Abstract
Niemann-Pick type C disease (NP-C) is a progressive lysosomal lipid storage disease caused by mutations in the NPC1 and NPC2 genes. NPC1 is essential for transporting cholesterol and other lipids out of lysosomes, but little is known about the mechanisms that control its cellular abundance and localization. Here we show that a reduction of TMEM97, a cholesterol-responsive NPC1-binding protein, increases NPC1 levels in cells through a post-transcriptional mechanism. Reducing TMEM97 through RNA-interference reduces lysosomal lipid storage and restores cholesterol trafficking to the endoplasmic reticulum in cell models of NP-C. In TMEM97 knockdown cells, NPC1 levels can be reinstated with wild type TMEM97, but not TMEM97 missing an ER-retention signal suggesting that TMEM97 contributes to controlling the availability of NPC1 to the cell. Importantly, knockdown of TMEM97 also increases levels of residual NPC1 in NPC1-mutant patient fibroblasts and reduces cholesterol storage in an NPC1-dependent manner. Our findings propose TMEM97 inhibition as a novel strategy to increase residual NPC1 levels in cells and a potential therapeutic target for NP-C.
Introduction
Niemann–Pick disease type C (NP-C, OMIM #257220 and #607625) is a severe autosomal-recessive lysosomal storage disease characterized by abnormal transport and storage of cholesterol and other lipids in cells. NP-C patients show a broad variety of visceral and neuropsychiatric manifestations, ranging from perinatal liver failure to cognitive decline in adulthood (1–3). Symptomatic treatments exist that focus on alleviating progression of neurodegeneration, but no curative therapy is available (4). The majority of NP-C patients carry mutations in the NPC1 gene (5), with a small minority harbouring mutations in NPC2 (6). NPC1, the protein encoded by NPC1, is a transmembrane glycoprotein that localizes to late endosomes and lysosomes. Here it interacts with the NPC2 protein to facilitate the export of cholesterol from lysosomes (7,8). NPC1 mutations either reduce NPC1 protein synthesis or stability, its transport to lysosomes or its activity at the lysosomal membrane (9–13). When NPC1 is dysfunctional or absent, transport of lipids from lysosomes is blocked and unesterified cholesterol and glycosphingolipids accumulate (9–11). Lysosomal lipid accumulation is considered the pathological hallmark of NP-C, and current treatment strategies focus on promoting the removal of excess lipids from lysosomes, e.g. with cholesterol-binding cyclodextrins (14–16). Alternative approaches aim at replacing dysfunctional with wild type NPC1 or NPC2 at the gene (17,18) or protein level (19). An emerging and potentially less challenging therapeutic concept is to increase the availability of mutant NPC1 protein with residual activity, since this has been shown to reduce cholesterol accumulation in NPC1-mutant cells in vitro (20,21). A better understanding of the distinct mechanisms that regulate NPC1 protein levels and availability may thus help to develop targeted NP-C therapies with improved efficacy and safety.
Through combining genome-wide expression profiling with unbiased RNA-interference screenings, we have previously identified the membrane protein TMEM97 as a modulator of cholesterol levels in cells and as a novel NPC1-interacting protein (22). Here we show that reducing TMEM97 increases NPC1 protein levels in NP-C cell models and in fibroblasts from NP-C patients with different NPC1 mutations. Importantly, decreasing TMEM97 also counteracts lysosomal lipid accumulation and ameliorates cholesterol storage in an NPC1-dependent manner. These findings suggest TMEM97 as a new target to increase residual NPC1 levels in NP-C.
Results
Reduction of TMEM97 in cells increases NPC1 protein levels
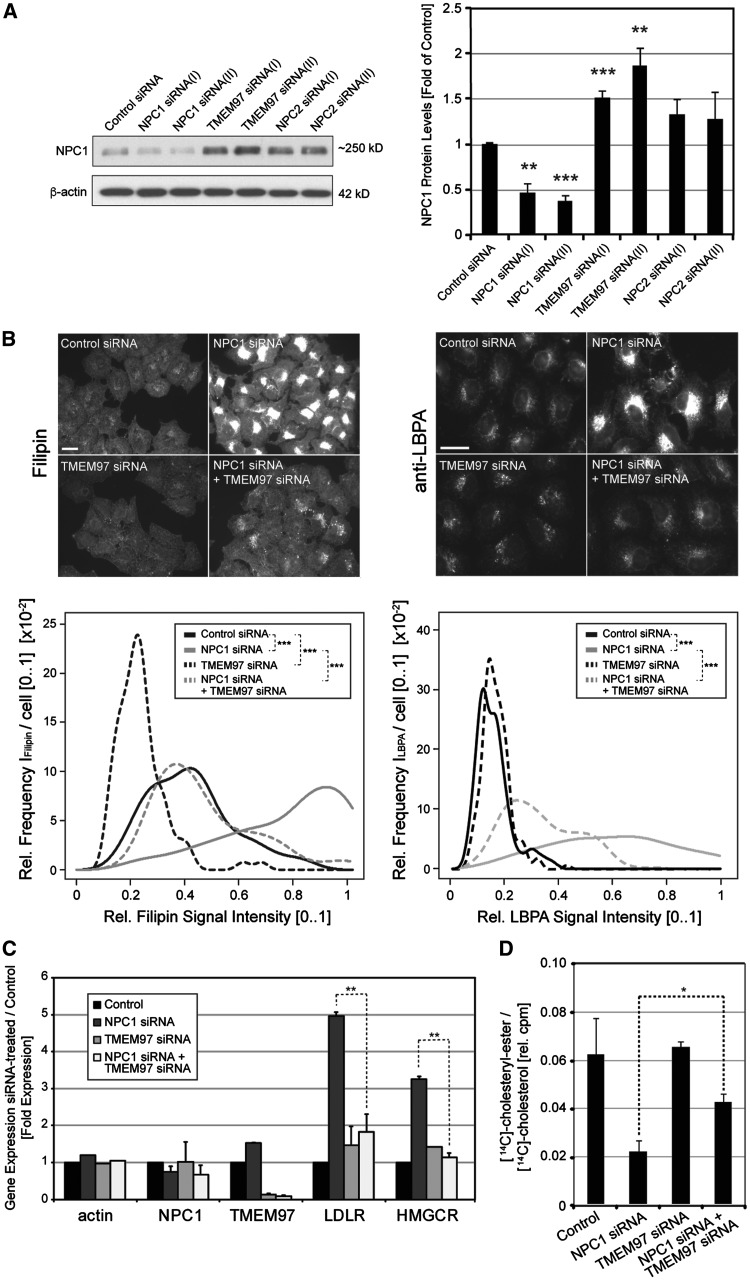
Our previous studies showed that TMEM97 and NPC1 co-immunoprecipitate (22). We thus hypothesized that reducing TMEM97 may impact NPC1 levels or function. To test this hypothesis, we first explored whether siRNA-mediated knockdown of TMEM97 would affect NPC1 levels in cultured HeLa cells. Indeed, while NPC1-siRNAs reduced NPC1 protein levels to 40–50% of control siRNA-treated cells, knockdown of TMEM97 with two independent siRNAs increased cellular NPC1 protein levels to ∼1.50 and ∼1.83-fold of controls, respectively (Fig. 1A) . This increase well exceeded the moderate increase in NPC1 protein levels upon knockdown of NPC2, as previously observed in NPC2-mutant fibroblasts (12,23). Unlike at the protein level, TMEM97-siRNAs did not impact NPC1 mRNA levels (Supplementary Material, Fig. S1A), suggesting that the increase in NPC1 upon TMEM97 knockdown is likely due to post-translational mechanisms. Conversely, and consistent with TMEM97 being a sterol-response element binding protein (SREBP) target gene (24) with stimulated expression in NPC1- deficient cells (25), NPC1-siRNAs increased TMEM97 mRNA levels to 1.5-fold of controls (Supplementary Material, Fig. S1B). Individual siRNAs reduced TMEM97 mRNA to ∼20% of baseline levels, but knockdown efficiency could be improved to <8% when both TMEM97-siRNAs where combined (Supplementary Material, Fig. S1A). Since this combination also reduced TMEM97 protein to <20% of controls (Supplementary Material, Fig. S1C), it was used in subsequent experiments to achieve a maximal reduction of TMEM97.
Figure 1.
SiRNA-mediated knockdown of TMEM97 increases NPC1 protein levels, ameliorates cholesterol accumulation and restores cholesterol delivery to the ER in NPC1-deficient HeLa cells. (A) Cultured HeLa cells were transfected for 48h with either control siRNA or indicated siRNAs targeting NPC1, TMEM97 or NPC2. Whole cell lysates were subjected to Western blotting and probed with antibodies against NPC1 and β-actin. NPC1 protein levels were quantified as a ratio to β-actin and normalized to levels of control siRNA treated cells (n = 3 independent experiments per condition; **P < 0.01; ***P < 0.001). (B) HeLa cells transfected with siRNAs targeting indicated genes for 48h were stained with either filipin dye (left panel) or a polyclonal antibody against LBPA (right panel) (scale bars = 10µm). Filipin and LBPA signal intensities in perinuclear areas were quantified as the relative intensity frequency distributions [0.1] (y-axis) of mean perinuclear filipin or LBPA signals per cell (x-axis) from up to 20 images per condition with an average of 25 cells per image (n = 2–3 independent experiments per condition, ***P < 0.001). (C) HeLa cells cultured in DMEM/5%FCS were transfected with siRNAs against the indicated genes. Eight hours after siRNA-transfection, medium was changed to DMEM/0.5%LDS for 16h. Next, cells were washed and medium was changed to DMEM/10%FCS/50μg/ml LDL for another 24h before mRNA was extracted. Mean mRNA-levels of indicated genes relative to RPL19 are shown (n = 2–3 independent experiments per condition; **P < 0.01). (D) Analysis of cholesteryl-ester formation from LDL-associated [14C]-cholesterol in HeLa cells transfected with indicated siRNAs. 24h after siRNA-transfection, medium was changed to DMEM/0.5%LDS and cells were cultured for another 24h. Next, pulse-labeled medium containing DMEM/0.5%LDS/50μg/ml LDL was added and [14C]LDL-cholesterol was internalized for 4.5h before cellular lipids were extracted and separated by thin-layer chromatography. Cell-associated free [14C]-cholesterol and [14C]-cholesteryl-ester signals (in counts per minute, cpm) were quantified by scintillation counting (n = 2 independent experiments; *P < 0.05).
Next, we tested whether TMEM97 knockdown would also impact NPC1 levels in a cellular model where we used NPC1 knockdown to induce a NP-C phenotype. To this end, we transfected HeLa cells with siRNAs against NPC1, TMEM97, or both proteins concomitantly. Interestingly, and similar to cells with normal NPC1 levels, NPC1-siRNA treated cells maintained higher steady-state NPC1 protein levels following TMEM97 knockdown (Supplementary Material, Fig. S2). This increase in NPC1 protein levels upon TMEM97 knockdown was also seen in cells cultured in the presence of 2-hydroxypropyl-β-cyclodextrin (Supplementary Material, Fig. S2). In summary, these results demonstrate that reducing TMEM97 through RNAi is associated with increased NPC1 protein levels in cells.
Reduction of TMEM97 counteracts lysosomal lipid accumulation and restores LDL-cholesterol transport in NPC1-deficient cells
We hypothesized that the increase in NPC1 protein upon TMEM97 knockdown might counteract the lysosomal lipid accumulation in NPC1-deficient cells. To test this hypothesis, we transfected HeLa cells with either control-siRNA, NPC1-siRNA alone, TMEM97-siRNA alone, or siRNAs against both NPC1 and TMEM97 and approximated cellular cholesterol content using the fluorescent dye filipin (12,26,27). NPC1 knockdown induced a robust cholesterol storage phenotype typical of NP-C, while, consistent with our previous observations (22), filipin signal was reduced in TMEM97-siRNA treated cells (Fig. 1B, left panel). Importantly, filipin signal in cells transfected with siRNAs against both proteins was significantly lower than in cells where only NPC1 was silenced (Fig. 1B, left panel). Quantitative image analysis confirmed that the distribution of lysosomal filipin signal intensity in cells treated with siRNAs against both proteins closely resembled that of control siRNA treated cells (Fig. 1B, left graph). To investigate whether TMEM97 knockdown also prevented the accumulation of other lipids, we measured levels of lysobisphosphatidic acid (LBPA), another major storage product in NP-C (28). Similar to filipin signal, knockdown of NPC1 strongly increased perinuclear LBPA signal (Fig. 1B, right panel). Again, this was significantly attenuated upon co-transfection of TMEM97-siRNA with NPC1-siRNA (Fig. 1B, right panel and graph). Taken together, these results strongly propose that the accumulation of cholesterol and LBPA in response to a reduction in NPC1 levels can be counteracted by knockdown of TMEM97.
NPC1 is crucial for delivering cholesterol from lysosomes to the ER, where cholesterol suppresses the activation of SREBP transcription factors and is esterified by acyl-CoA cholesterol-acyltransferase (22,25). In NPC1-deficient cells, LDL cholesterol does not reach the ER and SREBP target genes are constitutively expressed. To test whether TMEM97 levels would restore cholesterol transport to the ER, we measured mRNA levels of two well-established SREBP target genes, LDL-receptor (LDLR) and HMG-CoA reductase (HMGCR), under the four conditions described above. As expected, NPC1-siRNA, but not TMEM97-siRNA, significantly induced SREBP target gene expression (Fig. 1C). When TMEM97 was silenced together with NPC1, up-regulation of LDLR and HMGCR was suppressed and not significantly different from control cells (Fig. 1C). Likewise, the concomitant knockdown of TMEM97 and NPC1 was able to stimulate esterification of LDL-derived [14C]-cholesterol, which is suppressed in cells treated with NPC1-siRNAs alone (Fig. 1D). Taken together, these results show that a concomitant knockdown of NPC1 with TMEM97 counteracts lysosomal lipid accumulation and overcomes the transport block of LDL-derived cholesterol to the ER, most likely through increasing residual NPC1 protein levels.
Reduction of TMEM97 elevates residual mutant NPC1 protein levels and restores cholesterol trafficking in NPC1-mutant fibroblasts
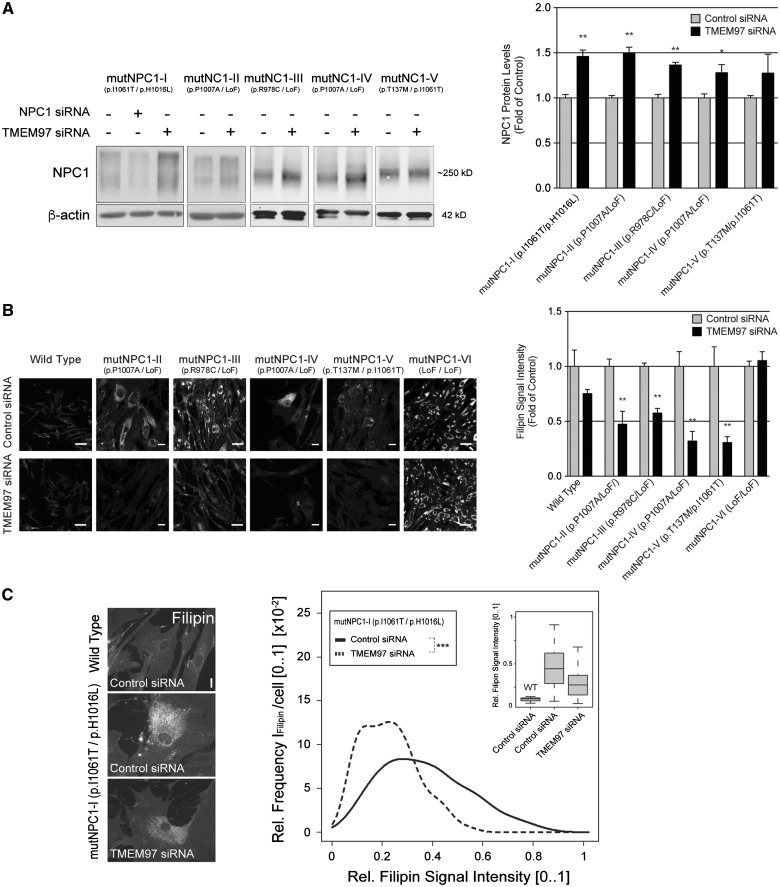
Mutations in NPC1 account for the vast majority of NP-C cases (29). To test whether knockdown of TMEM97 would increase levels not only of wild type NPC1, but also mutant NPC1 protein, we next studied primary fibroblasts from well-characterized NP-C patients with confirmed NPC1 mutations (Supplementary Material, Table S1). Indeed, in all five independent patient cell lines tested, knockdown of TMEM97 moderately increased levels of residual mutant NPC1 to 1.2–1.5-fold of control siRNA-treated patient cells (Fig. 2A). A previous report showing that even a moderate increase in mutant NPC1 may restore cholesterol export from lysosomes (20) prompted us to analyse whether knockdown of TMEM97 in NPC1-mutant fibroblasts would also reduce cholesterol storage. Quantitative analyses of filipin signals in NPC1-mutant fibroblasts showed that this was indeed the case (Fig. 2B). Transfecting TMEM97-siRNAs into four independent patient cell lines with residual levels of mutant NPC1 protein between 12 and 36% (relative to cells from a healthy control individual, Supplementary Material, Table S1) significantly reduced cholesterol storage to 38–56% of control-siRNA treated cells (Fig. 2B and C). Notably, this effect was dependent on the presence of residual mutant NPC1, since cholesterol storage remained unchanged in cells of patient mutNPC1-VI, who carries protein-truncating loss-of-function alleles on both copies of the NPC1 gene and thus shows a complete absence of NPC1 protein (Fig. 2B, Supplementary Material, Table S1). Taken together, these data propose that reducing TMEM97 in NP-C patient fibroblasts elevates residual mutant NPC1 levels and reduces cholesterol storage in an NPC1-dependent manner.
Figure 2.
Reduction of TMEM97 elevates residual mutant NPC1 protein levels and restores cholesterol trafficking in NPC1-mutant fibroblasts. (A) Primary human skin fibroblasts homozygous or compound-heterozygous for indicated NPC1 alleles (in brackets, see Supplementary Material, Table S1 for details) were transfected with NPC1 or TMEM97 siRNAs. Ninety-six hours after transfection, whole cell lysates were subjected to Western blotting and analyzed for NPC1 or β-actin. NPC1 protein levels were quantified as a ratio to β-actin and normalized to levels in control siRNA treated cells (n = 2 independent experiments; mean±SEM, *P < 0.05, **P < 0.01; LoF = NPC1 loss-of-function allele). (B) Images show filipin-stained fibroblasts from a healthy control (Wild Type) and NP-C patients treated with either control or TMEM97-siRNAs for 96h (scale bars = 10µm). The bar graph shows mean filipin signal intensities in TMEM97-siRNA treated cells relative to control-siRNA per cell line (see Supplementary Material, Table S1 for details; n = 3–4 independent experiments; mean±SEM; **P < 0.01). (C) Filipin signal in fibroblasts from NP-C patient mutNPC1-I (p.I1061T/p.H1016L) shown as relative intensity frequency distributions [0.1] (y-axis) of mean perinuclear filipin signals per cell (x-axis) from up to 30 images per condition with typically ∼3 fibroblasts per image (n = 2 independent experiment; ***P < 0.001). Box plots (inset) show medians (lines), lower and upper quartiles (boxes), and 10th and 90th percentiles (whiskers) of mean perinuclear filipin signals per cell from patient mutNPC1-I (middle and right box blot) relative to wild type (WT).
Knockdown of TMEM97 increases NPC1 protein levels in lysosomal and extra-lysosomal compartments
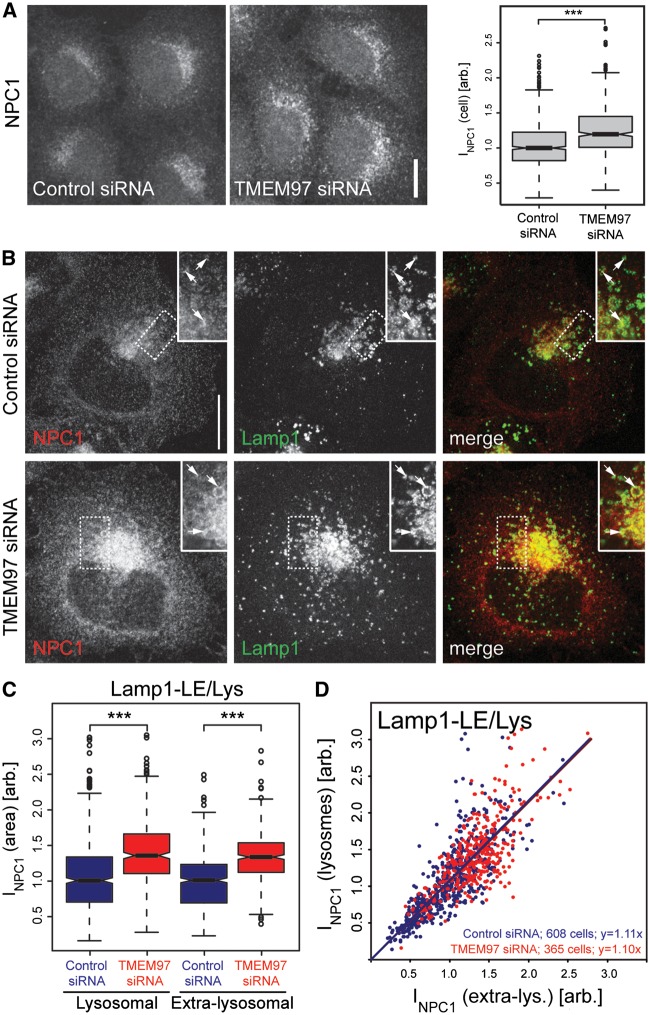
We next aimed to identify clues about the mechanisms by which reduction in TMEM97 may increase NPC1 protein levels. Since NPC1 has been well characterized as an integral membrane protein that resides in membranes of late endosomes and lysosomes (30,31), we tested whether TMEM97 knockdown alters NPC1 subcellular distribution. NPC1 signal per cell, as determined with a specific antibody (32), significantly increased upon TMEM97 knockdown (Fig. 3A), which further supports the Western blot results. Irrespective of TMEM97 knockdown, NPC1 localized to perinuclear vesicular compartments positive for Lamp1, Lamp2 and Rab7 (Figs 3B–D, Supplementary Material, Fig. S4 and S5), but consistent with the published literature, was also present in other areas of the cell. Automated quantification of signal intensities from stacks of confocal images revealed that TMEM97 siRNAs increased total NPC1 signal per cell, as well as NPC1 signal in perinuclear compartments that overlap with late endosomal and lysosomal markers (Fig. 3B–D). Ratiometric analyses at the level of single cells showed that the extent of this increase was similar in both lysosomal as well as extra-lysosomal compartments (Figs 3C–D and Supplementary Material, Fig. S4). In line with these findings, knockdown of NPC1 reduced NPC1 signal to a similar extent in both lysosomal and extra-lysosomal compartments (Supplementary Material, Fig. S5). These results suggest that knockdown of TMEM97 increases the overall abundance of NPC1 protein in cells, rather than shifting its subcellular distribution from extra-lysosomal compartments towards lysosomes, or vice versa.
Figure 3.
Knockdown of TMEM97 increases NPC1 protein levels in lysosomal and extra-lysosomal compartments. (A) Confocal images from HeLa cells treated with indicated siRNAs for 48h and stained for NPC1 with an NPC1-specific antibody (32) (scale bar = 10µm). Box plots show medians (lines), lower and upper quartiles (boxes), 10th and 90th percentiles (whiskers) and outliers (circles) of NPC1 signal intensities per cell as quantified automatically from stacks of confocal images using CellProfiler software (n = 5 independent experiments; ***P < 0.001). (B) Representative maximal intensity projections from confocal stacks of HeLa cells stained for NPC1 and lysosomal marker Lamp1 in TMEM97 and control siRNA treated cells. Arrows denote selected lysosomes (scale bar = 10µm). (C) NPC1 signal from lysosome-like areas overlapping with lysosomal marker Lamp1 or extra-lysosomal cellular regions were quantified from automatically acquired widefield images using CellProfiler. Box plots in left graph show medians (lines), lower and upper quartiles (boxes), 10th and 90th percentiles (whiskers) and outliers (circles) of mean NPC1 signal per indicated area from the number of cells given in (D). Median of control-siRNA treated cells/condition was set to 1 (n = 5 independent experiments per condition, ***P < 0.001). (D) Ratios of NPC1-signal within relative to outside of lysosome-like areas in indicated numbers of individual cells (dots) treated with either control (blue) or TMEM97-siRNAs (red). Formulas indicate linear regression curves.
Expression of wild type TMEM97, but not TMEM97 missing an ER-retention signal, reverses increased NPC1 levels in TMEM97-deficient cells
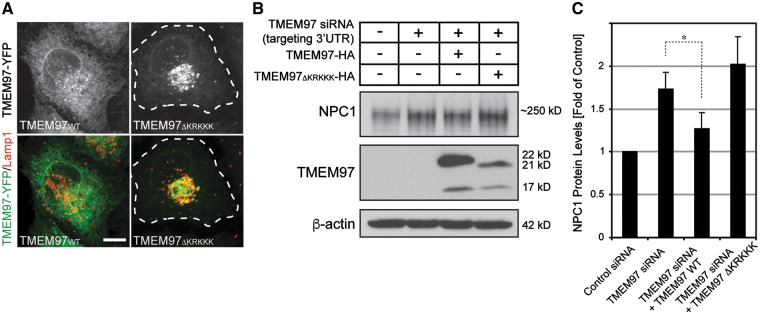
In addition to late endosomes and lysosomes, a significant proportion of NPC1 resides within the endoplasmic reticulum (ER) where wild type NPC1 is prepared for lysosomal targeting and mutant forms are selected for ER-based degradation (20). Likewise, we previously found that TMEM97 may associate with ER-like reticular organelles and the nuclear membrane (22). We therefore considered the ER as one potential site for the interaction between TMEM97 and NPC1. Analysis of the amino acid sequence of TMEM97 predicted a potential ER-retention motif at the carboxy-terminus. In contrast to wild type TMEM97, constructs missing this motif (TMEM97ΔKRKKK) failed to be retained in reticular organelles, but instead localized to late endosomes and lysosomes (Fig. 4A). To test whether ER-retention of TMEM97 could be relevant for regulating NPC1 levels, we expressed HA-tagged cDNA constructs encoding either wild type TMEM97 or TMEM97 ΔKRKKK in HeLa cells that had previously been transfected with an siRNA targeting the 3’ untranslated region (3’UTR) of TMEM97-mRNA. Expression of wild type TMEM97-HA in TMEM97 3’UTR-siRNA treated cells mitigated the increase in NPC1 protein levels observed in cells treated with TMEM97 3’UTR-siRNA alone. This confirmed that the increase in NPC1 following treatment with TMEM97 siRNA is indeed a consequence of a reduction in TMEM97 (Fig. 4B and C). In contrast, TMEM97 ΔKRKKK was unable to rescue increased NPC1 levels (Fig. 4B and C). Interestingly, TMEM97 knockdown appears to increase NPC1 protein through mechanisms independent of its ER-associated degradation (ERAD) via the proteasomal pathway, since blocking proteasomal degradation in TMEM97 siRNA-treated cells with the inhibitor clasto-lactacystin-β-lactone further increased residual NPC1 levels (Supplementary Material, Fig. S6), arguing that upon reduction of TMEM97 a substantial amount of NPC1 still undergoes proteasomal degradation. Taken together, these results suggest that the mechanisms by which a reduction in TMEM97 increases NPC1 protein levels are likely to occur outside of lysosomal compartments, most likely within ER-membranes.
Figure 4.
Expression of wild type TMEM97, but not TMEM97 missing an ER-retention signal, reverses increased NPC1 levels in TMEM97 knockdown cells. (A) HeLa cells transiently transfected for 24h with cDNA-plasmids encoding for YFP-tagged wild type TMEM97 (TMEM97WT) or TMEM97 missing an ER-retention signal (TMEM97ΔKRKKK), respectively, and counterstained for Lamp1. (B) Whole cell lysates from HeLa cells (co-)transfected for 48h with HA-tagged TMEM97WT (lane 3) or TMEM97ΔKRKKK (lane 4) as well as either control siRNA (lane 1) or siRNA targeting the TMEM97 3´untranslated region (UTR) (lanes 2-4) were subjected to Western blotting and probed for NPC1, HA and β-actin. (C) NPC1 protein levels were quantified as a ratio to β-actin and normalized to levels of control siRNA treated cells (n = 2 independent experiments per condition; *P < 0.05).
In vivo knockdown of Tmem97 in Npc1tm(I1061T)dso mice does not alter hepatic NPC1 or lipid levels
Our combined in vitro results provide strong evidence that reduction of TMEM97 by RNAi could be a viable strategy for the prevention or treatment of NP-C. To explore whether knockdown of TMEM97 could ameliorate NP-C in vivo, we conducted a proof-of-concept experiment using transgenic Npc1tm(I1061T)Dso mice in which the human NPC1 mutation p.I0161T has been introduced into the murine locus (33). Locked-nucleic acid antisense oligonucleotides (LNA-ASO) targeting murine Tmem97 or GFP were injected intraperitoneally into postnatal day 28 mice. After 4 weeks, LNA-ASO efficiently reduced hepatic Tmem97 mRNA levels to <6% of control-injected mice (Fig. 5A; n = 6 animals per group). However, no significant changes were observed in hepatic Npc1 mRNA (Fig. 5B) or protein levels (Fig. 5C) during this observation period. Also, Tmem97 LNA-ASO did not induce significant changes in any of the major lipid species typically altered in NP-C as determined by mass spectrometry (Fig. 5D, n ≥ 7 animals per group). An alternative strategy to reduce Tmem97 in vivo using adeno-associated virus 8 (AAV8) particles carrying a plasmid encoding murine Tmem97-shRNA yielded only an inconsistent reduction of Tmem97 mRNA in murine liver homogenates (Supplementary Material, Fig. S7). Taken together, additional animal studies will be required to assess a potential impact of Tmem97 inhibition on the manifestation or progression of NP-C in vivo.
Figure 5.
Targeting of Tmem97 by RNA interference in Npc1tm(I1061T)Dso mice fails to increase hepatic NPC1 levels or to improve lipid abnormalities. (A & B) Npc1tm(I1061T)Dsomice received intraperitoneal injections of locked nucleic acid antisense oligonucleotides (LNA-ASO) targeting murine Tmem97 or GFP starting at postnatal day 28. Vehicle treated mice served as a second control cohort. Livers were harvested on postnatal day 56. Graphs show mRNA levels of Tmem97 and Npc1 relative to untreated control. Tmem97 mRNA was significantly reduced in Tmem97 LNA-ASO treated mice relative to GFP LNA-ASO treated controls and vehicle treated control. No significant changes in Npc1 mRNA levels were observed (n = 8 Vehicle, 8 GFP LNA-ASO, 7 Tmem97 LNA-ASO, ***P < 0.001). (C) Npc1 protein levels in whole liver lysates harvested at postnatal day 56 from Npc1tm(I1061T)Dsomice treated with LNA-ASO targeting murine Tmem97 (n = 7, T1-7) or GFP (n = 8, GFP1-8), or vehicle (n = 8 Vehicle, Veh1-8) as described in (A). (D) Quantification of lipids from liver lysates (C) using tandem mass spectrometry. Graph shows the percent change in Npc1tm(I1061T)Dsomice treated with Tmem97 LNA-ASO (n = 7) relative to those receiving GFP LNA-ASO (n = 8). None of the changes reached statistical significance.
Discussion
In this study, we aimed to assess whether reducing TMEM97 could be a novel therapeutic strategy for the treatment or prevention of NP-C. We show that several independent approaches to reduce TMEM97 through RNA-interference in cultured cells increase NPC1 protein levels via a post-transcriptional mechanism. Reduction of TMEM97 ameliorates lipid accumulation in NP-C cell models and restores cholesterol trafficking, most likely through increasing cellular availability of NPC1. Importantly, knockdown of TMEM97 not only increased wild type, but also mutant NPC1, and this increase was accompanied by a mobilization of cholesterol from lysosomes in NPC1-mutant fibroblasts from different NP-C patients.
Excess lipid levels in lysosomes compromise endo-lysosomal homeostasis and result in degenerative changes in multiple tissues, including the central nervous system (9,10,34). Disease severity in NP-C, to some degree, correlates with the levels of residual NPC1 protein and the extent of cholesterol storage (3,12,23,26). Consistently, multiple lines of evidence in vitro and in vivo support the hypothesis that preventing lipid accumulation and reducing storage through increasing residual NPC1 is efficacious for the treatment of NP-C (20,21). Little is known about the mechanisms that control NPC1 protein levels. Unlike other genes that increase cholesterol availability in cells, NPC1 expression does not seem to strongly depend on regulation through SREBP (22,24). NPC1 is moderately increased in NPC2-deficient fibroblasts (12,23), presumably due to compensatory mechanisms secondary to impaired export of cholesterol from lysosomes (7).
TMEM97 is one of very few proteins described to interact with NPC1 (22), and to our knowledge our study is the first to report a protein that affects NPC1 cellular abundance, potentially through direct interaction. Certainly, the precise mechanisms by which TMEM97 impacts NPC1 levels remain to be determined. However, two lines of evidence suggest TMEM97 as a factor that controls the overall NPC1 availability to the cell rather than a regulator of specific lysosomal activities: First, knockdown of TMEM97 increases NPC1 to a similar extent in lysosomal and non-lysosomal compartments. And second, only expression of the full-length, but not a truncated TMEM97 lacking its ER-retention signal, was able to reverse the effect of TMEM97 knockdown on NPC1 levels. We speculate that TMEM97 could serve as a chaperone protein to NPC1 that may play a role in limiting the formation and/or export of NPC1 from ER membranes. Consequently, reducing TMEM97 would increase the amount of NPC1 available for delivery to post-ER compartments including lysosomes. An analogous, yet inverse regulatory function has been introduced for the Nogo-B receptor in controlling the entry of NPC2 into the secretory pathway (35).
Several strategies to increase residual mutant NPC1, including histone deacetylase (HDAC) inhibitors and HSP70 analogues, are currently being explored for clinical use (21,36,37). In principle, these strategies are believed to be efficacious since most NP-C patients still express substantial amounts of mutant NPC1 protein with preserved cholesterol-transporting function (12,23). For instance, increasing cellular levels of the most common NPC1 mutant p.I1061T, which is normally degraded by ER-based quality control mechanisms, allows a fraction of mutant NPC1 to fold correctly, localize to lysosomes and reduce cholesterol storage with an efficiency close to that of wild type protein (20). In order to explore potential therapeutic implications of TMEM97 inhibition, we used cultured fibroblasts from NP-C patients carrying different pathogenic NPC1 mutations. We found that knockdown of TMEM97 reduced cholesterol storage across a range of fibroblasts with common NPC1 mutations, but not in cells from a patient with a homozygous protein-truncating mutation that abrogates formation of NPC1 protein. Taken together, these findings suggest TMEM97 inhibition as a novel therapeutic strategy for NP-C. They further lend support to the notion that even a small increase in functional NPC1 levels may be sufficient to restore cholesterol metabolism and to prevent disease manifestations.
Despite these encouraging results, efforts to translate our in vitro findings into an in vivo model have thus far not met with success. At four weeks, TMEM97-ASO-mediated knockdown of Tmem97 failed to increase hepatic Npc1 levels and to modify hepatic lipid levels in mice carrying the pathogenic homozygous p.I1061T mutation. Potential therapies directed against TMEM97 will likely be required to very efficiently reduce TMEM97 expression for a prolonged period of time, and longer observation periods may be necessary to detect amelioration of pathologies. Given the failure to detect a response in hepatocytes in vivo, it also remains important to test if the mechanisms that control NPC1 levels could be tissue-specific. For this, more extensive animal studies including prolonged and sustained knockdown paradigms, additional time points, additional tissues, and more detailed analyses at the molecular and cellular level will be necessary. Future therapy development is further challenged by the fact that TMEM97 is a yet poorly characterized and highly hydrophobic membrane protein, features that make it a difficult target for small molecules or therapeutic antibodies. Somewhat surprisingly, however, a very recent study identified TMEM97 as a highly ligandable protein with the capacity to bind multiple small molecule chemotypes (38). The same study also proposed a lipid tool compound that binds TMEM97 with micromolar affinity (38), which provides a promising starting point for the development of a specific TMEM97 inhibitor. Alternatively, new strategies to tackle TMEM97 at the mRNA level or through targeted gene editing, in conjunction with appropriate delivery vehicles, might become available (39) and provide additional means to investigate TMEM97 as a therapeutic target for NP-C.
In summary, the experiments detailed here introduce TMEM97 as a novel factor that controls cellular and lysosomal levels of NPC1. We find that knockdown of TMEM97 increases NPC1 protein levels and ameliorates lysosomal lipid storage in cell models of NP-C and NPC1-mutant fibroblasts. Future work is needed to deepen our understanding of the molecular interplay between both proteins and the best strategies to advance insights into the development of novel mechanism-based therapies for NP-C.
Materials and Methods
Antibodies, plasmids and cell lines
The following antibodies were used: β-actin (mouse-monoclonal, clone AC-15, Sigma-Aldrich), GFP (mouse-monoclonal, Roche), HA (rabbit-polyclonal, clone MSA-106, Biomol), LBPA (mouse-monoclonal, clone 6c4, Echelon), Lamp1 (mouse-monoclonal, clone H4A3, DSHB, University of Iowa), Lamp2 (mouse-monoclonal, clone H4B4, DSHB, University of Iowa), NPC1 (rabbit-polyclonal, Novus Biologicals), NPC1 (rabbit-polyclonal, described in (32)) and TMEM97 (rabbit-polyclonal, Novus Biologicals). For RNA interference the following siRNAs were used: For knockdown of TMEM97: siRNAs #140160 (I) (Ambion) and #SI03198314 (II) (QIAGEN); for knockdown of NPC1: siRNAs #114041 (I) and #106017 (II) (both from Ambion); for knockdown of NPC2: siRNAs #135801 (I) and #135802 (II) (both from Ambion); as a non-silencing negative control: scrambled-siRNA #4611 (Ambion). Oligonucleotides sequences selected to generate plasmids for short-hairpin RNA encoding plasmids for knockdown of murine Tmem97 and Npc1 are provided in Supplementary Material, Table S2A. 21-nt shRNA consensus sequences targeting the 3’ untranslated regions of human or mouse Tmem97 and Npc1 were chosen from reference transcripts (www.ensembl.org) with the help of www.sirnawizard.com and ligated into vector pBSU6 (40). TMEM97 LNA-ASO were custom-designed and generated by Exiqon (#500125, Supplementary Material, Table S2B). To generate TMEM97ΔKRKKK, cDNA-plasmids encoding for HA- or YFP-tagged wild type human TMEM97 (ENSG00000109084) (22) were truncated from the carboxy-terminal five amino-acids by PCR using the primer pair 5’-TTTGCGGCCGCTCGGGGCTCCGGCAACCAG-3’ and 5’-CCCGG AATTCTCACTCTTCATACTTGTAGTAG-3’ and reinserted into respective source vectors (described in (22)). A431-cells stably expressing Rab7-GFP were a kind gift from M. Zerial (MPI-CBG, Dresden, Germany) and have been described previously (41). Fibroblasts were cultured from skin biopsies of NP-C patients or a healthy control and are summarized in Supplementary Material, Table S1. Written informed consent was obtained from patients and/or legal guardians. Experiments involving patient-derived cell lines were approved by the institutional review board at the Heidelberg University Faculty of Medicine (IRB approval no. S-032/2012).
Cell culture and transfection
HeLa, A431 and fibroblast cultures were maintained as described previously (12,22,27). Sterol-depletion by lipoprotein-depleted serum (LDS) and 2-hydroxypropyl-β-cyclodextrin (Sigma-Aldrich) were performed as described previously (12,22,27). HeLa and A431 cells were siRNA-transfected for 48h with Oligofectamine (Invitrogen) and cDNA-transfected for 24h with Lipofectamine 2000 (Invitrogen) according to manufacturerś instructions. Fibroblasts were transfected with silentFect (Biorad) or Oligofectamine (Invitrogen) for 96h according to manufacturerś instructions. SiRNA knockdowns were performed with 30µM siRNA/gene. TMEM97 knockdown to <8% of controls was achieved by combining both TMEM97-siRNAs at 15µM each. Consistent with recent reports in cancer cell lines (42,43), we found that TMEM97 knockdown moderately but significantly impacts cell proliferation, leading to a reduced cell number (p = 0.046) as well as a smaller nuclear area (p = 0.049), without any obvious changes in cellular morphology (p = 0.70, compared to siRNA-treated controls; n = 3). For the combined knockdown of TMEM97 and NPC1, TMEM97-siRNAs were mixed with 30µM of NPC1-siRNA (I) and compared to control-siRNA at 60µM concentration. For simultaneous knockdown and overexpression experiments, HeLa cells were Lipofectamine 2000-transfected with 500ng/ml cDNA and 30µM siRNA and cultured for 48h. Clasto-lactacystin-beta-lactone (Enzo Life Sciences) was used at 10μM for 6hr.
qPCR, western blotting and lipid assays
RNA isolation, qPCR, Western blotting, LDL isolation and analysis of cholesteryl-ester formation were performed as described previously (22,44). Total intensities of Western blot bands were quantified by NIH ImageJ software and normalized to β-actin. Esterification of LDL- [14C]cholesterol was determined from HeLa cells pulse-labeled for 4.5h at 37 °C, 5%CO2 with 0.25 mCi/ml total [14C]-activity. Bands representing free cholesterol and cholesteryl-esters were scraped and scintillation counted as described in (22).
Immunocytochemistry, image acquisition and analysis
Cells cultured on glass coverslips were fixed with 3% paraformaldehyde and either directly stained for cholesterol with 50 µg/ml filipin (Sigma-Aldrich) or permeabilized with 0.1% TritonX-100 for 2min at 4 °C. Cell nuclei were stained with 1µg/ml DAPI (Carl Roth) or 10µM Draq5 (Biostatus). Images from filipin- and LBPA-stained samples or cells expressing TMEM97-YFP were acquired on an Axiovert200 epifluorescence microscope (Carl Zeiss) using a 40x oil objective. Filipin and LBPA signal in whole cells was quantified from background-subtracted images within masks generated by ImageJ (22). Mean signal intensity per area was plotted with modules of the statistics software R (http://www.r-project.org). Stacks of images of cells expressing TMEM97-YFP or stained for immunocytochemistry were acquired on a Zeiss LSM Meta confocal microscope using a 63x oil objective. To quantify NPC1 levels and subcellular distribution, images were acquired on an automated epifluorescence microscope (Olympus ScanR) using a 40x air-immersion objective. NPC1 signal in whole cells and subcellular regions overlapping with Lamp1, Lamp2 or Rab7-GFP, respectively, were quantified automatically from background-subtracted images using image analysis procedures implemented as modules in a MATLAB-based version of CellProfiler (www.cellprofiler.org, (45)). In brief, area of single cells was approximated by stepwise dilation of masks generated from the images of DAPI-stained cell nuclei. NPC1 signal was quantified within and outside of late endosomal/lysosomal (LE/Lys) areas as determined by local adaptive thresholding according to pre-defined parameters for the size and shape of perinuclear vesicular structures (Supplementary Material, Fig. S4), as described previously in (46).
Npc1tm(I1061T)dso mice
Npc1tm(I1061T)Dso transgenic mice carrying the equivalent of the human pathogenic NPC1 mutation p.I1061T in the Npc1 locus have been recently described (33). The animals were held in a light-dark cycle, temperature and humidity controlled animal vivarium with ad libitum food and water. Experimental procedures were approved by the Washington University Animal Studies Committees and were conducted in accordance with the USDA Animal Welfare Act and the Public Health Service Policy for the Humane Care and Use of Laboratory Animals.
LNA ASO-mediated knockdown of Tmem97 in vivo
Male Npc1tm(I1061T)Dsomice received intraperitoneal injections of locked nucleic acid antisense oligonucleotides (LNA-ASO) targeting murine Tmem97 or GFP starting at postnatal day 28. Vehicle treated mice served as a second control cohort. Livers here harvested on postnatal day 56. Preparation of liver lysates and quantification of lipids was done using tandem mass spectrometry as described previously (33,47).
AAV vector generation and in vivo AAV8-shRNA mediated knockdown of Tmem97
Recombinant adeno-associated virus (AAV) vector plasmids to drive short-hairpin RNA expression under an H1 promoter were generated and purified as described previously (40). In brief, murine Tmem97-shRNA-1 (Supplementary Material, Table S2) or control shRNA targeting Renilla luciferase was packaged into recombinant AAV8 particles, and viral particles were purified from supernatant of HEK293T cells via caesium-chloride density gradient centrifugation. Purified AAV8 particles were injected intraperitoneally into Npc1 knock-in mice Npc1tm(I1061T)Dso (described in (33)) on postnatal day 28. 14 days after injection, mice were sacrificed, whole RNAs were extracted from mouse liver, and Tmem97 mRNA levels were quantified via qPCR.
Statistical analysis
All graphical data present mean ± standard deviation unless otherwise indicated. Differences between the mean values were tested for statistical significance (P < 0.05) using two-tailed Student t-tests for two groups, and either one-way or two-way ANOVA (with Bonferroni’s correction when appropriate) for three or more groups, followed by Bonferroni post-hoc test.
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
We thank R. Pepperkok, C. Tischer and the Advanced Light Microscopy Facility (ALMF, EMBL) for helpful discussions and supporting image acquisition and analysis. Lamp1 and Lamp2 antibodies (deposited by August, J.T./Hildreth, J.E.K.) were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City.
Conflict of Interest statement. None declared.
Funding
This work was supported by research grants from the Ara Parseghian Medical Research Foundation (to H.R.) and Dana’s Angels Research Trust (to D.O). D.E.-F. and L.W. acknowledge support from the Young Investigator Award Program at Ruprecht-Karls-University Heidelberg Faculty of Medicine, the Daimler and Benz Foundation (to D.E.-F.), and the Reinhard-Frank Foundation (to D.E.-F.).
D.E.-F. and L.W. report receiving travel grants from Actelion Pharmaceuticals for attending a scientific conference in 2014. H.R. and D.O. have received travel grants and honoraria from Actelion Pharmaceuticals and are members of the Scientific Advisory Board of Vtesse Inc. . H.R. is a full-time employee of Merck & Co. Inc. Funding to pay the Open Access publication charges for this article was provided by Fondation Leducq (Career Development Award 12CDA04).
Footnotes
Present address: Department of Genetics and Pharmacogenomics, Merck Research Laboratories, Boston, MA 02115, USA
#The authors wish it to be known that, in their opinion, the first three authors should be regarded as joint First Authors.
References
- 1.Mengel E., Klunemann H.H., Lourenco C.M., Hendriksz C.J., Sedel F., Walterfang M., Kolb S.A. (2013) Niemann-Pick disease type C symptomatology: an expert-based clinical description. Orphanet J. Rare Dis., 8, 166.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patterson M.C., Mengel E., Wijburg F.A., Muller A., Schwierin B., Drevon H., Vanier M.T., Pineda M. (2013) Disease and patient characteristics in NP-C patients: findings from an international disease registry. Orphanet J. Rare Dis., 8, 12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stampfer M., Theiss S., Amraoui Y., Jiang X., Keller S., Ory D.S., Mengel E., Fischer C., Runz H. (2013) Niemann-Pick disease type C clinical database: cognitive and coordination deficits are early disease indicators. Orphanet J. Rare Dis., 8, 35.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patterson M.C., Hendriksz C.J., Walterfang M., Sedel F., Vanier M.T., Wijburg F., Group N.C.G.W. (2012) Recommendations for the diagnosis and management of Niemann-Pick disease type C: an update. Mol. Genet. Metab., 106, 330–344. [DOI] [PubMed] [Google Scholar]
- 5.Carstea E.D., Polymeropoulos M.H., Parker C.C., Detera-Wadleigh S.D., O'Neill R.R., Patterson M.C., Goldin E., Xiao H., Straub R.E., Vanier M.T., et al. (1993) Linkage of Niemann-Pick disease type C to human chromosome 18. Proc. Natl. Acad. Sci. U. S. A., 90, 2002–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naureckiene S., Sleat D.E., Lackland H., Fensom A., Vanier M.T., Wattiaux R., Jadot M., Lobel P. (2000) Identification of HE1 as the second gene of Niemann-Pick C disease. Science, 290, 2298–2301. [DOI] [PubMed] [Google Scholar]
- 7.Wang M.L., Motamed M., Infante R.E., Abi-Mosleh L., Kwon H.J., Brown M.S., Goldstein J.L. (2010) Identification of surface residues on Niemann-Pick C2 essential for hydrophobic handoff of cholesterol to NPC1 in lysosomes. Cell Metab., 12, 166–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deffieu M.S., Pfeffer S.R. (2011) Niemann-Pick type C 1 function requires lumenal domain residues that mediate cholesterol-dependent NPC2 binding. Proc. Natl. Acad. Sci. U. S. A., 108, 18932–18936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Platt F.M., Wassif C., Colaco A., Dardis A., Lloyd-Evans E., Bembi B., Porter F.D. (2014) Disorders of cholesterol metabolism and their unanticipated convergent mechanisms of disease. Annu Rev Genomics Hum Genet, 15, 173–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schulze H., Sandhoff K. (2011) Lysosomal lipid storage diseases. Cold Spring Harb. Perspect. Biol., 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanier M.T. (2015) Complex lipid trafficking in Niemann-Pick disease type C. J. Inherit. Metab. Dis., 38, 187–199. [DOI] [PubMed] [Google Scholar]
- 12.Tangemo C., Weber D., Theiss S., Mengel E., Runz H. (2011) Niemann-Pick Type C disease: characterizing lipid levels in patients with variant lysosomal cholesterol storage. J. Lipid Res., 52, 813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Runz H., Dolle D., Schlitter A.M., Zschocke J. (2008) NPC-db, a Niemann-Pick type C disease gene variation database. Hum. Mutat., 29, 345–350. [DOI] [PubMed] [Google Scholar]
- 14.Ottinger E.A., Kao M.L., Carrillo-Carrasco N., Yanjanin N., Shankar R.K., Janssen M., Brewster M., Scott I., Xu X., Cradock J., et al. (2014) Collaborative development of 2-hydroxypropyl-beta-cyclodextrin for the treatment of Niemann-Pick type C1 disease. Curr. Top. Med. Chem., 14, 330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vance J.E., Peake K.B. (2011) Function of the Niemann-Pick type C proteins and their bypass by cyclodextrin. Curr. Opin. Lipidol., 22, 204–209. [DOI] [PubMed] [Google Scholar]
- 16.Vite C.H., Bagel J.H., Swain G.P., Prociuk M., Sikora T.U., Stein V.M., O'Donnell P., Ruane T., Ward S., Crooks A., et al. (2015) Intracisternal cyclodextrin prevents cerebellar dysfunction and Purkinje cell death in feline Niemann-Pick type C1 disease. Sci. Transl. Med., 7, 276ra226.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez M.E., Klein A.D., Dimbil U.J., Scott M.P. (2011) Anatomically defined neuron-based rescue of neurodegenerative Niemann-Pick type C disorder. J. Neurosci., 31, 4367–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loftus S.K., Erickson R.P., Walkley S.U., Bryant M.A., Incao A., Heidenreich R.A., Pavan W.J. (2002) Rescue of neurodegeneration in Niemann-Pick C mice by a prion-promoter-driven Npc1 cDNA transgene. Hum. Mol. Genet., 11, 3107–3114. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen G.K., Dagnaes-Hansen F., Holm I.E., Meaney S., Symula D., Andersen N.T., Heegaard C.W. (2011) Protein replacement therapy partially corrects the cholesterol-storage phenotype in a mouse model of Niemann-Pick type C2 disease. PLoS One, 6, e27287.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gelsthorpe M.E., Baumann N., Millard E., Gale S.E., Langmade S.J., Schaffer J.E., Ory D.S. (2008) Niemann-Pick type C1 I1061T mutant encodes a functional protein that is selected for endoplasmic reticulum-associated degradation due to protein misfolding. J. Biol. Chem., 283, 8229–8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helquist P., Maxfield F.R., Wiech N.L., Wiest O. (2013) Treatment of Niemann–pick type C disease by histone deacetylase inhibitors. Neurotherapeutics, 10, 688–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartz F., Kern L., Erz D., Zhu M., Gilbert D., Meinhof T., Wirkner U., Erfle H., Muckenthaler M., Pepperkok R., et al. (2009) Identification of cholesterol-regulating genes by targeted RNAi screening. Cell Metab., 10, 63–75. [DOI] [PubMed] [Google Scholar]
- 23.Millat G., Marcais C., Tomasetto C., Chikh K., Fensom A.H., Harzer K., Wenger D.A., Ohno K., Vanier M.T. (2001) Niemann-Pick C1 disease: correlations between NPC1 mutations, levels of NPC1 protein, and phenotypes emphasize the functional significance of the putative sterol-sensing domain and of the cysteine-rich luminal loop. Am. J. Hum. Genet., 68, 1373–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horton J.D., Shah N.A., Warrington J.A., Anderson N.N., Park S.W., Brown M.S., Goldstein J.L. (2003) Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. U. S. A., 100, 12027–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J.R., Coleman T., Langmade S.J., Scherrer D.E., Lane L., Lanier M.H., Feng C., Sands M.S., Schaffer J.E., Semenkovich C.F, et al. (2008) Niemann-Pick C1 protects against atherosclerosis in mice via regulation of macrophage intracellular cholesterol trafficking. J. Clin. Invest, 118, 2281–2290.18483620 [Google Scholar]
- 26.Vanier M.T., Latour P. (2015) Laboratory diagnosis of Niemann-Pick disease type C: The filipin staining test. Methods Cell Biol., 126, 357–375. [DOI] [PubMed] [Google Scholar]
- 27.Blattmann P., Schuberth C., Pepperkok R., Runz H. (2013) RNAi-based functional profiling of loci from blood lipid genome-wide association studies identifies genes with cholesterol-regulatory function. PLoS Genetics, 9, e1003338.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chevallier J., Chamoun Z., Jiang G., Prestwich G., Sakai N., Matile S., Parton R.G., Gruenberg J. (2008) Lysobisphosphatidic acid controls endosomal cholesterol levels. J. Biol. Chem., 283, 27871–27880. [DOI] [PubMed] [Google Scholar]
- 29.Vanier M.T. (2010) Niemann-Pick disease type C. Orphanet J. Rare Dis., 5, 16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neufeld E.B., Wastney M., Patel S., Suresh S., Cooney A.M., Dwyer N.K., Roff C.F., Ohno K., Morris J.A., Carstea E.D., et al. (1999) The Niemann-Pick C1 protein resides in a vesicular compartment linked to retrograde transport of multiple lysosomal cargo. J. Biol. Chem., 274, 9627–9635. [DOI] [PubMed] [Google Scholar]
- 31.Higgins M.E., Davies J.P., Chen F.W., Ioannou Y.A. (1999) Niemann-Pick C1 is a late endosome-resident protein that transiently associates with lysosomes and the trans-Golgi network. Mol. Genet. Metab., 68, 3588–3599.. [DOI] [PubMed] [Google Scholar]
- 32.Millard E.E., Srivastava K., Traub L.M., Schaffer J.E., Ory D.S. (2000) Niemann-pick type C1 (NPC1) overexpression alters cellular cholesterol homeostasis. J. Biol. Chem., 275, 38445–38451. [DOI] [PubMed] [Google Scholar]
- 33.Praggastis M., Tortelli B., Zhang J., Fujiwara H., Sidhu R., Chacko A., Chen Z., Chung C., Lieberman A.P., Sikora J., et al. (2015) A Murine Niemann-Pick C1 I1061T Knock-In Model Recapitulates the Pathological Features of the Most Prevalent Human Disease Allele. J. Neurosci., 35, 8091–8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ebrahimi-Fakhari D., Wahlster L., Hoffmann G.F., Kolker S. (2014) Emerging role of autophagy in pediatric neurodegenerative and neurometabolic diseases. Pediatr. Res., 75, 217–226. [DOI] [PubMed] [Google Scholar]
- 35.Harrison K.D., Miao R.Q., Fernandez-Hernando C., Suarez Y., Davalos A., Sessa W.C. (2009) Nogo-B receptor stabilizes Niemann-Pick type C2 protein and regulates intracellular cholesterol trafficking. Cell Metab., 10, 208–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirkegaard T., Gray J., Olsen O.D., Drndarski S., Ingemann L., Jørgensen S.H., Williams I., Wallom K.L., Priestman D.A., Begley D., et al. (2014), In 2014 Michael, Marcia, and Christa Parseghian Scientific Conference for Niemann-Pick Type C Research, Vol. Abstract #15, p. 26.
- 37.Alam M.S., Getz M., Haldar K. (2016) Chronic administration of an HDAC inhibitor treats both neurological and systemic Niemann-Pick type C disease in a mouse model. Sci. Transl. Med., 8, 326ra323.. [DOI] [PubMed] [Google Scholar]
- 38.Niphakis M.J., Lum K.M., Cognetta A.B., 3rd, Correia B.E., Ichu T.A., Olucha J., Brown S.J., Kundu S., Piscitelli F., Rosen H., et al. (2015) A Global Map of Lipid-Binding Proteins and Their Ligandability in Cells. Cell, 161, 1668–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S., et al. (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature, 520, 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grimm D., Pandey K., Kay M.A. (2005) Adeno-associated virus vectors for short hairpin RNA expression. Methods Enzymol., 392, 381–405. [DOI] [PubMed] [Google Scholar]
- 41.Runz H., Rietdorf J., Tomic I., de Bernard M., Beyreuther K., Pepperkok R., Hartmann T. (2002) Inhibition of intracellular cholesterol transport alters presenilin localization and amyloid precursor protein processing in neuronal cells. J. Neurosci., 22, 1679–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu G., Sun W., Zou Y., Cai Z., Wang P., Lin X., Huang J., Jiang L., Ding X., Hu G. (2015) RNA interference against TMEM97 inhibits cell proliferation, migration, and invasion in glioma cells. Tumour Biol., 36, 8231–8238. [DOI] [PubMed] [Google Scholar]
- 43.Xu X.Y., Zhang L.J., Yu Y.Q., Zhang X.T., Huang W.J., Nie X.C., Song G.Q. (2014) Down-regulated MAC30 expression inhibits proliferation and mobility of human gastric cancer cells. Cell. Physiol. Biochem., 33, 1359–1368. [DOI] [PubMed] [Google Scholar]
- 44.Ebrahimi-Fakhari D., Cantuti-Castelvetri I., Fan Z., Rockenstein E., Masliah E., Hyman B.T., McLean P.J., Unni V.K. (2011) Distinct roles in vivo for the ubiquitin-proteasome system and the autophagy-lysosomal pathway in the degradation of alpha-synuclein. J. Neurosci., 31, 14508–14520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamentsky L., Jones T.R., Fraser A., Bray M.A., Logan D.J., Madden K.L., Ljosa V., Rueden C., Eliceiri K.W., Carpenter A.E. (2011) Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics, 27, 1179–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thormaehlen A.S., Schuberth C., Won H.H., Blattmann P., Joggerst-Thomalla B., Theiss S., Asselta R., Duga S., Merlini P.A., Ardissino D., et al. (2015) Systematic cell-based phenotyping of missense alleles empowers rare variant association studies: a case for LDLR and myocardial infarction. PLoS Genetics, 11, e1004855.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fan M., Sidhu R., Fujiwara H., Tortelli B., Zhang J., Davidson C., Walkley S.U., Bagel J.H., Vite C., Yanjanin N.M., et al. (2013) Identification of Niemann-Pick C1 disease biomarkers through sphingolipid profiling. J. Lipid Res., 54, 2800–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.