Biobanks1 are well-organized repositories of biological material. They have become the fundamental resource for advancing medical research and constitute a major component of more generally understood bioresources. Yet they face a number of challenges to become more utilized on the national and global scale. These challenges range from fragmentation of data structure and sometimes even lack of availability of data,2–4 lack of consistent quality management and traceability5–8 to fragmentation of privacy protection regulations9–13 and technical, organizational, and legal aspects of scalable secure storage and processing of privacy-sensitive big data.14–16 To address the fragmentation and findability aspects, BBMRI-ERIC has released its Directory as a first IT service, providing aggregate information about the biobanks and bioresources. The Directory features a novel scalable distributed architecture, which enables updating data about changing resources in a long-term sustainable manner.
Inventory data about the bioresources, describing availability of various resource types such as biological material, data, expertise, and offered services, are the basis for any further interaction between the biobanks as resource/service providers and their users or collaborators. There have been various terms used for these types of services, including “catalogs” and “registries.” Inventory data cover various types of information that is not considered privacy sensitive and thus shareable in an open-access mode. The business model of a bioresource may impose access restrictions, however. From the users' perspective, it is important to achieve consistent or at least algorithmically harmonizable semantics of the information, so that it is possible to implement efficient search or filtering services.
There have been a number of attempts to improve the situation with availability and consistency of the inventory data in the past decade both internationally and nationally. Prominent international examples include P3G Observatory,17 BBMRI Preparatory Phase Catalogue,3 ISBER International Resource Locator,18 Maelstrom Repository,19 BBMRI-LPC catalogs,20,21 or RD-CONNECT Catalogue22,23 and the NIH/NCATS GRDR®24 on rare diseases. Although being very valuable for helping to organize biobanking and bioresources in projects with limited life spans, these tools also demonstrate the key deficiency of such centrally built and managed systems: because of the lack of automated data updates, the information becomes sooner or later obsolete and thus of limited use for the users.
In contrast, distributed information systems are well known in computer infrastructures, such as cloud and grid computing systems,25 where various architectures have been explored, ranging from client-server communication schemes26,27 to peer-to-peer systems.28–30 The biobanking community needs to learn from these endeavors and take a similar approach with (a) distributed architecture that allows for information flow from the original sources to the inventory services, (b) well-defined stable application programming interfaces (APIs) that allow for their implementation in the biobank information management systems, (c) clear component-based architecture that allows for simple implementation of relevant data extraction and harmonization components as close to the original information sources as possible to include in-depth knowledge of the data.
BBMRI-ERIC Directory
BBMRI-ERIC, the Biobanking and BioMolecular Resources Research Infrastructure-European Research Infrastructure Consortium, is a new form of legal organization for biobanking in Europe. BBMRI-ERIC has started to develop the BBMRI-ERIC Directory as its first information technology tool. Directory 1.0 was released in July 2015 with basic support for biobanks, and Directory 2.0 was released in December 2015, supporting biobanks, collections, and biobank networks. The BBMRI-ERIC Directory has been designed with the following two primary use cases in mind: biomedical and bioinformatics researchers seeking retrieval of samples/data, or for collecting/hosting services for their samples/data; biobank operators needing to identify similar biobanks (experience sharing, collaboration, etc.) and to promote their visibility. One can also imagine other use cases such as research participants (donors/patients) and their organizations interested in determining where their samples might be located, the purposes they are being used, and for funding and governance bodies looking into the extent and use statistics of funded infrastructures.
The first development step was designing an extensible data model, which covers all three key components of biobanks: (a) biological material and associated physical storage facilities, (b) data and associated data storage facilities, and (c) expertise of the biobankers. The core of the data model for Directory 2.0 relies on MIABIS 2.0,31 which is the evolution of the previously published MIABIS model.32 The Directory's data model includes biobanks as institutional units hosting collections of samples and data, as well as biobank networks used to further aggregate biobanks or their collections. Our current data model is highly aggregated and serves to primarily identify candidate biobanks that might have samples for the given purpose or biobanks that provide relevant services.
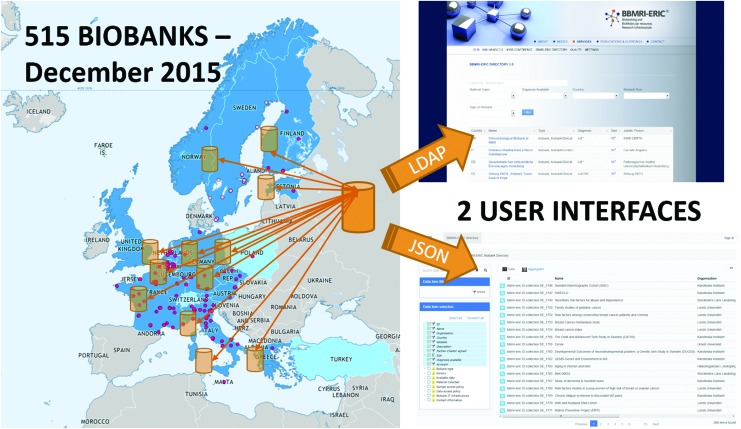
From the architectural perspective, the Directory is a distributed system using multilayer architecture as shown in Figure 1. Each layer uses clearly defined machine-readable APIs (LDAP, REST/JSON) and data formats, which enable automated propagation of updates, allows for building purpose-focused user interfaces, as well as integrates into larger automated workflows (e.g., currently developed BBMRI-ERIC Negotiator to facilitate access negotiation). As for Directory 2.0, there are two web-based user interfaces implemented: the main BBMRI-ERIC Directory interface33 and the BBMRI.nl interface,34 integrating BBMRI-ERIC data using the Molgenis platform.35
FIG. 1.
Distributed and modular BBMRI-ERIC Directory architecture. It typically comprises online or offline data flow from biobanks and other bioresources (not shown for sake of simplicity) → aggregating nodes (e.g., BBMRI-ERIC national nodes) → central BBMRI-ERIC Directory → user interfaces. Color images available online at www.liebertpub.com/bio
The current Directory 2.0 includes 515 biobanks and standalone collections, with an estimated number of samples exceeding 60,000,000. This covers 136 clinical or disease-specific biobanks and 189 population biobanks, based on the classification proposed in an article3 from the BBMRI Preparatory Phase. This is a conservative estimate based on the 10n order of magnitude attribute, which is mandatory for each collection in Directory 2.0, compared with optional exact size. For the largest biobanks, the estimate has been adjusted based on direct communication to avoid substantial bias. We consider these estimates sufficient, as exact counting would require consensus on sample and aliquot definition (expected to be clarified by ISO TC 276 between 2017 and 2018).*
It should also be noted that the access to the samples/data is controlled by the biobanks, which means biobanks may or may not allow access depending on the types of requests received by them. Based on Directory 2.0 data, ∼23% of biobanks provide access to samples/data based on a fee structure and ∼28% based on joint projects. Approximately 60% do not publish this information and need to be contacted directly to receive information on access conditions.
Future Work
There are two basic directions to improve the inventory services, and particularly the BBMRI-ERIC Directory. The first direction is to improve specificity in the responses. Although the biobanks are already urged to publish data that are as accurate as possible, many issues will only be resolved once we help biobanks fully implement online interfaces to their primary information systems. A particular example is the list of available diagnoses, which is among the most searched for parameters.36 Some biobanks do not have this information themselves and must either retrieve it using targeted questions to hospital information systems or must resort to consulting external registries.
The second direction for the future extensions is improving coverage of various aspects of the biobanks, such as availability of quality management systems, advertising additional services such as sample/data hosting, or providing semantic translation support for data that comes in different coding or semantics from different sources. The Directory service can also be used to map various types of identifiers and to publish persistent identifiers for sample sets and datasets once they are used for publishing, hence further supporting efforts toward reproducible biomedical research. These extensions are expected in Directory 3.0 (to be released in 2016) and onward. BBMRI-ERIC also works on extending geographical coverage of the Directory by merging with the validated RD-CONNECT Catalogue data during 2016 (e.g., data from the United States and Australia).
Last but not least, the communication of many users with many biobanks at the same time is not efficient, and tools for simplifying such communication are needed. BBMRI-ERIC will address these issues using the Negotiator tool integrated with the Directory, intended for cumulative communication between a user and multiple biobanks at the same time.
Because of its potential global impact, the Directory has been proposed as a tool for organizing bioresources' inventory information as a part of the BBMRI-ERIC application for the G7 Group of Senior Officials on Global Research Infrastructures.
Acknowledgments
This work is part of the ADOPT BBMRI-ERIC project, funded by the European Commission, topic H2020-INFRADEV-3-2015, Grant Agreement Number 676550.
The authors would like to thank the directors of BBMRI-ERIC National Nodes and their IT representatives for their involvement in designing and deploying the BBMRI-ERIC Directory. Particular thanks goes to Michael Hummel of BBMRI.de for facilitating data model discussions and Araceli Diez-Fraile of BBMRI.be for valuable contributions to the data structures and extensive beta testing of Directory 1.0 and Directory 2.0. We would also like to thank the developers at BBMRI.nl for their valuable contributions to implementing the Molgenis-based user interface for Directory 1.0. The authors would also like to thank the members of the MIABIS Working Group: Roxana Merino-Martinez, Loreana Norlin, Gabriele Anton, Simone Schuffenhauer, Kaisa Silander, Linda Mook, Raffael Bild, Martin Fransson, Roman Siddiqui, Klaus Kuhn, Linda Zaharenko, Helmut Spengler, Araceli Diez-Fraile, Joakim Geeraert, Ondřej Vojtíšek, Anita Nieminen, Kristjan Metsalu, Murat Sariyar, Michael Hummel, and Cathleen Ploetzand.
Footnotes
We would advise against “abandoning samples” and using only number of research participants (donors), as has already happened in some Nordic population biobanks, because such an approach does not allow differentiation between a biobank that collects one sample per participant and a time-consistent series of samples per participant.
Authors' Contributions
P.H. designed the distributed system, implemented the server infrastructure and connectors to national nodes, and coordinated writing the article; J.-E.L. contributed to the overall design of the system; M.S. and D.v.E. contributed Molgenis-based user interface integrated with BBMRI.nl National Node; and R.R. and H.M. contributed the user interface integrated on the BBMRI-ERIC Web pages. All the authors contributed to writing this article.
Author Disclosure Statement
No conflicting financial interests exist.
References
- 1. Harris JR, Burton P, Knoppers BM, et al. Toward a roadmap in global biobanking for health. Eur J Hum Genet 2012;20:1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Litton JE. Biobank informatics: Connecting genotypes and phenotypes. In: Dillner J, ed. Methods in Biobanking. Totowa, NJ: Humana Press/Springer; 2011:343–361 [DOI] [PubMed] [Google Scholar]
- 3. Wichmann HE, Kuhn KA, Waldenberger M, et al. Comprehensive catalog of European biobanks. Nat Biotechnol 2011;29:795–797 [DOI] [PubMed] [Google Scholar]
- 4. Yuille M, Ommen GJ, van Bréchot C, et al. Biobanking for Europe. Brief Bioinform 2008;9:14–24 [DOI] [PubMed] [Google Scholar]
- 5. Ioannidis JP, Allison DB, Ball CA, et al. Repeatability of published microarray gene expression analyses. Nat Genet 2009;41:149–155 [DOI] [PubMed] [Google Scholar]
- 6. Prinz F, Schlange T, Asadullah K. Believe it or not: How much can we rely on published data on potential drug targets? Nat Rev Drug Discov 2011;10:712. [DOI] [PubMed] [Google Scholar]
- 7. Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature 2012;483:531–533 [DOI] [PubMed] [Google Scholar]
- 8. Bissell M. Reproducibility: The risks of the replication drive. Nature 2013;503:333–334 [DOI] [PubMed] [Google Scholar]
- 9. Hansson MG, Dillner J, Bartram CR, et al. Should donors be allowed to give broad consent to future biobank research? Lancet Oncol 2006;7:266–269 [DOI] [PubMed] [Google Scholar]
- 10. Bäumen TSID, Paci D, Ibarreta D. Data protection and sample management in biobanking—A legal dichotomy. Genomics Soc Policy 2010;6:33–46 [Google Scholar]
- 11. Williams H, Spencer K, Sanders C, et al. Dynamic consent: A possible solution to improve patient confidence and trust in how electronic patient records are used in medical research. JMIR Med Inform 2015;3:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chadwick R, Strange H. Harmonisation and standardisation in ethics and governance: Conceptual and practical challenges. In: Widdows H, Mullen C, ed. The Governance of Genetic Information: Who Decides, vol. 9 Cambridge University Press; 2009: 201–213 [Google Scholar]
- 13. Abbott A. European medical research escapes stifling privacy laws. Nature News, December 16, 2015. DOI: 10.1038/nature.2015.19054 [DOI] [Google Scholar]
- 14. Verissimo PE, Bessani A. E-biobanking: What have you done to my cell samples? IEEE Security&Privacy 2013;11:62–65 [Google Scholar]
- 15. Gholami A, Lind AS, Reichel J, et al. Privacy threat modeling for emerging BiobankClouds. Procedia Comput Sci 2014;37:489–496 [Google Scholar]
- 16. Gholami A, Dowling J, Laure E. A security framework for population-scale genomics analysis. In: 2015 International Conference on High Performance Computing & Simulation (HPCS) IEEE; 2015: 106–114 [Google Scholar]
- 17. Knoppers B, Fortier I, Legault D, et al. The Public Population Project in Genomics (P3G): A proof of concept. Eur J Hum Genet 2008;16:664–665 [DOI] [PubMed] [Google Scholar]
- 18. ISBER Resource Locator, International Repository Locator Working Group. www.isber.org/?IRL (accessed November22, 2016)
- 19. Maelstrom Repository. www.maelstrom-research.org/repository (accessed November22, 2016)
- 20. Catalogue of BBMRI-LPC biobanks. www.bbmri-lpc-biobanks.eu/cataloque.html (accessed November22, 2016)
- 21. Catalog of variables in BBMRI-LPC biobanks. http://bbmri-lpc.iarc.fr/mica/?q=variable-search (accessed November22, 2016)
- 22. Thompson R, Johnston L, Taruscio D, et al. RD-Connect: An integrated platform connecting databases, registries, biobanks and clinical bioinformatics for rare disease research. J Gen Intern Med 2014;29:780–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. RD-CONNECT Catalogue. http://catalogue.rd-connect.eu (accessed November22, 2016)
- 24. NIH/NCATS Global Rare Diseases Patient Registry Data Repository (GRDR). https://ncats.nih.gov/grdr (accessed November22, 2016)
- 25. Foster I, Kesselman C, Tuecke S. The anatomy of the grid: Enabling scalable virtual organizations. Int J High Perform Comput Appl 2001;15:200–222 [Google Scholar]
- 26. Czajkowski K, Fitzgerald S, Foster I, et al. Grid information services for distributed resource sharing. In: 10th IEEE International Symposium on High Performance Distributed Computing, 2001 IEEE; 2001: 181–194 [Google Scholar]
- 27. Plale B, Dinda P, von Laszewski G. Key concepts and services of a grid information service. In: Proceedings of the 15th International Conference on Parallel and Distributed Computing Systems (PDCS 2002) CiteSeerX, 2002: 437–442 [Google Scholar]
- 28. Andrzejak A, Xu Z. Scalable, efficient range queries for grid information services. In: Second International Conference on Peer-to-Peer Computing (P2P 2002), 2002 IEEE; 2002: 33–40 [Google Scholar]
- 29. Cai M, Frank M, Chen J, et al. Maan: A multi-attribute addressable network for grid information services. J Grid Comput 2004;2:3–14 [Google Scholar]
- 30. Puppin D, Moncelli S, Baraglia R, et al. A grid information service based on peer-topeer. In: Euro-Par 2005 Parallel Processing Springer; 2005: 454–464 [Google Scholar]
- 31. Merino-Martinez R, Loreana Norlin DvE, Anton G, et al. Towards global biobank integration by implementation of the Minimum Information About Biobank data Sharing (MIABIS 2.0 Core). Biopreserv Biobank 2016;14:298–306 [DOI] [PubMed] [Google Scholar]
- 32. Norlin L, Fransson MN, Eriksson M, et al. A minimum data set for sharing biobank samples, information, and data: MIABIS. Biopreserv Biobank 2012;10:343–348 [DOI] [PubMed] [Google Scholar]
- 33. BBMRI-ERIC Directory. http://bbmri-eric.eu/bbmri-eric-directory (accessed November22, 2016)
- 34. MOLGENIS-based BBMRI-ERIC Directory user interface. http://directory-molgenis.bbmri-eric.eu/ (accessed November22, 2016)
- 35. Swertz MA, Dijkstra M, Adamusiak T, et al. The MOLGENIS toolkit: Rapid prototyping of biosoftware at the push of a button. BMC Bioinformatics 2010;11:S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Puchois P. General overview of industry needs and requirements. In: HandsOn: Biobanks 2015. Conference presentation Milan, Italy, July2015 [Google Scholar]