Abstract
Spinal muscular atrophy (SMA), the leading genetic cause of infant mortality, predominantly affects high metabolic tissues including motor neurons, skeletal muscles and the heart. Although the genetic cause of SMA has been identified, mechanisms underlying tissue-specific vulnerability are not well understood. To study these mechanisms, we carried out a deep sequencing analysis of the transcriptome of spinal motor neurons in an SMA mouse model, in which we unexpectedly found changes in many genes associated with mitochondrial bioenergetics. Importantly, functional measurement of mitochondrial activities showed decreased basal and maximal mitochondrial respiration in motor neurons from SMA mice. Using a reduction-oxidation sensitive GFP and fluorescence sensors specifically targeted to mitochondria, we found increased oxidative stress level and impaired mitochondrial membrane potential in motor neurons affected by SMA. In addition, mitochondrial mobility was impaired in SMA disease conditions, with decreased retrograde transport but no effect on anterograde transport. We also found significantly increased fragmentation of the mitochondrial network in primary motor neurons from SMA mice, with no change in mitochondria density. Electron microscopy study of SMA mouse spinal cord revealed mitochondria fragmentation, edema and concentric lamellar inclusions in motor neurons affected by the disease. Intriguingly, these functional and structural deficiencies in the SMA mouse model occur during the presymptomatic stage of disease, suggesting a role in initiating SMA. Altogether, our findings reveal a critical role for mitochondrial defects in SMA pathogenesis and suggest a novel target for improving tissue health in the disease.
Introduction
Spinal Muscular Atrophy (SMA) is an autosomal recessive neuromuscular disease that occurs in one of every 10,000 live births (1–3), ranking as the leading genetic cause of infant mortality. SMA is caused by mutation in survival motor neuron 1 (SMN1) gene (4). In addition to SMN1, humans have SMN2, which is different from SMN1 by a cytosine (C) to thymine (T) change in its exon 7. This single nucleotide change disrupts the efficient splicing of SMN2 gene. Consequently, only about 10% of SMN protein generated by SMN2 is full-length and functional. In SMA patients, genetic mutations cause a complete loss of SMN production from SMN1 (4), leading to reduced levels of SMN protein, which correlate inversely with disease severity (5). Patients with a severe form of disease exhibit disease onset around 5 months of age, with death from respiratory distress within 2 years (5). SMN is implicated in regulating a variety of biological functions, including small nuclear ribonucleoprotein (snRNP) biogenesis and pre-mRNA splicing (6–9). Although SMN protein is ubiquitously expressed, SMA disease is characterized by the predominant loss of lower alpha motor neurons in the spinal cord (10–12). Other tissues affected in SMA include skeletal muscles and the heart, all of which require high levels of energy supply (13–16). Mechanisms underlying tissue-specific vulnerability in SMA are poorly understood.
Tissues with high-energy demand are particularly enriched in mitochondria, whose primary function is to supply cells with ATP generated by oxidative phosphorylation (17). Depending on the cell type, hundreds and sometimes thousands of mitochondria populate a cell. Neurons and muscles, which expend more energy than other cell types, have a particularly high requirement of mitochondrial functions. Within a cell mitochondria are dynamically transported to and localized in regions that utilize more energy, such as the axon hillock and presynaptic terminal of neurons (18,19). Mutations in genes coding components of the respiratory chain of oxidative phosphorylation lead to mitochondrial diseases (18,20,21), which preferentially affect tissue types with high bioenergetic requirement. SMA share many common features with mitochondrial diseases including the specific vulnerability of tissues with high energy demand, suggesting that mitochondria might be functionally impaired in SMA. Clinical studies have indicated fatty acid metabolism defects in SMA patient plasma (22–24), and impaired myogenesis and mitochondrial biogenesis in SMA patient’s muscles (25). However, potential defects of mitochondrial structure and function in motor neurons of SMA mouse models have not been explored.
In this study, we report that mitochondria in motor neurons affected by SMA are functionally and structurally defective before the onset of disease symptoms. We found that mitochondria in motor neurons from SMA mice were functionally impaired and fragmented, with reduced mitochondrial respiration, decreased mitochondrial ATP synthesis, defective retrograde mitochondrial transport, decreased mitochondrial membrane potential and increased oxidative stress level. In addition, electron micrographs of lumbar level spinal cord motor neurons from SMA mice show significant mitochondrial fragmentation and edema at a presymptomatic stage, suggesting a role for mitochondrial defects in initiating SMA pathogenesis.
Results
Impaired mitochondrial bioenergetics
To better understand why motor neurons are specifically vulnerable in SMA, we decided to isolate spinal motor neurons from SMA and control mice and study the difference between their RNA expression profiles. We crossed the Hung-Li SMA mouse model with the Hb9:GFP mice, which specifically label motor neurons with GFP (26). After micro-dissection and enzymatic dissociation, GFP-expressing spinal motor neurons from SMA (SMA; Hb9:GFP) mice and control (Hb9:GFP) mice were isolated by fluorescence activated cell sorting (FACS). Then RNA deep sequencing (RNA-seq) was used to analyse RNA expression profiles in these purified motor neurons. Analysis of the RNA-seq data using GSEA (Gene Set Enrichment Analysis) and the hallmark gene sets showed that many mitochondria bioenergetics-related genes were significantly dysregulated in motor neurons affected by SMA (Fig. 1A and B, Supplementary Material, Table S1) . Therefore, we decided to test whether mitochondrial functions were changed in motor neurons from SMA mouse models.
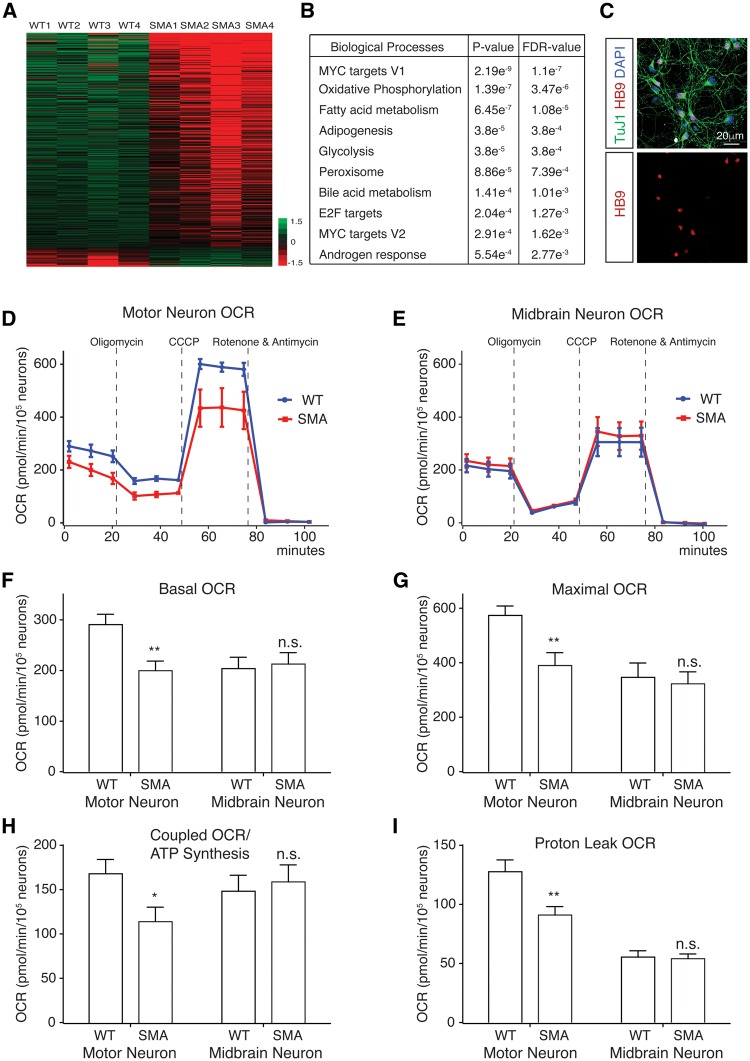
Figure 1.
Impaired mitochondrial bioenergetics in spinal motor neurons affected by SMA. (A) Heat map depicting differential gene expression in spinal motor neurons isolated by fluorescence activated cell sorting (FACS) from SMA; Hb9:GFP mice and littermate wild type (WT) mice (n = 4). (B) Gene ontology analysis of RNA-seq results using GSEA revealed that several categories of genes, whose expression changed most significantly in SMA motor neurons, were related to mitochondrial function and energy metabolism (fold change cutoff = 1.5, P < 0.05, FDR: False Discovery Rate). (C) Immunostaining of motor neurons cultured from E12.5 mouse spinal cords. Cells were stained with antibodies recognizing neuronal marker Tuj1 (green) and motor neuron marker HB9 (red). (D) Mitochondrial oxygen consumption rates (OCR) of motor neurons from Δ7 SMA and wild type control mice. OCR measured by a Seahorse XF24 Analyzer showed impaired mitochondrial bioenergetics in motor neurons affected by SMA. (E) Midbrain neuron mitochondrial OCR was not changed in Δ7 SMA mice. (F) Basal mitochondrial OCR was significantly decrease in spinal motor neurons (P = 0.0019, WT n = 27; SMA n = 28), but not in midbrain neurons (P = 0.78, WT n = 20; SMA n = 16), isolated from SMA mice compared to those from wild type (WT) control mice. (G) Maximal mitochondrial OCR measured in the presence of CCCP was significantly reduced in SMA spinal motor neurons (P = 0.0029) but not in midbrain neurons (P = 0.84). (H) Coupled mitochondrial OCR used for ATP synthesis was significantly reduced in SMA spinal motor neurons (P = 0.024) but not in midbrain neurons (P = 0.69). (I) Mitochondrial proton leak OCR was also specifically reduced in SMA spinal motor neurons (P = 0.0049) but not in midbrain neurons (P = 0.73). Results are mean ± SEM, from at least three independent experiments, were normalized to cell number. n.s.: not significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t test.
The primary function of mitochondria in cells is to generate ATP through oxidative phosphorylation. To test whether mitochondrial bioenergetics were compromised in SMA disease conditions, we isolated and cultured motor neurons from the spinal cord of Δ7 SMA mice and control littermates (Fig. 1C). Mitochondrial respiration of primary spinal motor neurons was measured using a Seahorse analyzer. Motor neurons affected by SMA showed significantly lower basal mitochondrial respiration rate/oxygen consumption rate (OCR) than non-disease motor neurons (Fig. 1D and F). In addition, maximal mitochondrial respiration rate/OCR induced by mitochondrial uncoupler carbonyl cyanide m-chlorophenyl hydrazone (CCCP), which is an indication of the highest mitochondrial functional capacity under stress conditions, was also significantly reduced (Fig. 1D and G). Importantly, coupled mitochondrial respiration rate/OCR used for ATP synthesis, as well as the OCR for counteracting proton leaking across the mitochondrial inner membrane were both significantly decreased (Fig. 1D,H,I). To test whether the observed mitochondrial respiration deficiency is specific to motor neurons, we examined primary mouse midbrain neurons from SMA and wild type mice. Midbrain neurons from SMA mice showed no change of mitochondrial basal, maximal, coupled or proton leak OCR, compared to those of midbrain neurons from wild type mice (Fig. 1E–I). These results suggest that mitochondrial bioenergetics are specifically impaired in motor neurons affected by SMA.
Increased mitochondrial oxidative stress and reduced mitochondrial membrane potential
Defective mitochondrial respiration and ATP synthesis often lead to increased mitochondrial oxidative stress (27). To determine whether this was the case in SMA motor neurons, we expressed a redox-sensitive variant of green fluorescent protein (roGFP) with a mitochondria targeting sequence (mito-roGFP) (28), which provides a reversible, quantitative means of monitoring mitochondrial oxidative stress level in motor neurons from Δ7 SMA mice (Fig. 2A and B). Because mitochondria are localized in both neuronal cell body and axon, we decided to measure mitochondrial oxidative stress in both regions using fluorescence live imaging. We found that the oxidation levels of mito-roGFP were significantly higher in both the soma and axon of SMA motor neurons (Fig. 2C and D) than those in wild type motor neurons, indicating increased mitochondrial oxidative stress in motor neurons affected by SMA.
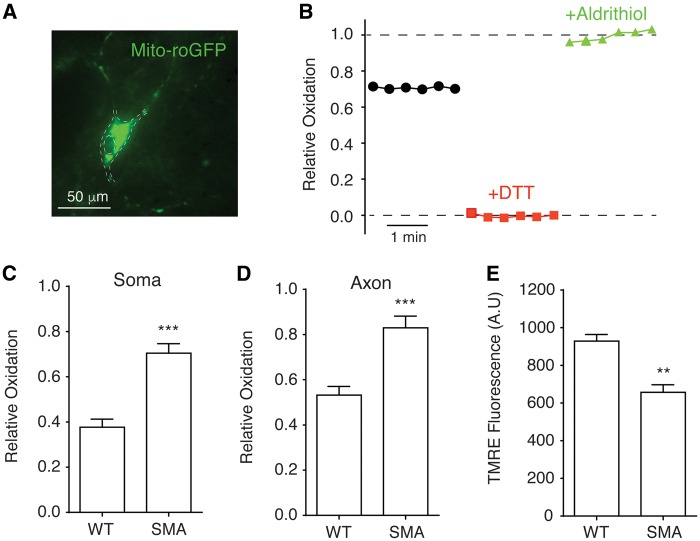
Figure 2.
Increased mitochondrial oxidative stress and compromised membrane potential in spinal motor neurons affected by SMA. (A) A primary mouse spinal motor neuron expressing redox-sensitive green fluorescent protein with mitochondria targeting sequence (mito-roGFP). The contour of motor neuron was marked by dashed lines. (B) Measurement of mitochondrial oxidation level. The intensities of mito-roGFP in motor neurons were measured with a live imaging fluorescent microscope. DTT were applied to fully reduce (red trace) and aldrithiol were used to fully oxidize (green trace) roGFP for calculating relative oxidation levels as described in Methods. (C, D) Mitochondrial oxidative stress was significantly increased in both the soma (C) (P = 0.00013) and axon (D) (P = 0.0003) of spinal motor neurons from Δ7 SMA mice. Mitochondrial roGFP oxidation levels in the soma and axon (at least 50 µm away from the motor neuron soma) were recorded in 5–7 experimental repeats for each group. (E) Mitochondrial membrane potential was significantly reduced (P = 0.0069) in SMA motor neurons. The membrane potentials of mitochondria on 42 samples of Δ7 SMA motor neurons and 42 wild type samples in three independent experiments were measured using fluorescent dye tetramethylrhodamine ethyl (TMRE) and normalized to cell number. **P < 0.01; ***P < 0.001, Student’s t test.
Increased mitochondrial oxidative stress is often associated with compromised mitochondrial membrane potential. To test this possibility, the mitochondria membrane potential of spinal motor neurons from SMA mice was measured with fluorescent dye tetramethylrhodamine ethyl (TMRE). Depolarized or inactive mitochondria have decreased membrane potentials that reduce the uptake of cationic TMRE. We found that the mitochondrial membrane potential of motor neurons from SMA mice was significantly compromised compared with non-disease motor neurons (Fig. 2E). Together, the increased mitochondrial oxidative stress level and reduced mitochondrial membrane potential suggest impaired health and function of mitochondria in motor neurons affected by SMA, consisting with defective mitochondrial bioenergetics we observed in SMA mice.
Defective mitochondrial transport
Mitochondria are dynamically transported in neurons to ensure sufficient energy supply and proper mitochondrial repairment. Thus, appropriate transport is an important indication of mitochondrial function. Mitochondria are transported anterogradely from the cell body towards the synaptic terminals and retrogradely from the terminals back along cytoskeleton microtubule tracks (29). We recently found that hyper-phosphorylation of the microtubule-associated protein Tau, a key regulator of cellular trafficking, contributed to the specific loss of motor neurons in SMA (30). This finding motivated us to explore the possibility that mitochondrial transport is impaired in SMA motor neurons. We used time-lapse confocal live imaging to measure mitochondrial transport in primary motor neurons transfected with mitochondria targeting sequence-tagged DsRed (mito-DsRed) (Fig. 3A) (31). We found that mitochondria travelled significantly shorter distance in axons of Δ7 SMA motor neurons compared to wild type neurons (Fig. 3B). Further analysis revealed that retrograde travel distance (Fig. 3E) of mitochondria was also decreased in motor neurons affected by SMA, which was caused by decreased speed (Fig. 3D) and time (Fig. 3C) spent on retrograde transport. There was also a significant increase in the time spent in the immobile state (Fig. 3C) by mitochondria in SMA motor neurons, but no significant effect on any aspects of anterograde transport. To test whether defective mitochondrial transport is unique to the Δ7 SMA mice, we performed these studies using a different SMA model, the Hung-Li SMA mice. Both of these mouse models were generated by expressing low levels of human SMN transgene in Smn knock-out mice to recapitulate hallmarks of SMA pathogenesis at the molecular, cellular and behaviour levels, with similar average lifespan of around 13 days (32,33). Similarly, mitochondria in spinal motor neurons from the Hung-Li SMA mice also showed significantly reduced total travel distance, decreased retrograde transport distance, speed and time, as well as increased percent of time in the immobile state (Supplementary Material, Fig. S1). By contrast, every aspect of mitochondrial transport appears to be normal in midbrain neurons isolated from SMA mice (Fig. 3F–J), suggesting defective mitochondrial transport is specific for motor neurons. Given the particularly long axons of lower alpha motor neurons, it is conceivable that detrimental consequences of defective mitochondrial transport may be exacerbated in motor neurons, thus contributing to motor neuron-specific degeneration in SMA pathogenesis.
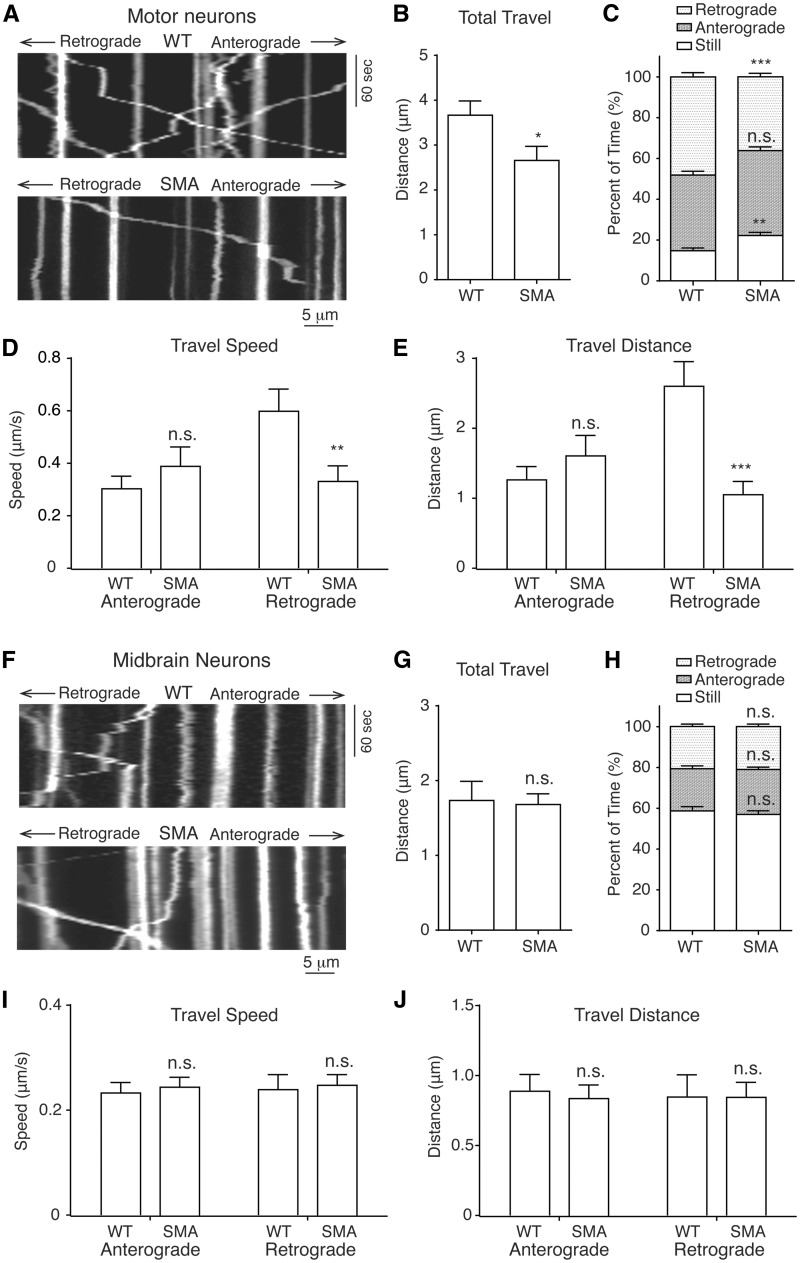
Figure 3.
Impaired mitochondrial transport in spinal motor neurons from Δ7 SMA mice. (A) Representative kymographs of mitochondrial transport in wild type and SMA mouse motor neuron axons. (B) The average total travel distance of all tracked mitochondria is significantly reduced in SMA motor neurons compared to wild type neurons (P = 0.03; WT: 174 mitochondria on 40 axons; SMA: 260 mitochondria on 58 axons). (C) The percent of time spent by all analysed mitochondria on retrograde movement was significantly reduced (P = 0.00002) in SMA motor neurons, while the percent of time spent in still was increased (P = 0.001), with no significant change of time in anterograde movement (P = 0.08). (D) The retrograde movement speed of mitochondria during their mobile phases was significantly reduced in SMA motor neurons (P = 0.008), but not the anterograde movement speed (P = 0.37). (E) The average total retrograde travel distance of all analysed mitochondria was significantly reduced in motor neurons affected by SMA (P = 0.00007), but not the anterograde travel distance (P = 0.37). (F) Representative kymographs of mitochondrial transport in wild type and SMA mouse midbrain neuron axons. (G) Analysis of mitochondrial transport showed that the average total travel distance of all tracked mitochondria in SMA mouse midbrain neurons was not significantly different from that in wild type mice (P = 0.84; WT: 316 mitochondria on 23 axons; SMA: 378 mitochondria on 33 axons). (H-J) No significant difference between SMA mouse and wild type mouse midbrain neurons in characteristics of mitochondrial movement including percent of time in immobility (P = 0.53), retrograde (P = 0.84) or anterograde (P = 0.47) movement, speed of retrograde (P = 0.80) or anterograde movement (P = 0.68), total anterograde (P = 0.69) or retrograde (P = 0.90) travel distance. n.s.: not significant; *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t test.
Increased mitochondria fragmentation
Mitochondrial functions are closely related to its size and morphology. Elongated mitochondria have higher density of cristae with better efficiency in ATP production; whereas short, fragmented mitochondria are more likely to have compromised membrane potential, with lower levels of the dimeric form of ATPase and decreased cristae density, both of which are associated with the impaired ATP synthesis (34–36). To test whether mitochondria fragmentation is associated with the functional defects observed in SMA disease conditions, we compared the mitochondria length in motor neurons from the Δ7 SMA mice and control mice transfected with mito-DsRed. Confocal live imaging of mitochondria located in motor neuron axons showed significantly reduced mitochondria length in SMA motor neurons, with no change of mitochondria density (Fig. 4A–C). In addition, we also tested mitochondria fragmentation in motor neurons from the Hung-Li SMA mice. The length of mitochondria was also significantly reduced in motor neurons from the Hung-Li SMA mice, without any change in mitochondrial density (Supplementary Material, Fig. S2). These data suggest that mitochondria fragmentation is likely associated with the general SMA pathology but not limited to a specific SMA mouse model. To test whether mitochondria fragmentation represents a common defect associated with all neuronal cell types, we also examined the mitochondria morphology in midbrain neurons isolated from SMA mice. We found that neither the length nor density of mitochondria in SMA midbrain neurons was different from that of wild type mice (Fig. 4D–F). This finding suggests that, similar to impaired mitochondrial bioenergetics, mitochondria fragmentation is also a defect specific to motor neurons, potentially contribute to the selective vulnerability of motor neurons in SMA disease conditions.
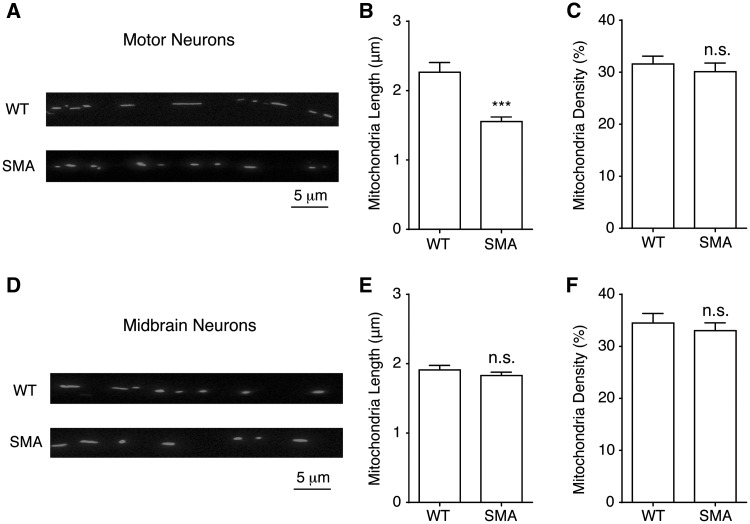
Figure 4.
Mitochondria fragmentation in spinal motor neurons from Δ7 SMA mice. (A) A representative image for measuring mitochondria length and density in wild type (WT) and SMA mouse motor neuron axons. Primary spinal motor neurons were transfected with mito-dsRed at 5-7 days in vitro and imaged using a confocal microscope 48 h later. Mitochondria length and density were analysed along axons at least 50 µm away from the soma. (B, C) Mitochondria length in spinal motor neurons from SMA mice was significantly shorter (P = 0.00001), than that of wild type motor neurons while mitochondria density was not changed (P = 0.51). Wild type motor neurons: 174 mitochondria from 40 axons; SMA motor neurons: 260 mitochondria from 58 axons. (D) A representative image of mitochondria in wild type and SMA mouse midbrain neuron axons. (E, F) Neither mitochondria length (E) (P = 0.33) nor density (F) (P = 0.53) was changed in SMA midbrain neurons. Wild type midbrain neurons: 316 mitochondria from 23 axons; SMA midbrain neurons: 378 mitochondria from 33 axons. Results are mean ± SEM, from at least three different cultures. n.s: not significant; ***P < 0.001, Student’s t test.
Ultrastructural mitochondrial abnormalities
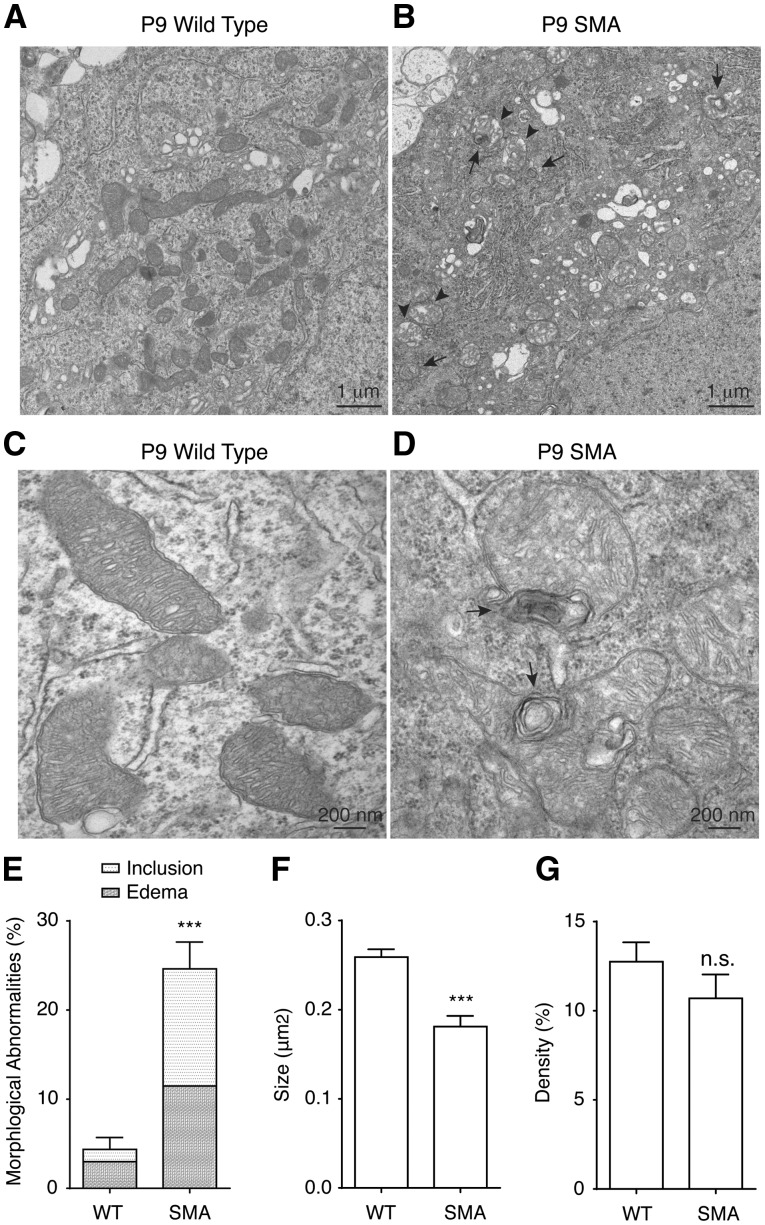
To further examine mitochondrial defects in vivo, we investigated the mitochondria ultrastructural morphology in SMA mice. Electron microscopy study of motor neurons from lumbar 1 (L1) level mouse spinal cord showed significantly reduced mitochondria size (Fig. 5A–D,F) in postnatal day 9 (P9) Δ7 SMA mice, whereas mitochondria density was not changed (Fig. 5G). Importantly, 25% of mitochondria in motor neurons from SMA mice show abnormal morphology, compared to only 4% in wild type motor neurons (Fig. 5A–E). These ultrastructural abnormalities include edema (in 12% mitochondria) and concentric lamellar inclusions (in 13% mitochondria) (Fig. 5E), which are commonly found in mitochondrial diseases (37,38). Furthermore, cristae density is decreased in motor neurons affected by SMA (Fig. 5A–D). Decreased cristae density has been linked to impaired ATP production efficiency (34,35). These findings not only are consistent with our observations of decreased mitochondrial bioenergetics, length and retrograde transport in cultured SMA motor neurons, but also provide additional solid evidence for impaired mitochondrial health and function in motor neurons affected by SMA in vivo.
Figure 5.
Ultrastructural abnormalities of mitochondria in motor neurons in Δ7 SMA mice. (A-D) Representative low-magnification (A, B) and high-magnification (C, D) electron micrographs of mitochondria from lumbar 1 (L1) level spinal cord motor neurons in wild type and SMA mice at postnatal day 9 (P9). Mitochondria in SMA mice displayed concentric lamellar inclusions (arrows in B, D) and edema (arrow heads in B) with decreased cristae density compared to mitochondria in wild type mice. (E) 25% mitochondria in L1 level motor neurons in P9 SMA mice exhibited ultrastructural abnormalities compared to 4% mitochondria in P9 wild type mouse L1 level motor neurons (P = 0.0001; WT: 200 mitochondria from three wild type mice, SMA: 230 mitochondria from three SMA mice). (F) Average size of mitochondria in L1 level motor neurons of P9 SMA mice was significantly smaller than mitochondria in P9 wild type L1 level motor neurons (P = 0.0009, WT: 200 mitochondria from three wild type mice, SMA: 230 mitochondria from three SMA mice). (G) Mitochondria density in L1 level motor neurons in P9 SMA mice was similar to that in P9 wild type mice (P = 0.26, WT: 200 mitochondria from three wild type mice, SMA: 230 mitochondria from three SMA mice). Results are mean ± SEM, from at least three independent experiments. n.s: not significant; ***P < 0.001, Student’s t test.
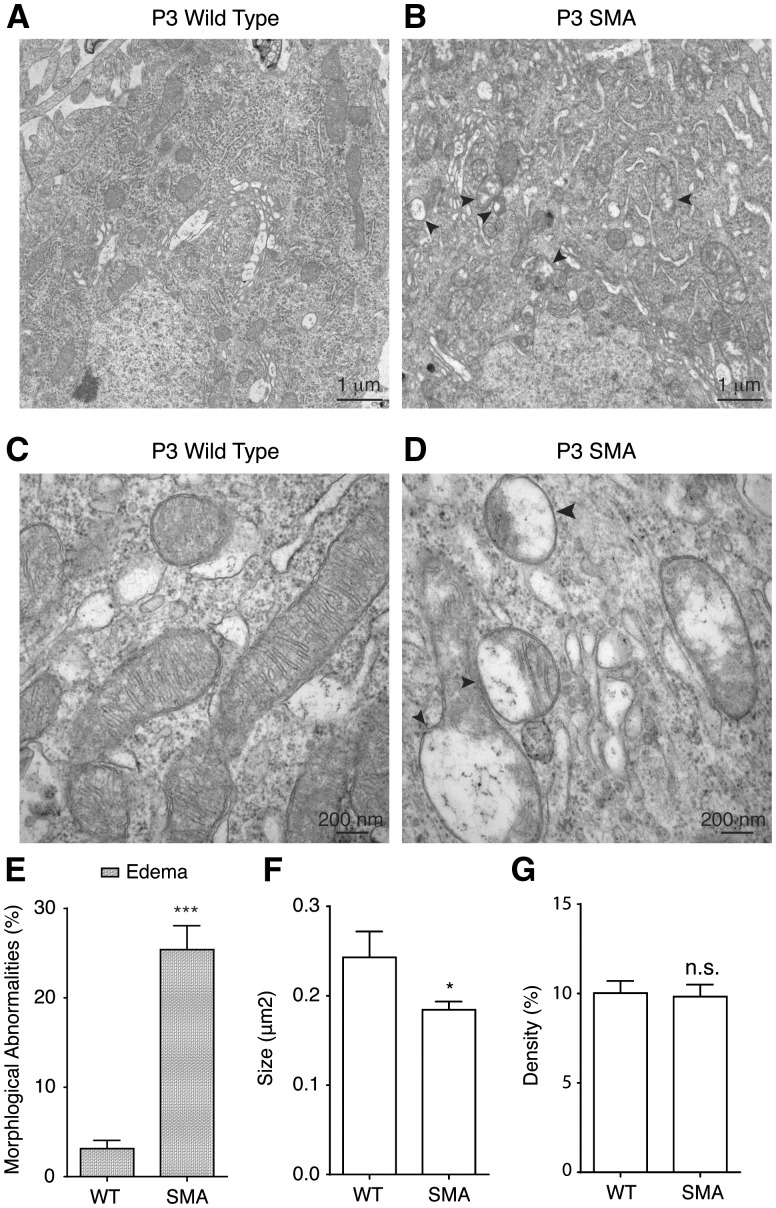
In Δ7 SMA mice at P9, some disease phenotypes including motor behavioural defects can already be detected, thus P9 is considered as a postsymptomatic stage. To test whether mitochondrial ultra-structural abnormalities exist in SMA mice before the onset of disease symptom, we examined the mitochondria ultrastructure at postnatal day 3 (P3), a presymptomatic stage when SMA mice are indistinguishable from their wild type littermate both at the molecular and behavioural levels. Intriguingly, structural abnormalities were detected in 25% of mitochondria in P3 SMA mouse motor neurons, compared to 3% in littermate control mice (Fig. 6A–E). All affected mitochondria at this stage displayed edema, with no sign of concentric lamellar inclusions (Fig. 6A–E). We found that even at presymptomatic P3, mitochondria size was reduced in SMA motor neurons (Fig. 6A–D,F), but not mitochondria density (Fig. 6G). In addition, P3 SMA mouse spinal motor neurons also showed reduced cristae density. These finding suggest that morphological and functional impairments of mitochondria are likely a cause, rather than an effect, of motor neuron degeneration in SMA pathogenesis.
Figure 6.
Mitochondria show ultrastructural abnormalities in Δ7 SMA mice before the onset of disease symptom. (A-D) Representative low-magnification (A, B) and high-magnification (C, D) electron micrographs of mitochondria from lumbar 1 (L1) level spinal cord motor neurons in wild type and SMA mice at presymptomatic stage postnatal day 3 (P3). Mitochondria in P3 SMA mice showed edema (arrow heads in B, D), but not concentric lamellar inclusions, with decreased cristae density. (E) 25% mitochondria in L1 level motor neurons in presymptomatic P3 SMA mice displayed edema compared to 3% mitochondria in P3 wild type mice (P = 0.00001; WT: 326 mitochondria from three wild type mice, SMA: 883 mitochondria from three SMA mice). (F) Average size of mitochondria in L1 level motor neurons in P3 SMA mice was significantly smaller than that in P3 wild type mice (P = 0.02, WT: 326 mitochondria from three wild type mice, SMA: 883 mitochondria from three SMA mice). (G) Mitochondria density in L1 level motor neurons in presymptomatic P3 SMA mice was similar to that in P3 wild type mice (P = 0.85, WT: 326 mitochondria from three wild type mice, SMA: 883 mitochondria from three SMA mice). Results are mean ± SEM, from at least three independent experiments. n.s: not significant; *P < 0.05; ***P < 0.001, Student’s t test.
Discussion
SMA is characterized by selective defects in tissues with high metabolic rate, such as spinal motor neurons and muscles, but mitochondrial dysfunction in motor neurons during SMA pathogenesis has not been systematically explored. Using mouse models of SMA, we found that mitochondria in SMA motor neurons were functionally impaired, with reduced respiration rate, decreased membrane potential, increased oxidative stress level, defective transport along axons and fragmented morphology. Electron microscopy analysis of motor neurons from SMA mice showed substantial mitochondrial fragmentation and edema, which occur prior to the onset of disease symptoms, suggesting a role for mitochondrial dysfunction in contributing to SMA pathogenesis.
Functional defects of mitochondria and motor neuron-specific vulnerability in SMA
Previous clinical studies of infants with severe forms of SMA showed abnormal fatty acid metabolism (22,23,39,40). Muscle biopsies from SMA patients also showed metabolic deficits (24). In addition, depletion of mitochondrial DNA and disintegrated mitochondria were found in muscles affected by SMA (25,41,42). However, all these works are primarily clinical studies focusing on metabolic or mitochondrial abnormalities in muscles. Here we systematically investigated functional defects of mitochondria in motor neurons using SMA mouse models. We found significantly reduced basal mitochondrial respiration and coupled respiration, which is used for ATP synthesis, in mouse motor neurons affected by SMA (Fig. 1D,F,H). In addition, the maximal mitochondrial respiration capacity, revealed when mitochondria were stressed by uncoupler CCCP, was also decreased (Fig. 1D and G). Spinal cord alpha motor neurons are characterized by large soma, long axon, elaborated dendritic branching and dynamic synaptic activities (43,44). It is conceivable that defects in mitochondria bioenergetics will predominantly affect these fast fatigable motor neurons, which have a high metabolic rate and are constantly under energy stress. The active transport of mitochondria, especially the retrograde transport, is slowed down in motor neurons affected by SMA (Fig. 3A–E). Given that motor neuron axons are particularly long, defects in the axonal transport of mitochondria may be specifically exacerbated in these neurons. Together, these functional impairments of mitochondria bioenergetics and transport could help explain why spinal motor neurons are particularly vulnerable in SMA disease.
Our new observation that mitochondrial axonal transport is defective in neurons affected by SMA is consistent with our recent finding that microtubule-associated protein tau is hyperphosphorylated in SMA conditions and contribute to disease pathogenesis (30). Mitochondria are actively transported by motor proteins kinesin and dynein along cytoskeleton microtubules (29,19). Microtubule-associated protein tau is enriched in axons and serves as an axonal marker (45). Hyperphosphorylated tau may interfere with the transport of mitochondria by impeding their movement along axons.
Mitochondria fragmentation and oxidative stress in SMA motor neurons
Mitochondria are highly dynamic organelles that constantly fuse and divide to maintain their normal structure and function. A balance between fusion and fission is essential for maintaining the proper mitochondria size and function; disruption of the balance is associated with a variety of neurodegenerative diseases (46). Fusion allows mitochondria to share and exchange content such as mitochondrial DNA, proteins and lipids, whereas fission facilitates apoptosis and the clearance of damaged mitochondria (47). Increased mitochondrial fusion protects against neurodegeneration (48), while disruption of fusion leads to smaller, fragmented mitochondria that display reduced membrane potential and impaired respiration (35,49). In this study, we found severely fragmented mitochondria with reduced size in motor neurons affected by SMA (Figs. 4–6, Supplementary Material, Fig. S2), suggesting potential defects in the balance between fusion and fission. In addition, our findings were confirmed in two widely used SMA mouse models, indicating that these defects are associated with general SMA pathology but not limited to a particular mouse model. A recent study found that mitochondria length is not changed but mitochondria density is decreased in human SMA induced pluripotent stem cell (iPSC)-derived motor neurons (50), different from our finding. Although this study also found impaired retrograde transport and less mobile mitochondria in SMA iPSC motor neurons, which is consistent with our observations, there are drawbacks associated with its use of mitoTracker Red to study mitochondria length and density. We used the mito-DsRed expression to label mitochondria and measure their size and transport; whereas Xu et al utilized mitochondrial membrane potential-dependent fluorescent dye MitoTracker Red in their assays. We have found that mitochondrial membrane potential was compromised in SMA motor neurons (Fig. 2E), consistent with studies using SMN knock-down in NSC34 cells (51) and embryonic stem cell-derived motor neurons (52). Therefore, mitochondria with reduced membrane potential in SMA motor neurons may not be effectively labelled by MitoTracker Red. In addition, MitoTracker Red is not entirely specific to mitochondria (53), further complicating the interpretation of results from using this dye.
Mitochondria fragmentation has been linked with reduced membrane potential and increased oxidative stress (49,54). This is consistent with our observation of dramatically increased mitochondrial oxidative stress level measured by mito-roGFP (Fig. 2) and mitochondrial fragmentation (Figs. 4–6, Supplementary Material, Fig. S2) in SMA motor neurons. Studies in SMA iPSC motor neurons generated conflicting results regarding oxidative stress (50,55). Patitucci et al found no change of oxidative stress while Xu et al observed decreased membrane potential and increased oxidative stress in mitochondria from SMA iPSC motor neurons. These discrepancies are likely due to different experimental conditions used by these laboratories or the intrinsic limitations of iPSC as a model system to study SMA pathogenesis (56). The increased mitochondrial oxidative stress we observed in SMA motor neurons may be caused by fragmented mitochondria or reduced retrograde transport of mitochondria for repairment and clearance. Whether disruption of fission/fusion balance or impaired mitophagy may contribute to these defects, requires further investigation.
Ultrastructural changes of mitochondria in SMA mice before the onset of disease
Do these profound mitochondrial impairments in primary spinal motor neurons reflect similar abnormalities in vivo? Ultrastructural analysis by electron microscope revealed decrease of mitochondria size but not density in motor neurons in SMA mice (Figs. 5 and 6), consistent with our findings in primary motor neurons. Decreased mitochondria size has been linked with reduced level of dimeric ATPase, and decreased efficiency of ATP synthesis (34,35). We also found that 25% of all mitochondria in SMA motor neurons had abnormal morphology, including edema and concentric lamellar inclusions (Fig. 5), which are characteristics of mitochondrial diseases (37,38). Edema was accompanied by a decrease in the density of cristae (Fig. 5). Decreased mitochondrial cristae density has been linked to impaired ATP production efficiency (34,35,57). These results are consistent with our finding of decreased mitochondrial bioenergetics in SMA. Besides edema, concentric lamellar inclusions were found in 13% of mitochondria in SMA motor neurons. These inclusions are characteristic of mitochondrial Leigh disease (37,38) and have been associated with mutations in mitochondrial gene ATP6 (58). SMA is caused by insufficient SMN protein, whose best characterized function is linked to pre-mRNA splicing. Deep sequencing with higher resolution than that in Fig. 1A is underway in our laboratory to test whether the splicing of mitochondria-related genes are altered in SMA disease.
To determine whether the mitochondrial ultrastructural abnormalities appear before the onset of disease symptoms, we examined spinal motor neurons in SMA mice at postnatal day 3 (P3), a presymptomatic stage of disease. Intriguingly, structural abnormalities were detected in 25% of mitochondria in P3 SMA mouse motor neurons, compare to 3% in littermate control mice. All affected mitochondria at this stage displayed edema, with no sign of concentric lamellar inclusions (Fig. 6A–E). Mitochondria in presymptomatic P3 SMA motor neurons also displayed decreased size and reduced cristae density (Fig. 6A–D, F). Decreased mitochondria size and cristae density has been linked to impaired ATP synthesis (34,35,57). Our findings imply that ATP production in SMA motor neurons is likely impaired at a presymptomatic stage. These findings strongly suggest that mitochondrial defects contribute to SMA disease pathogenesis, rather than being a secondary effect of motor neuron degeneration.
As another prevalent neuromuscular disease, amyotrophic lateral sclerosis (ALS) is also characterized by motor neuron degeneration and muscle atrophy. In ALS, retrograde movement of mitochondria is impaired (59–61). Ultrastructural abnormalities of mitochondria were also observed in ALS by electron microscopy. However, in ALS there were reports of enlarged mitochondria (60,62,63) whereas we found mitochondria of reduced size in motor neurons affected by SMA. In addition, lamellar inclusions, which are characteristic of mitochondria diseases and not reported in ALS, were observed by us in SMA motor neurons. These differences suggest that distinct aetiologies underlying SMA and ALS cause different mitochondrial impairments, but the high sensitivity of motor neuron to mitochondrial dysfunction leads to the similar symptom of motor neuron degeneration.
Mitochondrial health and function are crucial for maintaining cellular bioenergetic demand. Impairment of mitochondrial functions in spinal motor neurons may contribute to their specific vulnerability in SMA disease. Our findings reveal a critical role for mitochondrial defects in SMA pathogenesis and suggest a novel target for improving tissue health in the disorder.
Materials and Methods
SMA mouse models
The Δ7 SMA mouse model (Smn-/-;SMN2tg/tg;SMNΔ7tg/tg or Jackson Laboratory #005025), the Hung-Li SMA mouse model (Smn-/-;SMN2Hungtg/- or Jackson Laboratory #005058) and the Hb9:GFP transgenic mice (Hb9:GFP or Jackson Laboratory #005029) were obtained from the Jackson Laboratory. Genomic DNA extracted from tail samples was used for PCR-based genotyping as reported (26,32,33). Both male and female mice were used.
Motor neurons purification and RNA-seq analysis
Smn-/-;SMN2Hungtg/- SMA mice were crossed with the Hb9:GFP mice to label spinal motor neurons with GFP. Spinal cords from P9 SMA;Hb9:GFP mice and control Hb9:GFP mice were isolated and dissociated using Accutase (MP Biomedicals). GFP-expressing spinal motor neurons were purified using BD FacsAria SORP 4-Laser fluorescence-activated cell sorter (BD Biosciences) and lysed into Trizol LS (Thermo Fisher Scientific). Total RNA from sorted motor neurons was purified using RNeasy Mini kit (Qiagen). RNA-seq libraries were constructed using the strand specific dUTP method (64) with minor modifications. Briefly, 3 ug of DNAse treated RNA was depleted of rRNA using Ribozero (Epicentre). Two batches of rRNA-depleted samples were combined, cleaned by RiboMinus concentration module (Invitrogen) and fragmented at 90 °C for 3 min (NEB fragmentation buffer). First strand synthesis was followed by cleanup with RNAClean XP SPRI beads (Agencourt). Second strand synthesis incorporated dUTP, followed by sample clean up with MinElute PCR purification Kit (Qiagen). Fragment ends were repaired, adenylated, then ligated to True-Seq barcoded adaptors and cleaned up with AMPure XP SPRI beads (Agencourt). The libraries were then amplified by PCR for 12 cycles and cleaned up with AMPure XP SPRI beads. Illumina sequencing (1650 bp read length) was performed on a HiSeq 2000. Sequence analyses were carried out using Gene Set Enrichment Analysis (GSEA) with the hallmark gene sets and other software packages by the NUSeq Core Facility at Northwestern University Center for Genetic Medicine.
Mouse primary spinal motor neuron and midbrain neuron culture
Primary neurons from mouse spinal cord and ventral midbrain were cultured in Neurobasal (Life Technologies) supplemented with B27 (Life Technologies) as previously described (30). Briefly, spinal cords and midbrains from E12.5 mouse embryos were dissected out and dissociated with 0.25% trypsin. After enriching motor neurons with Optiprep (Sigma) density gradient centrifugation and BSA cushion, cells were seeded on glass cover slips coated with 20 μg/ml Poly-L-Lysine (PLL) (Sigma) and 8 μg/ml Laminin (Sigma), and grown in the presence of 50 μg/ml BDNF, 50 μg/ml CNTF and 25 μg/ml GDNF (PeproTech). Neurons were transfected with Lipofectamine 2000 (Life Technologies) following manufacturer’s instructions.
Mitochondrial oxygen consumption rate measurements
Mitochondrial oxygen consumption rates (OCR) were measured using a XF24 Seahorse Biosciences Extracellular Flux Analyzer. Mouse spinal motor neurons and midbrain neurons were plated onto a Seahorse 24-well plate at 180,000 neurons per well and grown for 7–9 days. Culture media were changed to 500 μl of fresh Neurobasal 30 min before the assay. Seahorse analyzer injection ports contained 1 μM ATP synthase inhibitor oligomycin A, or 5 μM mitochondrial uncoupler carbonyl cyanide m-chlorophenyl hydrazine (CCCP), or 1 μM mitochondrial respiratory complex I inhibitor rotenone plus 1 μM complex III inhibitor antimycin A. Basal mitochondrial oxygen consumption rate (OCR) was determined by subtracting the mitochondrial respiration following antimycin A and rotenone treatment from the base line OCR. Maximal mitochondrial OCR was calculated by subtracting the mitochondrial respiration following antimycin A and rotenone treatment from the CCCP-induced OCR. Coupled mitochondrial OCR was determined by subtracting oligomycin A-induced OCR from the basal mitochondrial OCR. After assays, neurons were immediately fixed for further immunofluorescence staining analysis. Oxygen consumption rates were normalized to cell number quantified by DAPI staining.
Transmission electron microscopy
SMA mice at 3 days and 9 days of age were euthanized and transcardially perfused with 2% paraformaldehyde and 2.5% glutaraldehyde (EM grade freshly prepared) in 0.05M sodium Phosphate buffer pH 7.4. Dissected lumber level spinal cord samples were fixed overnight in fresh fixative at 4 °C. A Pelco Biowave microwave with a Cold Spot and vacuum chamber was used for all processing steps. Second exchange of fixative was processed in the microwave at 250 Watts (all other steps were processed at 100 Watts). Spinal cord samples were washed in 0.05M sodium phosphate buffer, and post-fixed in reduced 1.5% osmium tetroxide, followed with three double-deionized water washes and an acetone dehydration series. EMBed 812 Resin was used for embedding. Blocks were polymerized in a 60 °C oven for 24 h. Thin sections (90 nm) were cut with a diamond knife on a Leica Ultracut S ultramicrotome and retrieved on 200 mesh copper grids. Grids were stained with 3% uranyl acetate and Reynold’s lead citrate at 4 °C. Micrographs were obtained using a Gatan Orius camera on a JEOL 1230 transmission electron microscope with an accelerating voltage of 80kV. Motor neurons were identified by their location, size and morphology on mouse spinal cord ventral horn ultrathin sections. The identity of motor neurons was confirmed by immunostaining of thick sections prepared from spinal cord tissue adjacent to EM ultrathin sections with antibodies recognizing motor neuron marker HB9. Motor neurons were photographed and examined in blinded experimental conditions. Mitochondria density was calculated by dividing the total area of mitochondria with the area of the cytoplasm in the same view field.
Mitochondrial oxidative stress measurement
Primary mouse neurons cultured on glass coverslips for 3–4 days were infected with adeno-associated virus expressing a redox-sensitive variant of green fluorescent protein (roGFP) with a mitochondria targeting sequence (AAV-mito-roGFP) from Dr. Jyothisri Kondapalli of the Surmeier laboratory (28). 96 h after transduction, cultured neurons were transferred to an imaging chamber with inverted epifluorescence microscope (Olympus IX71) using a 40X/NA 1.35 oil-immersion objective (Olympus). The imaging chamber was perfused with artificial cerebrospinal fluid (ACSF) containing 125 mM NaCl, 3 mM KCl, 1.25 mM NaH2PO4, 25 mM NaHCO3, 1 mM MgCl2, 2 mM CaCl2 and 25 mM D-glucose, pH 7.4. Cultures were kept in ACSF for 10 min to allow fluid environment reaching equilibrium before imaging. All experiments were performed at 35–36 °C. Two excitation wavelengths (410 nm and 470 nm) were used with emission monitored at 535 nm. Region of interests (ROIs) were selected using imaging software SlideBook (Intelligent Imaging Innovations) on the soma or axon (at least 50 µm from the neuronal cell body). Images were taken every 30 s using a cooled CCD camera (I-PentaMax, Princeton Instruments) and fluorescent intensities were measured ratiometrically. After initial basal measurements, cultures were treated with 2 mM dithiothreitol (DTT) to get fully reduced, followed by 100 µM aldrithiol treatment to reach maximal oxidation. Fluorescent intensities were measured correspondingly to determine the range of roGFP signal. The relative mitochondrial oxidative levels were then calculated as 1 − [(F − FAld)/(FDTT − FAld)], in which F, FDTT and FAld represent measured intensities at basal, reduced and oxidized states, respectively.
Mitochondrial membrane potential measurement
Mitochondrial membrane potential was measured with the potential-dependent fluorescent dye tetramethylrhodamine ethyl (TMRE) (Molecular Probes). 150,000 primary motor neurons were plated into each well of a BD-Falcon 96-well black plate (with clear bottom) and cultured for 7 days. Neurons were labelled with 200 nM TMRE for 30 min, followed by washing with PBS plus 0.2% BSA before measuring the fluorescence intensity with a Wallac Victor plate reader. Background fluorescence was measured after treating neurons with 40 µM FCCP for 15 min. Neurons were fixed immediately after the assay for immunofluorescence staining. TMRE intensity was normalized to cell number quantified by DAPI staining.
Live imaging and data analysis of mitochondria transport, density and fragmentation
Time-lapse live imaging by confocal microscope was used to measure axonal mitochondrial transport, density and fragmentation. After culturing for 5–7 days, primary mouse neurons were transfected with mitochondria targeting sequence-tagged DsRed (mito-DsRed). 48 h after transfection, images were acquired using a Zeiss LSM 700 confocal microscope equipped with a 63X/NA 1.15 water LD C-Apochromat objective lense and a temperature (37ºC) and CO2 (5%) controlled stage. Images were captured every 2 s for a period of 2 min using Zen 2009 software. The 561 nm laser intensity was set at 0.2 mW to minimize damage, and pinholes were opened maximally to allow the entire thickness of the axon to be imaged. Axon fragments of 50-100 µm in length located at least 50 µm away from the cell body were selected for analysis. A custom-made Image J plug-ins were used to generate kymographs and analyse mitochondria motility (31). Motility was assessed based on three parameters, Total Travel Distance, Travel Speed and Percent of Time in motion. Total Travel Distance was defined as the average of total distance travelled by each mitochondrion in 1 min; Travel speed was defined as the average of speed travelled in each direction; Percent of Time in motion was defined as the average of time spent mobile in each direction. Mitochondria that moved continuously in one direction were scored as 100% of Time in motion for that direction, while those that were entirely stationary or only moved in the opposite direction were scored as 0% Time in motion for that direction. Mitochondria length and density were measured by using the first frame of each time-lapse recording on selected axons, and analysed with Imaris software (Bitplane). Mitochondria density was calculated by dividing the total length of mitochondria with the length of axon in the same view field.
Immunohistochemistry
Primary motor neurons were fixed for 20 min in freshly made 4% PFA, followed by three washes with PBS and permeabilization in PBST buffer (PBS with 0.05% Tween-20). Samples were then blocked with 5% donkey serum and 5% goat serum in PBST before incubated with primary antibodies overnight at 4 °C, washed with PBST, incubated with secondary antibodies, washed with PBST and mounted in Aquamount (Fisher Scientific). All images were taken with a Zeiss LSM510 confocal microscope. Primary antibodies used in this study are as follows: HB9 (1:10000, Dr. Samuel Pfaff, rabbit polyclonal), Tuj1 (1:1000, Covance mouse monoclonal). Secondary antibodies are from Jackson ImmunoResearch and used at 1:500 dilution.
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
We thank Dr. Qinwen Mao for her help with analysing EM data, Drs. Benjamin W. Okaty and Jesse M. Gray for their help with RNA-seq and data analysis, Dr. Jyothisri Kondapalli for providing the AAV-mito-roGFP, Cathy Su for her help on analysing mitochondrial transport, Samuel Weinberg and Dr. Navdeep Chandel for their help with using the Seahorse analyzer.
Conflict of Interest statement. None declared.
Funding
This work was supported by National Institutes of Health [grant number R01NS094564] and grants from The Hartwell Foundation, Cure SMA and Whitehall Foundation to Y.C.M. Y.C.M. is Ann Marie and Francis Klocke M.D. Research Scholar supported by the Joseph and Bessie Feinberg Foundation.
References
- 1.Sugarman E.A., Nagan N., Zhu H., Akmaev V.R., Zhou Z., Rohlfs E.M., Flynn K., Hendrickson B.C., Scholl T., Sirko-Osadsa D.A., et al. (2012) Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: clinical laboratory analysis of > 72,400 specimens. Eur. J. Hum. Genet., 20, 27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prior T.W., Snyder P.J., Rink B.D., Pearl D.K., Pyatt R.E., Mihal D.C., Conlan T., Schmalz B., Montgomery L., Ziegler K., et al. (2010) Newborn and carrier screening for spinal muscular atrophy. Am. J. Med. Genet. A, 152A, 1608–1616. [DOI] [PubMed] [Google Scholar]
- 3.Pearn J. (1978) Incidence, prevalence, and gene frequency studies of chronic childhood spinal muscular atrophy. J. Med. Genet., 15, 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. (1995) Identification and characterization of a spinal muscular atrophy-determining gene. Cell, 80, 155–165. [DOI] [PubMed] [Google Scholar]
- 5.Faravelli I., Nizzardo M., Comi G.P., Corti S. (2015) Spinal muscular atrophy–recent therapeutic advances for an old challenge. Nat. Rev. Neurol., 11, 351–359. [DOI] [PubMed] [Google Scholar]
- 6.McWhorter M.L., Monani U.R., Burghes A.H., Beattie C.E. (2003) Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J. Cell Biol., 162, 919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossoll W., Jablonka S., Andreassi C., Kroning A.K., Karle K., Monani U.R., Sendtner M. (2003) Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J. Cell Biol., 163, 801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pellizzoni L., Kataoka N., Charroux B., Dreyfuss G. (1998) A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell, 95, 615–624. [DOI] [PubMed] [Google Scholar]
- 9.Akten B., Kye M.J., Hao le T., Wertz M.H., Singh S., Nie D., Huang J., Merianda T.T., Twiss J.L., Beattie C.E., et al. (2011) Interaction of survival of motor neuron (SMN) and HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits. Proc. Natl. Acad. Sci. U S A, 108, 10337–10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burghes A.H., Beattie C.E. (2009) Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat. Rev. Neurosci., 10, 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monani U.R. (2005) Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron, 48, 885–896. [DOI] [PubMed] [Google Scholar]
- 12.Murray L.M., Comley L.H., Thomson D., Parkinson N., Talbot K., Gillingwater T.H. (2008) Selective vulnerability of motor neurons and dissociation of pre- and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy. Hum. Mol. Genet., 17, 949–962. [DOI] [PubMed] [Google Scholar]
- 13.Hamilton G., Gillingwater T.H. (2013) Spinal muscular atrophy: going beyond the motor neuron. Trends Mol. Med, 19, 40–50. [DOI] [PubMed] [Google Scholar]
- 14.Rudnik-Schoneborn S., Heller R., Berg C., Betzler C., Grimm T., Eggermann T., Eggermann K., Wirth R., Wirth B., Zerres K. (2008) Congenital heart disease is a feature of severe infantile spinal muscular atrophy. J. Med. Genet., 45, 635–638. [DOI] [PubMed] [Google Scholar]
- 15.Shababi M., Habibi J., Yang H.T., Vale S.M., Sewell W.A., Lorson C.L. (2010) Cardiac defects contribute to the pathology of spinal muscular atrophy models. Hum. Mol. Genet., 19, 4059–4071. [DOI] [PubMed] [Google Scholar]
- 16.Lunn M.R., Wang C.H. (2008) Spinal muscular atrophy. Lancet, 371, 2120–2133. [DOI] [PubMed] [Google Scholar]
- 17.Mishra P., Chan D.C. (2014) Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol., 15, 634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schon E.A., Przedborski S. (2011) Mitochondria: the next (neurode)generation. Neuron, 70, 1033–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheng Z.H., Cai Q. (2012) Mitochondrial transport in neurons: impact on synaptic homeostasis and neurodegeneration. Nat. Rev. Neurosci., 13, 77–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DiMauro S., Schon E.A. (2003) Mitochondrial respiratory-chain diseases. N. Engl. J. Med., 348, 2656–2668. [DOI] [PubMed] [Google Scholar]
- 21.DiMauro S., Schon E.A., Carelli V., Hirano M. (2013) The clinical maze of mitochondrial neurology. Nat. Rev. Neurol., 9, 429–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tein I., Sloane A.E., Donner E.J., Lehotay D.C., Millington D.S., Kelley R.I. (1995) Fatty acid oxidation abnormalities in childhood-onset spinal muscular atrophy: primary or secondary defect(s)? Pediatr. Neurol., 12, 21–30. [DOI] [PubMed] [Google Scholar]
- 23.Crawford T.O., Sladky J.T., Hurko O., Besner-Johnston A., Kelley R.I. (1999) Abnormal fatty acid metabolism in childhood spinal muscular atrophy. Ann. Neurol., 45, 337–343. [DOI] [PubMed] [Google Scholar]
- 24.Harpey J.P., Charpentier C., Paturneau-Jouas M., Renault F., Romero N., Fardeau M. (1990) Secondary metabolic defects in spinal muscular atrophy type II. Lancet, 336, 629–630. [DOI] [PubMed] [Google Scholar]
- 25.Ripolone M., Ronchi D., Violano R., Vallejo D., Fagiolari G., Barca E., Lucchini V., Colombo I., Villa L., Berardinelli A., et al. (2015) Impaired Muscle Mitochondrial Biogenesis and Myogenesis in Spinal Muscular Atrophy. JAMA Neurol., 72, 666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wichterle H., Lieberam I., Porter J.A., Jessell T.M. (2002) Directed differentiation of embryonic stem cells into motor neurons. Cell, 110, 385–397. [DOI] [PubMed] [Google Scholar]
- 27.Nicholls D.G. (2008) Oxidative stress and energy crises in neuronal dysfunction. Ann. N Y Acad. Sci., 1147, 53–60. [DOI] [PubMed] [Google Scholar]
- 28.Dryanovski D.I., Guzman J.N., Xie Z., Galteri D.J., Volpicelli-Daley L.A., Lee V.M., Miller R.J., Schumacker P.T., Surmeier D.J. (2013) Calcium entry and alpha-synuclein inclusions elevate dendritic mitochondrial oxidant stress in dopaminergic neurons. J. Neurosci., 33, 10154–10164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Millecamps S., Julien J.P. (2013) Axonal transport deficits and neurodegenerative diseases. Nat. Rev. Neurosci., 14, 161–176. [DOI] [PubMed] [Google Scholar]
- 30.Miller N., Feng Z., Edens B.M., Yang B., Shi H., Sze C.C., Hong B.T., Su S.C., Cantu J.A., Topczewski J., et al. (2015) Non-aggregating tau phosphorylation by cyclin-dependent kinase 5 contributes to motor neuron degeneration in spinal muscular atrophy. J. Neurosci., 35, 6038–6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pekkurnaz G., Trinidad J.C., Wang X., Kong D., Schwarz T.L. (2014) Glucose regulates mitochondrial motility via Milton modification by O-GlcNAc transferase. Cell, 158, 54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le T.T., Pham L.T., Butchbach M.E., Zhang H.L., Monani U.R., Coovert D.D., Gavrilina T.O., Xing L., Bassell G.J., Burghes A.H. (2005) SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet., 14, 845–857. [DOI] [PubMed] [Google Scholar]
- 33.Hsieh-Li H.M., Chang J.G., Jong Y.J., Wu M.H., Wang N.M., Tsai C.H., Li H. (2000) A mouse model for spinal muscular atrophy. Nat. Genet., 24, 66–70. [DOI] [PubMed] [Google Scholar]
- 34.Strauss M., Hofhaus G., Schroder R.R., Kuhlbrandt W. (2008) Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. Embo J., 27, 1154–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomes L.C., Di Benedetto G., Scorrano L. (2011) During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol., 13, 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy M., Reddy P.H., Iijima M., Sesaki H. (2015) Mitochondrial division and fusion in metabolism. Curr. Opin. Cell. Biol., 33, 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pronicki M., Matyja E., Piekutowska-Abramczuk D., Szymanska-Debinska T., Karkucinska-Wieckowska A., Karczmarewicz E., Grajkowska W., Kmiec T., Popowska E., Sykut-Cegielska J. (2008) Light and electron microscopy characteristics of the muscle of patients with SURF1 gene mutations associated with Leigh disease. J. Clin. Pathol., 61, 460–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zick M., Rabl R., Reichert A.S. (2009) Cristae formation-linking ultrastructure and function of mitochondria. Biochim. Biophys. Acta, 1793, 5–19. [DOI] [PubMed] [Google Scholar]
- 39.Kelley R.I., Sladky J.T. (1986) Dicarboxylic aciduria in an infant with spinal muscular atrophy. Ann. Neurol., 20, 734–736. [DOI] [PubMed] [Google Scholar]
- 40.Bach J.R. (2007) Medical considerations of long-term survival of Werdnig-Hoffmann disease. Am. J. Phys. Med. Rehabil., 86, 349–355. [DOI] [PubMed] [Google Scholar]
- 41.Berger A., Mayr J.A., Meierhofer D., Fotschl U., Bittner R., Budka H., Grethen C., Huemer M., Kofler B., Sperl W. (2003) Severe depletion of mitochondrial DNA in spinal muscular atrophy. Acta Neuropathol., 105, 245–251. [DOI] [PubMed] [Google Scholar]
- 42.Voigt T., Meyer K., Baum O., Schumperli D. (2010) Ultrastructural changes in diaphragm neuromuscular junctions in a severe mouse model for Spinal Muscular Atrophy and their prevention by bifunctional U7 snRNA correcting SMN2 splicing. Neuromuscul. Disord., 20, 744–752. [DOI] [PubMed] [Google Scholar]
- 43.Kanning K.C., Kaplan A., Henderson C.E. (2010) Motor neuron diversity in development and disease. Annu. Rev. Neurosci., 33, 409–440. [DOI] [PubMed] [Google Scholar]
- 44.Cullheim S., Fleshman J.W., Glenn L.L., Burke R.E. (1987) Three-dimensional architecture of dendritic trees in type-identified alpha-motoneurons. J. Comp. Neurol., 255, 82–96. [DOI] [PubMed] [Google Scholar]
- 45.Kosik K.S., Finch E.A. (1987) MAP2 and tau segregate into dendritic and axonal domains after the elaboration of morphologically distinct neurites: an immunocytochemical study of cultured rat cerebrum. J. Neurosci., 7, 3142–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Itoh K., Nakamura K., Iijima M., Sesaki H. (2013) Mitochondrial dynamics in neurodegeneration. Trends Cell Biol., 23, 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pernas L., Scorrano L. (2016) Mito-morphosis: mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu Rev Physiol., 78:505–531. [DOI] [PubMed] [Google Scholar]
- 48.Chen H., McCaffery J.M., Chan D.C. (2007) Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell, 130, 548–562. [DOI] [PubMed] [Google Scholar]
- 49.Chen H., Chomyn A., Chan D.C. (2005) Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem., 280, 26185–26192. [DOI] [PubMed] [Google Scholar]
- 50.Xu C.C., Denton K.R., Wang Z.B., Zhang X., Li X.J. (2016) Abnormal mitochondrial transport and morphology as early pathological changes in human models of spinal muscular atrophy. Dis. Model Mech., 9, 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Acsadi G., Lee I., Li X., Khaidakov M., Pecinova A., Parker G.C., Huttemann M. (2009) Mitochondrial dysfunction in a neural cell model of spinal muscular atrophy. J. Neurosci. Res., 87, 2748–2756. [DOI] [PubMed] [Google Scholar]
- 52.Wang Z.B., Zhang X., Li X.J. (2013) Recapitulation of spinal motor neuron-specific disease phenotypes in a human cell model of spinal muscular atrophy. Cell Res., 23, 378–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song W., Song Y., Kincaid B., Bossy B., Bossy-Wetzel E. (2013) Mutant SOD1G93A triggers mitochondrial fragmentation in spinal cord motor neurons: neuroprotection by SIRT3 and PGC. Neurobiol. Dis., 51, 72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin M.T., Beal M.F. (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature, 443, 787–795. [DOI] [PubMed] [Google Scholar]
- 55.Patitucci T.N., Ebert A.D. (2015) SMN deficiency does not induce oxidative stress in SMA iPSC-derived astrocytes or motor neurons. Hum. Mol. Genet., 25:514–523. [DOI] [PubMed] [Google Scholar]
- 56.Edens B.M., Ajroud-Driss S., Ma L., Ma Y.C. (2015) Molecular mechanisms and animal models of spinal muscular atrophy. Biochim. Biophys. Acta, 1852, 685–692. [DOI] [PubMed] [Google Scholar]
- 57.Cogliati S., Frezza C., Soriano M.E., Varanita T., Quintana-Cabrera R., Corrado M., Cipolat S., Costa V., Casarin A., Gomes L.C., et al. (2013) Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell, 155, 160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rak M., Tetaud E., Godard F., Sagot I., Salin B., Duvezin-Caubet S., Slonimski P.P., Rytka J., di Rago J.P. (2007) Yeast cells lacking the mitochondrial gene encoding the ATP synthase subunit 6 exhibit a selective loss of complex IV and unusual mitochondrial morphology. J. Biol. Chem., 282, 10853–10864. [DOI] [PubMed] [Google Scholar]
- 59.Magrane J., Sahawneh M.A., Przedborski S., Estevez A.G., Manfredi G. (2012) Mitochondrial dynamics and bioenergetic dysfunction is associated with synaptic alterations in mutant SOD1 motor neurons. J. Neurosci., 32, 229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang W., Li L., Lin W.L., Dickson D.W., Petrucelli L., Zhang T., Wang X. (2013) The ALS disease-associated mutant TDP-43 impairs mitochondrial dynamics and function in motor neurons. Hum. Mol. Genet., 22, 4706–4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Magrane J., Cortez C., Gan W.B., Manfredi G. (2014) Abnormal mitochondrial transport and morphology are common pathological denominators in SOD1 and TDP43 ALS mouse models. Hum. Mol. Genet., 23, 1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fornai F., Longone P., Cafaro L., Kastsiuchenka O., Ferrucci M., Manca M.L., Lazzeri G., Spalloni A., Bellio N., Lenzi P., et al. (2008) Lithium delays progression of amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U S A, 105, 2052–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parone P.A., Da Cruz S., Han J.S., McAlonis-Downes M., Vetto A.P., Lee S.K., Tseng E., Cleveland D.W. (2013) Enhancing mitochondrial calcium buffering capacity reduces aggregation of misfolded SOD1 and motor neuron cell death without extending survival in mouse models of inherited amyotrophic lateral sclerosis. J. Neurosci., 33, 4657–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parkhomchuk D., Borodina T., Amstislavskiy V., Banaru M., Hallen L., Krobitsch S., Lehrach H., Soldatov A. (2009) Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res., 37, e123.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.