Abstract
CHD7 mutations are implicated in a majority of cases of the congenital disorder, CHARGE syndrome. CHARGE, an autosomal dominant syndrome, is known to affect multiple tissues including eye, heart, ear, craniofacial nerves and skeleton and genital organs. Using a morpholino-antisense-oligonucleotide-based zebrafish model for CHARGE syndrome, we uncover a complex spectrum of abnormalities in the neural crest and the crest-derived cell types. We report for the first time, defects in myelinating Schwann cells, enteric neurons and pigment cells in a CHARGE model. We also observe defects in the specification of peripheral neurons and the craniofacial skeleton as previously reported. Chd7 morphants have impaired migration of neural crest cells and deregulation of sox10 expression from the early stages. Knocking down Sox10 in the zebrafish CHARGE model rescued the defects in Schwann cells and craniofacial cartilage. Our zebrafish CHARGE model thus reveals important regulatory roles for Chd7 at multiple points of neural crest development viz., migration, fate choice and differentiation and we suggest that sox10 deregulation is an important driver of the neural crest-derived aspects of Chd7 dependent CHARGE syndrome.
Introduction
CHD7 is a chromodomain helicase DNA-binding domain (CHD) protein and is a member of the SWI-SNF superfamily of ATP-dependent chromatin remodelers that bind to DNA and modulate gene expression (1). CHD7 has been shown to bind to gene enhancer elements and promoters, functioning as a transcriptional co-regulator (2,3). In vitro assays have shown that CHD7, powered by ATP hydrolysis remodels chromatin and that disease-causing mutations identified in patients impair this function of the protein (4). CHD7 co-localizes with the histone acetyl transferase, P300 and the stem cell pluripotency factors OCT4, SOX2 and NANOG on enhancers to regulate transcription (5). The occupancy of CHD7 correlates both with activator as well as repressor functions on these enhancers (3,6), however, the functional relevance of these activities is still not completely clear.
Mutations in CHD7 cause CHARGE syndrome (OMIM ID: 214800), which is characterized by defects in multiple tissues ranging from eye, heart, craniofacial skeleton, vertebrae, genital organs, with neuronal disorders, deafness and general growth retardation (7–9). Mutations along the whole length of the protein have been associated with CHARGE syndrome (10) suggesting that most domains in the protein are functionally important. CHARGE syndrome models have been created using morpholino based antisense oligonucleotide knockdowns of chd7 in xenopus (11) and zebrafish (12,13), mutations and knock-outs in mouse (14,15) and drosophila (16) as well as by shRNA-based knockdown in human stem cells (11). A few of these models have identified defects in neural crest derived organs implying a conserved role for CHD7 in the neural crest.
Neural crest (NC) is a transient population of multipotent cells in the embryo that emerges from the ectoderm to migrate and populate a diverse set of tissues in the body (17). Abnormalities in many of these tissues, such as the craniofacial skeleton and peripheral nerves, are major contributors to CHARGE syndrome. In cultured human neural crest like cells (hNCLCs) CHD7 has been shown to be a part of the PBAF (polybromo-and BRG1-associated factor-containing) complex that binds to and activates enhancers of NC specific genes such as SOX9 and TWIST1 (11). CHD7 has been demonstrated to be important for maintaining multipotency of NC cells in mouse neural crest stem cells when complexed with BRG1 and other proteins (18). SOX2, a transcription factor with an important role in the epithelial to mesenchymal transition of NC cells (19), has been shown to physically interact with CHD7 to co-regulate gene expression (3). Microarray analysis of a mouse mutant for Chd7 revealed its role in the regulation of a number of genes that are important for NC migration (20). Also, NC-specific conditional deletion of Chd7 in mouse causes deformities in the craniofacial skeleton (21). However, the role of Chd7 in the various stages of neural crest development and differentiation has not yet been explored.
Here we investigate the effects of chd7 knockdown on the development and differentiation of zebrafish neural crest. We show that chd7 deficiency affects early NC gene expression as well as NC migration. We demonstrate that Chd7 is critical for fate determination of NC derivatives and that its deficiency adversely affects crest-derived precursor as well as differentiated populations of craniofacial cartilage, cranial ganglia and enteric neurons. We find that Chd7 is crucial for the differentiation of pigmented cells and myelinated Schwann cells, which has not been reported previously. In the chd7 morphants we also observed a deregulation of the transcription factor sox10, known for its role in neural crest. By reducing the levels of Sox10, we could rescue a number of different crest-derived phenotypes indicating the importance of sox10 deregulation in the pathophysiology of CHARGE.
Results
chd7 RNA is inherited from parents and ubiquitously expressed in early zebrafish embryos
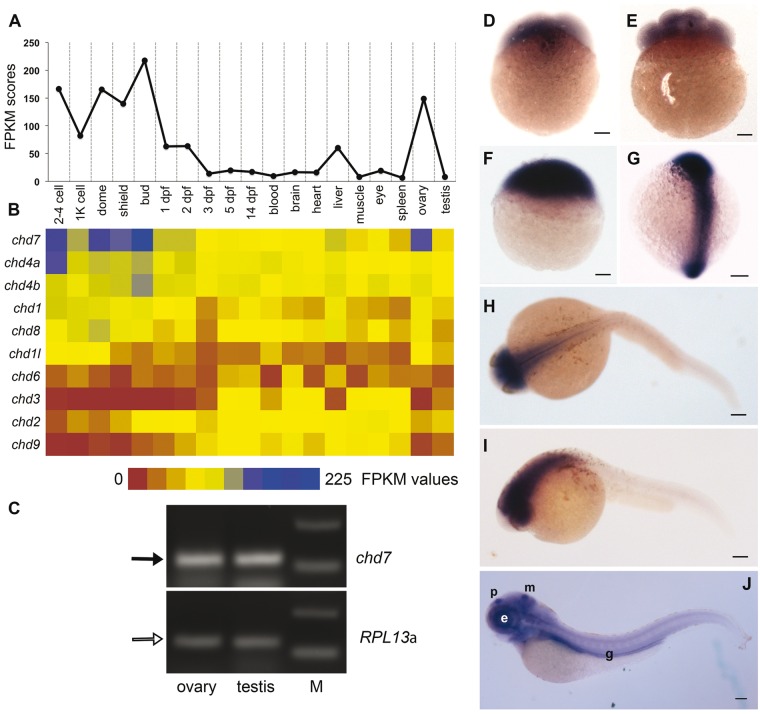
CHD proteins play important roles in development, stem cell function, adult physiology and diseases (1,22). To determine the expression of these genes in zebrafish, we analysed existing RNA sequencing data available in the public domain for different developmental stages and tissues of zebrafish [SRP009426 (23), SRP008845 (24), ERP000016 (Zebrafish transcriptome sequencing project, PRJEB1986), PRJNA207719 (25) and SRP017135 (26)] . We found expression data for 10 members of the chd family. Chd1-like, chd2, chd3, chd6 and chd9 had very low expression at most stages of zebrafish development, while chd1, chd8, chd4a and chd4b showed high expression in some tissues and stages (Fig. 1B). The most abundant chd mRNA in zebrafish was chd7 (Fig. 1B).
Figure 1.
Parental contribution and ubiquitous expression of chd7 in the early stages. (A) FPKM scores for chd7 plotted at different stages and tissues. Highest expression is seen in 2–4 cell, dome stage and bud stage. In adult tissues, ovary had the highest expression. (B) Heatmap depicting FPKM values from RNA sequencing analysis (from data available in the public domain) of 10 chd transcripts across different zebrafish developmental stages and tissues. chd7 is the most abundant mRNA among the Chd group of proteins. (C) Agarose gel electrophoresis showing amplification of chd7 transcript (black arrow) from adult ovary and testes, suggesting maternal and paternal contribution of chd7 to zygote. RPL13α (white arrow) is used to normalize the RNA levels. (D–G) chd7 is ubiquitously expressed in early stages. RNA in situ hybridization of chd7 in zebrafish embryos at 2-cell (D), 8-cell (E), dome (F) and 10-somite stage (G) shows ubiquitous expression. The animal pole is to the top (D–F). Dorsal view, anterior to the top (G). (H–I) At 24hpf, expression of chd7 is restricted to the head. (J) 4dpf larva expresses chd7 more in the eye, pineal gland-p, midbrain-hindbrain boundary-m and gut tube-g. Anterior is to the left. (H) dorsal view, (I,J) lateral view. The scale bar marks 100µm.
Chd7 expression was maximum in the early stages of development starting from 2 to 4 cells (Fig. 1A). In zebrafish embryos, the zygotic transcription starts between 1K cell stage and dome stage (3–4 hours post fertilization (hpf)) (27), thus mRNA present in 2–4 cell and 1K cell stages are indicative of parental contribution. The only adult tissue with comparable expression to the early embryonic stages was the ovary (Fig. 1A). Chd7 could be amplified from both adult zebrafish ovary and testis cDNA (Fig. 1C), suggesting that the contribution of the chd7 transcript to the zygote could be from both parents.
To confirm observations made from RNA sequencing analysis and to understand the spatial distribution of the chd7 RNA, we performed RNA in situ hybridization at various embryonic stages (Fig. 1D–J). Reinforcing the RNA sequencing data, chd7 was highly expressed from 2-cell stage onwards (Fig. 1D). Ubiquitous expression was observed at 2-cell, 8-cell, sphere and 10-somite stage (Fig. 1D–G). CHD7 is known as a positive regulator of ribosomal RNA biogenesis in stem cells (6) and might be essential in the first few hours when the translation of maternal RNA is essential for the rapid progression of embryonic development. By 24hpf, the expression becomes more restricted to the head, specifically in the eye and the mid-and hindbrain (Fig. 1H and I), as has also been shown previously (12,28). At 4 days post fertilization (dpf), chd7 expression was visible in the pineal complex, midbrain-hindbrain boundary, eye, jaw region and the gut tube (Fig. 1J) which supports previous studies showing the important roles for Chd7 in neurogenesis in the embryo as well as the adult (29,30).
Knockdown of CHD7 in Zebrafish
To address the role of Chd7 in zebrafish development, we designed a splice blocking antisense morpholino oligonucleotide (MO) against the exon8-intron8 boundary (MO1) (Supplementary Material, Fig. S1B). The MO1 would be predicted to cause retention of intron8 in the processed transcript (Supplementary Material, Fig. S1C). PCR analysis of the cDNA from embryos injected with 2.4ng of chd7 MO1 confirmed the presence of a defective splice product; (Supplementary Material, Fig. S1D). We amplified the aberrant splice-product from cDNA of both control MO and chd7 MO1 injected embryos using primers in the exon6 and intron8 (Supplementary Material, Fig. S1B and S1C). The control embryos had no detectable expression of the aberrant product while the chd7 morphant embryos had a robust expression (Supplementary Material, Fig. S1D). The product size matched retention of only intron8 and not intron6 or intron7 also confirming that the aberrant product was the result of splicing defects, not genomic DNA contamination in the cDNA. We further amplified the exon8-intron8 boundary by PCR from the morphant cDNA and sequenced this amplicon to confirm the retention of intron (Supplementary Material, Fig. S1E). The insertion of the intron8 would be predicted to cause a frame shift mutation in the chd7 transcript resulting in a 943 amino acid long protein, instead of the 3140 amino acid long full-length Chd7 protein (Supplementary Material, Fig. S1A). The truncated form of Chd7 is predicted to contain only the chromodomain-1 and to lack all other known domains of the Chd7 protein. Similar was the case when we injected a previously published chd7 morpholino (MO2) (13) that is predicted to result in a 1447 aa Chd7 protein product (Supplementary Material, Fig. S1A).
Injection of 2.4ng chd7 MO1 caused developmental defects without causing lethality and all following experiments in this study were performed at this concentration of the MO1, unless otherwise mentioned. From hereon, we refer to chd7 MO1 as chd7 MO (and where MO2 has been used, it is explicitly stated). All the control embryos were injected with equal amount of a control MO with random scrambled sequence from Gene Tools®.
Injection of chd7 MO caused multiple morphological defects including small head and eyes, missing or reduced jaw and pericardial edema (Fig. 7B). In order to determine if the phenotypes are due to downregulation of Chd7, we co-injected 1.5ng of full-length human CHD7 RNA (11) with 1.2ng of chd7 MO. The expression of the human CHD7 RNA was determined by PCR of 24hpf injected embryos (Supplementary Material, Fig. S1F). Craniofacial defects have been noted in chd7 morphant zebrafish previously (13). Four-day old Tg(sox10:eGFP) larvae had GFP expression in the craniofacial cartilage stacks (Supplementary Material, Fig. S1G). In larvae injected with chd7 MO severe defects in the craniofacial cartilage was evident. Co-injection of wildtype human CHD7 RNA with the chd7 MO partially rescued the craniofacial defects in the morphant larvae (Supplementary Material, Fig. S1G). No rescue was observed in larvae co-injected with chd7 MO and dsRed RNA. Thus, human CHD7 is able to replace the loss of zebrafish Chd7 suggesting a high degree of functional conservation between species. These observations also reinforce that the phenotypes observed in the chd7 MO are caused specifically due to reduction of Chd7 activity.
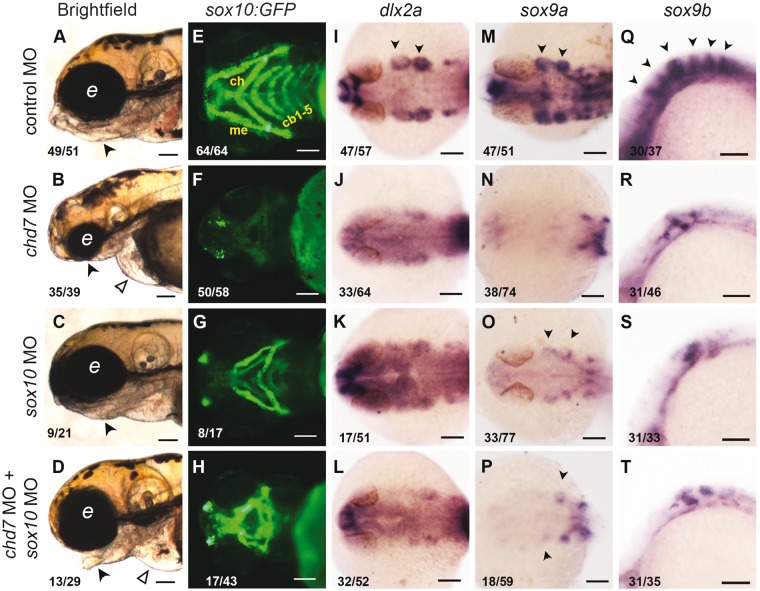
Figure 7.
chd7 knockdown leads to defects in craniofacial cartilage specification and differentiation in a Sox10-dependent manner. (A–D) 4dpf control larvae exhibited a well-formed lower jaw (black arrowhead, A) while chd7 morphants had a missing lower jaw (black arrowhead, B, smaller head and pericardial edema (open arrowhead). Embryos injected with sox10 MO had partial loss of lower jaw (black arrowhead, C). Larvae coinjected with chd7 MO and sox10 MO showed partial rescue of the jaw structures (black arrowhead, D). (E–H) Tg(Sox10:eGFP) marks chondrocyte stacks in the jaw of 4dpf control larvae highlighting the Meckel’s (me), Ceratohyal (ch) and Ceratobranchial (cb1-5) cartilages. All major cartilage elements were absent in the chd7 morphant larvae (F) while ceratobranchial arches were absent in Sox10 morphants (G). Larvae coinjected with chd7 MO and sox10 MO had recovered significant cartilage structures (H). (I–T) RNA in situ hybridization of control (I, M, Q), chd7 MO (J, N, R), sox10 MO (K, O, S) and chd7 and sox10 MO coinjected 24 hpf embryos (l, p, t). Dlx2a marks the migratory NC cells in the pharyngeal arches (black arrowheads, I), which was downregulated in chd7 morphants and sox10 morphants (K). Embryos coinjected with chd7 and sox10 MO did not show any recovery of dlx2a expression (L). (M–P) sox9a which marks the craniofacial cartilage precursors in the pharyngeal arches (black arrowheads, m) was completely lost in the chd7 morphants (N). sox10 morphants had much reduced expression of sox9a (O). Embryos coinjected with chd7 and sox10 MO did not look very different from chd7 morphants (P). (Q–T) sox9b marks the perichondrium bands in control embryos (black arrowheads, Q) and this expression was reduced and the bands disorganized in the chd7 morphants (R) and sox10 morphants (S). Embryos coinjected with chd7 and sox10 MO were similar to the chd7 morphants with no rescue apparent (T). All images have anterior to the left, lateral view (A–D, Q–T) dorsal view (I–P), ventral view (E–H). All scale bars are 100µm. The numbers in the bottom left corner indicate the actual number of embryos of the total represented by the image.
Chd7 Regulates Neural Crest Gene Expression and Migration
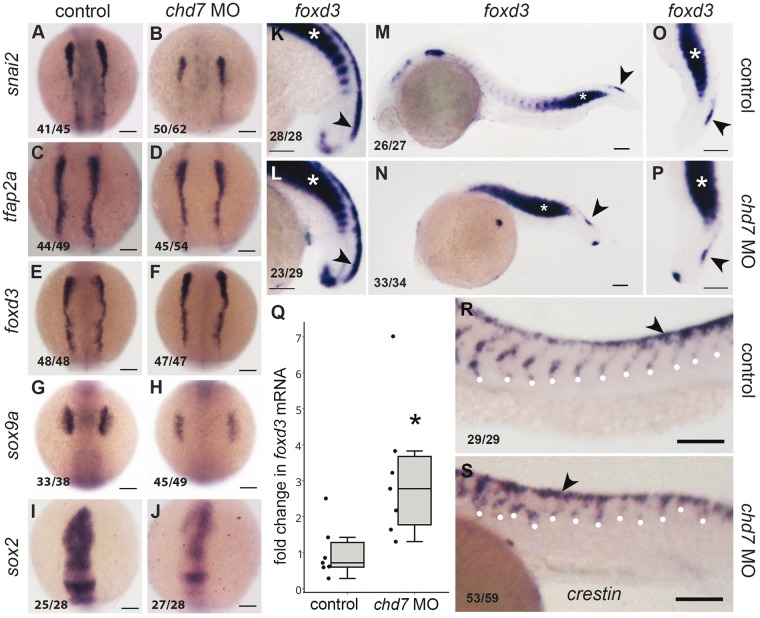
Many NC-derived cell types such as craniofacial cartilage and cranial motor and sensory neurons are affected in the CHARGE syndrome and Chd7 has been implicated in the development of NC derived cell types (11,18,21). However, no study has examined the role of Chd7 in the NC cells in detail. We set out to systematically dissect the effect of chd7 knockdown on NC in the zebrafish embryos, as a model for vertebrate development. We began by analysing 3-somite stage (ss) embryos; around the stage when NC cells are specified. Expression of one of the earliest NC specification markers snai2, is reduced in the chd7 morphants (Fig. 2A and B) while levels of tfap2a and foxd3, both early markers of NC cells, appear largely unperturbed in the morphants compared to the controls (Fig. 2C and 2F). sox9a, an SRY box gene that is crucial for NC specification and migration, is severely downregulated (Fig. 2G and H). Thus, the neural crest appears to be specified but not patterned correctly in the chd7 morphants, perhaps eventually influencing the downstream steps in NC development.
Figure 2.
chd7 knockdown causes differential effects on neural crest markers and impairs neural crest migration. RNA in situ hybridization of neural crest markers at 3 somite (A–J), 18 somite (K–L) and 24hpf (M–P and R–S) stages; control (A, C, E, G, I, K, M, O, R) and chd7 morphant (B, D, F, H, J, L, N, P, S) embryos. (A,B) snai2, the earliest marker of neural crest specification was downregulated in chd7 morphant, while early neural crest markers tfap2a (C,D) and foxd3 (E,F) appeared unaffected. (G, H) sox9a was downregulated in the chd7 morphants. (I, J) sox2, a marker of neural tube, was downregulated in the chd7 morphants. Dorsal views, with anterior to the top (A–J). (K,L) At 18hpf, the somitic (white asterisk) and crest expression (black arrowhead) of foxd3, was upregulated in morphants. (M–P) However at 24hpf, foxd3 was predominantly upregulated in somites (white asterisks) while the expression in caudal neural crest was unchanged (black arrowheads) in chd7 morphants. (O,P) magnified views of trunk region of m, n. (Q) Quantitative real time PCR analysis of foxd3 mRNA shows upregulation in 24hpf chd7 morphant embryos (3.13 fold, P < 0.05). (R,S) crestin-marked neural crest cells migrate robustly in controls but are stalled near the dorsal crest in chd7 morphants (white dots mark the point of migration). Lateral views with anterior to the left, dorsal to the top (K–P, R–S). All scale bars are 100µm. The numbers in the bottom left corner indicate the actual number of embryos of the total represented by the image.
Since NC cells delaminate from the neural tube, we analysed the expression of sox2, a neural tube marker. Expression of sox2, is significantly downregulated in chd7 MO injected embryos (Fig. 2I and J). Human fetuses with CHD7 mutation have been known to have neural tube defects (31). It is possible that some of the effects on NC could be secondary to the defect in neural tube patterning.
We followed the expression of foxd3 into the 18-somite stage embryos. foxd3 is expressed in pre-migratory NC cells and in the somites in the 18-somite stage embryos (Fig. 2K and L) and there is an upregulation of both domains of expression in the chd7 morphants. At 24hpf, the NC-specific expression of foxd3 remains only in the caudal-most region and this appeared unperturbed in the chd7 morphants (Fig. 2M and P). Foxd3 is also expressed robustly in the somites and this expression was significantly upregulated in the morphants (Fig. 2M and P). Foxd3 has been implicated in regulation of myf5 (32), the myogenesis factor expressed in the somites as they differentiate to myoblasts. The foxd3 deregulation may suggest possible skeletal muscle-related complications in CHARGE. We performed quantitative RT-PCR of 24hpf embryos and found an average 3.13 fold upregulation of foxd3 mRNA (Fig. 2Q). A recent study compared the RNA profiles of wildtype and whirlgig (chd7wg/wg) mouse embryos during NC migration (9.5dpc) using microarray and they found a 2.5 fold upregulation of foxd3 in the chd7wg/wg mouse embryos (20). Our whole organism analysis suggests that the overall upregulation evident in the chd7 mutant/morphant background may be primarily attributed to the somitic expression.
By 24hpf NC cells are rapidly migrating in a rostral-to-caudal chronology. Thus, the cranial and vagal cells have completed their migration while the trunk NC may be seen in different stages of the migration as visualized by crestin, a NC marker (33). Trunk migration was severely impaired in the chd7 morphants and cells appear to be still residing in the dorsal position when compared to the control embryos (Fig. 2R and S). This data supports previous studies on human NC like cells (hNCLC) and Xenopus embryos, which demonstrated that NC cell migration is impaired upon CHD7 knockdown (11).
Chd7 Deficiency Causes Deregulation of sox10 Expression
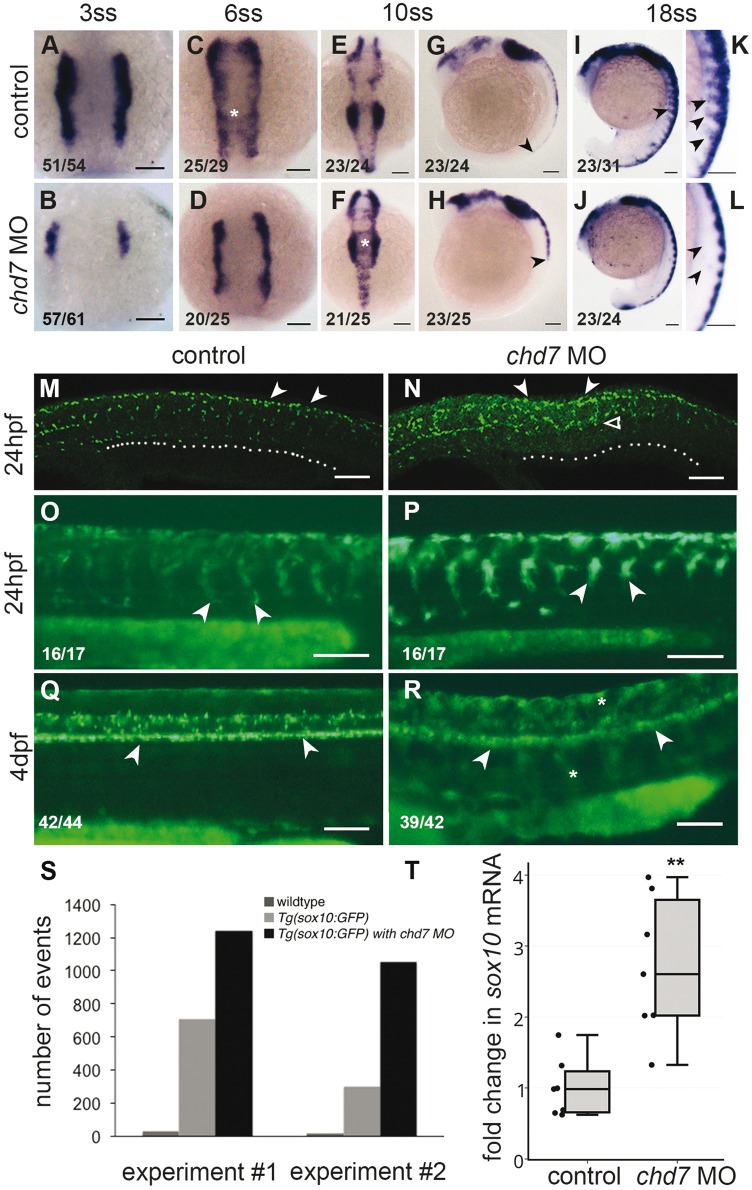
A critical regulator of neural crest cell specification, migration and fate choice, is Sox10. We analysed the expression of sox10 in chd7 morphants from 3ss, when the NC is first evident, to 4dpf, when many NC derived cells are undergoing differentiation, in zebrafish embryo. We mapped sox10 expression in the 3ss (11hpf), 6ss (12hpf), 10ss (14hpf), 14ss (16hpf), 18ss (18hpf) and 22ss (20hpf) embryos. The embryos were staged by counting somites (till 22ss) to confirm the developmental stage. The 3ss control embryos expressed sox10 in the neural plate border cells while in chd7 morphant embryos this expression was not very apparent (Fig. 3A and B). By 6ss the control NC cells were migrating medially (also described in (34), however the NC in the chd7 morphants were still farther apart and there was no evidence of medial migration (Fig. 3C and D). As the vagal and cranial NC condense at 10ss, the medial migration was no longer evident in the control, but appeared to continue in the chd7 morphants (Fig. 3E and F). In lateral views of the embryos the NC, marked by sox10 expression, stretches caudally to more than half the length of the embryo, while in the chd7 morphants the crest was seen only in the anterior trunk region (Fig. 3G and H). In the 14ss (Supplementary Material, Fig. S2A and S2B) and 18ss embryos (Fig. 3I and L), sox10 marks the ventrally streaming NC cells and these streams were absent or severely reduced in the chd7 morphants; this effect was also evident in 20hpf embryos (Supplementary Material, Fig. S2C and S2D). In summary, we observed that the sox10 expression pattern in the chd7 morphants was reminiscent of expression in the control embryos, but a few hours younger, indicating a delay specifically in the NC development.
Figure 3.
Chd7 knockdown causes deregulation of Sox10 expression in the neural crest. (A–L) RNA in situ hybridization of sox10 at different stages; control (A, C, E, G, I, K) and chd7 morphant (B, D, F, H, J, L). (A, B) At 3 somite stage (ss), Sox10, expressed in neural plate border cells is less in chd7 morphants. (C,D) At 6ss, the NC cells begin migration medially (asterisk) in control but not in chd7 morphants. (E–H) At 10ss, the medial migration concluded, cells condense in the vagal and cranial regions, while the chd7 morphants continue medial migration (asterisk) Dorsal views, with anterior to the top (A–F). (G, H) Lateral views of 10ss embryos shows that the NC stretches caudally to more than half the length of the control embryo, while in the chd7 morphants the crest was seen only in the anterior trunk region (black arrowheads). (I–L) At 18ss, Sox10 marks the ventrally migrating streaks of NC cells, which were absent or severely reduced in the chd7 morphants. (K, L) are magnified images of (I, J). Lateral views, with anterior to top (G–L). (M,N). At 24hpf, Sox10 antibody stained cells which have migrated to ventral region of embryo in control (white dots) while in chd7 morphant Sox10 positive cells have travelled only halfway (white open arrowhead) and are accumulated at the dorsal crest (white arrowheads). (O–P) At 24hpf, Tg(sox10:eGFP) marks NC cells that have migrated to the ventral-most region while chd7 morphant showed retention of GFP expression and abnormal migration of NC cells that have stuck midway (white arrowheads). (Q–R) At 4dpf, Tg(sox10:eGFP) shows sox10 expression in spinal cord (white arrowheads), however in chd7 morphant embryo spinal cord expression was not so evident but ectopic GFP positive cells appeared in the trunk region (white asterisks). Lateral views with anterior to left (M–R). (S) Flow cytometry analysis of sox10:eGFP positive cells showed a 1.7 fold to 3.5 fold expansion of GFP positive cell population in the chd7 morphants (T) Quantitative real time PCR analysis of sox10 mRNA showed upregulation in 24hpf chd7 morphant embryos (2.7 fold, P < 0.01). All scale bars are 100µm. The numbers in the bottom left corner indicate the actual number of embryos of the total represented by the image.
At 24hpf, as the NC completes their migration, sox10 is downregulated in these cells. In the control embryos, sox10 expression decreased rapidly and was found only in the occasional dispersed cells and in the yet-to-migrate caudal crest (Supplementary Material, Fig. S2I). We observed that at 24hpf, the chd7 morphant embryos have impaired NC migration and retain a robust expression of sox10 in the dorsal crest region (Supplementary Material, Fig. S2J). This effect on sox10 was confirmed independently in chd7 MO2 injected embryos (Supplementary Material, Fig. S2O and S2P). By quantitative RT-PCR, we observed an average 2.7 fold increase in sox10 gene expression in the chd7 morphant embryos (Fig. 3T). Schulz et al. as described in the previous section, in their microarray analysis on wildtype and whirlgig (chd7wg/wg) 9.5dpc mouse embryos also reported a 2.2 fold upregulation of sox10 RNA in the chd7wg/wg embryos (20).
To determine if the increase in sox10 RNA impacts the protein expression, we performed immunostaining with an anti-Sox10 antibody in 24hpf embryos. We found that the chd7 morphant embryos had more cells expressing Sox10 protein compared to the controls (Fig. 3M and N). We used the Tg(–4.9sox10:egfp)ba2 line to visualize the migrating NC in live embryos at 24hpf and observed a similar increase in the number of GFP positive cells in the morphants when compared to controls (Fig. 3O and P). We quantified this effect using flow cytometry and found a 1.7 to 3.5 fold increase in the number of GFP positive cells in the chd7 morphants (Fig. 3S).
The increase in sox10 expressing cells at 24hpf may be attributed to the delay in migration and the concomitant delay in downregulation of Sox10 in migrating NC cells. However, we observed that the elevation in Sox10 expression was not confined to the 24hpf morphant embryos. We observed ectopic and elevated expression of sox10 in 28hpf sox10:eGFP morphant embryos (Supplementary Material, Fig. S2K–S2N) and in 36hpf chd7 MO embryos by RNA in situ hybridization (Supplementary Material, Fig. S2M and S2N). Examination of 4dpf sox10:eGFP larvae by microscopy showed that the control larvae have a clear pattern of fluorescence in the spinal cord but little elsewhere (Fig. 3Q). The chd7 morphant larvae did not have a very strong fluorescence in the spinal cord, however there was ectopic fluorescence in the trunk region (Fig. 3R). Thus, we conclude that chd7 morphant animals have an overall deregulation of sox10 expression from the early NC stages to later.
chd7 Knockdown Affects Migration of Pigment Precursors and Inhibits Differentiation of Pigment Lineages
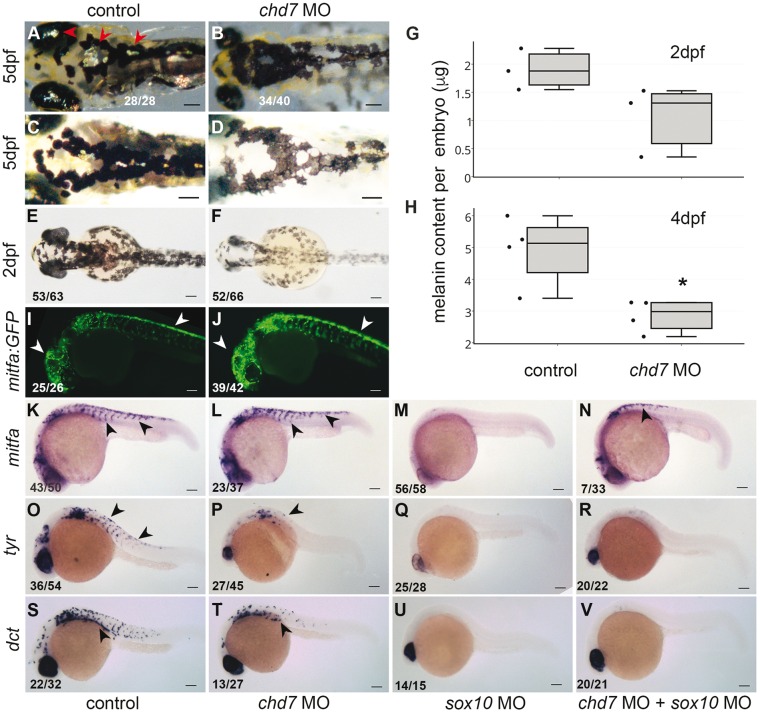
Sox10 is a crucial transcription factor for pigment development in vertebrates (35). Zebrafish has two different kinds of pigment cells melanophores and iridophores that are derived from same precursor pool of NC cells and regulated by Sox10 (36). Since sox10 is deregulated in the chd7 morphant fish, we analysed the status of these pigments in 5dpf zebrafish larvae. Iridophores produce the iridescent pigment in the eyes, top of the head, body and in discrete spots on the trunk in 5dpf control larvae (Fig. 4A). We found an absence of the iridophores (or rather the iridescent pigment) in the whole body of chd7 morphant larvae (Fig. 4B) (previously reported in the eye (12)). Melanophores that contain the black pigment melanin are evident by 30hpf in zebrafish embryos (37) and we observed that 2dpf and 5dpf chd7 morphant animals had decreased pigmentation. At 5dpf, the melanophore shape also appeared to be abnormal in the morphants (Fig. 4C and D). We quantified the melanin content per embryo in 2dpf and 4dpf animals and found a significant decrease in the chd7 morphants compared to controls (Fig. 4G and H).
Figure 4.
chd7 MO affects migration of pigment precursors and inhibits differentiation of pigment lineages. (A–B) At 5dpf, embryos had iridescent iridophores present in eye and body (red arrowheads, a) while iridophores were absent in chd7 morphants. (C–D) At 5dpf, embryos had condensed dark melanophores, while chd7 morphants had spread out dentritic shaped melanophores with overall less melanin. (E–F) 2dpf control embryos are darker than the chd7 morphants. (G–H) Relative melanin content was decreased mildly in 2dpf (1.8 fold, ns) and significantly in 4dpf (1.7 fold, P < 0.05) chd7 morpholino injected larvae. (I, J) Tg(mitfa-GFP) marks the melanoblasts at 24hpf, and these are distributed in the head and dorsal crest region (white arrowheads, I), while, chd7 morphants had expanded population of mitfa:GFP positive cells in both the regions (white arrowheads, J). (K,L) mitfa in situ hybridization at 24hpf marks the melanoblasts in crest and head region, however chd7 morphants had elevated mitfa expression mostly in the dorsal crest. (M,N) sox10 MO injected embryos had no mitf-a expression apparent, while embryos coinjected with sox10 MO and chd7 MO had less mitf-a expression than chd7 MO alone. (O–R) RNA in situ hybridization of tyrosinase marks differentiated melanophores in 24hpf embryos. chd7 morphants had severe downregulation of tyrosinase. Sox10 morphants also had complete absence of tyr, which was not recovered in embryos coinjected with chd7 and sox10 MO. (S–V) RNA in situ hybridization of dct also marks differentiated melanophores in 24hpf embryos. chd7 morphants had severe downregulation of dct as did sox10 morphants. The dct expression was not recovered in embryos coinjected with chd7 and sox10 MO. All images have anterior to the left; dorsal views (A–F) and lateral views (I–V). All scale bars are 100µm. The numbers in the bottom left corner indicate the actual number of embryos of the total represented by the image.
The differentiation of melanophores is initiated by the transcription factor mitf-a which is expressed in the precursors of melanophores as they migrate laterally from the crest (38). The mitf-a positive NC cells are marked by GFP in the Tg(mitf:GFP) transgenic line. At 24hpf, the chd7 morphant embryos have elevated levels of GFP (Fig. 4I and J) compared to controls. RNA in situ hybridization of mitf-a showed delayed migration and dorsal accumulation similar to that seen in sox10 (Fig. 4K and L). Melanophores express two enzymes that are important for melanin formation, tyrosinase (tyr) and dopachrome tautomerase (dct) (39). The expression of these genes is evident in distinct scattered cells throughout the embryo at 24hpf (Fig. 4O and S); this expression is severely downregulated in the chd7 morphants (Fig. 4P and T).
It is important to note that CHARGE patients have not been reported to have any pigmentation defects. Melanophore development is known to be very plastic and alternate routes of melanogenesis may become active even when the primary wave of melanogenesis is compromised (40). It is possible that the requirement for Chd7 by melanophores is species specific and is not necessary in the human melanocytes.
Knockdown of sox10 in Zebrafish Embryos
Since an upregulation of sox10 was noticed in the neural crest of the chd7 morphant (Fig. 3M and P) embryos and sox10 is a known regulator of mitf-a (41), we sought to downregulate Sox10 levels using a MO. We used two sox10 MOs in this study: MO1, a translation blocking MO (42) and MO2, a splice blocking MO (designed for this study, details in Supplementary material Table S1 and Supplementary Material, Fig. S3B). The sox10 MO1 (henceforth referred to as sox10 MO) is predicted to block the translation and immunostaining of sox10 MO injected embryos with anti-Sox10 antibody showed a dramatic reduction in the protein levels in 24hpf embryos (Supplementary Material, Fig. S3C). sox10 MO2 injection would be predicted to interfere with the splicing of exon2 and exon3 causing either retention of intron2 or loss of exon3 (Supplementary Material, Fig. S3D). We performed a PCR with primers on exon2 and exon4 and found that there was no detectable product in the morphant, as predicted if the 2kb long intron is retained in the morphant (Supplementary Material, Fig. S3E). We performed another PCR with primers in exon2 and exon3 and also found significant downregulation of the 382bp PCR product (Supplementary Material, Fig. S3D,S3E,S3F) confirming the aberrant splicing in the sox10 MO2.
Sox10 Downregulation Does Not Affect The Pigment Defect in chd7 Morphant Zebrafish
We examined the embryos after injection of the Sox10 MO for the pigmented melanocytes and pigment regulatory genes. As would be expected, Sox10 MO injected embryos exhibited a reduced pigment phenotype [as previously shown in (43); Supplementary Material, Fig. S4G]. Sox10 MO caused a complete loss of tyr, dct as well as mitf-a (Fig. 4M,Q,U). sox10 MO2 elicited a similar but weaker effect (Supplementary Material, Fig. S4N–S4S). Coinjection of chd7 MO and sox10 MO caused a downregulation of mitfa RNA, compared to the chd7 MO alone suggesting a role for Sox10 in the elevated levels of mitf-a (Supplementary Material, Fig. S4I). However, the double morphant embryos continued to have severe effects on the migration of mitf-a positive NC (Fig. 4N). The double morphants did not show any recovery of the expression of the melanophore differentiation factors tyr or dct (Fig. 4R and V, Supplementary Material, Fig. S4J and S4K). This indicated that chd7 may also play an independent role in the induction of differentiation genes in the melanophore precursors. Recent studies have shown that the MITF regulatory complex, which includes BRG1, that is responsible for induction of melanocytic genes, contains multiple chromatin modifying proteins, including CHD7 (44). Thus, although Sox10 along with Mitfa is known to activate the differentiation genes dct and tyr (45) chd7 may also be essential for the induction of differentiation in the pigment lineage, independent of the deregulation of Sox10.
Figure 5.
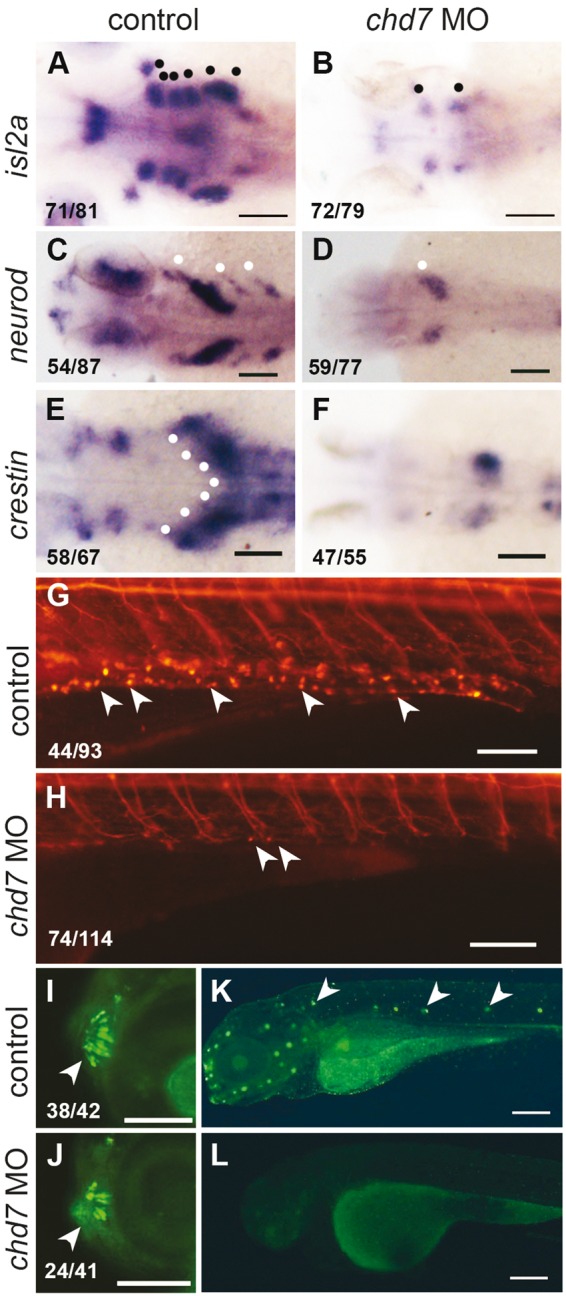
chd7 knockdown causes loss of peripheral neuronal lineages. (A, B) RNA in situ hybridization of islet 2a at 72hpf which marks the differentiated sensory and motor cranial neurons (black dots, a) was severely reduced in chd7 morphants. (C, D) The epibranchial neuronal precursors marked by neuroD in the head (white dots, C) was lacking in the chd7 morphants. (E, F) The vagal neural crest and precursors of enteric neurons marked by crestin in 36hpf control embryos (white dots, E) was reduced in chd7 morphants. (G,H) At 6dpf, Tg(NBT:dsRed) marks enteric neurons on the gut tube (white arrowheads, g) was severely decreased in chd7 morphants. (I, J) Tg(sox10:eGFP) marks well patterned and organised microvillous neurons at 4dpf (white arrowheads, i) and fewer and disorganized microvillous neurons were present in chd7 morphants. (K,L) DASPIE staining highlights the neuromasts cells along lateral line in 4dpf control embryos (white arrowheads, k) and these neurons were completely absent in the chd7 morphant embryos. All images have anterior to the left, dorsal views (A–F) and lateral views (G–L). Scale bars are 100µm (A–H, K–L) and 50µm (I–J). The numbers in the bottom left corner indicate the actual number of embryos of the total represented by the image.
Loss of CHD7 Causes Reduction of Peripheral Neuronal Lineages
Neuronal defects are a hallmark of CHARGE syndrome. CHARGE patients have multiple defects such as defects in facial muscles, xvestibular functions, hearing, olfaction and swallowing difficulties (9). The peripheral nervous system consists of neurons and glia derived from the neural crest. We probed the different peripheral neuronal lineages in our model for CHARGE syndrome. Cranial neurons include both sensory and motor neurons that control functions in the head and face. We detected differentiated cranial neurons in 72hpf embryos using islet-2a (isl2a)(46) as a marker and discovered that knockdown of chd7 leads to a dramatic reduction of these neurons (Fig. 5A and B). This is also substantiated by a previous study in zebrafish (12). Since the cranial neurons are reduced in chd7 morphants, we probed the status of the cranial neuron precursors in the epibranchial ganglia marked by neuroD (47) at 36 hpf; this expression was severely reduced in the chd7 morphant embryos (Fig. 5C and D) suggesting that the defects begin in the precursor populations. The chd7 MO2 had similar effects on neuroD and isl2a expression (Supplementary Material Fig. S5J and S5K and S5N–S5O).
Enteric nervous system innervates the gut. The enteric neurons are originally derived from the migratory vagal NC marked by crestin at 36hpf (48). Knockdown of chd7 reduced the expression of crestin in vagal NC cells (Fig. 5E and F) suggesting that the specification of these cells is compromised. This chd7 MO2 had similar effects on crestin expression (Supplementary Material, Fig. S5L and S5M). The differentiated enteric neurons in 6dpf zebrafish larvae can be visualized using the Tg(NBT:dsRed) line that expresses a red fluorescent protein in all neuronal cell bodies. Deficiency of chd7 led to a severe reduction in the number of enteric neurons (Fig. 5G and H). CHARGE patients have not been reported to have enteric neuronal defects, although there have been reports of chronic constipation in patients (49), which is a symptom of colonic agangliogenesis as seen in Hirschprung disease (50).
At 4dpf, the olfactory microvillous neurons are marked by sox10:eGFP expression (51). We visualized the effect of chd7 knockdown on these cells and found that the chd7 morphant larvae have a reduced number and structural disorganization of the olfactory microvillous neurons (Fig. 5I and J). Chd7 mutant mice also exhibit defects in the number and organization of olfactory sensory neurons (52). The lateral line hair cells are important mechanosensory neurons in the zebrafish. We used the fluorescent dye DASPEI to visualize the lateral line neuromasts and found that chd7 morphants had a significant reduction of these neurons (Fig. 5K and L). Thus, multiple sensory neuronal types are depleted in the chd7 morphant embryos.
To test whether the depletion of peripheral neurons is a result of of Sox10 deregulation, we co-injected embryos with chd7 MO and sox10 MO. We did not observe any rescue of the neurod, crestin or isl2a expression in the embryos co-injected with both chd7 MO and sox10 MO (Supplementary Material, Fig. S5A–S5I, S5P–S5R). Similarly, we analysed the co-injected embryos for olfactory and enteric neurons and did not find any significant rescue compared to chd7 morphants (Supplementary Material, Fig. S5S and S5T). Thus, peripheral neurons derived from the neural crest are dependent on Chd7 function and this function cannot be compensated for by downregulating sox10 mRNA.
Loss of chd7 Expands Glial Precursors But Inhibits Myelination of Schwann Cells in A Sox10-Dependent Manner
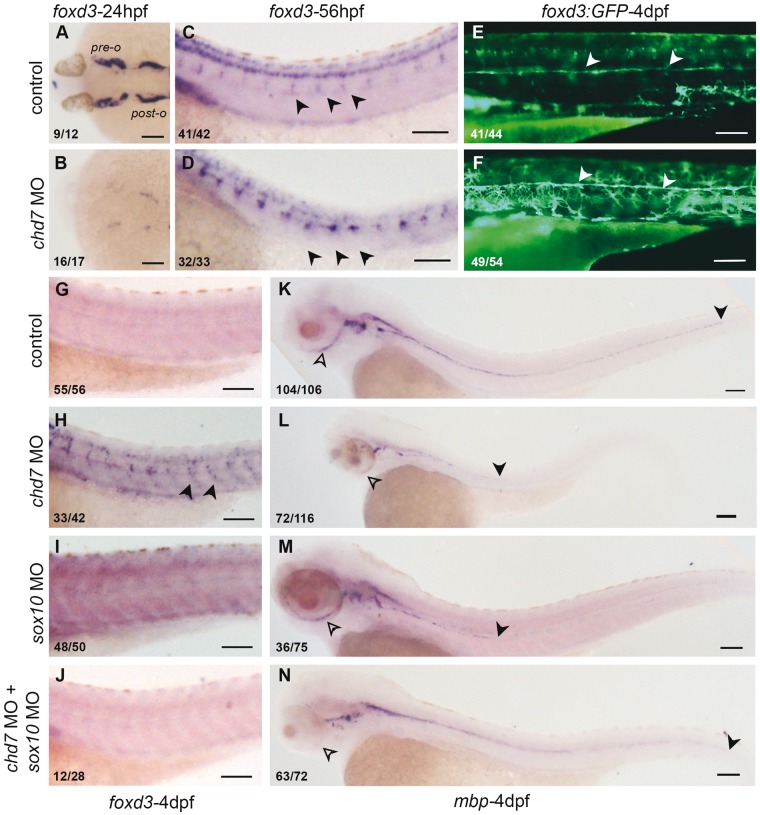
Neural crest cells give rise to a number of different kinds of glia associated with the peripheral neurons viz. satellite glia and myelinated and nonmyelinated Schwann cells (17). At 24hpf, foxd3 is expressed in cranial glia associated with the preotic and postotic ganglia (53) and this expression was completely abolished in the chd7 morphant embryos (Fig. 6A and B). However, at 56hpf foxd3 expression in the trunk satellite glia associated with the dorsal root ganglia (DRG) (54) showed an increase in chd7 morphants compared to the controls (Fig. 6C and D). We also observed an increase in the trunk glia of 4dpf chd7 morphant larvae (Fig. 6G and H) by foxd3 RNA in situ hybridization; this effect was dramatically illustrated in the Tg(foxd3:GFP) transgenic line (Fig. 6E and F) that marks the lateral line glia in 4dpf larvae with GFP (55).
Figure 6.
chd7 knockdown leads to expanded non-myelinating glia and reduced myelinating Schwann cells in a Sox10-dependent manner. (A, B) Cranial glia associated with pre-otic and post-otic ganglia in 24hpf embryos was completely absent in the chd7 morphant embryos. (C, D) In 56hpf embryos foxd3 positive satellite glia associated with the dorsal root ganglia are enhanced in the chd7 morphants. (E, F) Tg(foxd3:GFP) marks the lateral line glia in 4dpf control larvae (arrowheads, E), which shows increased fluorescence intensity with many more dentritic shaped cells present ectopically in the chd7 morphant embryos (injected with 0.8ng of chd7 MO) (arrowheads, F). (G–J) foxd3 RNA in situ hybridization of 4dpf larvae does not show any signal in control, while chd7 morphant embryos have enhanced signal. sox10 morphants and embryos coinjected with chd7 and sox10 MO are not different from the control. (K–N) RNA in situ hybridization for myelin basic protein (mbp) in 4dpf embryo marks the myelinated glia. The extend of myelinating Schwann cells on the lateral line are marked by the black arrowhead and the periocular ganglia are marked by black open arrowheads. .The chd7 morphant embryos (L) and the sox10 morphant embryos (N) have reduced extend of lateral line glia (black arrowhead) and periocular glia (open arrowhead). Emrbyos coinjected with chd7 MO and sox10 MO show a recovery in the posterior extend of the lateral line Schwann cells (black arrowheads N). All images have anterior to the left, dorsal views (A–B) and lateral views (C–N). All scale bars are 100µm. The numbers in the bottom left corner indicate the actual number of embryos of the total represented by the image.
To determine if this increase in foxd3 positive glial cells also results in more myelinated Schwann cells we performed RNA in situ hybridization for myelin basic protein (mbp) mRNA. mbp, a specific marker of myelinated Schwann cells is prominently expressed in the lateral line and in the periocular region at 4dpf (56). Deficiency of chd7 causes an overall decrease in the expression of mbp, around the eye and on the lateral line (Fig. 6K and L). This was further confirmed by quantitative RT-PCR (0.4 fold, Supplementary Material, Fig. S6H). Larvae injected with chd7 MO2 also had a similar downregulation of mbp in the lateral line and the periocular region (Supplementary Material, Fig. S6F and S6G). The expression of mbp and in turn myelination is induced by the transcription factor, krox20. Compared to control larvae, the expression of krox20 in the chd7 morphant larvae was also significantly reduced (Supplementary Material, Fig. S6A and S6B).
We have thus uncovered a novel aspect of CHARGE syndrome in our zebrafish model. The severe reduction in the myelinated Schwann cells of the peripheral nervous system was accompanied by an expansion of satellite and other non-myelinated glia marked by foxd3 in the morphant. We speculate that glial defects may contribute to many of the peripheral nervous system deficits in CHARGE patients. Micucci and colleagues have recently reported an increase in glia in the subventricular zone in the brain in Chd7 mutant mice resulting from a neuron-to-glia fate switch (57). Very recent studies by He et al. have shown that Chd7 and Sox10 interact to specify central nervous system glial cells in mouse (58). In our study, chd7 deficiency caused an expansion of glial cells in the trunk but was detrimental to myelination of these cells.
Sox10, overexpressed in 24hpf and older chd7 morphant embryos is a known regulator of foxd3 (59), the marker of glial precursors (55). Sox10 is also a crucial regulator of glial fate choice and myelination (60). We asked whether the glial cell phenotype in chd7 morphants is mediated by Sox10. We knocked down sox10 in the chd7 morphants and observed the effects on glial fate. The expansion of foxd3 expression in the trunk of 4dpf chd7 morphant larvae was reversed by the combined reduction of chd7 and sox10 (Fig. 6G and J, Supplementary Material, Fig. S6I). Further, we analysed the effect of the combined knockdown of chd7 and sox10 on myelination. We found that co-injection of chd7 MO and sox10 MO affected a dramatic rescue of the myelinated Schwann cells; 90% of double morphant embryos showed a nearly normal lateral line expression of mbp (Fig. 6K and N and Supplementary Material, Fig. S6J) and nearly half also rescued the periocular glia. This effect was also reproduced in embryos coinjected with chd7 MO and sox10 MO2 (Supplementary Material, Fig. S6C–S6E).
Sox10 is a known inducer of the myelination program in the glia; however a previous study in chick embryos has shown that the overexpression of Sox10 results in inhibition of differentiation of NC derivatives, including myelinated Schwann cells (61). We therefore suggest that the overexpression of Sox10 in the chd7 morphants is the cause of inhibition of myelination and that when this inhibition was relieved in double morphants, the foxd3 positive glial cells differentiated into myelinated Schwann cells efficiently.
chd7 Knockdown Leads to Defects in Craniofacial Cartilage Specification and Differentiation in a Sox10-Dependent Manner
Craniofacial skeletal abnormalities are a predominant feature of CHARGE syndrome (9). Our chd7 morphant larvae also exhibited a smaller head, smaller eyes and an absence of the lower jaw at 4dpf (Fig. 7A and B). Alcian blue staining of cartilage (Supplementary Material, Fig. S7B) and sox10:eGFP fluorescence in the chondrocyte stacks (Fig. 7F) in the 4dpf morphant larvae confirmed the severe reduction of craniofacial cartilage structures such as the Meckel’s, Ceratohyal, and Ceratobranchial cartilages. Craniofacial defects have been reported in zebrafish (13) and mouse models (21). chd7 MO2 caused similar but less severe jaw phenotypes (Supplementary Material, Fig. S7L).
The precursors of ectomesenchyme express transcription factors such as dlx2a, sox9a and sox9b that commit them to cartilage fate and promote the activation of cartilage-specific genes (62). We analysed the expression of dlx2a in 24hpf embryos, expressed in the migratory neural crest cells that form pharyngeal arches 1-4 (62,63), and found that the chd7 morphants showed a reduction of dlx2a in the arches (Fig. 7I and J). Sox9a and sox9b, two paralogs of SOX9 in zebrafish, regulate chondrogenesis in the head (62). The control embryos have robust sox9a and sox9b expression in the pharyngeal arches 1 and 2 and in the perichondrium (62) respectively. We found a severe reduction in the expression of both sox9a and sox9b genes in the chd7 deficient embryos (Fig. 7M,N,Q,R). It has been previously shown that the CHD7 protein can bind and activate the human SOX9 enhancer (11) and our present results suggest that sox9 might be an evolutionarily conserved target of CHD7 from zebrafish to humans.
We probed the precursor population of NC cells that give rise to the ectomesenchymal derivatives in the craniofacial region of the embryo. Migratory cranial NC cells are marked by crestin at 24hpf (64), and we observed an almost complete absence of crestin staining in the chd7 morphant embryos (Supplementary Material, Fig. S7I and S7J). On the other hand, as described earlier, more Sox10 positive cells were present in the cranial region in the chd7 morphants at 24hpf (Supplementary Material, Fig. S2E and S2F). This suggests that the defect in the derivatives of the neural crest in the head may be a defect of ectomesenchymal fate choice. So, could the increase in Sox10 expression contribute to a suppression of cartilage fate in the morphants? To address this, we co-injected the chd7 MO with sox10 MO and observed the effect on the jaw in sox10:eGFP larvae. Our analysis revealed that compared to 14% of chd7 morphants that have normal or partial cartilage structures, 72% of the double morphants have partial or normal cartilage (Fig. 7E and H and Supplementary Material, Fig. S7N). Using Alcian blue staining and sox10:eGFP fluorescence for the craniofacial cartilage we observed that both sox10 MO1 and sox10 MO2 caused defects and reduction in the calcification of craniofacial structures in the 4dpf larvae Supplementary Material, Fig. S7C, S7G, S7N and Fig. 7G). Upon co-injection of chd7 MO with sox10 MO1 or sox10 MO2 a partial recovery of the cartilage structures were evident in the larvae (Supplementary Material, Fig. S7D, S7H, S7N). Sox9a, sox9b and dlx2a did not show significant rescue in the double morphants (Fig. 7L,P,T and Supplementary Material, Fig. S7O,S7P,S7Q). This suggests that reducing the Sox10 levels can partially rescue the craniofacial cartilage defects in the chd7 morphants, however the mechanism remains unclear. Our results support the view that the craniofacial skeletal defects in CHARGE are due to defective fate specification in the ectomesenchymal lineage and that this is caused, at least in part, by the deregulation of Sox10.
Discussion
CHARGE syndrome is a debilitating and often fatal congenital disorder. 65–70% of patients have a mutation in the chromatin remodeler CHD7 (9). Many of the tissues affected in CHARGE syndrome such as heart, auditory neurons, facial neurons and craniofacial bones (9) have their origins in the neural crest lineage. Earlier studies have also implicated a role for Chd7 in the neural crest cells (11,18,21). In this study, we aimed to systematically understand the neural crest related defects in a chd7 compromised state. Since NC is a unique vertebrate tissue type (65), we chose zebrafish the highly amenable vertebrate model organism to perform this study.
Using morpholino antisense oligonucleotides to knockdown zebrafish chd7 we generated a model for CHARGE syndrome. Using this model we identified certain novel aspects of Chd7 function and perhaps CHARGE manifestation. We discovered defects in pigmentation, enteric neurons and myelination of Schwann cells, all previously undocumented in CHARGE patients or CHARGE models. We suggest that some of these phenotypes such as abnormalities in enteric ganglia or myelination defects may have relevance in patients and may have been overlooked just because they are not obvious during gross examination in the clinic.
The morphant zebrafish showed a selective delay in NC development with striking abnormalities in the trunk NC migration. We found that NC cells marked by sox10, migrate efficiently into the head NC regions but crestin positive cells were absent in the head. We interpret this as a change of fate in the cranial NC from crestin expressing cells to Sox10 expressing cells. We also discovered that the craniofacial cartilage, that form from the cranial NC are compromised in the chd7 morphants. Sox10 downregulation was able to rescue these craniofacial defects indicating a role for Sox10 in the fate determination of the cranial NC.
We also documented a persistence or upregulation of Sox10 in the NC cells in the chd7 morphant embryos. This could be a direct consequence of the delay or defect in migration of the NC in the chd7 morphants, since Sox10 downregulation in normal NC cells is linked to their ventral migration. However, our study indicates that timely downregulation of Sox10 is an important event in the specification and differentiation of NC cells into their derivatives and that a delay in the downregulation of Sox10 can have consequences on fate choice and differentiation of NC cells. This hypothesis is borne out by the apparent rescue of craniofacial cartilage and myelination defects in chd7 morphants by downregulation of Sox10 protein.
Thus, we conclude that Chd7 is an important player in the specification, migration, fate-choice and differentiation of NC. We also conclude that Sox10 is an important mediator of function of Chd7 in NC. We show that by external regulation of sox10 expression, we can rescue some aspects of the neural crest derived phenotypes in the CHARGE model suggesting possible avenues of intervention in the future.
Materials and Methods
Zebrafish lines and maintenance
Zebrafish (Danio rerio) were bred, raised and maintained at 28.5 °C under standard conditions as described (66). Embryo staging was done by using both timing (hours post fertilization, hpf) and morphological features as previously described (67). For fluorescence imaging and in situ hybridization analysis of embryos older than 24hpf, embryos were raised in 1-phenyl-2-thiourea (PTU) (0.003% in Embryo medium), to inhibit pigment formation (68). Zebrafish handling was in strict accordance with good animal practice as defined and all experiments were performed using protocols approved by the Institutional Animal Ethics Committee (IAEC) of the CSIR-Institute of Genomics and Integrative Biology, India. The zebrafish lines used in this study are Tuebingen (TU), used as the wild-type strain, Tg(–4.9Sox10:egfp)ba2(69), Tg(foxd3:GFP)(55), Tg(mitfa:GFP)w47(70), and Tg(NBT-dsRed) (71).
RNA sequencing analysis
The publicly available data for RNA-seq was obtained from NCBI SRA. Developmental timepoints RNA seq data was obtained from SRP009426 (23), SRP008845 (24), ERP000016 [Zebrafish transcriptome sequencing project, PRJEB1986] PRJNA207719 (25) and SRP017135 (26), corresponds to adult tissues. The SRA data was processed into fastq format, followed by preliminary quality check (QC) by FastQC and adapter trimming using Trimmomatic (72) The data comprised of 10 developmental stages viz., 2-4 cell, 1Kcell, dome, shield, bud, 1dpf, 2dpf, 3dpf, 5dpf, 14dpf and 9 adult tissues viz. blood, brain, heart, liver, muscle, eye, spleen, ovary and testis .
Once the preliminary QC was completed the reads for respective stages and tissues were aligned on the reference genome of zebrafish (zv10) using TopHat (73). The alignment was followed by analysing the differential expression of the annotated transcripts (ens84). Differential expression of the transcripts across the 10 stages and 9 tissues of zebrafish was obtained using Cuffdiff module (73) for which Binary alignment map (BAM) files served as input obtained from TopHat. The differential expression was in terms of Fragment per Kilo base Exon per Million Reads (FPKM) scores which is used to give the relative abundance of the transcripts across the data points. The FPKM scores were further fetched for the chd transcripts and the heatmap for the same was plotted so as to visualize the expression of them across the stages and tissues of zebrafish.
Morpholino and RNA injections
The endogenous expression of genes was perturbed by injection of morpholinos. All morpholino antisense oligonucleotides (MOs) were designed and synthesized by Gene-Tools, USA. For all experiments, the minimum effective concentration was used as determined by titration experiments. Two different chd7 splice blocking MOs have been used in this study. The first chd7 splice blocking MO (MO1) was designed against the exon8-intron8 boundary of chd7 transcript (ENSDART00000135230) (Supplementary Material, Fig. S1). For confirmation, other previously reported chd7 splice blocking MOs (termed here as, MO2) was also used (13).
To determine the specificity of the phenotypes observed full length human CHD7 RNA was co-injected with chd7 MO. pCDNA plasmid containing full length human CHD7 was linearized with AvrII. The linear fragment was gel-eluted and transcribed using T7-ultra mRNA synthesis kit (Ambion-Thermo Fisher). The full length mRNA was purified using oligodT based mRNA purification kit (Invitrogen), run on an agarose gel for confirmation, quantitated and frozen in small aliquotes. As truncated or partially degraded CHD7 mRNA can function as dominant negative, thawed aliquotes of mRNA were used only once. Each embryo was injected with 1.5ng of human CHD7 RNA and 1.2ng of chd7 MO.
sox10 knockdown was performed using a previously reported translation blocking morpholino (42) and a splice blocking morpholino designed across exon2 and intron2 hereafter termed as sox10 MO2. Concentrations of the various MOs used in injections are: 2.4ng for both chd7 MO1 and MO2 and sox10 MO1, and 12ng for sox10 MO2. A standard control scrambled sequence MO obtained from Gene Tools® was used in all experiments as a control. All injections were done at one-cell staged embryos. Morpholinos were suspended in nuclease free water to form a stock solution of 1mM. Working solutions were made by dissolving in nuclease free water and used within 2–3 days . The list of MO sequences is given in Supplementary Material, Table S1)
Polymerase chain reaction
Total RNA was extracted by homogenizing 15–20 pooled embryos using Trizol reagent (Invitrogen). RNA was further treated with DNase (Ambion) to remove genomic DNA contamination. Wildtype (zebrafish and human) as well as aberrant chd7 transcripts were detected using RT-PCRs. Quantitative real time PCR was carried out as described (74) on the Roche LightCycler® Real-Time PCR System. For quantification, the relative standard curve method was used (as described by the manufacturer) to generate raw values representing arbitrary units of RNA transcript. Each experiment was performed on three independent occasions in biological triplicates of pooled embryos (in every experiment). Statistical analyses on normalized data were performed using 2−ΔΔCT algorithm (known as the delta-delta-Ct or ddCt algorithm) (74,75). All genes were normalized against RPL13α unless mentioned otherwise. All primers used in this study are listed in Supplementary Materials Tables S4, S5 and S6.
Imaging
Embryos (staged between 1 and 6dpf) were monitored and scored for phenotypes using Zeiss (Stemi 2000C®) bright field microscope (with AxiocamICc1). For visualizing iridophores, reflected light with the black background was used. For fluorescent imaging, Zeiss AxioScope A1 microscope (with Axiocam HRc®) was used. Images were captured using Zeiss proprietary software and processed and analysed in Adobe® Photoshop®.
Wholemount RNA in situ hybridization and immunohistochemistry
RNA in situ hybridization was performed as described (76). Probes of the following genes were used: sox2, snai1b, Sox10, foxd3, tfap2a, sox9a, dlx2a, crestin, mitfa and dct (77); sox9b (62), neurod (78), isl2 (79), tyr (80), mbp (56) and krox20 (81).
To generate a probe for chd7 a 525bp fragment of the chd7 gene was PCR amplified from 24hpf zebrafish cDNA (Primer sequences in the Supplementary Material, Table S2). The fragment was cloned into pCR4-TOPO® vector and was used as template for in vitro transcription of antisense chd7 RNA probe. Images were processed as mentioned earlier.
Immunostaining for Sox10 was performed broadly as described (82). The embryos were fixed in 4% paraformaldehyde and were dehydrated through a graded methanol series. Further, the embryos were incubated with anti-Sox10 Ab (1:100 dilution) (GTX128374, Gene Tex®) for 24hrs and goat anti-rabbit IgG secondary antibody conjugated with Alexa fluor 488 was used (1:500 dilution) (A-11008, Thermo Fisher Scientific®).
Fluorescence Assorted Cell Sorting (FACS) Analysis
At 24hpf, live embryos were dechorionated with Pronase and de-yolked in ice-cold Ringers solution using a micropipette tip. Single cell suspension of the de-yolked embryos was prepared using TrypLeTM Express (Thermo Fisher Scientific®). The cells were collected, washed with ice-cold Ringers solution and passed through a 70µm filter to avoid cell clumping. The cells were then collected and re-suspended in DPBS with 2.5% FBS. The cells were analysed for GFP + ve cells in BD FACS Aria III®. Uninjected sox10:eGFP positive embryos were used as controls. FACS data were analysed using BD FACSDiva™ software.
Alcian blue staining
Alcian blue stain was used to visualize the structure of the craniofacial skeleton of zebrafish as described previously (83).
DASPEI staining
DASPEI (2-(4-(dimethylamino)styryl)-N-ethylpyridinium iodide) stains hair cells within lateral line neuromasts of live zebrafish embryos. The staining procedure was carried out as described (84).
Melanin content
Melanin content was determined as described previously in (85). Embryo lysates were prepared for both control embryos and chd7 morphants, at 2dpf and 4dpf. The sample pellet was dissolved in 1 N NaOH at 100 °C for 50 min. Optical density was measured at 490 nm, and the sample results were compared with a standard curve of known concentrations (0–300 µg/ml) of melanin (Sigma). The experiment was performed on three or more independent occasions; with sample size, i.e. number of embryos ≥50, and same for both control and chd7 MO, each time. For relative comparison, melanin content per embryo (µg) was plotted against sample.
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
We thank Alessandro Mongera, Prateek Mahalwar and Soundhar Ramsay for generous help and for reagents. We thank Archana Vats and D. Ayyappa Raja for help with FACS analysis and Manish Kumar for confocal microscopy. We thank Vivek Natarajan and Mageshi Kamaraj for comments and discussion.
Conflict of Interest Statement. None declared.
Funding
This work was supported by Council of Scientific and Industrial Research (CSIR), New Delhi [BSC0118 to C. S., research fellowships to Z.A. and S. K. and BSC0403 for the microscopy facility], National Institutes of Health-National Institute of Dental and Craniofacial Research [R01DE024584 to R. B.] and CHARGE Syndrome Foundation Grant [R.B.].
References
- 1.Marfella C.G.A., Imbalzano A.N. (2007) The Chd family of chromatin remodelers. Mutat Res-Fund Mol M, 618, 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schnetz M.P., Bartels C.F., Shastri K., Balasubramanian D., Zentner G.E., Balaji R., Zhang X., Song L., Wang Z., Laframboise T., et al. (2009) Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Research, 19, 590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engelen E., Akinci U., Bryne J.C., Hou J., Gontan C., Moen M., Szumska D., Kockx C., van IJcken W., Dekkers D.H.W., et al. (2011) Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat Genet, 43, 607. U153. [DOI] [PubMed] [Google Scholar]
- 4.Bouazoune K., Kingston R.E. (2012) Chromatin remodeling by the CHD7 protein is impaired by mutations that cause human developmental disorders. Proc. Natl Acad. Sci. U S A, 109, 19238–19243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schnetz M.P., Handoko L., Akhtar-Zaidi B., Bartels C.F., Pereira C.F., Fisher A.G., Adams D.J., Flicek P., Crawford G.E., Laframboise T., et al. (2010) CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. Plos Genet, 6, e1001023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zentner G.E., Hurd E.A., Schnetz M.P., Handoko L., Wang C.P., Wang Z.H., Wei C.L., Tesar P.J., Hatzoglou M., Martin D.M., et al. (2010) CHD7 functions in the nucleolus as a positive regulator of ribosomal RNA biogenesis. Hum. Mol. Genet., 19, 3491–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pagon R.A., Graham J.M., Jr., Zonana J., Yong S.L. (1981) Coloboma, congenital heart disease, and choanal atresia with multiple anomalies: CHARGE association. J. Pediatr., 99, 223–227. [DOI] [PubMed] [Google Scholar]
- 8.Vissers L.E.L.M., van Ravenswaaij C.M.A., Admiraal R., Hurst J.A., de Vries B.B.A., Janssen I.M., van der Vliet W.A., Huys E.H.L.P.G., de Jong P.J., Hamel B.C.J., et al. (2004) Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat. Genet., 36, 955–957. [DOI] [PubMed] [Google Scholar]
- 9.Zentner G.E., Layman W.S., Martin D.M., Scacheri P.C. (2010) Molecular and phenotypic aspects of CHD7 mutation in CHARGE syndrome. Am. J.Med. Genet. Part A, 152A, 674–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Legendre M., Gonzales M., Goudefroye G., Bilan F., Parisot P., Perez M.J., Bonniere M., Bessieres B., Martinovic J., Delezoide A.L., et al. (2012) Antenatal spectrum of CHARGE syndrome in 40 fetuses with CHD7 mutations. J. Med. Genet., 49, 698–707. [DOI] [PubMed] [Google Scholar]
- 11.Bajpai R., Chen D.A., Rada-Iglesias A., Zhang J., Xiong Y., Helms J., Chang C.P., Zhao Y., Swigut T., Wysocka J. (2010) CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature, 463, 958–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patten S.A., Jacobs-McDaniels N.L., Zaouter C., Drapeau P., Albertson R.C., Moldovan F. (2012) Role of Chd7 in Zebrafish: A Model for CHARGE Syndrome. Plos One, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balow S.A., Pierce L.X., Zentner G.E., Conrad P.A., Davis S., Sabaawy H.E., McDermott B.M., Scacheri P.C. (2013) Knockdown of fbxl10/kdm2bb rescues chd7 morphant phenotype in a zebrafish model of CHARGE syndrome. Dev. Biol., 382, 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bosman E.A., Penn A.C., Ambrose J.C., Kettleborough R., Stemple D.L., Steel K.P. (2005) Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum. Mol. Genet., 14, 3463–3476. [DOI] [PubMed] [Google Scholar]
- 15.Hurd E.A., Capers P.L., Blauwkamp M.N., Adams M.E., Raphael Y., Poucher H.K., Martin D.M. (2007) Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mamm Genome, 18, 94–104. [DOI] [PubMed] [Google Scholar]
- 16.Melicharek D.J., Ramirez L.C., Singh S., Thompson R., Marenda D.R. (2010) Kismet/CHD7 regulates axon morphology, memory and locomotion in a Drosophila model of CHARGE syndrome. Hum. Mol. Genet., 19, 4253–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trainor P. (2013) Neural Crest Cells: Evolution, Development and Disease. Academic Press. [Google Scholar]
- 18.Fujita K., Ogawa R., Kawawaki S., Ito K. (2014) Roles of chromatin remodelers in maintenance mechanisms of multipotency of mouse trunk neural crest cells in the formation of neural crest-derived stem cells. Mech. Dev., 133, 126–145. [DOI] [PubMed] [Google Scholar]
- 19.Mandalos N., Rhinn M., Granchi Z., Karampelas I., Mitsiadis T., Economides A., Dollé P.a, Remboutsika E. (2014) Sox2 acts as a rheostat of epithelial to mesenchymal transition during neural crest development. Front Physiol., 5, 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schulz Y., Wehner P., Opitz L., Salinas-Riester G., Bongers E.M.H.F., van Ravenswaaij-Arts C.M.A., Wincent J., Schoumans J., Kohlhase J., Borchers A., et al. (2014) CHD7, the gene mutated in CHARGE syndrome, regulates genes involved in neural crest cell guidance. Hum. Genet., 133, 997–1009. [DOI] [PubMed] [Google Scholar]
- 21.Sperry E.D., Hurd E.A., Durham M.A., Reamer E.N., Stein A.B., Martin D.M. (2014) The Chromatin Remodeling Protein CHD7, Mutated in CHARGE Syndrome, is Necessary for Proper Craniofacial and Tracheal Development. Dev. Dynam., 243, 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall J.A., Georgel P.T. (2007) CHD proteins: a diverse family with strong ties. Biochem. Cell Biol., 85, 463–476. [DOI] [PubMed] [Google Scholar]
- 23.Pauli A., Valen E., Lin M.F., Garber M., Vastenhouw N.L., Levin J.Z., Fan L., Sandelin A., Rinn J.L., Regev A., et al. (2012) Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res., 22, 577–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ulitsky I., Shkumatava A., Jan C.H., Sive H., Bartel D.P. (2011) Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell, 147, 1537–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaushik K., Leonard V.E., Kv S., Lalwani M.K., Jalali S., Patowary A., Joshi A., Scaria V., Sivasubbu S. (2013) Dynamic expression of long non-coding RNAs (lncRNAs) in adult zebrafish. Plos One, 8, e83616.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelkar D.S., Provost E., Chaerkady R., Muthusamy B., Manda S.S., Subbannayya T., Selvan L.D.N., Wang C.H., Datta K.K., Woo S. (2014) Annotation of the zebrafish genome through an integrated transcriptomic and proteomic analysis. Mol. Cell. Proteomics, 13, 3184–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Boyle S., Bree R.T., McLoughlin S., Grealy M., Byrnes L. (2007) Identification of zygotic genes expressed at the midblastula transition in zebrafish. Biochem. Biophys. Res. Commun., 358, 462–468. [DOI] [PubMed] [Google Scholar]
- 28.Jacobs-McDaniels N.L., Albertson R.C. (2011) Chd7 plays a critical role in controlling left-right symmetry during zebrafish somitogenesis. Dev Dyn., 240, 2272–2280. [DOI] [PubMed] [Google Scholar]
- 29.Kim K.H., Roberts C.W. (2013) CHD7 in charge of neurogenesis. Cell Stem Cell, 13, 1–2. [DOI] [PubMed] [Google Scholar]
- 30.Feng W., Khan M.A., Bellvis P., Zhu Z., Bernhardt O., Herold-Mende C., Liu H.K. (2013) The chromatin remodeler CHD7 regulates adult neurogenesis via activation of SoxC transcription factors. Cell Stem Cell, 13, 62–72. [DOI] [PubMed] [Google Scholar]
- 31.Haldipur P., Millen K.J. (2013) Deficits in early neural tube identity found in CHARGE syndrome. Elife, 2, e01873.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee H.C., Huang H.Y., Lin C.Y., Chen Y.H., Tsai H.J. (2006) Foxd3 mediates zebrafish myf5 expression during early somitogenesis. Dev. Biol., 290, 359–372. [DOI] [PubMed] [Google Scholar]
- 33.Honjo Y., Kniss J., Eisen J.S. (2008) Neuregulin-mediated ErbB3 signaling is required for formation of zebrafish dorsal root ganglion neurons. Development, 135, 2615–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drerup C.M., Wiora H.M., Topczewski J., Morris J.A. (2009) Disc1 regulates foxd3 and sox10 expression, affecting neural crest migration and differentiation. Development, 136, 2623–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silver D.L., Hou L., Pavan W.J. (2006), In Saint-Jeannet, Jean-Pierre (eds) Neural Crest Induction and Differentiation. Springer, US, pp. 155–169. [Google Scholar]
- 36.Curran K., Lister J.A., Kunkel G.R., Prendergast A., Parichy D.M., Raible D.W. (2010) Interplay between Foxd3 and Mitf regulates cell fate plasticity in the zebrafish neural crest. Dev. Biol., 344, 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tryon R.C., Johnson S.L. (2012) Clonal and lineage analysis of melanocyte stem cells and their progeny in the zebrafish. Methods Mol. Biol, 916, 181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mort R.L., Jackson I.J., Patton E.E. (2015) The melanocyte lineage in development and disease. Development, 142, 620–632.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quigley I.K., Parichy D.M. (2002) Pigment pattern formation in zebrafish: a model for developmental genetics and the evolution of form. Microsc. Res. Tech., 58, 442–455. [DOI] [PubMed] [Google Scholar]
- 40.Hultman K.A., Johnson S.L. (2010) Differential contribution of direct-developing and stem cell-derived melanocytes to the zebrafish larval pigment pattern. Dev. Biol., 337, 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elworthy S., Lister J.A., Carney T.J., Raible D.W., Kelsh R.N. (2003) Transcriptional regulation of mitfa accounts for the sox10 requirement in zebrafish melanophore development. Development, 130, 2809–2818. [DOI] [PubMed] [Google Scholar]
- 42.Dutton K., Dutton J., Pauliny A., Kelsh R. (2001) A morpholino phenocopy of the colourless mutant. Genesis, 30, 188–189. [DOI] [PubMed] [Google Scholar]
- 43.Whitlock K.E., Smith K.M., Kim H., Harden M.V. (2005) A role for foxd3 and sox10 in the differentiation of gonadotropin-releasing hormone (GnRH) cells in the zebrafish Danio rerio. Development, 132, 5491–5502. [DOI] [PubMed] [Google Scholar]
- 44.Laurette P., Strub T., Koludrovic D., Keime C., Le Gras S., Seberg H., Van Otterloo E., Imrichova H., Siddaway R., Aerts S., et al. (2015) Transcription factor MITF and remodeller BRG1 define chromatin organisation at regulatory elements in melanoma cells. Elife, 4, doi: 10.7554/eLife.06857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hou L., Pavan W.J. (2008) Transcriptional and signaling regulation in neural crest stem cell-derived melanocyte development: do all roads lead to Mitf? Cell Res., 18, 1163–1176. [DOI] [PubMed] [Google Scholar]
- 46.LoÌ p̂ez B.C., Nieto-Sampedro M. (2003) Glial cell function. Gulf Professional Publishing. [Google Scholar]
- 47.Holzschuh J., Wada N., Wada C., Schaffer A., Javidan Y., Tallafuss A., Bally-Cuif L., Schilling T.F. (2005) Requirements for endoderm and BMP signaling in sensory neurogenesis in zebrafish. Development, 132, 3731–3742. [DOI] [PubMed] [Google Scholar]
- 48.Reichenbach B., Delalande J.M., Kolmogorova E., Prier A., Nguyen T., Smith C.M., Holzschuh J., Shepherd I.T. (2008) Endoderm-derived sonic hedgehog and mesoderm Hand2 expression are required for enteric nervous system development in zebrafish. Dev. Biol., 318, 52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arvedson J., Hartshorne T., Hefner M., Davenport S., Thelin J. (2011) Feeding issues. CHARGE Syndrome, 113–138. in press., [Google Scholar]
- 50.Southard-Smith E.M., Kos L., Pavan W.J. (1998) Sox10 mutation disrupts neural crest development in DOM Hirschsprung mouse model. Nat. Genet., 18, 60–64. [DOI] [PubMed] [Google Scholar]
- 51.Saxena A., Peng B.N., Bronner M.E. (2013) Sox10-dependent neural crest origin of olfactory microvillous neurons in zebrafish. Elife, 2, e00336.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Layman W.S., McEwen D.P., Beyer L.A., Lalani S.R., Fernbach S.D., Oh E., Swaroop A., Hegg C.C., Raphael Y., Martens J.R., et al. (2009) Defects in neural stem cell proliferation and olfaction in Chd7 deficient mice indicate a mechanism for hyposmia in human CHARGE syndrome. Hum. Mol. Genet., 18, 1909–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kelsh R.N., Eisen J.S. (2000) The zebrafish colourless gene regulates development of non-ectomesenchymal neural crest derivatives. Development, 127, 515–525. [DOI] [PubMed] [Google Scholar]
- 54.Kelsh R.N., Dutton K., Medlin J., Eisen J.S. (2000) Expression of zebrafish fkd6 in neural crest-derived glia. Mech. Develop., 93, 161–164. [DOI] [PubMed] [Google Scholar]
- 55.Gilmour D.T., Maischein H.M., Nusslein-Volhard C. (2002) Migration and function of a glial subtype in the vertebrate peripheral nervous system. Neuron, 34, 577–588. [DOI] [PubMed] [Google Scholar]
- 56.Brosamle C., Halpern M.E. (2002) Characterization of myelination in the developing zebrafish. Glia, 39, 47–57. [DOI] [PubMed] [Google Scholar]
- 57.Micucci J.A., Layman W.S., Hurd E.A., Sperry E.D., Frank S.F., Durham M.A., Swiderski D.L., Skidmore J.M., Scacheri P.C., Raphael Y., et al. (2014) CHD7 and retinoic acid signaling cooperate to regulate neural stem cell and inner ear development in mouse models of CHARGE syndrome. Hum. Mol. Genet., 23, 434–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He D., Marie C., Zhao C., Kim B., Wang J., Deng Y., Clavairoly A., Frah M., Wang H., He X. (2016) Chd7 cooperates with Sox10 and regulates the onset of CNS myelination and remyelination. Nat. Neurosci., 19, 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nelms B.L., Labosky P.A. (2010) Transcriptional control of neural crest development. Dev. Biol, 1, 1–227. [PubMed] [Google Scholar]
- 60.Britsch S., Goerich D.E., Riethmacher D., Peirano R.I., Rossner M., Nave K.A., Birchmeier C., Wegner M. (2001) The transcription factor Sox10 is a key regulator of peripheral glial development. Gene Dev., 15, 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McKeown S.J., Lee V.M., Bronner-Fraser M., Newgreen D.F., Farlie P.G. (2005) Sox10 overexpression induces neural crest-like cells from all dorsoventral levels of the neural tube but inhibits differentiation. Dev. Dynam., 233, 430–444. [DOI] [PubMed] [Google Scholar]
- 62.Yan Y.L., Willoughby J., Liu D., Crump J.G., Wilson C., Miller C.T., Singer A., Kimmel C., Westerfield M., Postlethwait J.H. (2005) A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development, 132, 1069–1083. [DOI] [PubMed] [Google Scholar]
- 63.Akimenko M.A., Ekker M., Wegner J., Lin W., Westerfield M. (1994) Combinatorial Expression of 3 Zebrafish Genes Related to Distal-Less - Part of a Homeobox Gene Code for the Head. J. Neurosci., 14, 3475–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tu C.T., Yang T.C., Huang H.Y., Tsai H.J. (2012) Zebrafish arl6ip1 is required for neural crest development during embryogenesis. Plos One, 7, e32899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Le Douarin N.M., Dupin E. (2012) The neural crest in vertebrate evolution. Curr. Opin. Genet. Dev., 22, 381–389. [DOI] [PubMed] [Google Scholar]
- 66.Westerfield M. (2000) The zebrafish book: a guide for the laboratory use of zebrafish (Danio rerio). 4th edn University of Oregon Press. [Google Scholar]
- 67.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. (1995) Stages of embryonic development of the zebrafish. Dev. Dynam., 203, 253–310. [DOI] [PubMed] [Google Scholar]
- 68.Karlsson J., von Hofsten J., Olsson P.E. (2001) Generating transparent zebrafish: a refined method to improve detection of gene expression during embryonic development. Marine Biotechnol., 3, 522–527. [DOI] [PubMed] [Google Scholar]
- 69.Carney T.J., Dutton K.A., Greenhill E., Delfino-Machin M., Dufourcq P., Blader P., Kelsh R.N. (2006) A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development, 133, 4619–4630. [DOI] [PubMed] [Google Scholar]
- 70.Curran K., Raible D.W., Lister J.A. (2009) Foxd3 controls melanophore specification in the zebrafish neural crest by regulation of Mitf. Dev. Biol., 332, 408–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peri F., Nusslein-Volhard C. (2008) Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell, 133, 916–927. [DOI] [PubMed] [Google Scholar]
- 72.Bolger A.M., Lohse M., Usadel B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols, 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pfaffl M.W. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res., 29, e45.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Livak K.J., Schmittgen T.D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 76.Aerne B., Ish-Horowicz D. (2004) receptor tyrosine phosphatase psi is required for Delta/Notch signalling and cyclic gene expression in the presomitic mesoderm. Development, 131, 3391–3399. [DOI] [PubMed] [Google Scholar]
- 77.Stewart R.A., Arduini B.L., Berghmans S., George R.E., Kanki J.P., Henion P.D., Look A.T. (2006) Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Dev. Biol., 292, 174–188. [DOI] [PubMed] [Google Scholar]
- 78.van der Velden Y.U., Wang L.Q., Cano L.Q., Haramis A.P.G. (2013) The Polycomb Group Protein Ring1b/Rnf2 Is Specifically Required for Craniofacial Development. Plos One, 8, e73997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Appel B., Korzh V., Glasgow E., Thor S., Edlund T., Dawid I.B., Eisen J.S. (1995) Motoneuron fate specification revealed by patterned LIM homeobox gene expression in embryonic zebrafish. Development, 121, 4117–4125. [DOI] [PubMed] [Google Scholar]
- 80.Yang C.T., Johnson S.L. (2006) Small molecule-induced ablation and subsequent regeneration of larval zebrafish melanocytes. Development, 133, 3563–3573. [DOI] [PubMed] [Google Scholar]
- 81.Lister J.A., Cooper C., Nguyen K., Modrell M., Grant K., Raible D.W. (2006) Zebrafish Foxd3 is required for development of a subset of neural crest derivatives. Dev Biol, 290, 92–104. [DOI] [PubMed] [Google Scholar]
- 82.Solnica-Krezel L., Driever W. (1994) Microtubule arrays of the zebrafish yolk cell: organization and function during epiboly. Development, 120, 2443–2455. [DOI] [PubMed] [Google Scholar]
- 83.Dingerkus G., Uhler L.D. (1977) Enzyme clearing of alcian blue stained whole small vertebrates for demonstration of cartilage. Stain Technol., 52, 229–232. [DOI] [PubMed] [Google Scholar]
- 84.Harris J.A., Cheng A.G., Cunningham L.L., MacDonald G., Raible D.W., Rubel E.W. (2003) Neomycin-induced hair cell death and rapid regeneration in the lateral line of zebrafish (Danio rerio). J. Assoc. Res. Otolaryngol., 4, 219–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Choi T.Y., Kim J.H., Ko D.H., Kim C.H., Hwang J.S., Ahn S., Kim S.Y., Kim C.D., Lee J.H., Yoon T.J. (2007) Zebrafish as a new model for phenotype‐based screening of melanogenic regulatory compounds. Pigm. Cell Res., 20, 120–127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.