Abstract
The Archaeorhizomycetes are recently discovered fungi with poorly resolved ecology. Even their abundance in soil fungal communities is currently disputed. Here we applied a PCR-independent, RNA-based metatranscriptomic approach to determine their abundance among fungi in eleven different soils across Europe. Using small subunit (SSU) ribosomal RNA transcripts as marker, we detected Archaeorhizomycetes in 17 out of 28 soil metatranscriptomes. They had average relative SSU rRNA abundance of 2.0% with a maximum of 9.4% among fungal SSU rRNAs. Network analysis revealed that they co-occur with arbuscular mycorrhizal Glomerales, which is in line with their previously suggested association with plant roots. Moreover, Archaeorhizomycetes ranked among the potential keystone taxa. This metatranscriptomic survey exemplifies the usage of non-targeted molecular approaches for the study of soil fungi. It provides PCR- and DNA-independent evidence for the low abundance of Archaeorhizomycetes in soil fungal communities, although they might be non-negligible players despite their low abundance.
The Archaeorhizomycetes are an enigmatic fungal group that has been until recently without cultured representatives. They represent a deeply-branching class in the Ascomycota and have been detected in many soil ecosystems worldwide, indicating their ubiquity in soils1. As their name suggests, they show a strong association with the root environment. However, very limited information is available about their life style and ecology, with direct associations with roots through symbiotic or parasitic interactions and even secondary associations via interactions with other root-associated fungi being discussed2.
Several DNA-based PCR studies targeting the fungal internally transcribed spacer (ITS) of ribosomal RNA genes had suggested this group to be ubiquitous and numerically dominant in some soil fungal communities3. However, a recent global study of soil fungi could not confirm their high abundance4. In response, there arose a discussion about possible PCR primer biases and the true abundance of Archaeorhizomycetes in the latter study and generally in soils5,6.
The bias in PCR studies brought by primer mismatches and differing amplicon length can be avoided by targeting small subunit ribosomal RNA (SSU rRNA) transcripts instead of genes. This is facilitated by random hexamer-primed reverse transcription in metatranscriptomics approaches7. Total RNA extracted from soils is dominated by rRNA fragments (usually more than 90% is rRNA) and thus naturally enriched in target molecules, which enables PCR independent SSU rRNA profiling via direct sequencing of reversely transcribed cDNA. Furthermore, the obtained random-hexamer primed sequence fragments originate from different regions of the SSU rRNA molecule and are therefore insensitive to the presence of introns or PCR primer mismatches. Since ribosomal RNA is quickly degraded, it is a better marker for living cells than DNA used in metagenomics or targeted amplicon analysis8. The fact that metatranscriptomics does not suffer from PCR-introduced biases and thus does not discriminate any taxonomic group and on top of that gives information about the composition of the transcriptionally active part of the fungal community distinguishes such approach from standard amplicon sequencing. On the other hand, SSU rRNA profiling of fungi provides not the same high taxonomic resolution as the fungal ITS region used for amplicon sequencing.
This study aimed to assess the abundance of Archaeorhizomycetes in 28 metatranscriptomes7,9,10,11,12 from 11 soils belonging to different terrestrial biomes across Europe (for details see Table 1). Furthermore, network analysis identified possible fungal interactions partners of Archaeorhizomycetes.
Table 1. General site and sampling description.
| Site | Peatland soil “Knudsenheia” | Peatland soil “Solvatn” | Mofette | Mofette reference | Rothamsted grassland | Rotböhl | Forest Litter | Forest Soil | Mine L | Mine M | Mine H |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Abbreviation | PsK | PsS | MO | MR | RS | RB | FL | FS | MiL | MiM | MiH |
| Location | Ny-Ålesund, | Ny-Ålesund, | Hartoušov, | Hartoušov, | Rothamsted, | Darmstadt, | Vienna woods, | Vienna woods, | Coto Txomin, | Coto Txomin, | Coto Txomin, |
| Norway | Norway | Czech Republic | Czech Republic | United Kingdom | Germany | Austria | Austria | Spain | Spain | Spain | |
| (Svalbard) | (Svalbard) | ||||||||||
| Climatic zone | Arctic | Arctic | Temperate | Temperate | Temperate | Temperate | Temperate | Temperate | Temperate | Temperate | Temperate |
| Biome | Fen wet land | Fen wet land | Floodplain | Floodplain | Grassland | Grassland | Temperate deciduous forest | Temperate deciduous forest | Shrubland | Shrubland | Shrubland |
| Dominant vegetation | Mosses | Mosses | Filipendula ulmaria | Deschampsia cespitosa, Eriophorum vaginatum | N.A. | N.A. | Fagus sylvatica | Fagus sylvatica | Ulex europaeus | Festuca rubra | Festuca rubra |
| Substrate type / Horizon | Organic peat (Top layer) | Organic peat (Top layer) | Organic soil | Gleic fluvisol | Mineral soil | Mineral soil | Litter horizon | Mineral soil (A horizon) | Mineral soil | Mineral soil | Mineral soil |
| pH | 7.3 | 7.6 | 4.7 | 5.3 | 4.9 | 7.1 | N.A. | 4.5-5.1 | 3.9 | 5.6 | 5.9 |
| Moisture (% soil dry weight) | 1010 | 900 | N.A | N.A. | 33 | 32 | 18 | 43-64 | 52 | 49 | 30 |
| # of replicates | 2 | 2 | 3 | 3 | 2 | 1 | 2 | 4 | 3 | 3 | 3 |
| Sampling time | August 2009 | August 2009 | July 2013 | July 2013 | July 2009 | January 2006 | May 2008 | May 2008 | March 2011 | March 2011 | March 2011 |
| Sequencing method | 454 GS FLX Titanium | 454 GS FLX Titanium | Illumina HiSeq 2500 | Illumina HiSeq 2500 | 454 GS FLX Titanium | 454 GS 20 | 454 GS FLX | 454 GS FLX | Illumina HiSeq 2000 | Illumina HiSeq 2000 | Illumina HiSeq 2000 |
| Average eukaryotic SSU rRNA transcripts length | 369 | 372 | 166 | 167 | 314 | 105 | 218 | 271 | 171 | 171 | 178 |
| Average fungal SSU rRNA transcripts analysed | 514 | 1632 | 1102 | 5086 | 2164 | 3287 | 28640 | 2187 | 4445 | 3014 | 2125 |
| Average proportion of Archaeorhizomycetes SSU rRNA transcripts (%) | 0.00 | 0.00 | 0.00 | 0.95 | 1.05 | 0.14 | <0.01 | <0.01 | 4.03 | 7.28 | 5.74 |
| Reference | [9] | [9] | [12] | [12] | [10] | [7] | [10] | [10] | [11] | [11] | [11] |
Results and Discussion
Between 514 and 28640 fungal SSU rRNAs were on average analysed per soil, stemming from metatranscriptomes generated by 454 pyrosequencing and Illumina Hiseq sequencing (see Table 1). Generally, the fungal communities were dominated by Ascomycota (57.6% of all fungal rRNA transcripts), followed by Basidiomycota (23.2%), Glomeromycota (6.4%), Chytridiomycota (3.9%), Mortierellomycotina (2.5%) and Mucoromycotina (0.6%).
Archaeorhizomycetes were not among the abundant fungal classes (Fig. 1). The highest abundant were the Agaricomycetes (18.4%), which represented the vast majority of Basidiomycota. The other highly abundant classes were all ascomycetous: Leotiomycetes (10.5%), Eurotiomycetes (8.4%), Dothideomycetes (8.2%) and Sordariomycetes (7.3%).
Figure 1. Relative abundance of the 16 most abundant fungal classes (mean + SE) as average of all datasets (black columns) and in respective soils (open columns).
Archaeorhizomycetes SSU rRNAs were detected in 17 out of 28 metatranscriptomes and in 8 out of 11 soils (Fig. 1 and Table 1). The three sites where no Archaeorhizomycetes were detected were predominantly hypoxic, i.e. peatland and mofette soils. These metatranscriptomes had on average the lowest number of fungal reads, thus the detection limit could have been too high for Archaeorhizomycetes.
The overall relative abundance of Archaeorhizomycetes transcripts across all samples was 2.0% (Fig. 1). Exploring only metatranscriptomes containing Archaeorhizomycetes, this class represented on average 3.3% of SSU rRNAs. The maximum proportion recorded was 9.4%. Thus, these PCR-independent metatranscriptome results support the view that Archaeorhizomycetes are rather a low abundant fungal class in soils and that reports of their high abundance might be derived from a preferential amplification of their ITS or rRNA genes in PCR assays13. However, a particularly high abundance of Archaeorhizomycetes in PCR based studies was reported in alpine tundra14 and coniferous forest soils3, which were not included in our study. Furthermore, metatranscriptomics is not unbiased and shares some biases with DNA methods, such as protocol-dependent preferential nucleic acid extraction for certain taxonomic groups.
The relative abundance of Archaeorhizomycetes varied strongly between sites, with no SSU rRNA transcripts being detected in the predominantly hypoxic arctic peatland (PsS, PsK) and mofette (MO) soils (Fig. 1). Common to these soils was the low abundance of vascular plant roots, supporting once more the association of Archaeorhizomycetes with plant roots, although an obligate aerobic lifestyle could also result in such a distribution pattern. Remarkably, the highest relative abundance of Archaeorhizomycetes (average 5.6%) was found in soils of a former lead and zinc mining site (Fig. 1); with increasing heavy metal concentrations (MiL < MiM < MiH) having no effect on their relative abundance. These data indicate that Archaeorhizomycetes are tolerant to high concentrations of these metals, adding one additional piece of knowledge to their autecology. In the other surveyed sites, Archaeorhizomycetes abundance was minimal – up to ~1% for grassland (MR, RS, RB) and negligible <0.01% for beech forest (FL, FS) soils (Fig. 1).
Exploring seasonal patterns in the abundance of Archaeorhizomycetes SSU rRNAs, we observed a high relative abundance in samples collected in spring, which is in agreement with a study conducted in a Colorado alpine tundra that found a peak in abundance of Archaeorhizomycetes during spring14. Nevertheless, this conclusion is rather speculative as there was no seasonal sampling done for any of the soils surveyed in our study. To provide a well-founded description of the Archaeorhizomycetes seasonal variability more research is needed.
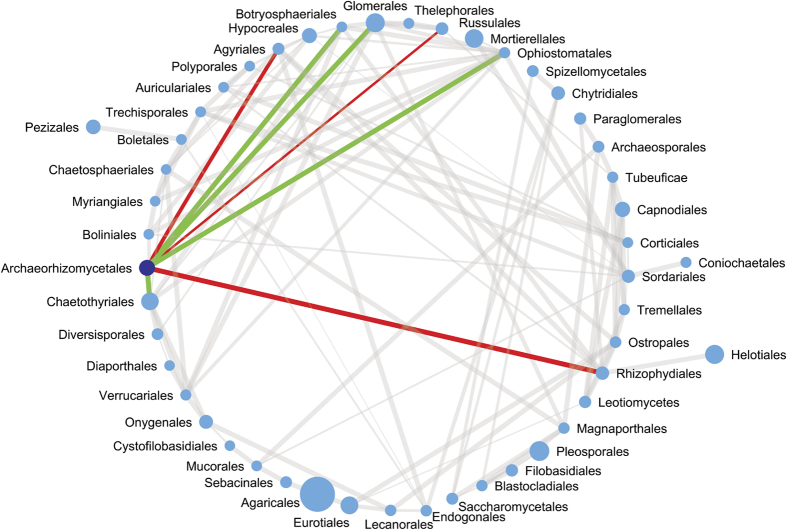
To identify possible interactions of Archaeorhizomycetes with other fungi we conducted network analysis (Fig. 2). We analysed the co-occurrence patterns of the 52 most abundant fungal orders in the metatranscriptomes using the CoNET algorithm15 in Cytoscape 3.0.216 (for details see Methods section). Archaeorhizomycetes had a strong co-presence relationship with the arbuscular mycorrhizal Glomerales (Fig. 2; Table 2). However, we are not able to resolve whether both fungal groups interact directly (share of mycorrhizal niche or parasitism) or only share the affinity to root environment without explicit relationship. Also other positively interacting fungal orders have representatives with strong association to plants as either pathogens or endophytes (Table 2). The results of network analysis therefore further supported the hypothesis that Archaeorhizomycetes prefer to live in close vicinity of plant roots.
Figure 2. Co-occurrence network of significantly interacting fungal orders.
Interactions with Archaeorhizomycetales are highlighted: positively (co-occurrence) interacting fungal orders are connected with green lines, negatively (mutual exclusion) interacting with red lines. The thickness of lines is proportional to significance of the interaction (q-value). The size of circle is proportional to the average relative abundance of fungal order in all datasets.
Table 2. Fungal orders significantly interacting with Archaeorhizomycetales, their assignment to phylum, prevailing lifestyle acc. to Tedersoo et al. 4, interaction q-value and mean relative abundance in all datasets.
| Interacting order | Phylum | Prevailing lifestyle | q-value | Mean proportion (%) |
|---|---|---|---|---|
| Co-presence | ||||
| Botryosphaeriales | Ascomycota | Plant pathogens | 1.2E-06 | 0.2 |
| Chaetothyriales | Ascomycota | Saprotrophs, plant endophytes | 6.4E-09 | 2.6 |
| Glomerales | Glomeromycota | Arbuscular mycorrhiza | 4.1E-07 | 3.1 |
| Ophiostomatales | Ascomycota | Plant pathogens | 7.4E-06 | 0.4 |
| Mutual exclusion | ||||
| Agyriales | Ascomycota | Lichenicolous | 1. E-02 | 0.6 |
| Russulales | Basidiomycota | Ectomycorrhiza and saprotrophs | 2.5E-02 | 0.9 |
| Rhizophydiales | Chytridiomycota | Pathogens, saprotrophs | 1.3E-06 | 1.2 |
Co-occurrence analysis placed Archaeorhizomycetes among the top 10 potential keystone taxa (Table 3), thus Archaeorhizomycetes seem to play a non-negligible role in soil fungal communities despite their low relative abundance.
Table 3. Potential keystone orders, their assignment to phyla, degree, betweenness centrality and closeness centrality.
| Potential keystone order | Phylum | Degree | Betweenness centrality | Closeness centrality |
|---|---|---|---|---|
| Ophiostomatales | Ascomycota | 12 | 0.13 | 0.45 |
| Glomerales | Glomeromycota | 9 | 0.12 | 0.42 |
| Chaetothyriales | Ascomycota | 9 | 0.05 | 0.41 |
| Sordariales | Ascomycota | 9 | 0.15 | 0.42 |
| Trechisporales | Basidiomycota | 8 | 0.11 | 0.42 |
| Ostropales | Ascomycota | 8 | 0.07 | 0.39 |
| Rhizophydiales | Chytridiomycota | 8 | 0.11 | 0.40 |
| Botryosphaeriales | Ascomycota | 7 | 0.03 | 0.41 |
| Hypocreales | Ascomycota | 7 | 0.03 | 0.41 |
| Archaeorhizomycetales | Ascomycota | 7 | 0.04 | 0.42 |
Orders are sorted according their degree (i.e. number of direct interactions with other fungal orders).
This study concludes Archaeorhizomycetes as fungal class with minor representation in transcriptionally active part of soil fungal communities in different terrestrial biomes. Yet, it highlights the need for more studies to elucidate more aspects of their ecology, as Archaeorhizomycetes were identified as putative key players in soil fungal communities.
Methods
Metatranscriptome sequences were filtered using PRINSEQ17 discarding short and low quality sequences (<150 bp, min average quality <0.25; exception dataset RB: <80 bp, min average quality <0.25). Metatranscriptomes generated with Illumina HiSeq were subsampled to 1 million reads and screened for eukaryotic SSU rRNA transcripts using SortMeRNA18 with default settings and the Silva SSUref 111 database19. Metatranscriptomes generated with 454 pyrosequencing were not subsampled and not subjected to SortMeRNA. For fungal community profiling, whole metatranscriptome datasets (454 pyrosequencing) or eukaryotic SSU rRNA transcripts (Illumina) were subsequently compared with BLASTN against the Silvamod SSU rRNA reference database20 and taxonomically classified with CREST20 and MEGAN521 using the lowest common ancestor algorithm (LCA parameters: minimum bit score 100, top percent 2). In addition, correctness of reads identified as belonging to the class Archaeorhizomycetes was manually verified with BLASTN searches of all identified reads against Archaeorhizomycetes SSU rRNA reference sequences in NCBI nt database (minimum query length 150 bp, minimum similarity with Archaeorhizomycetes reference sequences >92%; reference sequences: EU179934.1, EU179935.1, EU179933.1, EU179936.1, KF993708.1, JF836023.1, JF836020.1). The 92% cut off was chosen to be relaxed enough to allow acceptance of possible Archaeorhizomycetes transcripts slightly different to available Archaeorhizomycetes reference sequences, but on the other hand to avoid the false positive assignment of SSU rRNAs from related taxa to Archaeorhizomycetes.
The network analysis was done in Cytoscape 3.0.216 with CoNET algorithm15. Only those fungal orders whose sum of sequences in all datasets was higher than 200 were included into the network analyses. The parameters and settings for network analyses in CoNET algorithm were: -parent_child_exclusion, -row_minocc 10, -correlations (Spearman, Pearson, mutual information, Bray-Curtis dissimilatory and Kullback-Leibler dissimilatory), threshold for edge selection was set to 1000 top and bottom. In randomization step, 100 iterations were calculated for edge scores. In following bootstrap step 100 iterations were calculated, unstable edges were filtered out, Brown method was chosen as P-value merging method and Benjaminihochberg method for multiple test correction. Resulting network for fungal community was visualized and analysed (i.e. degree of nodes, betweenness centrality, closeness centrality) in Cytoscape 3.0.2 and nodes tending to have high mean degree, low betweenness centrality, and high closeness centrality were identified as potential keystone orders22.
Additional Information
How to cite this article: Choma, M. et al. Low abundance of Archaeorhizomycetes among fungi in soil metatranscriptomes. Sci. Rep. 6, 38455; doi: 10.1038/srep38455 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
We thank Thomas Rattei (University of Vienna, Austria) for access to the computer cluster and Andrea Söllinger (University of Vienna, Austria) for support with scripts. MC and HS were supported by Ministry of Education, Youth and Sport of the Czech Republic (LM2015075), JB and TU were supported by the Czech Science Foundation (31-16-18453 S).
Footnotes
Author Contributions T.U. designed the research. M.C. and T.U. analysed the metatranscriptome datasets. J.B. conducted the co-occurrence network analysis. All authors wrote the manuscript. M.C. prepared the figures.
References
- Rosling A. et al. Archaeorhizomycetes: Unearthing an Ancient Class of Ubiquitous Soil Fungi. Science (80-.). 333, 876–879 (2011). [DOI] [PubMed] [Google Scholar]
- Rosling A., Timling I. & Taylor D. L. In Genomics of Soil- and Plant-Associated Fungi (eds Horwitz B. A., Mukherjee P. K., Mukherjee M. & Kubicek C. P.) 36, 333–349 (Springer-Verlag Berlin Heidelberg, 2013). [Google Scholar]
- Porter T. M. et al. Widespread occurrence and phylogenetic placement of a soil clone group adds a prominent new branch to the fungal tree of life. Mol. Phylogenet. Evol. 46, 635–644 (2008). [DOI] [PubMed] [Google Scholar]
- Tedersoo L. et al. Global diversity and geography of soil fungi. Science (80-.). 346, 1078 (2014). [DOI] [PubMed] [Google Scholar]
- Schadt C. W. & Rosling A. Comment on ‘Global diversity and geography of soil fungi’. Science (80-.). 348, 1438–1438 (2015). [DOI] [PubMed] [Google Scholar]
- Tedersoo L. et al. Response to Comment on ‘Global diversity and geography of soil fungi’. Science (80-.). 349, 936 (2015). [DOI] [PubMed] [Google Scholar]
- Urich T. et al. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One 3 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urich T. & Schleper C. In Handbook of Molecular Microbial Ecology I (ed de Brujin F.) 587–596 (John Wiley & Sons, Inc., 2011). [Google Scholar]
- Tveit A., Schwacke R., Svenning M. M. & Urich T. Organic carbon transformations in high-Arctic peat soils: key functions and microorganisms. ISME J. 7, 299–311 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisen S. et al. Metatranscriptomic census of active protists in soils. ISME J. doi: 10.1038/ismej.2015.30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epelde L., Lanzén A., Blanco F., Urich T. & Garbisu C. Adaptation of soil microbial community structure and function to chronic metal contamination at an abandoned Pb-Zn mine. FEMS Microbiol. Ecol. 91, 1–11 (2015). [DOI] [PubMed] [Google Scholar]
- Beulig F. et al. Altered carbon turnover processes and microbiomes in soils under long-term extremely high CO2 exposure. Nat. Microbiol. 1, 15025 (2016). [DOI] [PubMed] [Google Scholar]
- Tedersoo L. et al. Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. MycoKeys 10, 1–43 (2015). [Google Scholar]
- Schadt C. W., Martin A. P., Lipson D. a. & Schmidt S. K. Seasonal dynamics of previously unknown fungal lineages in tundra soils. Science 301, 1359–1361 (2003). [DOI] [PubMed] [Google Scholar]
- Faust K. et al. Microbial co-occurrence relationships in the Human Microbiome. PLoS Comput. Biol. 8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R. & Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopylova E., Noé L. & Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–7 (2012). [DOI] [PubMed] [Google Scholar]
- Quast C. et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzén A. et al. CREST - Classification Resources for Environmental Sequence Tags. PLoS One 7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D. H., Auch A. F., Qi J. & Schuster S. C. MEGAN analysis of metagenomic data. Genome Res. 17, 377–86 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D. & Widder S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 5, 1–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]