Abstract
Multiple missense mutations in Leucine–rich repeat kinase 2 (LRRK2) have been linked to Parkinson’s disease (PD), the most common degenerative movement disorder. LRRK2 is expressed by both neurons and microglia, the residential immune cells in the brain. Increasing evidence supports a role of LRRK2 in modulating microglial activity, of which Lrrk2-null rodent microglia display less inflammatory response to endotoxin lipopolysaccharide (LPS). The underlying molecular mechanism, however, remains elusive. Chemokine (C–X3–C) receptor 1 (CX3CR1), predominantly expressed by microglia, suppresses microglial inflammation while promotes migration. Using whole-genome microarray screening, we found that Cx3cr1 mRNA levels were substantially higher in microglia derived from Lrrk2 knockout (Lrrk2–/–) mice. The total and cell surface levels of CX3CR1 proteins were also remarkably increased. In correlation with the enhanced CX3CR1 expression, Lrrk2-null microglia migrated faster and travelled longer distance toward the source of fractalkine (CX3CL1), an endogenous ligand of CX3CR1. To investigate the impact of CX3CR1 elevation in vivo, we compared LPS-induced inflammation in the striatum of Lrrk2–/– knockout mice with Cx3cr1 heterozygous and homozygous knockout background. We found that a complete loss of Cx3cr1 restored the responsiveness of Lrrk2–/– microglia to LPS stimulation. In conclusion, our findings reveal a previously unknown regulatory role for LRRK2 in CX3CR1 signalling and suggest that an increase of CX3CR1 activity contributes to the attenuated inflammatory responses in Lrrk2-null microglia.
Introduction
As the sole resident immune cell in the central nervous system (CNS), microglia actively survey their surrounding brain parenchyma through continuous extension and retraction of their multiple long and ramified processes under healthy condition (1). In response to stimulation, microglia undergo rapid morphological and functional alterations that include enlargement of cell bodies with shorter and thicker processes and enhancement of migration and phagocytosis (2). Previously, we have shown that inhibition of PD-related Lrrk2 suppresses microglial activation in a line of α-synuclein transgenic mice (3). It has been further documented that Lrrk2 inhibition attenuates the inflammatory responses of microglia induced by LPS exposure and α-synuclein overexpression (4,5). LRRK2 is expressed in many different cell types, including macrophagic and monocytic cells and microglia (6,7). However, the molecular mechanism of LRRK2 in regulating microglial activation remains poorly understood.
Fractalkine and its cognate receptor (CX3CR1) form a unique one-to-one ligand-receptor chemokine pair that play an important role in microglial activation (8–10). Notably, Fractalkine is highly expressed in neurons, while CX3CR1 is exclusively expressed in microglia in the CNS (11,12). Genetic disruption of Cx3cr1 causes excessive microglial activation, resulting in neurotoxicity in mouse models of systemic inflammation, PD, and amyotrophic lateral sclerosis (8). On the other hand, exogenous administration of Fractalkine is neuroprotective in models of neuroinflammation (9,13). Together, these previous studies support an inhibitory role of Fractalkine/CX3CR1 signalling in control of microglial inflammatory response (8,14).
Starting from genome–wide microarray screening, we found substantial increase of Cx3cr1 mRNA and protein expression in Lrrk2-null microglia. We further revealed a functional correlation between Lrrk2–deficiency and CX3CR1 augmentation in cultured microglia and mouse models. Our results suggest an enhancement of CX3CR1 signalling contributes to the attenuated inflammatory responses observed in Lrrk2–null microglia.
Results
Lrrk2–null microglia are less responsive to LPS exposure
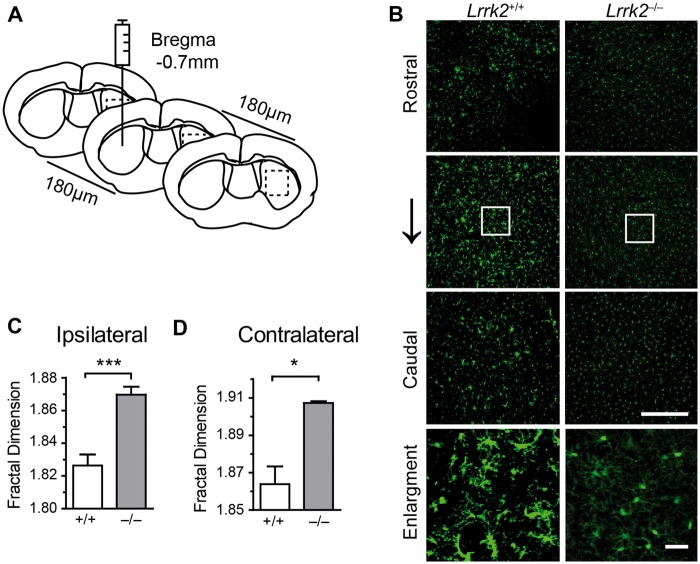
LPS activates microglia, resulting in enlarged cell bodies with shorter and thicker processes (15,16). Fractal dimension has been used to calculate the complexity of microglial morphology and activated microglia show the reduced value of fractal dimension (10). To investigate the role of LRRK2 in microglial activation, we stereotaxically injected LPS into the striatum of Lrrk2+/+ and Lrrk2–/– mice (Fig. 1A). Microglia were visualized by staining with an antibody against ionized calcium-binding adapter molecule 1 (Iba1) (17). More enlarged microglia were observed at both the ipsilateral and contralateral of LPS injection sites in the dorsal striatum of Lrrk2+/+ mice compared to the littermate Lrrk2–/– animals (Fig. 1B). The fractal dimension of Lrrk2–/– microglia was significantly lower than that of Lrrk2+/+ cells at both injection sites (Fig. 1C and D). By contrast, no difference in microglial morphology was observed at the striatum of vehicle-injected Lrrk2+/+ and Lrrk2–/– mice (data not shown). These results confirm the previous reports that Lrrk2-null microglia are less responsive to LPS exposure (4).
Figure 1.
Lrrk2–null microglia are less responsive to LPS intrastriatal injection. (A) A cartoon illustrates the sites of LPS intrastriatal injection. (B) Representative images show IBA1 staining in a series of coronal sections outlined at the boxed areas in (A). Scale bars, 100 μm. (C, D) Bar graphs summarize the calculation of microglial fractal dimension at both ipsilateral (C) and contralateral (D) injection sites of Lrrk2+/+ and Lrrk2–/– mice. Values represent mean ± S.E.M. Significant differences between the groups are determined by Student’s t-test and expressed as follows: *P<0.05; ***P<0.001. N = 5 per group.
Lrrk2–null microglia show higher levels of Cx3cr1 mRNA and protein expression
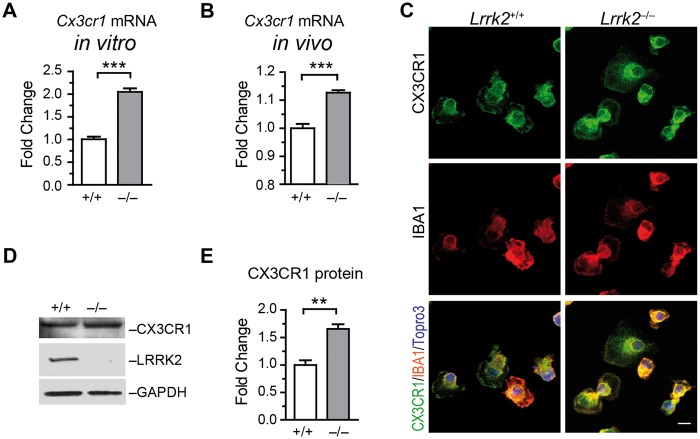
To understand the molecular mechanism of differential immunity of Lrrk2–/– microglia, we performed microarray analysis on primary microglial cultures. We found multiple alterations of gene expression between Lrrk2+/+ and Lrrk2–/– cells (GEO accession number: GSE83562). Noticeably, the mRNA levels of Cx3cr1 in Lrrk2–/– microglia were substantially higher than that in Lrrk2+/+ cells (Supplementary Material, Fig. S1). We further confirmed this finding in microglial cultures and mouse brains by quantitative RT-PCR (Fig. 2A and B). Using immunocytochemistry in cultured microglia, we found that CX3CR1 localized at the cell periphery and around the nucleus of both Lrrk2+/+ and Lrrk2–/– cells (Fig. 2C). Western blot analysis revealed the expression level of CX3CR1 proteins was substantially increased in Lrrk2–/– microglia compared to controls (Fig. 2D and E). These data demonstrate higher levels of Cx3cr1 mRNA and protein expression in Lrrk2–null microglia.
Figure 2.
Lrrk2–null microglia show increased Cx3cr1 mRNA and protein expression. (A, B) Bar graphs depict Cx3cr1 mRNA expression in cultured microglia (A) and mouse brains (B). Values represent mean ± S.E.M. Significant differences between the groups are determined by Student’s t-test and expressed as follows: ***P<0.001. N = 5 per group. (C) Representative images show CX3CR1and IBA1 co-staining at cultured microglia. Scale bars, 10 μm. (D) Western blot analyses the expression of CX3CR1 and LRRK2 proteins in cultured microglia. GAPDH was used as the loading control. (E) Bar graph shows CX3CR1 protein levels in cultured Lrrk2+/+ and Lrrk2–/– microglia. Values represent mean ± S.E.M. Significant differences between the groups were determined by Student’s t-test and expressed as follows: **P<0.01. N = 3 per group.
Lrrk2–null microglia show higher cell surface presentation of Cx3CR1
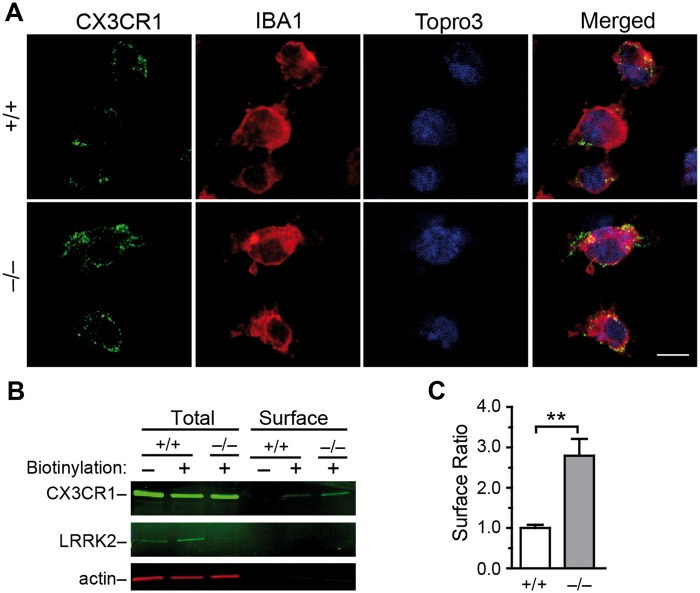
CX3CR1 binds to its ligand Fractalkine at the cell surface to initiate the intracellular signalling (11). We examined the cell surface localization of CX3CR1 by staining the non–permeabilized microglia using an antibody against the extracellular domain of CX3CR1. We found more CX3CR1 staining in Lrrk2–/– microglia (Fig. 3A). We further labelled surface proteins by biotin and immunoprecipitated the biotinylated proteins for western blot analysis of CX3CR1 levels. The ratio of cell surface presentation of CX3CR1 was substantially increased in Lrrk2–/– microglia (Fig. 3B and C). Collectively, these results show more CX3CR1 receptors at the cell surface of Lrrk2–/– microglia.
Figure 3.
Lrrk2–null microglia show increased presentation of CX3CR1 at the cell surface. (A) Representative images show CX3CR1 staining of impermeabilized microglia. The cells were counterstained with IBA1 and Topro3. Scale bar, 10 μm. (B) Western blot analyses the levels of CX3CR1. LRRK2 and actin at cell surface and in total lysates. (C) Bar graph depicts that the ratio of CX3CR1 proteins at cell surface in cultured Lrrk2+/+ and Lrrk2–/– microglia. Values represent means ± SEM. Significant differences between the groups are expressed as follows: **P < 0.01. The experiments were repeated three times independently.
Lrrk2–null microglia display increased motility in response to fractalkine stimulation
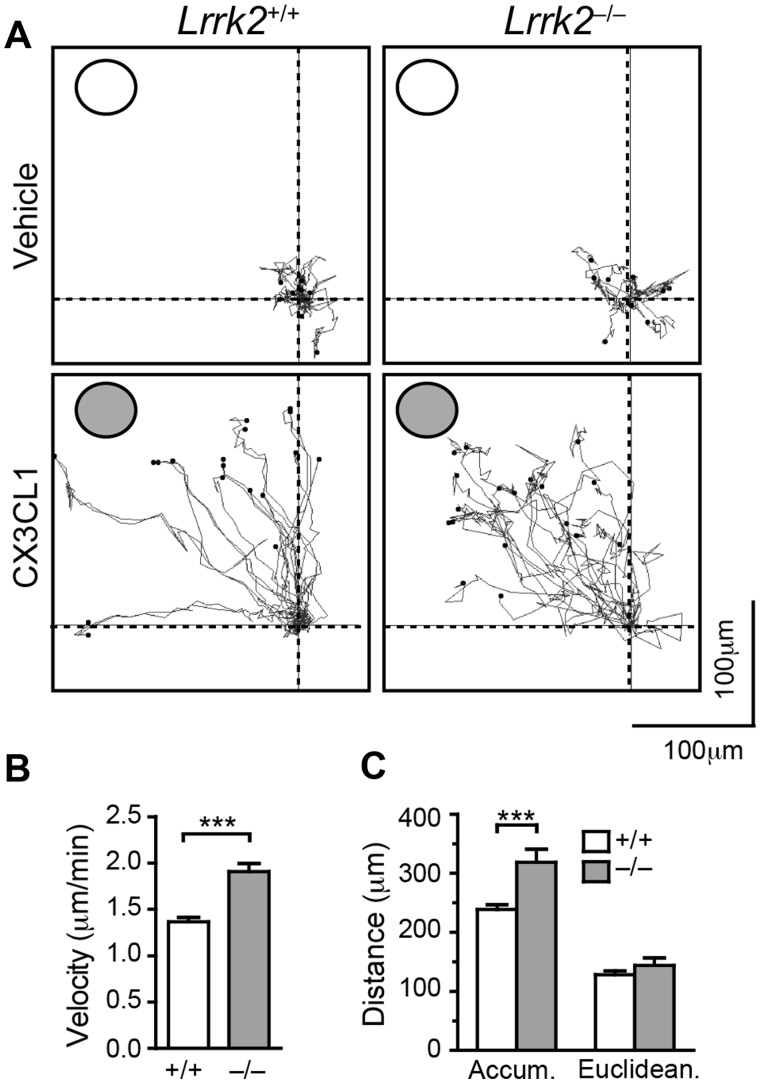
To test the functional significance of increased CX3CR1 expression in Lrrk2–/– microglia, we examined the responses of Lrrk2+/+ and Lrrk2–/– cells to Fractalkine stimulation. Using DUNN chamber, we found that Lrrk2–/– microglia migrated remarkably faster than Lrrk2+/+ cells in the presence of 5 nM Fractalkine (Fig. 4A and B). Although there was no difference in the Euclidean distance (the straight-line distance between the start point and end point), time-lapse recoding showed that Lrrk2–/– microglia travelled for longer distance compared to Lrrk2+/+ cells (accumulated distance) (Fig. 4C). It appeared that Lrrk2–/– microglia moved faster and made more changes of direction than the control cells in the presence of Fractalkine. The increased motility of Lrrk2–/– microglia correlates with the increased CX3CR1 expression in those cells.
Figure 4.
Lrrk2–null microglia show enhanced motility toward to the source of Fractalkine application. (A) Representative traces show the movement of microglia in the absence or presence of 5 nM Fractalkine in the outer well. Each dot represents an individual microglial cell. The outer wells are marked with circles. (B, C) Bar graphs show the velocity (B), as well as the accumulative (accum) and Euclidean distance (C) of microglia during the tracing period. The data were derived from three independent experiments.
Increased CX3CR1 activity contributes to the attenuated inflammatory responses in Lrrk2-null microglia
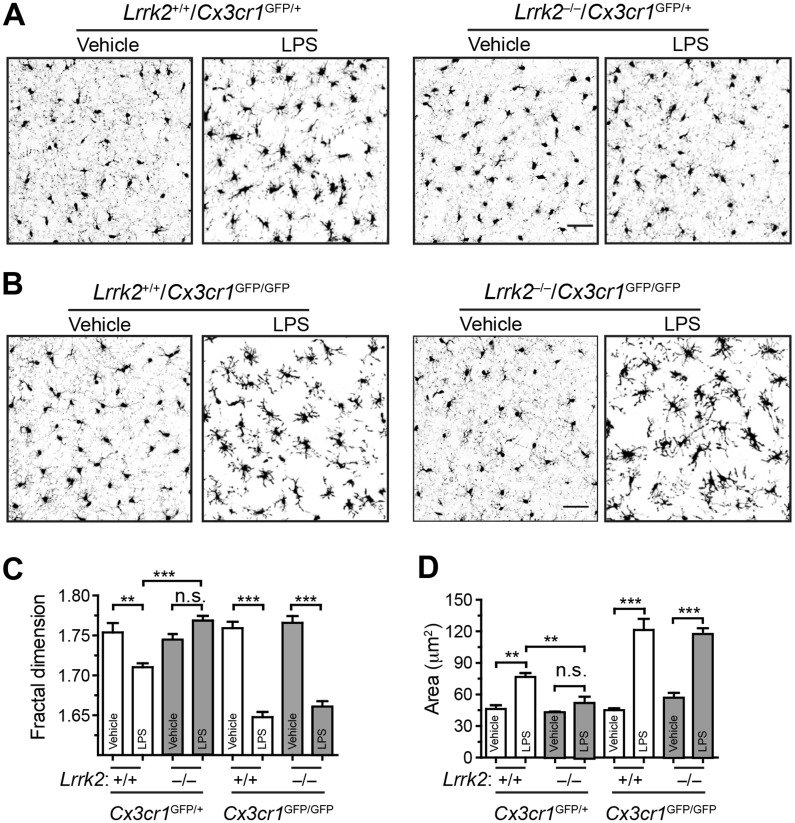
To investigate whether the enhanced CX3CR1 expression attenuates the response of Lrrk2–/– microglia to LPS stimulation, we generated a cohort of Lrrk2+/+ and Lrrk2–/– mice in Cx3cr1 heterozygous (Cx3cr1GFP/+) and homozygous knockout (Cx3cr1GFP/GFP) background. In this line of Cx3cr1 knockout mice, the coding exons of Cx3cr1 are replaced with a gene encoding green fluorescent protein (GFP)(18). The mice were administrated with LPS or vehicle by intraperitoneal injection. Both microglial fractal dimension and soma size were analysed in striatum (Fig. 5A–D). In the Cx3cr1 heterozygous background, Lrrk2+/+ microglia showed a decreased fractal dimension and increased soma size after treated with LPS compared to vehicle, while Lrrk2–/– microglia displayed no obvious changes of morphology (Fig. 5A, C and D). By contrast, in the Cx3cr1 homozygous knockout background, Lrrk2–/– microglia showed a very similar response to LPS administration as Lrrk2+/+ cells, including reduction of fractal dimension and enlargement of cell bodies (Fig. 5B–D). These findings thereby demonstrate an increase of CX3CR1 signalling attenuates the inflammatory responses of Lrrk2–/– microglia in vivo.
Figure 5.
Inhibition of CX3CR1 restores the responsiveness of Lrrk2–null microglia to LPS stimulation. (A, B) Representative images show GFP signals at the dorsal striatum of Lrrk2+/+/Cx3cr1GFP/+ and Lrrk2–/–/Cx3cr1GFP/+ (A), as well as Lrrk2+/+/Cx3cr1GFP/GFP and Lrrk2–/–/Cx3cr1GFP/GFP mice. Scale bars, 50 μm. (C, D) Bar graphs depict the measurements of fractal dimension (C) and area (D) of Lrrk2+/+/Cx3cr1GFP/+, Lrrk2–/–/Cx3cr1GFP/+, Lrrk2+/+/Cx3cr1GFP/GFP, Lrrk2–/–/Cx3cr1GFP/GFP microglia. Values represent means ± SEM. Significant differences between the groups are expressed as follows: **P<0.01; ***P<0.001. One-way ANOVA followed by Newman-Keuls multiple comparison tests. N = 4 per group.
Discussion
Using high-through microarray screening, we found that Cx3cr1 mRNA expression was substantially increased in Lrrk2–/– microglia. The amount of CX3CR1 proteins in total microglial lysates and on the cell surface was also remarkably elevated. Correlated with an augmentation of CX3CR1 expression, Lrrk2–/– microglia displayed enhanced motility in response to CX3CR1 ligand fractalkine. More importantly, an elevation of CX3CR1 expression contributes to the attenuated immune responses of Lrrk2–/– microglia to endotoxin LPS. Therefore, our findings reveal a previously unknown mechanism of LRRK2 in regulating CX3CR1 signalling in immune cells.
Fractalkine and its cognate receptor (CX3CR1) form a unique one-to-one ligand-receptor chemokine pair that has been demonstrated to play an important role in neuroinflammation and neuroprotection (11). The interaction between fractalkine and CX3CR1 contributes to maintain microglia in a surveillance phase. An activation of CX3CR1 signalling in microglia reduces the overproduction of proinflammatory molecules, such as inducible nitric oxide synthase (iNOS), interleukin (IL)-1β, tumour necrosis factor-α (TNF-α), and IL-6 (11,19,20). On the other hand, inhibition of LRRK2 kinase activity or genetic knockdown of LRRK2 protein attenuates TNFα secretion and iNOS induction (4,5). Based on these early findings, we propose that a lack of LRRK2 promotes CX3CR1 signalling in microglia, resulting in less inflammatory response.
LRRK2 could affect the expression and function of CX3CR1 at multiple levels. A previous study reports that the transcription of Cx3cr1 mRNA is regulated in part by nuclear factor of activated T-cells (NFAT), of which the expression of Cx3cr1 mRNA can be suppressed by NFAT inhibitors cyclosporine A and VIVIT (21). Our early work shows that LRRK2 is a potent negative regulator of NFAT transcriptional activity through retaining NFAT in the cytosol, while a lack of LRRK2 promotes NFAT activity by allowing NFAT to be more efficiently transported into the nucleus (22). Therefore, we speculate that LRRK2 may negatively regulate the expression of Cx3cr1 mRNA through preventing the nuclear translocation of NFAT. Previous studies have also demonstrated that LRRK2 regulates protein synthesis (23–25), ER-Golgi export (26), cytoskeleton dynamics (27), endocytosis (28), and autophagy (29). Potentially, LRRK2 could be involved in the translation of CX3CR1, exportation of newly synthesized CX3CR1 from ER to Golgi and cell surface, and recycling and degradation of CX3CR1 from the cell surface. LRRK2 also regulates protein kinase A (PKA)-dependent signalling pathway in neurons and microglia, in which a lack of Lrrk2 increases PKA activity (30,31). Since CX3CR1 activation inhibits PKA signalling (32), an increase of CX3CR1 in Lrrk2–/– microglia may serve as a negative feedback mechanism to prevent the excessive activation of PKA and related downstream pathways. The exact underlying molecular mechanism remains to determine.
Microglial cells are highly motile (1). The presence of Fractalkine increases the migration of microglia (8,33), which may explain the why Lrrk2–/– microglia moved faster toward the source of Fractalkine in cultures, since Lrrk2–/– cell possess more Cx3CR1. Additionally, Lrrk2–/– microglia tended to make more turns and travelled a longer distance. We suspect this irregular movement pattern of Lrrk2–null cells could be attributed to the dysfunction of LRRK2 in regulating the dynamics of actin and microtubule reorganization in cells (34–36). Previous studies suggest spleen tyrosine kinase (Syk) plays an important role in fractalkine/CX3CR1–induced chemotaxis (37,38). The binding of fractalkine with CX3CR1 activates Syk, which promotes cell motility through regulating actin dynamics via activation of both Rac1/WAVE2 and Cdc42/WASP pathways. Alternatively, LRRK2 can also regulate the movement of microglia by modulating the activity of focal adhesion kinase (FAK) in the organization of the dynamic actin network underneath cell protrusions, which allows Lrrk2–knockdown microglia more motile compared to control cells (39). Moreover, this study also demonstrates that PD-related LRRK2 G2019S missense mutation attenuates microglial motility by inhibiting FAK activity, while LRRK2 inhibitor restores the movement (39), suggesting an involvement of microglial abnormalities in the pathogenesis of PD. In addition, LRRK2 also regulates the activity of actin binding proteins ERM and cofilin during neuron development (27,30,35). However, we did not observe substantial alterations of ERM and cofilin activities in microglia (unpublished data). Therefore LRRK2 seems to participate multiple signalling pathways that actively regulate actin dynamics in microglia and neurons during cell migration and morphogenesis.
Increasing evidence suggests that neuronal death can be induced not only by defects in neurons but also by impairments in glia, including microglia, which provide critical support for neuron survival and function. Our findings on the modulatory role of LRRK2 in CX3CR1 signalling shed new light on the emerging pathogenic mechanisms of neuroinflammation in PD.
Materials and Methods
Mice
The Lrrk2−/− mice were generated through deletion of the second coding exon of LRRK2 (35). Cx3cr1GFP/+ mice (18) were obtained from the Jackson Laboratory. All mouse work follows the guidelines approved by the Institutional Animal Care and Use Committees of the National Institute of Child Health and Human Development, and the Johns Hopkins University School of Medicine.
LPS intrastriatal injection
Surgical procedures were carried out under gas anaesthesia (isoflurane:oxygen:nitrous oxide, 1:33:66) and aseptic conditions. One microliter LPS (5 mg/ml, TLR-grade, E.coli, Serotype O111:B4 S-form; Enzo Life Sciences) were slowly (2 min) injected by 10 µl hamilton syringe with 33 gauge needle into striatum of 1–month–old Lrrk2+/+ and Lrrk2–/– mice (n = 5). The coordinates of injection sites in striatum are: 0.7mm posterior to Bregma, 1.9mm lateral to midline, and 3.5mm ventral to skull surface. After injection, the needle was held in the injection place for two more minutes. Chloramphenicol (sodium succinate) was injected (i.m. 50 mg/kg) to avoid possible bacterial infection during surgery. The animals were then put back to the cages for full recovery. The mice were sacrificed at 72 h after injection, and the brains were fixed by 4% paraformaldehyde and processed for neuropathological analysis.
LPS intraperitoneal injection
Vehicle (phosphate-buffered saline; PBS solution) or LPS (TLR-grade, E.coli, Serotype O111:B4 S-form; Enzo Life Sciences) at the concentration (2 mg/kg body weight, intraperitoneal; two doses) was daily injected into 2-month-old C57BL/6 and transgenic mice (n = 4). Forty-eight hours after the first injection, the mice were sacrificed, and the brains were fixed by 4% paraformaldehyde and processed for neuropathological analysis.
Primary microglia culture and immunocytochemistry
Primary microglia culture was prepared as described previously (17). In brief, the mixed cell culture was prepared from postnatal 4 day–old mice and maintained at 37 °C and 5% CO2 for 14 days in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% heat-inactivated foetal bovine serum (FBS), 100U/ml penicillin and 100 µg/ml streptomycin. Microglia cells were collected as floating cells by gentle shaking for the following experiments.
Microglia were fixed in 4% PFA as described previously (17). Briefly, they were permeabilized with Triton X-100 and incubated with 10% donkey serum (Sigma-Aldrich) for 1h to block unspecific binding and incubated overnight with the primary antibody. Antibodies specific to CX3CR1 (1:500, Abcam), and ionized calcium binding adaptor molecule-1 (IBA1; 1:500, Wako Chemicals; 1:500, Abcam) were used for immunocytochemistry. Alexa 488- or Alexa 568-conjugated secondary antibody (1:500; Invitrogen) was used to visualize the staining.
For extracellular membrane staining of CX3CR1, live microglia were incubated with the primary CX3CR1 antibody (1:100, Abcam) on ice for 30 min in BWD buffer (20 mM HEPES, 125 mM NaCl, 5 mM KCl, 5 mM dextrose, 10 mM NaHCO3, 1 mM KH2PO4, 1 mM CaCl2, and 1 mM MgCl2, pH 7.4), containing mouse Fc Block (BD Biosciences) (37). Then the microglia were fixed and performed IBA1 immunocytochemistry as described above. Fluorescence images were captured using a laser scanning confocal microscope (LSM 510; Zeiss) at the same gain and offset settings. The images of staining were presented as a single optical layer after being acquired in z-series stack scans at 0.8 μm intervals.
Microarray
The primary microglia cells were lysed in the lysate buffer (included in the RNeasy kit, Qiagen, Hilden, Germany) and homogenized completely. The total RNA was extracted from the cell lysate with the RNeasy mini kit in accordance with the protocols prescribed by the manufacture (Qiagen). Biotinylated cRNA were prepared from 250 ng of total RNA in each sample with the Illumina Total Prep RNA Amplification kit (Ambion) and hybridized to the BeadChip Array Mouse WG-6 v2 (Illumina) following the standard hybridization protocol. The chip was scanned by iScan (Illumina) with the standard scanning protocol. The data were analysed with the Illumina BeadStudio software suite.
Real-time reverse-transcription PCR
Primary microglial cells were seeded on 6-well plates and washed with ice-cold PBS three times, and total RNA was isolated with RNeasy mini kit (Qiagen). The mouse brain samples were collected and isolated RNA by Trizol reagent, followed by clean-up using the RNeasy mini kit. The cDNA was then synthesized from 100 ng of RNA with the RT2 First Strand kit (Qiagen). The SYBR Green real-time PCR detection method was used to quantitate the transcript levels of the Cx3cr1, and β-actin using commercially prepared primers (Qiagen). The gene expression was calculated as fold changes normalized with the β-actin.
Immunohistochemistry and light microscopy
As described previously (40), mice were perfused via cardiac infusion with 4% paraformaldehyde in cold PBS. To obtain frozen sections, brain tissues were removed and submerged in 30% sucrose for 24 h and sectioned at 30 μm thickness with cryostat (Leica CM1950). Antibodies specific to Iba1 (1:500, Wako Chemicals) were used as suggested by the manufacturers. Alexa 488 or Alexa 568-conjugated secondary antibody (1:500, Invitrogen) was used to visualize the staining. Fluorescence images were captured using a laser scanning confocal microscope (LSM 510; Zeiss). The paired images in all of the figures were collected at the same gain and offset settings. When post collection processing was done, it was applied uniformly to all paired images. The images of staining were presented as a single optical layer after being acquired in z-series stack scans at 0.8 μm intervals.
Morphometric analysis of microglial morphology
The fractal dimension and cellular areas analysis was performed on images acquired by 25X objective using the NIH Image J software as described before (41,42). Briefly, the images were background subtracted, normalized (segmentation and threshold setting) and the readouts of images scanning were determined using a specific plugin “Fractal Dimension and Lacunarity”. For quantification, three sections at Bregma 0.28, 0 and −0.28 mm were acquired per animal, and three digital images at striatum were acquired per section.
Chemotaxis assay using the Dunn chemotaxis chamber
Chemotaxis was assessed using a Dunn chemotaxis chamber (Weber Scientific International Ltd., Teddington, UK) according to the method described previously (43). In brief, the cells were attached to square coverslips for 2 h, and washed three times with serum free DMEM. Then a coverslip was placed over the chamber, whose outer and inner wells were filled with DMEM. The medium in the outer well was exchanged for DMEM containing 5 nM Fractalkine (recombinant mouse CX3CL1/Fractalkine full length, R&D Systems) through the slit, and the chamber was set on the stage of a microscope (ECLIPSE TE300; Nikon), which was maintained at 37°C.
Western blot
Primary microglial cells were analysed by quantitative immunoblotting with anti-CX3CR1 (1:500, abcam), LRRK2 (1:1,000, University of California Davis/US National Institutes of Health NeuroMab Facility 73-188), and β-actin (1:1,000; Sigma) antibodies. All of the immunoblots were visualized by enhanced chemiluminescence development (Pierce) and quantified by the NIH Image J software.
Plasma membrane protein biotinylation assay
The surface biotinylation assay was performed as reported previously (26,44). Total protein and EZ-Link Sulfo-NHS-SS-Biotin (ThermoFisher) isolated biotinylated protein were analysed by quantitative immunoblotting with anti-CX3CR1 (1:500, Abcam), LRRK2 (1:1,000, University of California Davis/US National Institutes of Health NeuroMab Facility 73-188) and β-actin (1:1,000; Sigma) antibodies. All of the immunoblots were visualized by IRDye secondary antibodies and quantified on an Odyssey CLx Imaging System (LI-COR).
Statistical analysis
Statistical analysis was performed using the GraphPad Prism software (GraphPad Software). Data are presented as mean ± S.E.M. Statistical significance was determined by comparing means of different groups using the Student’s t-test or ANOVA followed by the post hoc test (*P < 0.05; **P < 0.01; ***P < 0.001).
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
We thank Cai lab members for their constructive suggestions.
Conflict of Interest statement. None declared.
Funding
This work was supported in part by the intramural research programs of the National Institutes of Health (NIH)–National Institute on Aging (Grants AG000928 and AG000944 to H.C.), and National Natural Science Grant of China (No. 81571257 to XD P.).
Footnotes
Present address: Department of Anesthesiology, Center for Translational Research in Neurodegenerative Disease, University of Florida, College of Medicine, 1275 Center Drive, Biomedical Sciences J448, Gainesville, FL 32610, USA.
Present address: Department of Neurology, Fujian Medical University Union Hospital, 29 Xinquan Road, Fuzhou 350001, China.
References
- 1.Nimmerjahn A., Kirchhoff F., Helmchen F. (2005) Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science, 308, 1314–1318. [DOI] [PubMed] [Google Scholar]
- 2.Kreutzberg G.W. (1996) Microglia: a sensor for pathological events in the CNS. Trends Neurosci., 19, 312–318. [DOI] [PubMed] [Google Scholar]
- 3.Lin X., Parisiadou L., Gu X.L., Wang L., Shim H., Sun L., Xie C., Long C.X., Yang W.J., Ding J., et al. (2009) Leucine-rich repeat kinase 2 regulates the progression of neuropathology induced by Parkinson's-disease-related mutant alpha-synuclein. Neuron, 64, 807–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moehle M.S., Webber P.J., Tse T., Sukar N., Standaert D.G., DeSilva T.M., Cowell R.M., West A.B. (2012) LRRK2 inhibition attenuates microglial inflammatory responses. J. Neurosci., 32, 1602–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daher J.P., Volpicelli-Daley L.A., Blackburn J.P., Moehle M.S., West A.B. (2014) Abrogation of alpha-synuclein-mediated dopaminergic neurodegeneration in LRRK2-deficient rats. Proc. Natl Acad. Sci. USA, 111, 9289–9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Z., Lee J., Krummey S., Lu W., Cai H., Lenardo M.J. (2011) The kinase LRRK2 is a regulator of the transcription factor NFAT that modulates the severity of inflammatory bowel disease. Nat Immunol., 12, 1063–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thevenet J., Pescini Gobert R., Hooft van Huijsduijnen R., Wiessner C., Sagot Y.J. (2011) Regulation of LRRK2 expression points to a functional role in human monocyte maturation. PLoS One, 6, e21519.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardona A.E., Pioro E.P., Sasse M.E., Kostenko V., Cardona S.M., Dijkstra I.M., Huang D., Kidd G., Dombrowski S., Dutta R., et al. (2006) Control of microglial neurotoxicity by the fractalkine receptor. Nature Neuroscience, 9, 917–924. [DOI] [PubMed] [Google Scholar]
- 9.Pabon M.M., Bachstetter A.D., Hudson C.E., Gemma C., Bickford P.C. (2011) CX3CL1 reduces neurotoxicity and microglial activation in a rat model of Parkinson's disease. J. Neuroinflammation, 8, 9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhaskar K., Konerth M., Kokiko-Cochran O.N., Cardona A., Ransohoff R.M., Lamb B.T. (2010) Regulation of tau pathology by the microglial fractalkine receptor. Neuron, 68, 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrison J.K., Jiang Y., Chen S., Xia Y., Maciejewski D., McNamara R.K., Streit W.J., Salafranca M.N., Adhikari S., Thompson D.A., et al. (1998) Role for neuronally derived fractalkine in mediating interactions between neurons and CX3CR1-expressing microglia. Proc Natl Acad Sci U S A., 95, 10896–10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes P.M., Botham M.S., Frentzel S., Mir A., Perry V.H. (2002) Expression of fractalkine (CX3CL1) and its receptor, CX3CR1, during acute and chronic inflammation in the rodent CNS. Glia, 37, 314–327. [PubMed] [Google Scholar]
- 13.Morganti J.M., Nash K.R., Grimmig B.A., Ranjit S., Small B., Bickford P.C., Gemma C. (2012) The soluble isoform of CX3CL1 is necessary for neuroprotection in a mouse model of Parkinson's disease. J. Neuroscience, 32, 14592–14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ransohoff R.M., Cardona A.E. (2010) The myeloid cells of the central nervous system parenchyma. Nature, 468, 253–262. [DOI] [PubMed] [Google Scholar]
- 15.Chow J.C., Young D.W., Golenbock D.T., Christ W.J., Gusovsky F. (1999) Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J. Biol. Chem., 274, 10689–10692. [DOI] [PubMed] [Google Scholar]
- 16.Lee S.J., Lee S. (2002) Toll-like receptors and inflammation in the CNS. Curr. Drug. Targets Inflamm. Allergy, 1, 181–191. [DOI] [PubMed] [Google Scholar]
- 17.Ma B., Yu J., Xie C., Sun L., Lin S., Ding J., Luo J., Cai H. (2015) Toll-Like Receptors Promote Mitochondrial Translocation of Nuclear Transcription Factor Nuclear Factor of Activated T-Cells in Prolonged Microglial Activation. J. Neurosci., 35, 10799–10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung S., Aliberti J., Graemmel P., Sunshine M.J., Kreutzberg G.W., Sher A., Littman D.R. (2000) Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell Biol., 20, 4106–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maciejewski-Lenoir D., Chen S., Feng L., Maki R., Bacon K.B. (1999) Characterization of fractalkine in rat brain cells: migratory and activation signals for CX3CR-1-expressing microglia. J. Immunol., 163, 1628–1635. [PubMed] [Google Scholar]
- 20.Re D.B., Przedborski S. (2006) Fractalkine: moving from chemotaxis to neuroprotection. Nat. Neurosci., 9, 859–861. [DOI] [PubMed] [Google Scholar]
- 21.Barlic J., McDermott D.H., Merrell M.N., Gonzales J., Via L.E., Murphy P.M. (2004) Interleukin (IL)-15 and IL-2 reciprocally regulate expression of the chemokine receptor CX3CR1 through selective NFAT1- and NFAT2-dependent mechanisms. J. Biol. Chem., 279, 48520–48534. [DOI] [PubMed] [Google Scholar]
- 22.Liu Z., Lee J., Krummey S., Lu W., Cai H., Lenardo M.J. (2011) The kinase LRRK2 is a regulator of the transcription factor NFAT that modulates the severity of inflammatory bowel disease. Nat. Immunol., 12, 1063–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imai Y., Gehrke S., Wang H.Q., Takahashi R., Hasegawa K., Oota E., Lu B. (2008) Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J, 27, 2432–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gehrke S., Imai Y., Sokol N., Lu B. (2010) Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature, 466, 637–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin I., Kim J.W., Lee B.D., Kang H.C., Xu J.C., Jia H., Stankowski J., Kim M.S., Zhong J., Kumar M., et al. (2014) Ribosomal protein s15 phosphorylation mediates LRRK2 neurodegeneration in Parkinson's disease. Cell, 157, 472–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho H.J., Yu J., Xie C., Rudrabhatla P., Chen X., Wu J., Parisiadou L., Liu G., Sun L., Ma B, et al. (2014) Leucine-rich repeat kinase 2 regulates Sec16A at ER exit sites to allow ER-Golgi export. EMBO J, 33(20):2314–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parisiadou L., Cai H. (2010) LRRK2 function on actin and microtubule dynamics in Parkinson disease. Commun. Integr. Biol, 3, 396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dodson M.W., Zhang T., Jiang C., Chen S., Guo M. (2012) Roles of the Drosophila LRRK2 homolog in Rab7-dependent lysosomal positioning. Hum. Mol. Genet., 21, 1350–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alegre-Abarrategui J., Christian H., Lufino M.M., Mutihac R., Venda L.L., Ansorge O., Wade-Martins R. (2009) LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum. Mol. Genet., 18, 4022–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parisiadou L., Yu J., Sgobio C., Xie C., Liu G., Sun L., Gu X.L., Lin X., Crowley N.A., Lovinger D.M, et al. (2014) LRRK2 regulates synaptogenesis and dopamine receptor activation through modulation of PKA activity. Nat Neurosci., 17, 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russo I., Berti G., Plotegher N., Bernardo G., Filograna R., Bubacco L., Greggio E. (2015) Leucine-rich repeat kinase 2 positively regulates inflammation and down-regulates NF-kappaB p50 signaling in cultured microglia cells. J. Neuroinflam., 12, 230.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Felouzis V., Hermand P., de Laissardiere G.T., Combadiere C., Deterre P. (2016) Comprehensive analysis of chemokine-induced cAMP-inhibitory responses using a real-time luminescent biosensor. Cell. Signal., 28, 120–129. [DOI] [PubMed] [Google Scholar]
- 33.Fuhrmann M., Bittner T., Jung C.K., Burgold S., Page R.M., Mitteregger G., Haass C., LaFerla F.M., Kretzschmar H., Herms J. (2010) Microglial Cx3cr1 knockout prevents neuron loss in a mouse model of Alzheimer's disease. Nat. Neurosci, 13, 411–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaleel M., Nichols R.J., Deak M., Campbell D.G., Gillardon F., Knebel A., Alessi D.R. (2007) LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson's disease mutants affect kinase activity. Biochem. J., 405, 307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parisiadou L., Xie C., Cho H.J., Lin X., Gu X.L., Long C.X., Lobbestael E., Baekelandt V., Taymans J.M., Sun L., et al. (2009) Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. J Neurosci., 29, 13971–13980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meixner A., Boldt K., Van Troys M., Askenazi M., Gloeckner C.J., Bauer M., Marto J.A., Ampe C., Kinkl N., Ueffing M. (2011) A QUICK screen for Lrrk2 interaction partners–leucine-rich repeat kinase 2 is involved in actin cytoskeleton dynamics. Mol. Cell. Proteomics, 10, M110 001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gevrey J.C., Isaac B.M., Cox D. (2005) Syk is required for monocyte/macrophage chemotaxis to CX3CL1 (Fractalkine). J. Immunol., 175, 3737–3745. [DOI] [PubMed] [Google Scholar]
- 38.Park H., Cox D. (2011) Syk regulates multiple signaling pathways leading to CX3CL1 chemotaxis in macrophages. J. Biol. Chem., 286, 14762–14769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi I., Kim B., Byun J.W., Baik S.H., Huh Y.H., Kim J.H., Mook-Jung I., Song W.K., Shin J.H., Seo H., et al. (2015) LRRK2 G2019S mutation attenuates microglial motility by inhibiting focal adhesion kinase. Nat. Commun., 6, 8255.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai H., Lin X., Xie C., Laird F.M., Lai C., Wen H., Chiang H.C., Shim H., Farah M.H., Hoke A., et al. (2005) Loss of ALS2 function is insufficient to trigger motor neuron degeneration in knock-out mice but predisposes neurons to oxidative stress. J. Neurosci., 25, 7567–7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varvel N.H., Bhaskar K., Kounnas M.Z., Wagner S.L., Yang Y., Lamb B.T., Herrup K. (2009) NSAIDs prevent, but do not reverse, neuronal cell cycle reentry in a mouse model of Alzheimer disease. J. Clin. Invest., 119, 3692–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soltys Z., Ziaja M., Pawlinski R., Setkowicz Z., Janeczko K. (2001) Morphology of reactive microglia in the injured cerebral cortex. Fractal analysis and complementary quantitative methods. J. Neurosci. Res., 63, 90–97. [DOI] [PubMed] [Google Scholar]
- 43.Honda S., Sasaki Y., Ohsawa K., Imai Y., Nakamura Y., Inoue K., Kohsaka S. (2001) Extracellular ATP or ADP induce chemotaxis of cultured microglia through Gi/o-coupled P2Y receptors. J. Neurosci., 21, 1975–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lai C., Xie C., McCormack S.G., Chiang H.C., Michalak M.K., Lin X., Chandran J., Shim H., Shimoji M., Cookson M.R., et al. (2006) Amyotrophic lateral sclerosis 2-deficiency leads to neuronal degeneration in amyotrophic lateral sclerosis through altered AMPA receptor trafficking. J. Neurosci., 26, 11798–11806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.