Abstract
Huntington’s disease (HD) is an autosomal dominant neurodegenerative disorder caused by a CAG expansion in the gene-encoding Huntingtin (HTT). Transcriptome dysregulation is a major feature of HD pathogenesis, as revealed by a large body of work on gene expression profiling of tissues from human HD patients and mouse models. These studies were primarily focused on transcriptional changes affecting steady-state overall gene expression levels using microarray based approaches. A major missing component, however, has been the study of transcriptome changes at the post-transcriptional level, such as alternative splicing. Alternative splicing is a critical mechanism for expanding regulatory and functional diversity from a limited number of genes, and is particularly complex in the mammalian brain. Here we carried out a deep RNA-seq analysis of the BA4 (Brodmann area 4) motor cortex from seven human HD brains and seven controls to systematically discover aberrant alternative splicing events and characterize potential associated splicing factors in HD. We identified 593 differential alternative splicing events between HD and control brains. Using two expanded panels with a total of 108 BA4 tissues from patients and controls, we identified four splicing factors exhibiting significantly altered expression levels in HD patient brains. Moreover, follow-up molecular analyses of one splicing factor PTBP1 revealed its impact on disease-associated splicing patterns in HD. Collectively, our data provide genomic evidence for widespread splicing dysregulation in HD brains, and suggest the role of aberrant alternative splicing in the pathogenesis of HD.
Introduction
Huntington’s disease (HD) is an autosomal dominant neurodegenerative disorder caused by an expansion in the glutamine-coding CAG tract in the Huntingtin (HTT) gene (1–3). Normal individuals have less than 36 CAG repeats (on average 15–25) in exon 1 of HTT, while affected individuals exhibit repeat lengths greater than 39 (2,3). The elongated polyglutamine tract at the amino terminus results in HTT protein aggregation and a gain-of-function toxicity (4). HD primarily affects neurons in the striatum and cortex, and patients can display emotional symptoms, cognitive deficits, motor impairment, sleep disturbance and weight loss (5–8). Currently, no disease modifying therapies are available, and treatment is limited to the symptomatic management of chorea and emotional disorders (2). Thus, efforts to clarify disease pathogenesis and identify potential therapeutic targets are of high priority.
Huntingtin’s role in part is as a transcription regulator (9), and while the functional targets of wild-type HTT are not fully understood, expanded polyglutamine HTT exhibits an altered DNA interaction profile (10) and causes transcriptional dysregulation of multiple genes (2,3,11). Some of the earliest studies looking into the molecular pathophysiology of HD revealed that neuronal genes, such as neurotransmitter receptors and neuropetides, are differentially expressed in patient brains (12–15). The importance of transcriptional dysregulation in the pathology of HD was also demonstrated in mouse models by nuclear-restricted variants of mutant mouse Htt transgenes that reproduced major aspects of the mouse phenotype (16,17).
Transcriptome dysregulation is widespread in HD, as revealed by a large body of work on gene expression profiling of RNA samples from human HD patients and mouse models. HD was among the first neurological diseases to which microarray technology was applied. Studies included HD cells and brains harvested from mouse models and postmortem HD brain tissues (6,18–33). Several studies have characterized transcriptional dysregulation in HD from the network perspective (34–39). More recently, changes in the microRNA class of noncoding RNAs have been evaluated (29,36,40–43). The major conclusion from these studies is that a multitude of gene expression changes occur before an overt cell loss is observed, and that this dysregulation contributes to regional vulnerability. Gene expression changes are remarkably similar in HD postmortem brains and HD mouse models (31), reinforcing the idea that they are important for pathogenesis. Brain regions studied in HD are generally striatum and cerebral cortex. The latter is clearly affected, but with limited cell loss, so changes in gene expression profiles likely reflect dysregulated transcription rather than merely altered cell type composition between HD brains and healthy controls. While these studies have provided much information about the HD transcriptome, they are primarily based on conventional gene expression microarrays designed for measuring overall gene expression levels. A significant missing component is a comprehensive survey of the HD transcriptome where information extends beyond changes in overall gene expression levels to include changes in transcript isoforms arising from alternative RNA processing such as alternative splicing (AS).
AS is a major mechanism for expanding the regulatory and functional complexity in higher eukaryotes. RNA sequencing (RNA-seq) studies suggest that over 90% of multi-exon human genes are alternatively spliced (44,45). AS is important in neural development and plays a critical role in regulating neuronal gene expression in a tissue and cell-type specific manner (45–47). The precise regulation of alternative splicing patterns requires complex interactions between cis-regulatory splicing elements and trans-acting splicing factors (48). Mutations that disrupt splicing factors can produce aberrant gene products and cause disease (49,50). A classic example of this as related to repeat expansion diseases is myotonic dystrophy type 1 (DM1). In DM1 the expanded trinucleotide repeat CUG in the 3’ UTR of DMPK sequesters the muscleblind-like splicing regulator 1 (MBNL1), disrupting many MBNL1-dependent splicing events (51). Similar mechanisms of “RNA toxicity” have been suggested for fragile X tremor ataxia syndrome (FXTAS) (52), spinocerebellar ataxia 3 (SCA3) (53), and spinocerebellar ataxia 8 (SCA8) (52). The role of alternative splicing has also been implicated in various neurodegenerative and psychiatric diseases such as Alzheimer’s disease, amyotrophic lateral sclerosis (ALS), and schizophrenia (46,47,54–56).
Emerging evidence suggests splicing defects in HD. HTT neuritic and cytoplasmic aggregates, which may reflect HD pathogenesis, co-localize with the ALS-linked splicing factor transactivation-responsive DNA-binding protein 43 (TDP-43) (57). Proteomics studies show that mutant and wildtype HTT proteins differentially interact with many RNA binding proteins including splicing factors, suggesting that mutant HTT may disrupt the splicing regulatory networks via protein–protein interactions (58). Several genes important for other neurodegenerative diseases have been found to undergo mis-splicing in HD brains – one example being tau, whose mis-splicing is known to cause frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17) (59). Finally, new data suggest that the CAG repeat expansion in HTT may induce HTT exon 1 missplicing to produce a premature polyadenylated mRNA coding for a toxic N-terminal truncated protein (60). Despite these interesting observations, there is limited knowledge about the transcriptome-wide landscape of AS in HD.
The recent advent of the RNA-seq technology has enabled high-throughput analyses of alternative splicing at an unprecedented resolution (44,45). In this work we carried out a deep RNA-seq analysis of human HD brains and healthy controls to systematically discover and characterize aberrant alternative splicing events and their potential associated splicing factors in HD. We selected BA4 motor cortex for our RNA-seq study, based on the early symptoms of motor dysfunction and limited neuronal cell loss in this region, as well as an abundance of gene expression changes documented from earlier microarray studies (6). Based on the RNA-seq data, we performed detailed molecular analyses of the splicing factor PTBP1 in an expanded panels of postmortem brains from patients and controls to assess its role in disease-associated splicing patterns in HD.
Results
RNA-seq of postmortem brains from patients with HD and healthy controls
To characterize transcriptome changes, especially alternative splicing changes in HD, we conducted a deep RNA-seq analysis of 14 Brodmann area 4 (BA4) primary motor cortex samples from HD (Grade 2–4) and control brains (n = 7 each, Table 1, Supplementary Material, Table S1). We generated 2.1 billion 100bp × 2 paired-end reads, with 128 million to 175 million pairs of reads per sample. We calculated RNA-seq based gene expression levels represented by FPKM values and performed unsupervised hierarchical clustering to assess similarities in global gene expression profiles between the postmortem brain samples (see Materials and Methods). Though an absolute separation between disease and normal groups was not observed, likely reflecting biological variability or limited power, the two groups displayed a tendency to segregate, with HD brain samples mostly clustered into one sub-group (5 HD and 2 control samples), suggesting common transcriptome signatures while control brain samples showed a more scattered distribution in the clustering dendrogram (Supplementary Material, Figure S1).
Table 1.
Sample summary
| RNA-Seq |
Set 1 qRT-PCR/RT-PCR |
Set 2 qRT-PCR |
||||
|---|---|---|---|---|---|---|
| Control | HD | Control | HD | Control | HD | |
| n = 7 | n = 7 | n = 17 | n = 37 | n = 27 | n = 27 | |
| Age (Year)* | 52.3±7.9 | 56.3±8.5 | 55.0±12.9 | 59.7±9.5 | 65.7±11.6 | 66.3±11.9 |
| Sex | M6:F1 | M4:F3 | M10:F7 | M21:F16 | M20:F6:?1 | M18:F9 |
| PMI (Hour) | 16.0±6.0 | 11.6±7.5 | 13.5±6.4 | 14.6±8.3 | N/A | N/A |
| HD Grade (1:2:3:4) | N/A | 0:2:4:1 | N/A | 6:10:12:9 | N/A | 0:12:14:1 |
Abbreviation: PMI, postmortem interval; M, male; F, female
Age was indicated by mean ± standard deviation
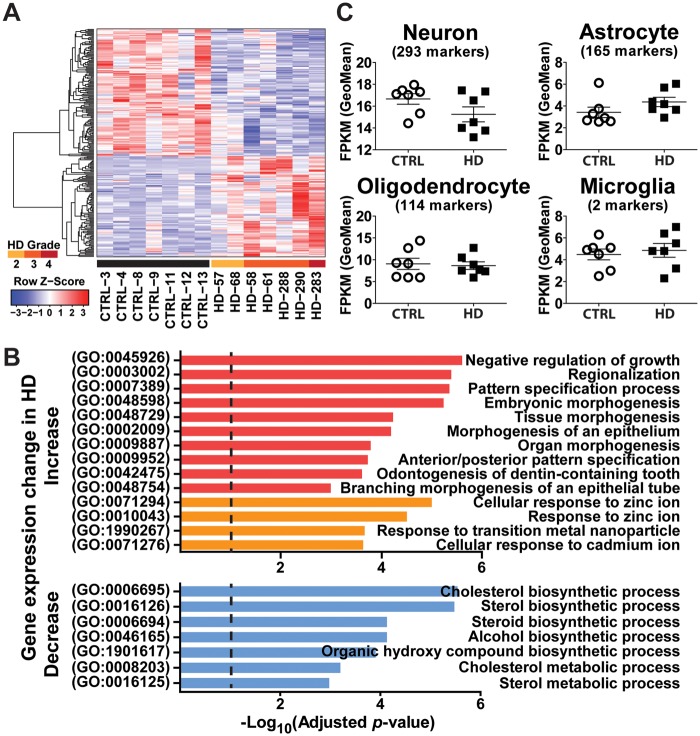
We identified 222 differentially expressed genes (Material and Methods) (Figure 1A, Supplementary Material, Table S2). Gene ontology (GO) analysis using Enrichr (61) revealed significantly enriched GO terms among the differentially expressed genes between HD and control brains (Material and Methods, Figure 1B, Supplementary Material, Table S3). Among genes upregulated in HD, the most significantly enriched GO terms fall almost exclusively into two categories: morphogenesis/growth (Figure 1B, red bars) and response to transition metal ions (Figure 1B, orange bars). All significantly enriched GO terms in downregulated genes in HD are biosynthetic/metabolic processes (cholesterol, sterol, steroid, alcohol and organic hydroxyl compound) (Figure 1B, blue bars).
Figure 1.
Gene expression analysis of HD and control motor cortex (BA4) samples using RNA-seq. (A) Heat map of 222 genes differentially expressed between HD and control motor cortex (BA4) samples. Color bars above sample IDs indicate the disease status: black, control; yellow, HD grade 2; orange, HD grade 3; red, HD grade 4. Each row of the heat map represents the Z-score transformed FPKM values of one differentially expressed gene across all samples. (B) Diagram depicting gene ontology (GO) terms that are significantly enriched in the differentially expressed genes. Categories in red and orange are enriched in up-regulated genes in HD. Categories in blue are enriched in down-regulated genes in HD. (C) Cell type composition analysis of human BA4 motor cortex samples. Geometric mean of the FPKM values of cell type marker genes of each cell type is used to indicate the change in specific cell type.
We also evaluated the cell type composition of these brain samples, using cell-type-specific marker genes for major brain cell types collected from the literature (Material and Methods). We did not observe statistically significant changes in the geometric mean of FPKM values of marker genes for four major cell types (neuron, astrocyte, oligodendrocyte and microglia, Figure 1C), consistent with the expectation that there is no overt neuronal cell loss in the BA4 region (6).
Finally, we assessed the concordance of our differential gene expression analysis based on RNA-seq data, with prior studies of HD transcriptomes using gene expression microarray. Specifically, we compared our results to the study by Hodges and colleagues, which provides the most comparable dataset with microarray profiling of BA4 motor cortex (6). Potentially due to the difference in experimental design including gene expression profiling platform (microarray vs RNA-seq) and sample size (31 vs 14), the numbers of differentially expressed genes identified between HD and control brains were quite different between these two datasets (1,374 by Hodges et al., and 222 in our study). 39 differentially expressed genes overlapped between the two datasets, representing a significant overlap over random expectation (P = 0.0002, Fisher exact test). We also performed a Gene Ontology analysis of the differentially expressed genes in the Hodges data set using Enrichr (61), and obtained similar enriched GO terms in HD-upregulated genes as in the current study using the same adjusted p-value and combined score cut-off (Supplementary Material, Table S3), indicating similarities between these two studies at the functional level.
Transcriptome-wide analysis of aberrant alternative splicing in HD brains
To discover and characterize differential alternative splicing between HD and control brains, we applied our RNA-seq splicing analysis software rMATS (62) to the RNA-seq data with a cutoff at FDR < 5% and |ΔPSI| ≥5% (see Materials and Methods). A total of 593 significant differential AS events were identified between the HD and control brains (Supplementary Material, Table S4), covering five basic types of AS patterns with cassette exon skipping (skipped exon, SE) being the majority of the AS events affected (Figure 2A–C), indicating widespread changes in AS in HD brains.
Figure 2.
Aberrant splicing program in Huntington's disease. (A) Differential alternative splicing events between HD and control motor cortex (BA4) samples. (B) Heat map of 316 differentially skipped exons in HD based on PSI (Percent Spliced In) values in HD and control motor cortex (BA4) samples. Color bars above sample IDs indicate the disease status: black, control; yellow, HD grade 2; orange, HD grade 3; red, HD grade 4. Each row of the heat map represents the Z-score transformed PSI values of one differentially spliced exon across all samples. (C) Examples of differential AS events in genes with neuronal functions. Box plots of PSI values of each alternative splicing event in 7 CTRL vs 7 HD samples. PSI values are represented by dots and dark horizontal lines represent the mean, with the box representing the 25th and 75th percentiles, the whiskers representing the 5th and 95th percentiles. (D) Significantly enriched binding sites of splicing factors and other RNA binding proteins in differentially skipped exons between HD and control samples. Gene symbols of splicing factors are followed by the consensus binding motifs and their Fisher’s exact test (one-sided) p-values (adjusted for multiple testing). IUPAC Ambiguity Codes were used to indicate motif patterns.
We found numerous events of differential AS in genes with important neuronal functions (see Table 2 and Figure 2C for a selected list). Here we briefly describe three examples with potential functional implications. DPF2 (D4, zinc and double PHD fingers family 2) is a transcription factor required for the apoptotic response following deprivation of survival factors. An alternative exon (exon 7) inserting 14 amino acids (aa) after aa position 212 showed decreased inclusion levels in HD brains. The position of the insertion falls within the C2H2-type zinc finger domain (aa 209-232), which may potentially change the binding property of the protein (63). Interestingly and with relevance to HD, DPF2 antagonizes PGC1α interaction with ERRα (64). Earlier work showed that the induction of PGC1α expression is intact in the N171-82Q mouse model of HD, but its transcriptional activation activity is impaired, as evidenced from decreased levels of target genes (65). It is possible that the elevated levels of the exon 7 skipping isoform of DPF2 may contribute to impaired PGC1α activity in HD brains. A second example is the gene Dystonin (DST). DST belongs to the plakin protein family of adhesion junction plague proteins. Mice deficient for DST develop neurodegeneration (66). In this gene exon 90 encodes 24aa near the C terminus of a neuronal variant of DST also known as BPAG1eA (67). Exon 90 was significantly reduced in HD brains. A third example is in SORBS1 (sorbin and SH3 domain containing 1), a gene encoding a CBL-associated protein that functions in the signalling and stimulation of insulin. Splice variants of SORBS1 have been reported to interact with ataxin-7 (68) and Huntingtin (69). The inclusion level of an alternatively spliced exon (exon 3) encoding a 32aa peptide (aa 26–57) is elevated in HD brain. The exon 3 inclusion isoform is designated as a foetal brain specific isoform when compared to the brain specific exon 3 skipping isoform (70). Decreased glucose metabolism and insulin signaling has been indicated in Huntington’s disease (71). It remains to be determined if the increase in foetal brain specific SORBS1 contributes to altered activities of the insulin signalling pathway in HD.
Table 2.
Examples of differential AS events in genes with neuronal functions.
| Gene Symbol | Ensembl Transcript ID | PSIa HD | PSI CTRL | Delta PSIb | Notes |
|---|---|---|---|---|---|
| (Exon number) | |||||
| DPF2 | ENST00000252268 (7) | 68% | 79% | −11% | Antagonize PGC1 alpha function |
| DST | ENST00000421834 (90) | 63% | 76% | −13% | Mice deficient for DST develop neurodegeneration |
| SORBS1 | ENST00000277982 (3) | 30% | 14% | 15% | A CBL-associated protein that functions in the signaling and stimulation of insulin |
| PTPRF | ENST00000359947 (12) | 15% | 6% | 9% | Function in the regulation of epithelial cell-cell contacts at adherens junctions, as well as in the control of beta-catenin signaling |
| PTPRS | ENST00000262963 (15) | 25% | 15% | 10% | Molecular control of adult nerve repair |
| ARAP2 | ENST00000303965 (24) | 89% | 98% | −9% | Associates with focal adhesions and functions downstream of RhoA to regulate focal adhesion dynamics |
aPSI: Percent Spliced In; bDelta PSI: PSI(HD-CTRL).
To investigate the potential mechanisms for widespread changes in AS in HD brains, we next sought to identify trans-acting splicing factors potentially responsible for these changes. Splicing factors are RNA-binding proteins that interact with splicing regulatory elements in exons or introns to regulate AS (48). We performed motif scan (Material and Methods) and identified 15 motifs in the exon bodies and one motif within 250nt downstream intronic sequences that were enriched among the differentially spliced exons as compared to background (control) exons (Figure 2D). For example, the motif of the splicing factor SRSF1 (also known as ASF/SF2) is significantly enriched in the exon bodies of the exons with elevated inclusion levels in HD, and in the downstream intronic sequences of the exons with reduced inclusion levels in HD. The motif of the splicing factor polypyrimidine tract binding protein 1 (PTBP1) is significantly enriched in the exon bodies of exons with elevated inclusion levels in HD. However, none of these splicing factors with enriched binding motifs showed significant changes in gene expression in the RNA-seq data of the seven controls and seven HD brain samples (Supplementary Material, Table S2), which could be due to the limited sample size as well as measurement noise in the RNA-seq gene expression data. For example, PTBP1 had higher average RNA-seq FPKM values in HD brains as compared to controls, but the change was not called significant (P = 0.1).
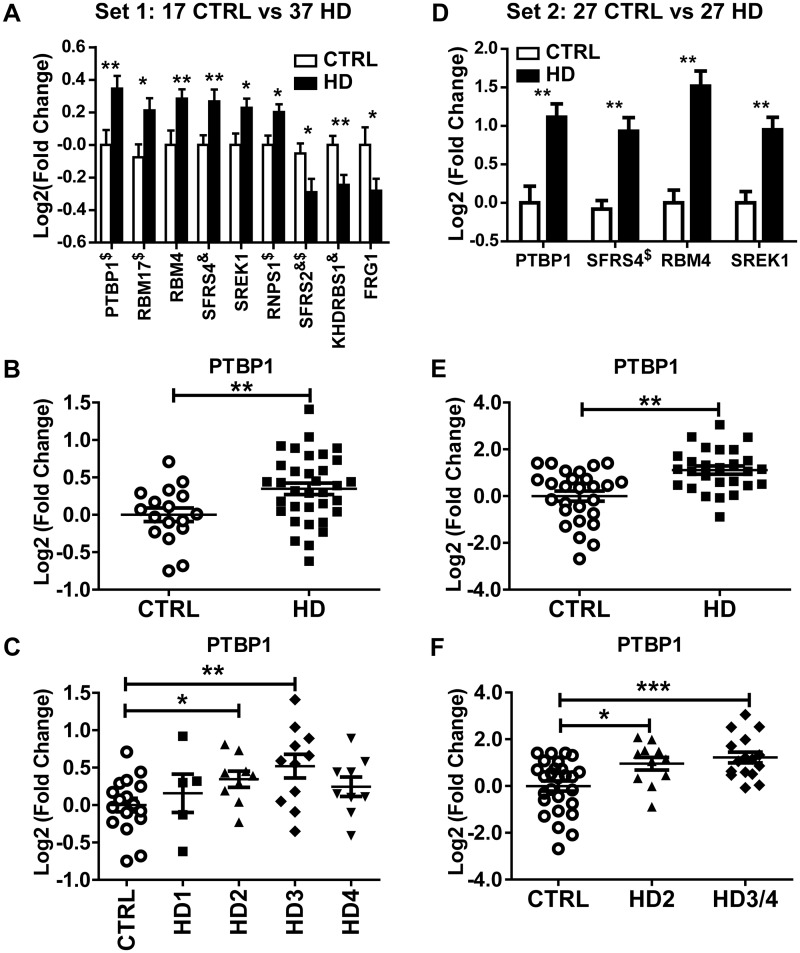
To further test the hypothesis that alternative splicing changes in the HD brains are associated with the gene expression changes of splicing factors, we performed real-time qPCR quantification of the expression levels of 64 splicing factors/RNA-binding proteins (Supplementary Material, Table S5) in a much more expanded patient cohort of 17 healthy controls and 37 patients with HD covering all stages of the disease (Sample set 1; Table 1, Supplementary Material, Table S1). Nine RNA binding proteins exhibited significantly altered expression level in HD patients (nominal P < 0.05), with six significantly upregulated and three significantly downregulated (Figure 3A). PTBP1 has one of the most significant changes between control and HD brains (Figure 3A–C) and is the only one with enriched binding motif in differentially spliced exons between HD and control brains (Figure 2D). To further validate these results, we repeated the qRT-PCR analysis of the nine significantly changed RNA-binding proteins in an independent set of human BA4 samples of 27 control and 27 HD brains (Sample set 2, mostly grade 2 and 3) (Table 1, Supplementary Material, Table S1) without any overlap with the Sample set 1. Four RNA binding proteins were validated to be also significantly changed (upregulated) in this new sample set (Figure 3D), including PTBP1 (Figure 3E–F). We also noticed that PTBP1 expression level showed significant increase as early as in grade 2 HD (Figure 3C, F).
Figure 3.
qRT-PCR analysis of splicing factors and RNA binding proteins in two independent sets of control and HD motor cortex (BA4) samples: Set 1 (17 control and 37 HD) (A-C); Set 2 (27 control and 27 HD) (D-F). (A) and (D) Splicing factors and RNA binding proteins with significantly altered gene expression. (B) and (E) PTBP1 expression comparison between control and HD brains. (C) and (F) PTBP1 expression comparison among control and different HD grades. Lines indicate group means. Error bars indicate ±SEM. ∗ Nominal P < 0.05; ∗∗ P <0.01, ∗∗∗ P < 0.001. $: Genes with outlier data point detected and excluded from analysis; &: Genes with unequal variance and p-value calculated using unpaired t-test with Welch's correction.
Quantitative analyses of PTBP1-regulated alternative splicing events in HD brains
Multiple lines of evidence including motif enrichment analysis and qRT-PCR analysis pointed to the splicing factor PTBP1 as potentially playing an important role in AS changes in HD brains. PTBP1 is a tissue-specific splicing factor that generally acts as a suppressor of neuronal-specific alternative splicing events (46,72). A prior gene expression microarray study of the BA4 brain region indicated that PTBP1 has elevated mRNA levels in HD patients (6). We also noticed that exon 7 of DPF2 (Table 2, Figure 2C), which had reduced inclusion levels in HD brains, is a known exon target repressed by PTBP1 (73).
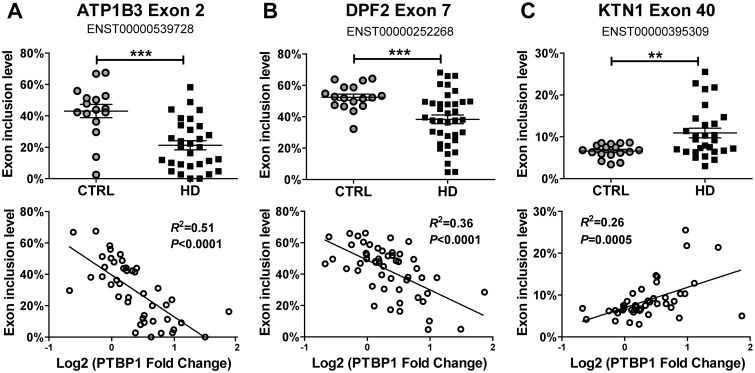
To further assess the downstream transcriptome impact of elevated PTBP1 expression in HD brains, we utilized a fluorescently labelled RT-PCR protocol to quantitatively assay for exon inclusion levels of known PTBP1 target exons. Since no global profiling studies have been done to determine exon targets of PTBP1 in human brains, we selected the comprehensive list generated earlier by Llorian and colleagues (73). Their study used a high-density exon and splice junction microarray to identify AS events regulated upon PTBP1 knockdown in HeLa cells. We selected 32 exons with a varying range of prediction confidence scores for PTBP1 targeting (8 highest, 11 high, 2 medium high, 2 medium, 8 medium low, and 1 low, Table 3) and designed RT-PCR primers to quantify splicing levels of these exons. For each exon, we tested if exon inclusion levels showed a strong correlation with PTBP1 expression levels across the 54 brain samples (Sample set 1, 17 control and 37 HD brains), which would suggest regulation by PTBP1, and secondly if splicing levels were significantly different between the control and HD brains. Of the 32 exons we assayed, 25 showed a significant correlation (nominal P < 0.05) with PTBP1 gene expression levels, among which 11 events showed a significant change (nominal P < 0.05) in transcript inclusion levels between HD patients and controls. Three examples of these findings are presented in Figure 4. Exon 2 of ATP1B3 has reduced splicing levels in HD brains, consistent with the elevated expression of PTBP1 in HD patients (Figure 4A). The splicing levels of this exon had a strong negative correlation with PTBP1 gene expression levels across all samples (R2 = 0.51, P < 0.0001). A similar pattern was also observed for the known PTBP1-repressed exon in DPF2 (Figure 4B).
Table 3.
Splicing changes of putative PTBP1 target exons in HD brains measured by fluorescently labeled quantitative RT-PCR.
| Gene Symbol | Exon Genome Coordinates UCSC hg19 (Feb 2009) | Exon Inclusion Change in HD |
Correlation Between Target Exon Inclusion Level And PTBP1 Gene Expression Level |
||||
|---|---|---|---|---|---|---|---|
| Direction | Nominal p-Value | Correlation direction | Nominal p-Value | r2 | Confidence in Llorian 2010 | ||
| CLASP1 | chr2:122203025-122203072 | Up | 0.0123** | Positive | <0.0001 | 0.30 | Med low |
| KTN1 | chr14:56139890-56139973 | Up | 0.0011** | Positive | 0.0005 | 0.26 | High |
| ATP1B3 | chr3:141620978-141621069 | Down | <0.0001 | Negative | <0.0001 | 0.51 | High |
| RPN2 | chr20:35866805-35866852 | Down | 0.0071 | Negative | <0.0001 | 0.48 | Highest |
| DPF2 | chr11:65112051-65112092 | Down | 0.0001** | Negative | <0.0001 | 0.36 | Highest |
| USP5 | chr12:6972473-6972541 | Down | 0.0015 | Negative | <0.0001 | 0.35 | High |
| AP2B1 | chr17:33997876-33997917 | Down | 0.0022** | Negative | < 0.0001 | 0.33 | Highest |
| EHBP1 | chr2:63215066-63215173 | Down | 0.0335 | Negative* | <0.0001 | 0.32 | Med low |
| MACF1 | chr1:39935219-39935287 | Down | 0.0295 | Negative | <0.0001 | 0.31 | Med low |
| ACTN1 | chr14:69345175-69345240 | Down | 0.0092 | Negative | <0.0001 | 0.31 | High |
| ABLIM1 | chr10:116213138-116213242 | Down | 0.0135 | Negative | 0.001 | 0.21 | Highest |
| PC | chr11:66721720-66721907 | Trending down | 0.0618 | Negative | 0.0005 | 0.26 | Med low |
| SNX14 | chr6:86248556-86248582 | Trending down | 0.0629 | Negative | 0.0003 | 0.25 | High |
| ATP2B4 | chr1:203702351-203702528 | NC | 0.1078 | Negative | < 0.0001 | 0.34 | Highest |
| BCLAF1 | chr6:136588167-136588313 | NC | 0.2021 | Negative | <0.0001 | 0.29 | Med low |
| CNNM2 | chr10:104831531-104831596 | NC | 0.3289 | Negative | 0.0002 | 0.26 | Med |
| ROD1 | chr9:115092721-115092754 | NC | 0.1173 | Negative | 0.0003 | 0.24 | Highest |
| ABI2 | chr2:204261508-204261690 | NC | 0.2816 | Negative | 0.0005 | 0.22 | High |
| APP | chr21:27369675-27369731 | NC | 0.1914 | Negative | 0.0009 | 0.21 | Highest |
| SYNJ1 | chr21:34051112-34051120 | NC | 0.4163 | Negative | 0.0065 | 0.16 | Med low |
| FMNL2 | chr2:153499933-153500058 | NC | 0.1496 | Negative | 0.0061 | 0.14 | Highest |
| KIF1B | chr1:10386169-10386417 | NC | 0.6374 | Negative | 0.0223 | 0.12 | Med low |
| SMARCC2 | chr12:56566721-56566813 | NC | 0.2357 | Negative | 0.0129 | 0.12 | Med high |
| LIMCH1 | chr4:41689857-41689934 | NC | 0.9682 | Negative | 0.0235 | 0.12 | High |
| EXOC7 | chr17:74087224-74087316 | NC | 0.2418 | Negative | 0.0405 | 0.09 | Med |
| DCUN1D5 | chr11:102953477-102953568 | NC | 0.1194** | No correlation | 0.214 | 0.03 | Low |
| HRB | chr2:228396854-228396925 | NC | 0.51** | No correlation | 0.371 | 0.02 | High |
| GANAB | chr11:62401782-62401847 | NC | 0.8245 | No correlation | 0.5411 | 0.01 | High |
| FAM21C | chr10:51886920-51886982 | NC | 0.1878 | No correlation | 0.6085 | 0.01 | Med low |
| PTBP2 | chr1:97271975-97272008 | NC | 0.2335 | No correlation | 0.7785 | 0.00 | Med high |
| FMNL2 | chr2:153501948-153502041 | NC | 0.2281 | No correlation | 0.7795 | 0.00 | High |
| TBC1D1 | chr4:38053520-38053681 | NC | 0.2787 | No correlation | 0.9883 | 0.00 | High |
Opposite direction from Llorian 2010
Unpaired t-test with Welch's correction, data with significantly different variances
NC: No Change
Figure 4.
Splicing change of putative PTBP1 target exons in HD and the correlation between exon inclusion level and PTBP1 gene expression level (log2(PTBP1 fold change)). (A) ATP1B3 exon 2 inclusion level is negatively correlated with PTBP1 gene expression level. (B) DPF2 exon 7 inclusion level is negatively correlated with PTBP1 gene expression level. (C) KTN1 exon 40 inclusion level is positively correlated with PTBP1 gene expression level. Error bars indicate ±SEM. ∗∗ Nominal P < 0.01; ∗∗∗ P < 0.001.
Although PTBP1 generally acts as a splicing repressor, it should be noted that certain exons can be positively regulated by PTBP1 (73). For example, exon 40 of KTN1 was previously identified to be up-regulated by PTBP1 (73). We found that this exon had elevated splicing levels in HD brains (Figure 4C), and the splicing levels of this exon showed a significant positive correlation with PTBP1 gene expression levels across all samples (R2 = 0.25, P = 0.0005). Although the confidence of target prediction in HeLa cells (73) does not necessarily correlate with changes of exon inclusion levels in human motor cortex (BA4 region), it is interesting to note that a greater proportion of the differential exon splicing events identified by RT-PCR had high or highest prediction confidence scores (8 out of 11) as compared to non-differential splicing events (11 in 21). However, not all previously confirmed PTBP1 target exons in HeLa cells showed differential splicing in HD brains. This discrepancy could be due to cell type-specific splicing regulation by PTBP1, or the dosage-dependent effects of PTBP1 on its target exons, which reflects the caveats of using putative PTBP1 target exons identified via knockdown in HeLa cells.
Discussion
Many cellular dysfunctions in HD have been attributed to alterations in gene expression (33). The last 15 years have seen multiple HD animal models, inducible cell-based systems, and postmortem tissues from patients with HD subject to transcriptome profiling experiments (6,18–33). Barring a few exceptions, the majority has relied extensively on microarray technology with the goal of discerning gene expression differences. This has led to a wealth of knowledge and a better understanding of disease pathogenesis. In mice, profiling studies have identified significant transcriptional differences at minimally symptomatic stages at the phenotypical and morphological level that worsen with disease progression (18,19,23,36,74–76). Additionally, there is substantial evidence that transcriptomic differences occur well before cell loss, mutant polyglutamine HTT aggregation, or mitochondrial dysfunction (20,77–79), and that individual neurons from early symptomatic brain samples mimic the altered transcriptome obtained from whole homogenates (6). These studies and others that correlate gene expression with phenotype (80) suggest that transcriptional dysregulation is a major contributor of HD pathogenesis (19,79,81,82).
In this work, we sought to identify alternative splicing changes in HD by RNA-seq. We identified 593 differential AS events between HD and control samples. This number was comparable to the number of differential AS events in our previous RNA-seq studies with perturbation of master splicing factors (62,83–89), suggesting widespread dysregulation of AS in HD. We also identified splicing factors whose RNA motifs were significantly enriched within or around these differential AS events, suggesting that they played important roles in aberrant splicing in HD. This could be due to differential expression of the splicing factors in HD brains (as is the case for PTBP1), or differential activity of the splicing factors at the protein level. We know that compared to wildtype HTT, mutant HTT exhibits a significantly different protein-protein interaction profile with a number of RNA binding proteins (90). It is possible that the mutant HTT may affect the protein activity or availability of certain splicing factors via protein–protein interactions.
Using two expanded panels of postmortem brains from HD patients and healthy controls, we quantitatively measured the mRNA levels of 64 RNA binding proteins including many well-known splicing factors. We identified 4 RNA binding proteins with significant differential expression (P < 0.05) in HD brains in both sample sets. Among these, the tissue-specific splicing factor PTBP1 showed one of the most significant changes in differential expression even in early stages (grade 2) of the disease (see Figure 3), a pattern also corroborated by prior microarray studies in human HD brains (6). Quantitative splicing profiling of 32 putative PTBP1 target exons in HD and control brains further confirmed the role of PTBP1 in driving aberrant AS patterns in HD. Interestingly, increased activity of PTBP1 in the brain has been associated with aging and observed in other neurodegenerative diseases such as frontotemporal lobar degeneration (FTLD) and Alzheimer's disease (AD) (91). Collectively, these data suggest PTBP1 may underlie a common transcriptome and splicing signature of neurodegenerative diseases.
Our study is a transcriptome-wide RNA-seq evaluation of splicing dysregulation in human HD brains. While our RNA-seq analysis was conducted on a cohort of limited sample size (7 HD and 7 control), our sequencing coverage on individual samples is high (128-175 million reads per sample). The small sample size, however, limits power and as such the data is more prone to RNA-seq noise or influence by outlier measurements. A study on a larger sample size may capture additional relevant AS events in HD that extend beyond those emerging from this important initial data set. We should also note that our analysis was conducted on bulk brain tissues, in which cell type composition always represents a potential confounding issue. In our RNA-seq data set, between HD and control brains we did not find any significant change in the overall expression levels of cell-type-specific marker genes for four major cell types (neuron, astrocyte, oligodendrocyte and microglia), consistent with the previous report that there is no overt neuronal cell loss in the BA4 region (6). However, we did observe a trend for neuron marker genes to go down slightly and astrocyte marker genes to go up slightly in HD, although neither was significant, thus, it remains possible that some of the identified splicing changes are at least in part contributed by shifts in cell-type composition. In the future, the use of single cell transcriptome analysis technologies such as Drop-seq (92) may allow us to better interrogate cell-type-specific HD transcriptome changes. It would also be useful to assess the regional differences in HD associated alternative splicing changes, using RNA-seq data from other brain regions (93). Future studies should also investigate the functional roles of the differentially expressed splicing factors and altered splicing events in HD pathogenesis.
Materials and Methods
Brain tissues
Postmortem brain samples were obtained from the New York Brain Bank (Columbia University, New York, NY), the Harvard Brain Tissue Resource Center (Belmont, MA), the New Zealand Neurological Foundation Brain Bank (University of Auckland, Auckland, New Zealand), and The Johns Hopkins University (Baltimore, MD). Detailed sample information for Brodmann area 4 (BA4) motor cortex is described in Table 1 and Supplementary Material, Table S1.
Total RNA preparation and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Single-pass cDNA was synthesized using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) according to manufacturer’s instructions. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA). qRT- PCR primers were designed using PRIMER3 (94). Primer sequences are described in Supplementary Material, Table S6. A panel of nine housekeeping endogenous control genes (Supplementary Material, Table S6) were used to normalize tested gene expression level. Gene expression levels were pre-analyzed using REST 2009 software (Qiagen, Valencia, CA) and a subset of 6 reference genes, TFRC, PPIA, HMBS, GAPDH, ALAS1 and ACTB were selected to be used in the final analysis. Average gene expression level in all control individuals were set to be 1. Log2 (expression fold change) were plotted and analyzed using GraphPad software (La Jolla, CA). Outliers were detected using outlier-labelling rule described in (95). Specifically, any data point outside of the interval (Q1-g(Q3-Q1), Q3 + g(Q3-Q1)) (Q1: 25th percentile, Q3: 75th percentile, g = 1.5) was marked as outlier and excluded from further analysis. When outlier was detected and excluded, the average of log2 (expression fold change) of the control sample was not reset to 0. Instead, a new re-calculated value was kept to reflect the exclusion of outlier data points. Two-tailed unpaired t-test was used to determine the P-value of log2 (expression fold change) between the control group (CTRL) and the HD group (HD). If F test detected the variance to be significantly different between the two sample groups, P-value was recalculated using the two-tailed unpaired t-test with Welch’s correction to account for the unequal variances.
RNA-seq analysis
RNA-seq libraries were prepared using TruSeq Stranded mRNA Sample Prep Kit (Illumina) and sequenced on an Illumina HiSeq 2000. The entire RNA-seq dataset is available at the Gene Expression Omnibus (GEO) database with the accession number GSE79666. We mapped RNA-seq reads to the human genome (hg19) and transcriptome (Ensembl, release 72) using the software TopHat (v1.4.1) allowing up to 3 bp mismatches per read and up to 2 bp mismatches per 25 bp seed. We used Cuffdiff (v2.2.0) to calculate RNA-seq based gene expression levels using the FPKM metric (fragments per kilobase of exon per million fragments mapped) (96). We also used Cuffdiff to identify 222 differentially expressed genes between control and HD samples (Supplementary Material, Table S2) using the cutoff of Cuffdiff FDR ≤ 25%, an extra filter of p-value ≤0.05 from two-sided t-test on FPKM values, and maximum average FPKMs ≥ 0.1 in at least one of the two sample groups. We then used rMATS (version3.0.8) (62) to identify differential alternative splicing (AS) events between the two sample groups corresponding to all five basic types of AS patterns. Briefly, rMATS uses a modified version of the generalized linear mixed model to detect differential AS from RNA-seq data with replicates, while controlling for changes in overall gene expression levels. It accounts for exon-specific sequencing coverage in individual samples as well as variation in exon splicing levels among replicates. For each AS event, we used both the reads mapped to the splice junctions and the reads mapped to the exon body as the input for rMATS. We ran rMATS with -c 0.0001 parameter and then detected significant splicing events using a cutoff at FDR < 5% and |deltaPSI| ≥5% (Supplementary Material, Table S4).
Cell type composition analysis
A list of ∼6,000 cell-type-specific genes for astrocytes, neurons, and oligodendrocytes were collected from (97). From this list of genes, we selected 173 astrocyte specific genes, 300 neuron-specific genes, and 119 oligodendrocyte-specific genes that showed at least 10-fold enrichment in this study. Since this study was done in mouse, we assigned corresponding human orthologs before computing the FPKMs of these genes. We also used two marker genes from Kuhn et al. (98) for the microglia cell population. We computed the geometric mean of FPKM values of the selected cell-type-marker genes for each cell type and used it as the indicator to detect changes in specific cell populations. Genes with FPKM = 0 in at least one sample was excluded due to the requirement for calculating geometric mean. Final numbers of cell type marker genes used are shown in Figure 1C. Data were plotted and analyzed using GraphPad software (La Jolla, CA). No outlier is detected using outlier-labelling rule described in (95). Specifically, any data point outside of the interval (Q1-g(Q3-Q1), Q3 + g(Q3-Q1)) (Q1: 25th percentile, Q3: 75th percentile, g = 2.2 for small sample size) was marked as outlier and excluded from further analysis. Two-tailed unpaired t-test was used to determine the p-value of the geometric mean of FPKM values between the control group (CTRL) and the HD group (HD). No unequal variances between the two sample groups were detected using F test.
Enrichment analysis of gene ontology (GO) terms
Enrichr (http://amp.pharm.mssm.edu/Enrichr/) was used to generate lists of Gene Ontology terms enriched in the upregulated, downregulated or differentially spliced gene lists (61). Default setting of Enrichr was used. Among all GO terms with adjusted p-value <0.05 (corrected for multiple hypothesis testing), only the ones with “Combined score” (c = log(p)*z; c, combined score; p, p-value; z, rank score or z-score, deviation from an expected rank) of 15 and greater from the differentially expressed genes were listed.
Motif enrichment analysis
We sought to identify binding sites of splicing factors and other RNA binding proteins that were significantly enriched in differential exon skipping events between HD and CTRL samples as compared to background (non-regulated) alternative exons. We collected 112 known binding sites (motifs) of human RNA binding proteins including many well-characterized splicing factors from the literature (77–81). The full list of motifs and their regular expression patterns we used can be found in (99) (http://rmaps.cecsresearch.org/Help/RNABindingProtein). The motif analysis followed our previously described procedure (83,84). Briefly, for each motif, we scanned based on a regular expression match for its occurrence in three separate regions: intronic regions 250 bp upstream of an exon, exon regions, and intronic regions 250 bp downstream of an exon. For intronic sequences, we excluded the 20 bp sequence within the 3’ splice site and the 6 bp sequence within the 5’ splice site. To determine motif enrichment, we counted the number of occurrences in the differentially spliced exons and the background exons for each motif in three separate regions. The background exons consisted of 6,241 exons without splicing changes (rMATS FDR > 50%) in highly expressed genes (FPKM > 5.0). Each motif would have its counts in target AS exons and in background exons in three separate regions. Using these counts, we calculated a P-value for motif enrichment via Fisher’s exact test (one-sided, target exons vs. background exons) in three separate regions per motif. A Benjamini-Hochberg FDR correction was used to adjust for multiple testing for exons, upstream introns, and downstream introns separately.
Fluorescently labelled RT–PCR
For each tested exon, we designed a pair of forward and reverse PCR primers targeting flanking constitutive exons using PRIMER3 (100,101) (Supplementary Material, Table S7). A 22 nt universal tag sequence (5′-CGTCGCCGTCCAGCTCGACCAG-3′) was added to the 5′ end of the gene-specific forward primer during oligo synthesis. A fluorescently labelled universal primer (5′-FAM-CGTCGCCGTCCAGCTCGACCAG-3′) was used as the third primer in PCR (102). Then, PCR reaction was carried out for 29-35 cycles (optimized for each exon). The reaction products were resolved on 5% TBE urea-PAGE gels. The fluorescent signal was captured by Typhoon 9200 (Molecular Dynamics, Sunnyvale, CA, USA) and quantified using the Quantity One 4.6.2 software (Bio-Rad, Hercules, CA, USA). Alternatively, the reaction products were ran on a 3730 capillary DNA analyzer (Applied Biosystems, Foster City, CA) and analyzed using GeneMapper software (Applied Biosystems, Foster City, CA). HD vs control comparison of exon inclusion levels was carried out using the same procedure as the qRT-PCR analysis of splicing factor expression level. The correlation between exon inclusion level and log2 (PTBP1 expression fold change) was plotted using GraphPad software (La Jolla, CA) and statistics were done using the built-in linear regression analysis. R2 represented the goodness of fit for the linear regression and p-value was for the deviation from zero of the slope.
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
We thank Mallory Stroik, Sara Miller, and Jennifer Dozier for technical assistance.
Conflict of Interest statement. None declared.
Funding
This work was supported by the National Institutes of Health (R01NS076631 to Y.X., B.L.D., and C.A.R., P50 AG005146 to O.P. and J.T.), CHDI Foundation (to Y.X. and B.L.D.), New Zealand Neurological Foundation (to R.L.F), the BrightFocus Foundation (to O.P. and J.T.), and the Roy J. Carver Trust (to B.L.D.). Y.X. is supported by an Alfred Sloan research fellowship. S.S. is supported by an HDSA Human Biology fellowship.
Footnotes
Present address: Department of Computer Engineering and Computer Science, KBRIN Bioinformatics Core, University of Louisville, Louisville, KY, USA
References
- 1.(1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell, 72, 971–983. [DOI] [PubMed] [Google Scholar]
- 2.Ross C.A., Shoulson I. (2009) Huntington disease: pathogenesis, biomarkers, and approaches to experimental therapeutics. Parkinsonism Relat Disord., 15 Suppl 3, S135–S138. [DOI] [PubMed] [Google Scholar]
- 3.Ross C.A., Tabrizi S.J. (2011) Huntington's disease: from molecular pathogenesis to clinical treatment. Lancet Neurol., 10, 83–98. [DOI] [PubMed] [Google Scholar]
- 4.Williams A.J., Paulson H.L. (2008) Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci., 31, 521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo Z., Rudow G., Pletnikova O., Codispoti K.E., Orr B.A., Crain B.J., Duan W., Margolis R.L., Rosenblatt A., Ross C.A., et al. (2012) Striatal neuronal loss correlates with clinical motor impairment in Huntington's disease. Mov Disord., 27, 1379–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hodges A., Strand A.D., Aragaki A.K., Kuhn A., Sengstag T., Hughes G., Elliston L.A., Hartog C., Goldstein D.R., Thu D., et al. (2006) Regional and cellular gene expression changes in human Huntington's disease brain. Hum Mol Genet., 15, 965–977. [DOI] [PubMed] [Google Scholar]
- 7.Thu D.C., Oorschot D.E., Tippett L.J., Nana A.L., Hogg V.M., Synek B.J., Luthi-Carter R., Waldvogel H.J., Faull R.L. (2010) Cell loss in the motor and cingulate cortex correlates with symptomatology in Huntington's disease. Brain, 133, 1094–1110. [DOI] [PubMed] [Google Scholar]
- 8.Unschuld P.G., Joel S.E., Pekar J.J., Reading S.A., Oishi K., McEntee J., Shanahan M., Bakker A., Margolis R.L., Bassett S.S., et al. (2012) Depressive symptoms in prodromal Huntington's Disease correlate with Stroop-interference related functional connectivity in the ventromedial prefrontal cortex. Psychiatry Res., 203, 166–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benn C.L., Sun T., Sadri-Vakili G., McFarland K.N., DiRocco D.P., Yohrling G.J., Clark T.W., Bouzou B., Cha J.H. (2008) Huntingtin modulates transcription, occupies gene promoters in vivo, and binds directly to DNA in a polyglutamine-dependent manner. J. Neurosci., 28, 10720–10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kegel K.B., Meloni A.R., Yi Y., Kim Y.J., Doyle E., Cuiffo B.G., Sapp E., Wang Y., Qin Z.H., Chen J.D., et al. (2002) Huntingtin is present in the nucleus, interacts with the transcriptional corepressor C-terminal binding protein, and represses transcription. J. Biol. Chem., 277, 7466–7476. [DOI] [PubMed] [Google Scholar]
- 11.Labbadia J., Morimoto R.I. (2013) Huntington's disease: underlying molecular mechanisms and emerging concepts. Trends Biochem. Sci., 38, 378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Augood S.J., Faull R.L., Emson P.C. (1997) Dopamine D1 and D2 receptor gene expression in the striatum in Huntington's disease. Ann. Neurol., 42, 215–221. [DOI] [PubMed] [Google Scholar]
- 13.Cha J.H., Kosinski C.M., Kerner J.A., Alsdorf S.A., Mangiarini L., Davies S.W., Penney J.B., Bates G.P., Young A.B. (1998) Altered brain neurotransmitter receptors in transgenic mice expressing a portion of an abnormal human huntington disease gene. Proc. Natl. Acad. Sci. U S A, 95, 6480–6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emson P.C., Arregui A., Clement-Jones V., Sandberg B.E., Rossor M. (1980) Regional distribution of methionine-enkephalin and substance P-like immunoreactivity in normal human brain and in Huntington's disease. Brain Res., 199, 147–160. [DOI] [PubMed] [Google Scholar]
- 15.Young A.B., Greenamyre J.T., Hollingsworth Z., Albin R., D'Amato C., Shoulson I., Penney J.B. (1988) NMDA receptor losses in putamen from patients with Huntington's disease. Science, 241, 981–983. [DOI] [PubMed] [Google Scholar]
- 16.Benn C.L., Landles C., Li H., Strand A.D., Woodman B., Sathasivam K., Li S.H., Ghazi-Noori S., Hockly E., Faruque S.M., et al. (2005) Contribution of nuclear and extranuclear polyQ to neurological phenotypes in mouse models of Huntington's disease. Hum. Mol. Genet., 14, 3065–3078. [DOI] [PubMed] [Google Scholar]
- 17.Schilling G., Savonenko A.V., Klevytska A., Morton J.L., Tucker S.M., Poirier M., Gale A., Chan N., Gonzales V., Slunt H.H., et al. (2004) Nuclear-targeting of mutant huntingtin fragments produces Huntington's disease-like phenotypes in transgenic mice. Hum. Mol. Genet., 13, 1599–1610. [DOI] [PubMed] [Google Scholar]
- 18.Luthi-Carter R., Strand A., Peters N.L., Solano S.M., Hollingsworth Z.R., Menon A.S., Frey A.S., Spektor B.S., Penney E.B., Schilling G., et al. (2000) Decreased expression of striatal signaling genes in a mouse model of Huntington's disease. Hum. Mol. Genet., 9, 1259–1271. [DOI] [PubMed] [Google Scholar]
- 19.Luthi-Carter R., Strand A.D., Hanson S.A., Kooperberg C., Schilling G., La Spada A.R., Merry D.E., Young A.B., Ross C.A., Borchelt D.R., et al. (2002) Polyglutamine and transcription: gene expression changes shared by DRPLA and Huntington's disease mouse models reveal context-independent effects. Hum. Mol. Genet., 11, 1927–1937. [DOI] [PubMed] [Google Scholar]
- 20.Sipione S., Rigamonti D., Valenza M., Zuccato C., Conti L., Pritchard J., Kooperberg C., Olson J.M., Cattaneo E. (2002) Early transcriptional profiles in huntingtin-inducible striatal cells by microarray analyses. Hum. Mol. Genet., 11, 1953–1965. [DOI] [PubMed] [Google Scholar]
- 21.Nucifora F.C., Jr., Sasaki M., Peters M.F., Huang H., Cooper J.K., Yamada M., Takahashi H., Tsuji S., Troncoso J., Dawson V.L., et al. (2001) Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science, 291, 2423–2428. [DOI] [PubMed] [Google Scholar]
- 22.Dunah A.W., Jeong H., Griffin A., Kim Y.M., Standaert D.G., Hersch S.M., Mouradian M.M., Young A.B., Tanese N., Krainc D. (2002) Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science, 296, 2238–2243. [DOI] [PubMed] [Google Scholar]
- 23.Luthi-Carter R., Hanson S.A., Strand A.D., Bergstrom D.A., Chun W., Peters N.L., Woods A.M., Chan E.Y., Kooperberg C., Krainc D., et al. (2002) Dysregulation of gene expression in the R6/2 model of polyglutamine disease: parallel changes in muscle and brain. Hum. Mol. Genet., 11, 1911–1926. [DOI] [PubMed] [Google Scholar]
- 24.Lieberman A.P., Harmison G., Strand A.D., Olson J.M., Fischbeck K.H. (2002) Altered transcriptional regulation in cells expressing the expanded polyglutamine androgen receptor. Hum. Mol. Genet., 11, 1967–1976. [DOI] [PubMed] [Google Scholar]
- 25.Thompson L.M. (2003) An expanded role for wild-type huntingtin in neuronal transcription. Nat. Genet., 35, 13–14. [DOI] [PubMed] [Google Scholar]
- 26.Ross C.A., Thompson L.M. (2006) Transcription meets metabolism in neurodegeneration. Nat. Med., 12, 1239–1241. [DOI] [PubMed] [Google Scholar]
- 27.Cui L., Jeong H., Borovecki F., Parkhurst C.N., Tanese N., Krainc D. (2006) Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell, 127, 59–69. [DOI] [PubMed] [Google Scholar]
- 28.Chen-Plotkin A.S., Sadri-Vakili G., Yohrling G.J., Braveman M.W., Benn C.L., Glajch K.E., DiRocco D.P., Farrell L.A., Krainc D., Gines S., et al. (2006) Decreased association of the transcription factor Sp1 with genes downregulated in Huntington's disease. Neurobiol. Dis., 22, 233–241. [DOI] [PubMed] [Google Scholar]
- 29.Marti E., Pantano L., Banez-Coronel M., Llorens F., Minones-Moyano E., Porta S., Sumoy L., Ferrer I., Estivill X. (2010) A myriad of miRNA variants in control and Huntington's disease brain regions detected by massively parallel sequencing. Nucleic Acids Res., 38, 7219–7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bithell A., Johnson R., Buckley N.J. (2009) Transcriptional dysregulation of coding and non-coding genes in cellular models of Huntington's disease. Biochem. Soc. Trans., 37, 1270–1275. [DOI] [PubMed] [Google Scholar]
- 31.Cha J.H. (2007) Transcriptional signatures in Huntington's disease. Prog. Neurobiol., 83, 228–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas E.A., Coppola G., Tang B., Kuhn A., Kim S., Geschwind D.H., Brown T.B., Luthi-Carter R., Ehrlich M.E. (2011) In vivo cell-autonomous transcriptional abnormalities revealed in mice expressing mutant huntingtin in striatal but not cortical neurons. Hum. Mol. Genet., 20, 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valor L.M. (2015) Transcription, epigenetics and ameliorative strategies in Huntington's Disease: a genome-wide perspective. Mol. Neurobiol., 51, 406–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oldham M.C., Langfelder P., Horvath S. (2012) Network methods for describing sample relationships in genomic datasets: application to Huntington's disease. BMC Syst. Biol., 6, 63.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langfelder P., Cantle J.P., Chatzopoulou D., Wang N., Gao F., Al-Ramahi I., Lu X.H., Ramos E.M., El-Zein K., Zhao Y., et al. (2016) Integrated genomics and proteomics define huntingtin CAG length-dependent networks in mice. Nat. Neurosci., 19, 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin J., Cheng Y., Zhang Y., Wood W., Peng Q., Hutchison E., Mattson M.P., Becker K.G., Duan W. (2012) Interrogation of brain miRNA and mRNA expression profiles reveals a molecular regulatory network that is perturbed by mutant huntingtin. J. Neurochem., 123, 477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narayanan M., Huynh J.L., Wang K., Yang X., Yoo S., McElwee J., Zhang B., Zhang C., Lamb J.R., Xie T., et al. (2014) Common dysregulation network in the human prefrontal cortex underlies two neurodegenerative diseases. Mol. Syst. Biol., 10, 743.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neueder A., Bates G.P. (2014) A common gene expression signature in Huntington's disease patient brain regions. BMC Med. Genomics, 7, 60.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ni Q., Su X., Chen J., Tian W. (2015) Prediction of Metabolic Gene Biomarkers for Neurodegenerative Disease by an Integrated Network-Based Approach. Biomed. Res. Int., 2015, 432012.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee S.T., Chu K., Im W.S., Yoon H.J., Im J.Y., Park J.E., Park K.H., Jung K.H., Lee S.K., Kim M., et al. (2011) Altered microRNA regulation in Huntington's disease models. Exp. Neurol., 227, 172–179. [DOI] [PubMed] [Google Scholar]
- 41.Soldati C., Bithell A., Johnston C., Wong K.Y., Stanton L.W., Buckley N.J. (2013) Dysregulation of REST-regulated coding and non-coding RNAs in a cellular model of Huntington's disease. J. Neurochem., 124, 418–430. [DOI] [PubMed] [Google Scholar]
- 42.Packer A.N., Xing Y., Harper S.Q., Jones L., Davidson B.L. (2008) The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J. Neurosci., 28, 14341–14346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J.H., Tecedor L., Chen Y.H., Monteys A.M., Sowada M.J., Thompson L.M., Davidson B.L. (2015) Reinstating aberrant mTORC1 activity in Huntington's disease mice improves disease phenotypes. Neuron, 85, 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet., 40, 1413–1415. [DOI] [PubMed] [Google Scholar]
- 45.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. (2008) Alternative isoform regulation in human tissue transcriptomes. Nature, 456, 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Black D.L., Grabowski P.J. (2003) Alternative pre-mRNA splicing and neuronal function. Prog. Mol. Subcell. Biol., 31, 187–216. [DOI] [PubMed] [Google Scholar]
- 47.Mazin P., Xiong J., Liu X., Yan Z., Zhang X., Li M., He L., Somel M., Yuan Y., Phoebe Chen Y.P., et al. (2013) Widespread splicing changes in human brain development and aging. Mol. Syst. Biol., 9, 633.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z., Burge C.B. (2008) Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA, 14, 802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cooper T.A., Wan L., Dreyfuss G. (2009) RNA and disease. Cell, 136, 777–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anthony K., Gallo J.M. (2010) Aberrant RNA processing events in neurological disorders. Brain Res., 1338, 67–77. [DOI] [PubMed] [Google Scholar]
- 51.Osborne R.J., Lin X., Welle S., Sobczak K., O'Rourke J.R., Swanson M.S., Thornton C.A. (2009) Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum. Mol. Genet., 18, 1471–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ranum L.P., Cooper T.A. (2006) RNA-mediated neuromuscular disorders. Annu. Rev. Neurosci., 29, 259–277. [DOI] [PubMed] [Google Scholar]
- 53.Li L.B., Yu Z., Teng X., Bonini N.M. (2008) RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature, 453, 1107–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clinton S.M., Haroutunian V., Davis K.L., Meador-Woodruff J.H. (2003) Altered transcript expression of NMDA receptor-associated postsynaptic proteins in the thalamus of subjects with schizophrenia. Am. J. Psychiatry, 160, 1100–1109. [DOI] [PubMed] [Google Scholar]
- 55.Dredge B.K., Polydorides A.D., Darnell R.B. (2001) The splice of life: alternative splicing and neurological disease. Nat. Rev. Neurosci., 2, 43–50. [DOI] [PubMed] [Google Scholar]
- 56.Licatalosi D.D., Darnell R.B. (2006) Splicing regulation in neurologic disease. Neuron, 52, 93–101. [DOI] [PubMed] [Google Scholar]
- 57.Schwab C., Arai T., Hasegawa M., Yu S., McGeer P.L. (2008) Colocalization of transactivation-responsive DNA-binding protein 43 and huntingtin in inclusions of Huntington disease. J. Neuropathol. Exp. Neurol., 67, 1159–1165. [DOI] [PubMed] [Google Scholar]
- 58.Ratovitski T., Chighladze E., Arbez N., Boronina T., Herbrich S., Cole R.N., Ross C.A. (2012) Huntingtin protein interactions altered by polyglutamine expansion as determined by quantitative proteomic analysis. Cell Cycle, 11, 2006–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fernandez-Nogales M., Cabrera J.R., Santos-Galindo M., Hoozemans J.J., Ferrer I., Rozemuller A.J., Hernandez F., Avila J., Lucas J.J. (2014) Huntington's disease is a four-repeat tauopathy with tau nuclear rods. Nat. Med., 20, 881–885. [DOI] [PubMed] [Google Scholar]
- 60.Sathasivam K., Neueder A., Gipson T.A., Landles C., Benjamin A.C., Bondulich M.K., Smith D.L., Faull R.L., Roos R.A., Howland D., et al. (2013) Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc. Natl. Acad. Sci. U S A, 110, 2366–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. (2013) Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics, 14, 128.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen S., Park J.W., Lu Z.X., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y. (2014) rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U S A, 111, E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang W., Xu C., Bian C., Tempel W., Crombet L., MacKenzie F., Min J., Liu Z., Qi C. (2011) Crystal structure of the Cys2His2-type zinc finger domain of human DPF2. Biochem. Biophys. Res. Commun., 413, 58–61. [DOI] [PubMed] [Google Scholar]
- 64.Nilsson S., Makela S., Treuter E., Tujague M., Thomsen J., Andersson G., Enmark E., Pettersson K., Warner M., Gustafsson J.A. (2001) Mechanisms of estrogen action. Physiol. Rev., 81, 1535–1565. [DOI] [PubMed] [Google Scholar]
- 65.Weydt P., Pineda V.V., Torrence A.E., Libby R.T., Satterfield T.F., Lazarowski E.R., Gilbert M.L., Morton G.J., Bammler T.K., Strand A.D., et al. (2006) Thermoregulatory and metabolic defects in Huntington's disease transgenic mice implicate PGC-1alpha in Huntington's disease neurodegeneration. Cell. Metab., 4, 349–362. [DOI] [PubMed] [Google Scholar]
- 66.Guo L., Degenstein L., Dowling J., Yu Q.C., Wollmann R., Perman B., Fuchs E. (1995) Gene targeting of BPAG1: abnormalities in mechanical strength and cell migration in stratified epithelia and neurologic degeneration. Cell, 81, 233–243. [DOI] [PubMed] [Google Scholar]
- 67.Okumura M., Yamakawa H., Ohara O., Owaribe K. (2002) Novel alternative splicings of BPAG1 (bullous pemphigoid antigen 1) including the domain structure closely related to MACF (microtubule actin cross-linking factor). J. Biol. Chem., 277, 6682–6687. [DOI] [PubMed] [Google Scholar]
- 68.Jiang Y.J., Zhou C.J., Zhou Z.R., Wu M., Hu H.Y. (2013) Structural basis for recognition of the third SH3 domain of full-length R85 (R85FL)/ponsin by ataxin-7. FEBS Lett., 587, 2905–2911. [DOI] [PubMed] [Google Scholar]
- 69.Kaltenbach L.S., Romero E., Becklin R.R., Chettier R., Bell R., Phansalkar A., Strand A., Torcassi C., Savage J., Hurlburt A., et al. (2007) Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet., 3, e82.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lebre A.S., Jamot L., Takahashi J., Spassky N., Leprince C., Ravise N., Zander C., Fujigasaki H., Kussel-Andermann P., Duyckaerts C., et al. (2001) Ataxin-7 interacts with a Cbl-associated protein that it recruits into neuronal intranuclear inclusions. Hum. Mol. Genet., 10, 1201–1213. [DOI] [PubMed] [Google Scholar]
- 71.Lopes C., Ribeiro M., Duarte A.I., Humbert S., Saudou F., Pereira de Almeida L., Hayden M., Rego A.C. (2014) IGF-1 intranasal administration rescues Huntington's disease phenotypes in YAC128 mice. Mol. Neurobiol., 49, 1126–1142. [DOI] [PubMed] [Google Scholar]
- 72.Wagner E.J., Garcia-Blanco M.A. (2001) Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell Biol., 21, 3281–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Llorian M., Schwartz S., Clark T.A., Hollander D., Tan L.Y., Spellman R., Gordon A., Schweitzer A.C., de la Grange P., Ast G., et al. (2010) Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat. Struct. Mol. Biol., 17, 1114–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abou-Sleymane G., Chalmel F., Helmlinger D., Lardenois A., Thibault C., Weber C., Merienne K., Mandel J.L., Poch O., Devys D., et al. (2006) Polyglutamine expansion causes neurodegeneration by altering the neuronal differentiation program. Hum. Mol. Genet., 15, 691–703. [DOI] [PubMed] [Google Scholar]
- 75.Nguyen H.P., Metzger S., Holzmann C., Koczan D., Thiesen H.J., von Horsten S., Riess O., Bonin M. (2008) Age-dependent gene expression profile and protein expression in a transgenic rat model of Huntington's disease. Proteomics Clin. Appl., 2, 1638–1650. [DOI] [PubMed] [Google Scholar]
- 76.Valor L.M., Guiretti D., Lopez-Atalaya J.P., Barco A. (2013) Genomic landscape of transcriptional and epigenetic dysregulation in early onset polyglutamine disease. J. Neurosci., 33, 10471–10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kita H., Carmichael J., Swartz J., Muro S., Wyttenbach A., Matsubara K., Rubinsztein D.C., Kato K. (2002) Modulation of polyglutamine-induced cell death by genes identified by expression profiling. Hum. Mol. Genet., 11, 2279–2287. [DOI] [PubMed] [Google Scholar]
- 78.Wyttenbach A., Swartz J., Kita H., Thykjaer T., Carmichael J., Bradley J., Brown R., Maxwell M., Schapira A., Orntoft T.F., et al. (2001) Polyglutamine expansions cause decreased CRE-mediated transcription and early gene expression changes prior to cell death in an inducible cell model of Huntington's disease. Hum. Mol. Genet., 10, 1829–1845. [DOI] [PubMed] [Google Scholar]
- 79.Runne H., Regulier E., Kuhn A., Zala D., Gokce O., Perrin V., Sick B., Aebischer P., Deglon N., Luthi-Carter R. (2008) Dysregulation of gene expression in primary neuron models of Huntington's disease shows that polyglutamine-related effects on the striatal transcriptome may not be dependent on brain circuitry. J. Neurosci., 28, 9723–9731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hodges A., Hughes G., Brooks S., Elliston L., Holmans P., Dunnett S.B., Jones L. (2008) Brain gene expression correlates with changes in behavior in the R6/1 mouse model of Huntington's disease. Genes Brain Behav., 7, 288–299. [DOI] [PubMed] [Google Scholar]
- 81.Desplats P.A., Kass K.E., Gilmartin T., Stanwood G.D., Woodward E.L., Head S.R., Sutcliffe J.G., Thomas E.A. (2006) Selective deficits in the expression of striatal-enriched mRNAs in Huntington's disease. J. Neurochem., 96, 743–757. [DOI] [PubMed] [Google Scholar]
- 82.Kuhn A., Goldstein D.R., Hodges A., Strand A.D., Sengstag T., Kooperberg C., Becanovic K., Pouladi M.A., Sathasivam K., Cha J.H., et al. (2007) Mutant huntingtin's effects on striatal gene expression in mice recapitulate changes observed in human Huntington's disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage. Hum. Mol. Genet., 16, 1845–1861. [DOI] [PubMed] [Google Scholar]
- 83.Bebee T.W., Park J.W., Sheridan K.I., Warzecha C.C., Cieply B.W., Rohacek A.M., Xing Y., Carstens R.P. (2015) The splicing regulators Esrp1 and Esrp2 direct an epithelial splicing program essential for mammalian development. Elife, 4, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cieply B., Park J.W., Nakauka-Ddamba A., Bebee T.W., Guo Y., Shang X., Lengner C.J., Xing Y., Carstens R.P. (2016) Multiphasic and Dynamic Changes in Alternative Splicing during Induction of Pluripotency Are Coordinated by Numerous RNA-Binding Proteins. Cell Rep., 15, 247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Damianov A., Ying Y., Lin C.H., Lee J.A., Tran D., Vashisht A.A., Bahrami-Samani E., Xing Y., Martin K.C., Wohlschlegel J.A., et al. (2016) Rbfox Proteins Regulate Splicing as Part of a Large Multiprotein Complex LASR. Cell, 165, 606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dittmar K.A., Jiang P., Park J.W., Amirikian K., Wan J., Shen S., Xing Y., Carstens R.P. (2012) Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing. Mol. Cell Biol., 32, 1468–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ji X., Park J.W., Bahrami-Samani E., Lin L., Duncan-Lewis C., Pherribo G., Xing Y., Liebhaber S.A. (2016) alphaCP binding to a cytosine-rich subset of polypyrimidine tracts drives a novel pathway of cassette exon splicing in the mammalian transcriptome. Nucleic Acids Res., 44, 2283–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lu Z.X., Huang Q., Park J.W., Shen S., Lin L., Tokheim C.J., Henry M.D., Xing Y. (2015) Transcriptome-wide landscape of pre-mRNA alternative splicing associated with metastatic colonization. Mol. Cancer Res., 13, 305–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang Y., Park J.W., Bebee T.W., Warzecha C.C., Guo Y., Shang X., Xing Y., Carstens R.P. (2016) Determination of a comprehensive alternative splicing regulatory network and the combinatorial regulation by key factors during the epithelial to mesenchymal transition. Mol. Cell Biol., 36(11):1704–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Culver B.P., Savas J.N., Park S.K., Choi J.H., Zheng S., Zeitlin S.O., Yates J.R, 3rd., Tanese N. (2012) Proteomic analysis of wild-type and mutant huntingtin-associated proteins in mouse brains identifies unique interactions and involvement in protein synthesis. J. Biol. Chem., 287, 21599–21614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tollervey J.R., Wang Z., Hortobagyi T., Witten J.T., Zarnack K., Kayikci M., Clark T.A., Schweitzer A.C., Rot G., Curk T., et al. (2011) Analysis of alternative splicing associated with aging and neurodegeneration in the human brain. Genome Res., 21, 1572–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M., et al. (2015) Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell, 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Labadorf A., Hoss A.G., Lagomarsino V., Latourelle J.C., Hadzi T.C., Bregu J., MacDonald M.E., Gusella J.F., Chen J.F., Akbarian S., et al. (2015) RNA Sequence Analysis of Human Huntington Disease Brain Reveals an Extensive Increase in Inflammatory and Developmental Gene Expression. PLoS One, 10, e0143563.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rozen S., Skaletsky H. (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol., 132, 365–386. [DOI] [PubMed] [Google Scholar]
- 95.Hoaglin D.C., Iglewicz B., Tukey J.W. (1986) Performance of Some Resistant Rules for Outlier Labeling. J. Amer. Statist. Assoc., 81, 991–999. [Google Scholar]
- 96.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol., 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cahoy J.D., Emery B., Kaushal A., Foo L.C., Zamanian J.L., Christopherson K.S., Xing Y., Lubischer J.L., Krieg P.A., Krupenko S.A., et al. (2008) A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci., 28, 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kuhn A., Thu D., Waldvogel H.J., Faull R.L., Luthi-Carter R. (2011) Population-specific expression analysis (PSEA) reveals molecular changes in diseased brain. Nat. Methods, 8, 945–947. [DOI] [PubMed] [Google Scholar]
- 99.Park J.W., Jung S., Rouchka E.C., Tseng Y.T., Xing Y. (2016) rMAPS: RNA map analysis and plotting server for alternative exon regulation. Nucleic Acids Res., pii: gkw410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Koressaar T., Remm M. (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics, 23, 1289–1291. [DOI] [PubMed] [Google Scholar]
- 101.Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. (2012) Primer3–new capabilities and interfaces. Nucleic Acids Res., 40, e115.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lu Z.X., Jiang P., Cai J.J., Xing Y. (2011) Context-dependent robustness to 5' splice site polymorphisms in human populations. Hum. Mol. Genet., 20, 1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.