulvestrant treatment (FASR). HOTAIR expression levels were increased in MCF7X and TAMR cells (Figure 5H). In contrast, FASR cells had significantly less HOTAIR RNA. Together with HOTAIR, expression of ESR1, FOXM1 and FOXA1 were also altered, particularly in TAMR cells, which have decreased expression of ESR1 while maintaining expression of the FOX genes (Figure 5H). These data indicate that HOTAIR expression increases in cell line models of endocrine therapy resistance.
HOTAIR regulators enhance the power of HOTAIR as a biomarker in HER2-enriched tumours
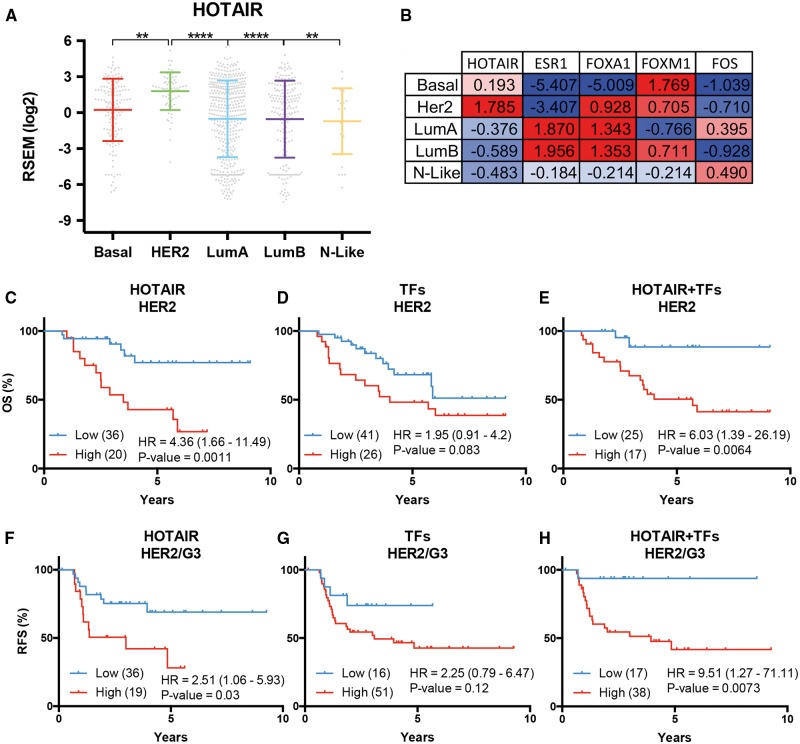
To determine whether the prognostic potential of HOTAIR and FOXM1 coexpression applies to other breast cancer subtypes, HOTAIR levels were assessed across all of the intrinsic molecular subtypes of breast cancer. This analysis revealed significant enrichment of expression in the HER2-enriched subtype (Figure 6A). Both FOXA1 and FOXM1 are also highly expressed in this subtype, whilst ESR1 expression is depleted (Figure 6B). Interestingly, both HOTAIR and FOXM1 (HOTAIR + FOXM1) expression is significantly enriched in the basal-like and HER2-enriched subtype (Supplementary Material, Figure S8).
Figure 6.
HOTAIR regulators improve utility of HOTAIR in stratifying HER2-Enriched tumours. (A and B) Relative expression of HOTAIR and transcription factors across the PAM50 intrinsic breast cancer subtypes. Referring to A, horizontal bars are the mean and error bars standard deviation. Significance was found via one-way ANOVAs with Tukey’s corrected multiple comparisons for HER2 tumours against all others. P-values are <0.05 (*), <0.01 (**), <0.001 (***) and <0.0001 (****). Tumour numbers are as follow Basal-like (140, Basal), HER2-enriched (67, HER2), Luminal A (420, LumA) Luminal B (194, LumB) and Normal-like (24, N-Like). (C-E) Kaplan-Meier curves stratifying the overall survival of patients with HER2-enriched tumours by HOTAIR alone, the transcription factors or their combination. Logrank hazards ratios with their 95% confidence intervals (CI) and P-values indicated b elow. Tumour number in the low or high expression groups are indicated in each graph. (F to G) As above, except for the relapse free survival (RFS) of patients with HER2-enriched Grade 3 tumours.
HOTAIR expression significantly stratified the RFS of patients with luminal B, HER2-enriched and ER+ tumours, with high expression associating with poor outcome (Supplementary Material, Table S5). High expression of HOTAIR, FOXA1, FOXM1 and FOS combined with low expression of ESR1, (HOTAIR + TFs), as a functional network, enhanced the stratification overall survival (OS) for patients with HER-enriched tumours (Figures 6C-E). This enhanced stratification is most evident in the more aggressive Grade 3 HER2-enriched tumours where HOTAIR + TFs expression significantly stratified the RFS greater than either HOTAIR or the TFs alone (Figures 6F–H). This stratification indicated that patients with low expression of HOTAIR + TFs, at the end of the studies, had a 93.75% chance of survival compared to 41.705% for the high expression group. These patients with dramatically worse survival are likely to respond poorly to therapies.
Collectively, we have identified several transcription factors that regulate the expression of HOTAIR and enhance the capacity of HOTAIR to predict the therapeutic response of patients with ER+ breast tumours and survival in breast cancer patients with HER2-enriched tumours.
Discussion
Transcriptional enhancers enable precise regulation of gene expression, are frequently disrupted in cancer, and are emerging as the next generation of prognostic biomarkers for a range of human diseases. This study describes the long-range regulatory elements and factors controlling the transcription of HOTAIR in breast cancer. Further, we show that transcription factors correlated with HOTAIR expression in breast tumours are associated with poor prognosis.
In this paper, we have identified several novel transcriptional control elements regulating the expression of the lncRNA HOTAIR. These include an alternate promoter situated approximately 8 kb upstream of the previously described HOTAIR promoter, and a distal enhancer (HDE) located approximately 150 kb downstream of the HOTAIR gene. These elements are distinct from the core promoter and CpG elements described previously (29–31,34). We also show that the HOTAIR enhancer regulates HOTAIR expression by DNA looping and that this enhancer can augment the transcriptional activity of both the canonical and alternative promoter elements. Importantly we demonstrate that HOTAIR and its regulators, as identified through mapping of the HDE, combine to significantly stratify survival of breast cancer patients, supportive of previous work also demonstrating that enhancer activity and transcription factor binding align closely with clinical outcome (35). It would be worth exploring this aspect for the HDE, by utilizing ChIP-Seq, DNA accessibility assay or 3C data from tumours samples, it may be possible to stratify clinical response through activity of this enhancer.
Several studies have highlighted the promiscuous nature of promoter-enhancer interactions (36,37). To further the research presented in this study, it would be worth exploring the interaction landscape of the HOXC locus through 5C, a technique that looks at all interactions within a large genomic region (38). In addition to these experiments, mutagenesis of the HDE using a CRISPR/Cas9 approach would be valuable for further establishing the role of this enhancer (39,40). Such studies may shed more light on the interaction of the HOXC distal enhancer, the HOTAIR promoter and potentially other targets of this enhancer.
Reporter mutagenesis, ChIP-seq and siRNA experiments show that HOTAIR transcription is regulated by a number of breast cancer-associated transcription factors, including FOXA1 and FOXM1. FOXA1 is a crucial pioneer factor for ERα binding (41). FOXM1 has a wide variety of functions including regulating G2/M phase transition, mammary gland luminal progenitor cell maintenance, co-binding with ERα in cell lines, and is strongly implicated in the progression of multiple cancer types including breast cancer (42–43). FOXM1 expression has previously been linked with ERα activity and resistance to endocrine therapies, where elevated expression is correlated with poor prognosis (44, 45). Whilst the tumour expression data for HOTAIR, ESR1 and FOXM1 was consistent with in vitro results, this was not the case for HOTAIR and FOXA1. The latter may represent differences between the molecular environment in cell lines and tumours. The data presented here support an association between forkhead box proteins and HOTAIR expression, likely through direct transcriptional regulation.
Re-analyses of published ChIP-Seq data and oestrogen treatment in vitro demonstrate that oestrogen represses HOTAIR transcription. Our observations are consistent with recent finding by Xue et al. (46) indicating a direct association between ER and HOTAIR repression following oestrogen treatment in MCF7 cells. In support of our findings, there is a strong inverse correlation between HOTAIR and ESR1 expression in breast tumours, arguing against the notion of oestrogen enhancing HOTAIR expression. Our findings, and those of others (46), conflict with a previous report showing induction of HOTAIR expression by oestrogen (31). This discrepancy may reflect differences in experimental design, including time of oestrogen treatment and the method of transcript detection, or the use of a different sub-line of MCF7 cells.
We hypothesized that the association between oestrogen and HOTAIR may play a role in endocrine resistance. To explore this, we used the well-documented tamoxifen resistant, fulvestrant resistant and oestrogen independent sub-lines of MCF7 cells (48–51). HOTAIR expression increased in both the oestrogen-deprived MCF7X, and tamoxifen-resistant TAMR cells, concordant with a repressive role of oestrogen/ER signalling. Importantly, our previous work (Zhuang et al. 2014 (52)) also demonstrated that HOTAIR expression was significantly increased in a cell line model of resistance to induced cell death via TNF, which displays loss of ER and altered oestrogen signalling. These findings may be of clinical significance as they suggest therapies blocking oestrogen signalling may induce HOTAIR, which could in turn promotes breast cancer progression.
Endocrine therapy (e.g. tamoxifen, aromatase-inhibitors) is a standard of care for patients with HR+ breast tumours, but deciding which patients should be managed more aggressively by the addition of chemotherapy can be difficult (53). These patients are therefore a primary focus of prognostic and predictive biomarker development (54). It is highly significant that FOXM1 and HOTAIR co-expression significantly enhances the prognostic power of either factor individually in predicting relapse-fee survival in patients with ER+ and ER+/N+ breast cancer.
There are several pathways in which HOTAIR and FOXM1 are likely to collaborate in tumourigenesis, explaining their prognostic power. Although we have shown that FOXM1 regulates the expression of HOTAIR through the HDE, we acknowledge that their link in tumourigenesis may be through other means, such as a common functional pathway. FOXM1 is involved in cell cycle regulation (55,56) and the cyclin-dependent kinases CDK1 and CDK2 are known to phosphorylate EZH2 during S and G2 phases of the cell cycle (57), actively promoting EZH2-HOTAIR interactions (57). HOTAIR over-expression has also been implicated in cell cycle progression in pancreatic cancer (27). It is possible that FOXM1 and HOTAIR corroborate to regulate the aspects of the cell cycle, further work should aim to understand this, and if this functionality explains the prognostic potential of FOXM1 and HOTAIR in breast cancer.
FOXM1 has been implicated in chromosome instability (CIN), a process associated with aneuploidy, aggressive tumours and poor response to therapy (58–60). CIN, together with ER signalling, has recently been described as a powerful predictor of survival in ER+ patients (61). It is tempting to speculate that HOTAIR may play a role in the relationship between CIN and ER signalling. Reduced ER signalling (e.g. reduced oestrogen) may drive the expression of HOTAIR (via FOXA1 or FOXM1) possibly in association with increased expression of the CIN genes. This could explain the poor prognosis of subsets of ER+ tumours that appear genomically unstable and have reduced response to endocrine therapy (tamoxifen) alone or in combination with chemotherapy. It also raises the possibility that patients with ER+/N+ tumours who are resistant to endocrine therapy may benefit from anti-aneuploidy drugs that target molecules in the CIN module, such as TTK, AURK and PLK1 (62). Indeed, we have previously shown that oestrogen-deprived or deficient cells are more sensitive to CIN inhibitors (Stone et al. 2013 (50); Al-Ejeh et al. 2014 (61)).
We demonstrate that the expression of HOTAIR in concert with its transcriptional regulators, are informative in predicting outcome in HER2 breast cancer. Our findings support an earlier study indicating that HOTAIR is most highly expressed in the HER2-enriched intrinsic molecular subtype of breast cancer (63). The primary treatment for a patient with a HER2+ tumour is chemotherapy and Herceptin, a targeted monoclonal antibody to the HER2 receptor (64,65). Herceptin resistance is a significant clinical problem, resulting in lower RFS, and leading to poor disease outcome (66). The combination of HOTAIR and its long-range transcriptional regulators can better predict outcome in patients with high-grade HER2-enriched tumours. Use of these biomarkers, could immediately facilitate better targeting of expensive Herceptin treatment and improve patient surveillance. Longer term, inhibitors of this functional network could be investigated as targeted therapies.
The results presented describe a novel regulatory pathway controlling the expression of the lncRNA HOTAIR and show that the associated elements and factors can illuminate the mechanisms underlying the role of HOTAIR in breast cancer. This information can be used to identify novel biomarker combinations to better predict response to therapy and survival of breast cancer patients. Continuation of this work should aim at understanding the molecular mechanisms that explain the prognostication potential of HOTAIR and its regulatory elements, with particular relevance to endocrine therapy resistant tumours that have aberrant ER signalling and the relationship this may have to chromosome instability.
Materials and Methods
Cell culture
Cell lines; MCF7, T47D, ZR-751, MDA-MB-453 and MDA-MB-468; were cultured as per recommendations from ATCC. MCF7 cells were a kind gift from Chris Ormandy at the Garvan Institute of Medical Research (NSW, Australia) whilst T47D, ZR-751, MDA-MB-453 and MDA-MB-468 cells were sourced from ATCC. Estradiol (E2) treatment experiments followed a time course of 10 or 1 nM E2 across indicated time points within the text. Positive controls for the RNA-Seq and qRT-PCR experiments are shown in Figure S6. MCF7 cells, from which the endocrine resistant sublines were derived, were originally obtained from AstraZeneca. MCF7-derived acquired endocrine resistant sub-line RNA was sourced through A. Stone and J. M. W. Gee see author list for details. Tamoxifen-resistant (TAMR), fulvestrant-resistant (FASR), and oestrogen-deprived (MCF7X) derivatives were cultured as previously described (47,48,51). All cell lines were authenticated by short-tandem repeat (STR) profiling (Cell Bank, Australia) and cultured for less than 6 months after authentication.
Chromosome conformation capture (3C)
Chromosome conformation capture was performed as previously described by in Tan-Wong et al. 2008 (67–69). Briefly, cells were grown to 60–80% confluence and fixed with 1% formaldehyde. Libraries were generated for each cell line using HindIII and or BglII with control libraries undigested and unligated, representing native DNA without chromosome conformation. Any 3C-qPCR products were excluded if they amplified within the control libraries. GAPDH primers (amplified fragment contains no cut sites for these restriction enzymes) were used to determine the digestion and ligation efficiency for each library by comparing 3C-qPCR values to primers that amplifed a fragment containing a HindIII or BglII cut sites (Table S6). For each 3C-qPCR, primers were designed between 100–250 bp up or downstream of each HindIII cut site with the primer across the putative HOTAIR distal enhancer used as bait in each 3C-qPCR (Supplementary Material, Table S6). BglII primers were sourced from Ferraiuolo et al. (70). 3C-qPCR conditions 50 °C, 2 min, 95 °C 10 min, (95 °C 15 sec and 60 °C 1 min) for 45 cycles. With a final melt starting at 50 °C 90 sec rising to 99 °C in 1 °C increments and 5 sec at each step. Each 3C-qPCR was performed on a pool of at least three independent libraries for each cell line and the qPCR done in duplicate. BAC controls were constructed following the protocol from (69) and primer pairs assayed for appropriate efficiency.
Cloning and luciferase reporter assays
HOTAIR enhancers and promoters were cloned into the luciferase reporter plasmid pGL3-Basic; see Supplementary Material, Table S7 for primers. The canonical HOTAIR promoter used in this study incorporated the previously described promoter region and E-box with additional bases either side (29). MCF7, T47D and MDA-MB-453 cells were transfected in antibiotic-free media with 700 ng of modified pGL3 promoter less reporter plasmids, 20 ng of Renilla and with 1μL of Lipofectamine® 2000 (Life Technologies, Grand Island, NY, USA). 48hours post transfection luciferase readings were measured using a DTX-880 luminometer and Dual-Glo® luciferase reporter kit (Promega, Madison, WI, USA), following the manufacturer’s recommended protocol. For oestrogen treatment and luciferase reporter levels of HOTAIR promoters and the HDE we followed the previously published method by Tan-Wong et al. (67).
Gene expression analysis
RNA for gene expression analysis was extracted using TRIzol® reagent (Life Technologies) and phenol-chloroform purified. RNA was then DNase I (Ambion, ThermoFisher) treated and cDNA generated using SuperScript® III (Invitrogen, ThermoFisher) using random hexamers supplemented with RNaseOUT™ (Invitrogen, ThermoFisher. Gene expression was assayed with TaqMan® probes (Life Technologies, ThermoFisher) following the manufacturer’s recommended protocol, probe list in Supplementary Material, Table S8. RNA-Seq in MCF7 cells was performed as previously described by K. Nephew (see author list) (71). Analysis of HOTAIR and FOXM1 expression in response to MCF7 cells treated with thiostreptin was sourced from GSE40767 (44). The probe sequence for HOTAIR expression on the array was as follows: TACACGCCTCTCCAAG ACAC AGTGGCACCGCTTTT CTAACTGGC AGCACA and FOXM1: GCCA CCTCCCCGTGTT TCCAAGTCAGCTTTC CTGCAAGAA GAA A T CCT GG. HOTAIR isoforms were sourced from public repositories for the RefSeq (72) and Broad (73) data sets. For analysis of HOTAIR and FOXM1 in the PAM50 subtypes, TCGA RSEM data was log2 transformed and mean-center normalized for all tumours, tumuor numbers as follows Basal-like (140, Basal), HER2-enriched (67, HER2), Luminal A (420, LumA) Luminal B (194, LumB) and Normal-like (24, N-Like).
RNAi
Short-interfering RNA-mediated knockdown was performed using Flexitube siRNA (Qiagen). SiRNAs utilized in this study were FOXA1; SI04311888 and SI04217038 and FOXM1; SI04261831 and SI04166309 which were compared to the AllStars Negative Control (Qiagen). MCF7 cells were transfected using 3L of RNAi-Max (Life Technologies, ThermoFisher) and with a final concentration of 10nM for each siRNA. RNA was extracted and conversion into cDNA and subsequent qRT-PCR for gene expression as described above.
Bioinformatic analysis of publically available data
All data was sourced from public repositories and analyzed through the IGV browser and the human genome assembly Hg18 and/or Hg19, as indicated. Publically available data were sourced from the Gene Expression Omnibus (GEO). Accession codes for each dataset are listed in Supplementary Material, Table S9. ChIP-Seq data that was not available as processed mapped profiles was done so utilizing standard mapping software. Briefly, data were downloaded in .sra format and converted to .fastq using fastq-dump and subsequently mapped with Bowtie where any multi-mapping reads were disregarded. Output .sam files were then converted to .bam files, using SamTools, .bed files using BedTools and finally converted to .bedGraphs that were tiled by IGV-Tools for IGV browser analysis (74).
The Cancer Genome Atlas (TCGA) (75) was accessed and analyzed through the University of California Santa Cruz (UCSC) Cancer Browser (75,76). HOTAIR + TFs were determined based on the average expression of HOTAIR, FOXM1, FOXA1, FOS together with the inverted average expression of ESR1. HOTAIR + FOXM1 is the combined average expression for these two genes.
Kaplan-meier analysis
The online tool, Kaplan-Meier Plotter (http://kmplot.com/analysis/) was used to produce survival curves based on gene expression with a maximum follow-up time of 10-years post diagnosis (77). High- and low-expression cutoffs were determined by the ‘Autoselect best cutoff,’ feature that determines the most significant stratification based on median, tertile or quartile splits. Kaplan-Meier Plotter uses logrank hazard’s ratios with corresponding P-values to determine significance. Probe IDs for each gene utilized in this analysis: HOTAIR (239153_at), ESR1 (205225_at), FOXA1 (204667_at), FOXM1 (202580_x_at) and FOS (209189_at).
Tumour clustering and correlations
Normalized TCGA (78) microarray expression data were used to cluster HOTAIR expression with ERα and ERα cofactors. The heat-map was produced using hierarchical average-linkage Manhattan-based clustering performed in Multiple experiment Viewer (MeV (79)) and Pearson correlation coefficients calculated for each comparison. The dendrogram tree cut point is indicated in Figure 5A, which creates 5 clusters of tumours, clusters 1-4 were grouped together based on the negative correlation between HOTAIR and ESR1 expression.
Western blot analysis
Total protein from MCF7 cells was extracted using RIPA buffer with 15 μg total per well and analyzed using Novex™ NuPAGE® gels from Life Technologies. Antibodies for FOXA1 and FOXM1 were ab23738 (Abcam, Cambridge, MA, USA) and sc-502 (Santa Cruz, Dallas, TX, USA) respectively.
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
We thank Jonathan Harris for the provision of DHT and David Miller for technical assistance and Kyle Upton for his critical review of the manuscript.
Conflict of Interest statement. None declared.
Funding
Research was funded by the National Breast Cancer Foundation (NBCF: 2007003445 and CG-12-07) of Australia, the Australian Research Council (ARC: DP0985025), the Cancer Council of Queensland (CCQ: 1026095) and The University of Queensland. DHD was supported by a Collaborative Program Grant from the, National Breast Cancer Foundation [NBCF; CG-08-03]. Institute, Integrative Cancer Biology Program U54CA1113001 (KPN). SLE, JDF, AMS and ED are supported by Fellowships from the NBCF [ID# ECF-10-05, ECF-12-04, ECF-12-12 and ECF-13-04 respectively]. FA is supported by Future Fellowship from the Australian Research Council [ID: FT130101417]. JMWG is funding by a Scientific Fellowship from Breast Cancer Now. MJM and JAB are supported by an Australian Postgraduate Award (APA). AS was supported from NBCF program grant. SJC supported by NHMRC fellowship. Funding to pay the Open Access publication charges for this article was provided by The University of Queensland.
References
- 1.Vuong D., Simpson P.T., Green B., Cummings M.C., Lakhani S.R. (2014) Molecular classification of breast cancer. Virchows Arch, 465, 1–14. [DOI] [PubMed] [Google Scholar]
- 2.Early Breast Cancer Trialists' Collaborative, G.(2005) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet, 365, 1687–1717. [DOI] [PubMed] [Google Scholar]
- 3.Osborne C.K., Schiff R. (2011) Mechanisms of endocrine resistance in breast cancer. Annu Rev Med, 62, 233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung A., Cui X., Audeh W., Giuliano A. (2013) Current status of anti-human epidermal growth factor receptor 2 therapies: predicting and overcoming herceptin resistance. Clin. Breast Cancer, 13, 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dos Anjos Pultz B., da Luz F.A., de Faria P.R., Oliveira A.P., de Araujo R.A., Silva M.J. (2014) Far beyond the usual biomarkers in breast cancer: a review. J. Cancer, 5, 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maston G.A., Evans S.K., Green M.R. (2006) Transcriptional regulatory elements in the human genome. Annu. Rev. Genomics Hum. Genet., 7, 29–59. [DOI] [PubMed] [Google Scholar]
- 7.Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M., et al. (2011) Mapping and analysis of chromatin state dynamics in nine human cell types. Nature, 473, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kron K.J., Bailey S.D., Lupien M. (2014) Enhancer alterations in cancer: a source for a cell identity crisis. Genome Med., 6, 77.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herz H.M., Hu D., Shilatifard A. (2014) Enhancer malfunction in cancer. Mol. Cell, 53, 859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cowper-Sal lari R., Zhang X., Wright J.B., Bailey S.D., Cole M.D., Eeckhoute J., Moore J.H., Lupien M. (2012) Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat. Genet., 44, 1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.French J.D., Ghoussaini M., Edwards S.L., Meyer K.B., Michailidou K., Ahmed S., Khan S., Maranian M.J., O'Reilly M., Hillman K.M., et al. (2013) Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am. J. Hum. Genet., 92, 489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akhtar-Zaidi B., Cowper-Sal-lari R., Corradin O., Saiakhova A., Bartels C.F., Balasubramanian D., Myeroff L., Lutterbaugh J., Jarrar A., Kalady M.F., et al. (2012) Epigenomic enhancer profiling defines a signature of colon cancer. Science, 336, 736–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loven J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R., Bradner J.E., Lee T.I., Young R.A. (2013) Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell, 153, 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozsolak F., Song J.S., Liu X.S., Fisher D.E. (2007) High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol., 25, 244–248. [DOI] [PubMed] [Google Scholar]
- 15.Marson A., Levine S.S., Cole M.F., Frampton G.M., Brambrink T., Johnstone S., Guenther M.G., Johnston W.K., Wernig M., Newman J., et al. (2008) Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell, 134, 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corcoran D.L., Pandit K.V., Gordon B., Bhattacharjee A., Kaminski N., Benos P.V. (2009) Features of mammalian microRNA promoters emerge from polymerase II chromatin immunoprecipitation data. PLoS One, 4, e5279.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang G., Wang Y., Shen C., Huang Y.W., Huang K., Huang T.H., Nephew K.P., Li L., Liu Y. RNA polymerase II binding patterns reveal genomic regions involved in microRNA gene regulation. PLoS One, 5, e13798.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sati S., Ghosh S., Jain V., Scaria V., Sengupta S. (2012) Genome-wide analysis reveals distinct patterns of epigenetic features in long non-coding RNA loci. Nucleic Acids Res., 40, 10018–10031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venkatesh S., Workman J.L. (2012) Non-coding transcription SETs up regulation. Cell Res., 23(3), 311–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., Huarte M., Zuk O., Carey B.W., Cassady J.P., et al. (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature, 458, 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A. (2013) Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell, 153, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraser P. (2006) Transcriptional control thrown for a loop. Curr. Opin. Genet. Dev., 16, 490–495. [DOI] [PubMed] [Google Scholar]
- 23.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., et al. (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell, 129, 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morey L., Helin K. (2010) Polycomb group protein-mediated repression of transcription. Trends Biochem. Sci., 35, 323–332. [DOI] [PubMed] [Google Scholar]
- 25.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. (2010) Long noncoding RNA as modular scaffold of histone modification complexes. Science, 329, 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L., et al. (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature, 464, 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim K., Jutooru I., Chadalapaka G., Johnson G., Frank J., Burghardt R., Kim S., Safe S. (2013) HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene, 8, 32(13):1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hajjari M., Salavaty A. (2015) HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol. Med., 12, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma M.Z., Li C.X., Zhang Y., Weng M.Z., Zhang M.D., Qin Y.Y., Gong W., Quan Z.W. (2014) Long non-coding RNA HOTAIR, a c-Myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer. Mol. Cancer, 13, 156.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang G., Zhang S., Gao F., Liu Z., Lu M., Peng S., Zhang T., Zhang F. (2014) Osteopontin enhances the expression of HOTAIR in cancer cells via IRF1. Biochim. Biophys. Acta., 1839, 837–848. [DOI] [PubMed] [Google Scholar]
- 31.Bhan A., Hussain I., Ansari K.I., Kasiri S., Bashyal A., Mandal S.S. (2013) Antisense Transcript Long Noncoding RNA (lncRNA) HOTAIR is Transcriptionally Induced by Estradiol. J Mol. Biol., 425(19), 3707–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X., Zhou L., Fu G., Sun F., Shi J., Wei J., Lu C., Zhou C., Yuan Q., Yang M. (2014) The identification of an ESCC susceptibility SNP rs920778 that regulates the expression of lncRNA HOTAIR via a novel intronic enhancer. Carcinogenesis, 35, 2062–2067. [DOI] [PubMed] [Google Scholar]
- 33.Bayram S., Sumbul A.T., Batmaci C.Y., Genc A. (2015) Effect of HOTAIR rs920778 polymorphism on breast cancer susceptibility and clinicopathologic features in a Turkish population. Tumour Biol., 36, 3863–3870. [DOI] [PubMed] [Google Scholar]
- 34.Lu L., Zhu G., Zhang C., Deng Q., Katsaros D., Mayne S.T., Risch H.A., Mu L., Canuto E.M., Gregori G., et al. (2012) Association of large noncoding RNA HOTAIR expression and its downstream intergenic CpG island methylation with survival in breast cancer. Breast Cancer Res. Treat., 136, 875–883. [DOI] [PubMed] [Google Scholar]
- 35.Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R, et al. (2012) Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature, 481, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Consortium, E.P. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Affer M., Chesi M., Chen W.D., Keats J.J., Demchenko Y.N., Tamizhmani K., Garbitt V.M., Riggs D.L., Brents L.A., Roschke A.V., et al. (2014) Promiscuous MYC locus rearrangements hijack enhancers but mostly super-enhancers to dysregulate MYC expression in multiple myeloma. Leukemia, 28, 1725–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dostie J., Richmond T.A., Arnaout R.A., Selzer R.R., Lee W.L., Honan T.A., Rubio E.D., Krumm A., Lamb J., Nusbaum C., et al. (2006) Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res., 16, 1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo Y., Xu Q., Canzio D., Shou J., Li J., Gorkin D.U., Jung I., Wu H., Zhai Y., Tang Y., et al. (2015) CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell, 162, 900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wright A.V., Nunez J.K., Doudna J.A. (2016) Biology and Applications of CRISPR Systems: Harnessing Nature's Toolbox for Genome Engineering. Cell, 164, 29–44. [DOI] [PubMed] [Google Scholar]
- 41.Jozwik K.M., Carroll J.S. (2012) Pioneer factors in hormone-dependent cancers. Nat. Rev. Cancer, 12, 381–385. [DOI] [PubMed] [Google Scholar]
- 42.Carr J.R., Kiefer M.M., Park H.J., Li J., Wang Z., Fontanarosa J., DeWaal D., Kopanja D., Benevolenskaya E.V., Guzman G., et al. (2012) FoxM1 regulates mammary luminal cell fate. Cell Rep., 1, 715–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koo C.Y., Muir K.W., Lam E.W. (2012) FOXM1: From cancer initiation to progression and treatment. Biochim. Biophys. Acta, 1819, 28–37. [DOI] [PubMed] [Google Scholar]
- 44.Sanders D.A., Ross-Innes C.S., Beraldi D., Carroll J.S., Balasubramanian S. (2013) Genome-wide mapping of FOXM1 binding reveals co binding with estrogen receptor alpha in breast cancer cells. Genome Biol., 14, R6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bergamaschi A., Madak-Erdogan Z., Kim Y., Choi Y.L., Lu H., Katzenellenbogen B.S. (2014) The forkhead transcription factor FOXM1 promotes endocrine resistance and invasiveness in estrogen receptor-positive breast cancer by expansion of stem-like cancer cells. Breast Cancer Res., 16, 436.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xue X., Yang Y.A., Zhang A., Fong K.W., Kim J., Song B., Li S., Zhao J.C., Yu J. (2015) LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene, 35(21), 2746–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McClelland R.A., Barrow D., Madden T.A., Dutkowski C.M., Pamment J., Knowlden J.M., Gee J.M., Nicholson R.I. (2001) Enhanced epidermal growth factor receptor signaling in MCF7 breast cancer cells after long-term culture in the presence of the pure antiestrogen ICI 182,780 (Faslodex). Endocrinology, 142, 2776–2788. [DOI] [PubMed] [Google Scholar]
- 48.Knowlden J.M., Hutcheson I.R., Jones H.E., Madden T., Gee J.M., Harper M.E., Barrow D., Wakeling A.E., Nicholson R.I. (2003) Elevated levels of epidermal growth factor receptor/c-erbB2 heterodimers mediate an autocrine growth regulatory pathway in tamoxifen-resistant MCF-7 cells. Endocrinology, 144, 1032–1044. [DOI] [PubMed] [Google Scholar]
- 49.Hurtado A., Holmes K.A., Ross-Innes C.S., Schmidt D., Carroll J.S. (2011) FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet., 43, 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stone A., Cowley M.J., Valdes-Mora F., McCloy R.A., Sergio C.M., Gallego-Ortega D., Caldon C.E., Ormandy C.J., Biankin A.V., Gee J.M., et al. (2013) BCL-2 hypermethylation is a potential biomarker of sensitivity to antimitotic chemotherapy in endocrine-resistant breast cancer. Mol. Cancer. Ther., 12, 1874–1885. [DOI] [PubMed] [Google Scholar]
- 51.Staka C.M., Nicholson R.I., Gee J.M. (2005) Acquired resistance to oestrogen deprivation: role for growth factor signalling kinases/oestrogen receptor cross-talk revealed in new MCF-7X model. Endocr. Relat. Cancer, 12 Suppl 1, S85–S97. [DOI] [PubMed] [Google Scholar]
- 52.Zhuang Y., Nguyen H.T., Burow M.E., Zhuo Y., El-Dahr S.S., Yao X., Cao S., Flemington E.K., Nephew K.P., Fang F, et al. (2014) Elevated expression of long intergenic non-coding RNA HOTAIR in a basal-like variant of MCF-7 breast cancer cells. Mol Carcinog., 54(12), 1656–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ali S., Coombes R.C. (2002) Endocrine-responsive breast cancer and strategies for combating resistance. Nat. Rev. Cancer, 2, 101–112. [DOI] [PubMed] [Google Scholar]
- 54.Eccles S.A., Aboagye E.O., Ali S., Anderson A.S., Armes J., Berditchevski F., Blaydes J.P., Brennan K., Brown N.J., Bryant H.E., et al. (2013) Critical research gaps and translational priorities for the successful prevention and treatment of breast cancer. Breast Cancer Res., 15, R92.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carter S.L., Eklund A.C., Kohane I.S., Harris L.N., Szallasi Z. (2006) A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet., 38, 1043–1048. [DOI] [PubMed] [Google Scholar]
- 56.Sadasivam S., Duan S., DeCaprio J.A. (2012) The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev., 26, 474–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaneko S., Li G., Son J., Xu C.F., Margueron R., Neubert T.A., Reinberg D. (2010) Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev., 24, 2615–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rajagopalan H., Lengauer C. (2004) Aneuploidy and cancer. Nature, 432, 338–341. [DOI] [PubMed] [Google Scholar]
- 59.Cheng W.Y., Ou Yang T.H., Anastassiou D. (2013) Biomolecular events in cancer revealed by attractor metagenes. PLoS Comput. Biol., 9, e1002920.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ricke R.M., van Ree J.H., van Deursen J.M. (2008) Whole chromosome instability and cancer: a complex relationship. Trends Genet., 24, 457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Al-Ejeh F., Simpson P.T., Sanus J.M., Klein K., Kalimutho M., Shi W., Miranda M., Kutasovic J., Raghavendra A., Madore J., et al. (2014) Meta-analysis of the global gene expression profile of triple-negative breast cancer identifies genes for the prognostication and treatment of aggressive breast cancer. Oncogenesis, 3, e100.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Al-Ejeh F., Shi W., Miranda M., Simpson P.T., Vargas A.C., Song S., Wiegmans A.P., Swarbrick A., Welm A.L., Brown M.P., et al. (2013) Treatment of triple-negative breast cancer using anti-EGFR-directed radioimmunotherapy combined with radiosensitizing chemotherapy and PARP inhibitor. J. Nucl. Med., 54, 913–921. [DOI] [PubMed] [Google Scholar]
- 63.Su X., Malouf G.G., Chen Y., Zhang J., Yao H., Valero V., Weinstein J.N., Spano J.P., Meric-Bernstam F., Khayat D., et al. (2014) Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget., 5, 9864–9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baselga J., Norton L., Albanell J., Kim Y.M., Mendelsohn J. (1998) Recombinant humanized anti-HER2 antibody (Herceptin) enhances the antitumor activity of paclitaxel and doxorubicin against HER2/neu overexpressing human breast cancer xenografts. Cancer Res., 58, 2825–2831. [PubMed] [Google Scholar]
- 65.Anampa J., Makower D., Sparano J.A. (2015) Progress in adjuvant chemotherapy for breast cancer: an overview. BMC Med., 13, 195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thery J.C., Spano J.P., Azria D., Raymond E., Penault Llorca F. (2014) Resistance to human epidermal growth factor receptor type 2-targeted therapies. Eur. J. Cancer, 50, 892–901. [DOI] [PubMed] [Google Scholar]
- 67.Tan-Wong S.M., French J.D., Proudfoot N.J., Brown M.A. (2008) Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc. Natl. Acad. Sci. U S A, 105, 5160–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vakoc C.R., Letting D.L., Gheldof N., Sawado T., Bender M.A., Groudine M., Weiss M.J., Dekker J., Blobel G.A. (2005) Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol. Cell, 17, 453–462. [DOI] [PubMed] [Google Scholar]
- 69.Hagege H., Klous P., Braem C., Splinter E., Dekker J., Cathala G., de Laat W., Forne T. (2007) Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc., 2, 1722–1733. [DOI] [PubMed] [Google Scholar]
- 70.Ferraiuolo M.A., Rousseau M., Miyamoto C., Shenker S., Wang X.Q., Nadler M., Blanchette M., Dostie J. (2010) The three-dimensional architecture of Hox cluster silencing. Nucleic Acids Res., 38, 7472–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miller D.F., Yan P.S., Buechlein A., Rodriguez B.A., Yilmaz A.S., Goel S., Lin H., Collins-Burow B., Rhodes L.V., Braun C., et al. (2013) A new method for stranded whole transcriptome RNA-seq. Methods, 63, 126–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M., et al. (2014) RefSeq: an update on mammalian reference sequences. Nucleic Acids Res., 42, D756–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L. (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev., 25, 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol., 10, R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cline M.S., Craft B., Swatloski T., Goldman M., Ma S., Haussler D., Zhu J. (2013) Exploring TCGA Pan-Cancer data at the UCSC Cancer Genomics Browser. Sci. Rep., 3, 2652.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rosenbloom K.R., Armstrong J., Barber G.P., Casper J., Clawson H., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M., et al. (2015) The UCSC Genome Browser database: 2015 update. Nucleic Acids Res., 43, D670–D681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gyorffy B., Lanczky A., Eklund A.C., Denkert C., Budczies J., Li Q., Szallasi Z. (2010) An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat., 123, 725–731. [DOI] [PubMed] [Google Scholar]
- 78.Cancer Genome Atlas, N. (2012) Comprehensive molecular portraits of human breast tumours. Nature, 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques, 34, 374–378. [DOI] [PubMed] [Google Scholar]
- 80.Joseph R., Orlov Y.L., Huss M., Sun W., Kong S.L., Ukil L., Pan Y.F., Li G., Lim M., Thomsen J.S., et al. (2010) Integrative model of genomic factors for determining binding site selection by estrogen receptor-alpha. Mol. Syst. Biol., 6, 456.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hah N., Danko C.G., Core L., Waterfall J.J., Siepel A., Lis J.T., Kraus W.L. (2011) A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell, 145, 622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.