Abstract
The diversity of non-canonical amino acids (ncAAs) endows proteins with new features for a variety of biological studies and biotechnological applications. The genetic code expansion strategy, which co-translationally incorporates ncAAs into specific sites of target proteins, has been applied in many organisms. However, there have been only few studies on pathogens using genetic code expansion. Here, we introduce this technique into the human pathogen Salmonella by incorporating p-azido-phenylalanine, benzoyl-phenylalanine, acetyl-lysine, and phosphoserine into selected Salmonella proteins including a microcompartment shell protein (PduA), a type III secretion effector protein (SteA), and a metabolic enzyme (malate dehydrogenase), and demonstrate practical applications of genetic code expansion in protein labeling, photocrosslinking, and post-translational modification studies in Salmonella. This work will provide powerful tools for a wide range of studies on Salmonella.
Non-canonical amino acids (ncAAs) are powerful tools for protein studies. In the past few years, more than 150 different ncAAs have been incorporated into proteins in both prokaryotic and eukaryotic organisms using varied approaches. One of the most powerful methods of ncAA incorporation is genetic code expansion1,2,3,4,5,6,7,8. This approach uses an orthogonal aminoacyl-tRNA synthetase (AARS)/tRNA pair, which does not cross-react with host AARSs and tRNAs, to direct the incorporation of an ncAA at an assigned codon (usually the amber stop codon). This allows the introduction of ncAAs with novel functional groups at a precise position in a protein of interest. Scientists have already used this strategy to insert photocrosslinkers for mapping weak, transient, or pH sensitive protein interactions, to install post-translational modifications for regulating biological processes or identifying modifying enzymes, to incorporate photo-caged amino acids for controlling signaling and channeling by light, and to introduce biophysical probes and labels for providing exquisite insights into protein dynamics. These approaches have been combined with imaging systems, single-molecule studies, biophysical techniques, structural biology, and mass spectrometry to answer biological questions that are difficult or impossible to address by most classical methods9,10,11,12,13,14.
As the key components of genetic code expansion, a number of orthogonal AARS/tRNA pairs have been developed. In Escherichia coli, three pairs have been most successful: (1) an evolved orthogonal pair based on the Methanocaldococcus jannaschii tyrosyl-tRNA synthetase (mjTyrRS) and its cognate tRNA, which has been used to install a diverse array of tyrosine and phenylalanine derivatives15; (2) a natural orthogonal pair of pyrrolysyl-tRNA synthetase (PylRS) and its cognate tRNA from Methanosarcinaceae species, which facilitates the incorporation of a variety of lysine and phenylalanine analogs16,17; and (3) the phosphoseryl-tRNA synthetase (SepRS) and its cognate tRNA from methanogenic archaea, which were engineered for phosphoserine incorporation18,19.
Salmonella infects both human and animals, causing millions of illnesses every year. It is also an important model organism for gastrointestinal and extraintestinal pathogenesis studies. Due to its significance in public health, we sought to expand the genetic code in Salmonella with a variety of ncAAs, increasing the diversity of approaches that can be used in Salmonella studies. We utilized the mjTyrRS, PylRS, and SepRS systems to incorporate p-azido-phenylalanine, benzoyl-phenylalanine, acetyl-lysine, and phosphoserine in Salmonella, individually. For validation, we incorporated these ncAAs into Salmonella native proteins including a microcompartment shell protein, a type III secretion effector protein, and a TCA cycle enzyme respectively to demonstrate practical applications of genetic code expansion in protein labeling, photocrosslinking, and post-translational modification studies in Salmonella.
Results
Expression of target proteins and efficiency of orthogonal pairs in Salmonella
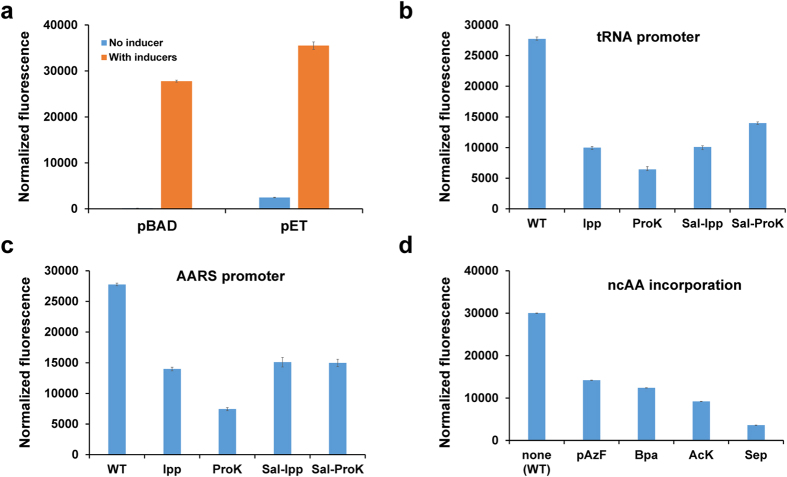
First, we compared the expression of target proteins from different vectors in Salmonella. We used superfolder green fluorescent protein (sfGFP) as a reporter, which was engineered to have improved tolerance of circular permutation, greater resistance to chemical denaturants, and enhanced folding kinetics20, and put its gene in either a modified pBAD vector with the arabinose promoter or a modified pET vector with the lac promoter. For using the arabinose promoter, we removed the araBAD gene cluster which encodes arabinose metabolic enzymes from the genome. Although the pET vector had higher expression of sfGFP, the background of the pBAD vector was much lower (Fig. 1a). Furthermore, the expression of sfGFP in the pBAD vector can be modulated by the concentration of arabinose in the media (Supplementary Fig. S1). So we chose the pBAD vector to express target proteins in later experiments.
Figure 1. Expression of target proteins and noncanonical amino acid incorporation efficiency in Salmonella.
(a) Comparison of the Para and Plac promoters to determine optimal superfolder green fluorescent protein (sfGFP) expression; (b) Comparison of the promoters to determine optimal tRNA expression for p-azido-phenylalanine (pAzF) incorporation: WT represents full-length wild-type sfGFP; (c) Comparison of the promoters to determine optimal aminoacyl-tRNA synthetase expression for pAzF incorporation: WT represents full-length wild-type sfGFP; (d) Comparison of the incorporation of noncanonical amino acids (ncAAs) at the position 151 of sfGFP: None (WT) represents full-length wild-type sfGFP without any ncAA supplied in the growth media. The normalized fluorescence was the absolute fluorescence readings at 12 h normalized by the corresponding cell densities. The mean values and standard deviations were calculated from three replicates.
To allow varied applications, we chose four orthogonal pairs of AARS/tRNA that are widely used in E. coli: (1) a recently evolved mjTyrRS variant which is highly specific for p-azido-phenylalanine (pAzF) incorporation21; (2) an optimized mjTyrRS variant for incorporating a photocrosslinker, benzoyl-phenylalanine (Bpa); (3) an optimized PylRS system for the introduction of an important post-translational modification, acetyl-lysine (AcK)22; and 4) an optimized SepRS system for the incorporation of phosphoserine (Sep), which is another major post-translational modification23.
There are considerable genetic differences between E. coli and Salmonella which may affect the incorporation efficiency of ncAAs in Salmonella by these orthogonal pairs which were developed in E. coli. To modify these pairs for use in Salmonella, we first optimized their expression. In E. coli, the genes of orthogonal AARSs and tRNAs are constructed with the lpp promoter before AARS genes and the ProK promoter before tRNA genes22. Sequence alignments showed that the lpp promoter in Salmonella is highly similar to that in E. coli, but the ProK promoters are quite different between these two organisms (Supplementary Fig. S2). Thus, we compared the effect of tRNA promoters on the incorporation of pAzF by using a sfGFP-based fluorescence assay24. By site-directed mutagenesis, a stop codon (UAG) was introduced at position 151 of the sfGFP gene which was shown to be permissive at that site22,24,25. Higher pAzF incorporation results in better readthrough of the stop codon thus generating higher fluorescence. The results showed that the Salmonella ProK promoter (Sal-ProK) gave the highest ncAA incorporation (Fig. 1b). A similar approach was applied to test the effect of AARS promoters, and the results showed the Salmonella lpp promoter (Sal-lpp) had the best efficiency (Fig. 1c). To further optimize AARS and tRNA expression based on varying promoter strength, we made a matrix of 16 combinations of four promoters for tRNA expression and four promoters for AARS expression. The results also showed that the combination of Sal-lpp for AARS expression and Sal-ProK for tRNA expression was the best pair (Supplementary Table S1). Thus, we put the AARS genes after Sal-lpp promoter and the tRNA genes after the Sal-ProK promoter. With the same fluorescence assay, we tested the incorporation of the other three ncAAs mentioned above. Promisingly, the incorporation efficiency of pAzF, Bpa, and AcK was about 30% in average compared to the expression of the wild-type sfGFP (Fig. 1d). The incorporation of Sep was relatively low at ~15%, similar to that in E. coli, but it could still produce ~50 mg protein in 1 L media which was sufficient for further studies. Finally, all the ncAA incorporation was confirmed with mass spectrometry (MS) analyses (Supplementary Fig. S3).
Incorporation of p-azido-phenylalanine (pAzF) as a handle for protein labeling by click chemistry
Currently, the most popular tags to label proteins for imaging purposes are fluorescent proteins (FPs)26. However, there are also disadvantages of this method. First, the relatively large size of FPs may perturb the structures and functions of target proteins. Second, FP fusions are limited to the N- or C- termini of target proteins. Moreover, FPs need oxygen to produce fluorescence. Here, we applied a protein labeling method based on the incorporation of pAzF, which is then reacted with an alkyne-fluorescent dye by the click reaction to produce fluorescence without oxygen. Moreover, due to its small size, pAzF can be incorporated at essentially any site within a protein by using a UAG codon and the optimized pAzF incorporation system21.
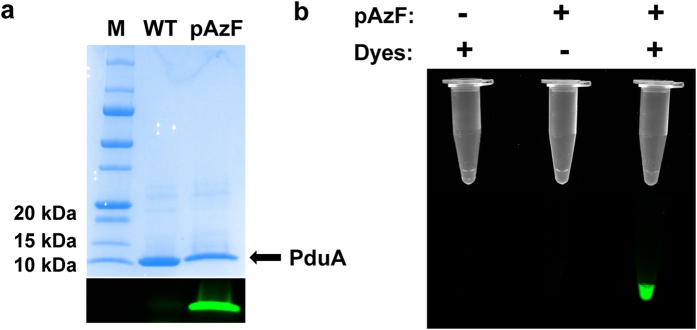
As for the target protein for validation, we chose a native Salmonella protein, PduA, one of the major shell proteins of 1,2-propanediol utilization (Pdu) microcompartments (MCPs) which are large multi-protein complexes, functioning as organelles for 1,2-propanediol degradation27. Based on the crystal structure of PduA28, residue N67 which is at the outer surface of Pdu MCPs was selected for pAzF incorporation. First, we inserted an amber stop codon (UAG) at the position 67 of PduA and overexpressed PduA from the pBAD plasmid. The purified pAzF-containing PduA was labeled by a fluorescent dye with an alkyne group through click chemistry29. With a 5-minute reaction, the pAzF-containing PduA protein gave a clear and bright green band, while the wild-type PduA protein had a very low background (Fig. 2a). Then, we used the same approach to label Pdu MCPs in living cells of Salmonella. We mutated the position 67 of PduA gene to an amber stop codon in the genome, expressed the entire pdu operon to produce pAzF-containing Pdu MCPs, which were then purified by previous protocols30, and labeled with fluorescent dyes. After washing away excess fluorescent dyes, only pAzF-containing MCPs had fluorescence (Fig. 2b). The SDS-PAGE gel also indicated the specific labeling of PduA in the whole Pdu MCP protein profile (Supplementary Fig. S4). The pAzF incorporation in PduA was confirmed by LC-MS/MS analysis (Supplementary Fig. S5).
Figure 2. Incorporation of p-azido-phenylalanine (pAzF) in Salmonella PduA protein.
(a) The SDS-PAGE gel of PduA labeling. The upper panel is the gel stained with Coomassie Blue. The lower panel was captured with fluorescent filters for Alexa Fluor 488. (b) The fluorescence labeling of purified Pdu microcompartments in vitro. The upper panel was captured without filters. The lower panel was captured with fluorescent filters for Alexa Fluor 488. The left lane was wild-type Pdu MCPs with dyes. The middle lane only contained pAzF-containing Pdu MCPs. The right lane included both pAzF-containing Pdu MCPs and dyes.
Incorporation of benzoyl-phenylalanine (Bpa) for photocrosslinking
Understanding how proteins interact is one of the most common questions to be solved for any biological processes. However, it can be difficult when the interactions are weak, transient, pH-dependent, or when the interactions are at particular subcellular locations such as membranes. The covalent nature of photocrosslinking enables detection of low-affinity interactions. Moreover, it can be used in living cells to identify specific, direct protein-protein interactions31. Benzoyl-l-phenylalanine (Bpa) is a widely used photocrosslinker excited by UV at the wavelength of 365 nm. Here, we utilized an optimized mjTyrRS variant32,33 to incorporate Bpa into proteins in Salmonella.
As for the target protein for validation, we chose a native Salmonella protein, SteA, a type III secretion effector which contributes to the control of membrane dynamics of Salmonella-containing vacuoles34, globally affecting host cell proliferation, morphology, adhesion and migration35. For minimal perturbation to the protein structure, we selected two tyrosine residues at positions 40 and 155 for Bpa incorporation, individually. First, we mutated these two positions to the amber stop codon separately, and overexpressed SteA from the pBAD plasmid. Then, we exposed the purified Bpa-containing SteA proteins to the long wavelength 365 nm UV light and analyzed the products by SDS-PAGE. Interestingly, the SteA (155-Bpa) formed a covalent dimer, while the SteA (40-Bpa) showed no crosslinking (Fig. 3). This indicated that residue Tyr155 may be at the dimer interface, so the SteA (155-Bpa) variant may affect further experiments to identify SteA-interacting proteins in vivo, while SteA (40-Bpa) variant should be ideal for this purpose. The Bpa incorporation in SteA was confirmed by LC-MS/MS analyses (Supplementary Fig. S5).
Figure 3. Incorporation of benzoyl-phenylalanine (Bpa) in Salmonella SteA protein.
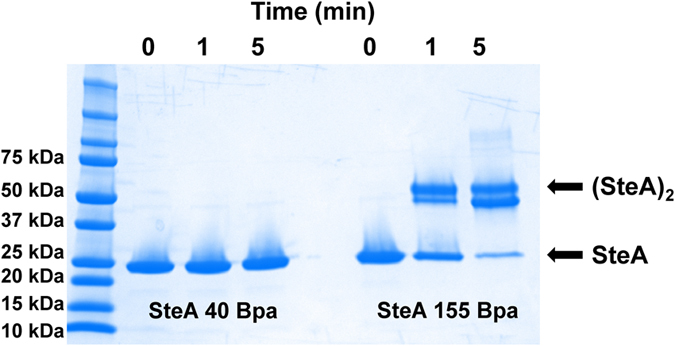
The SAS-PAGE gel for photocrosslinking of SteA proteins with Bpa incorporated at position 40 and 155, individually. The 365 nm UV irritation time was 0, 1 and 5 minutes. With 1-minute exposure, about 60% of total proteins were cross-linked, while 5-minute exposure made more than 95% of total protein cross-linked. The expected SteA monomer and SteA dimer were indicated at their corresponding molecular weights.
Incorporation of acetyl-lysine (AcK) and phosphoserine (Sep) for studying protein post-translational modifications
Acetylation and phosphorylation are two of the most common post-translational modifications in natue36,37. Previous studies have shown that lysine acetylation regulates the functions of many proteins in Salmonella38,39,40. Although proteomic analyses revealed that more than 20% of proteins in bacteria are modified by acetylation or phosphorylation41,42, limited validation has been done to confirm these modifications. One of the challenges is that it is difficult to synthesize proteins that are fully modified at desired positions by most classical methods. Therefore, we applied the genetic code expansion strategy to directly incorporate AcK and Sep site-specifically into proteins in Salmonella to overcome such challenge.
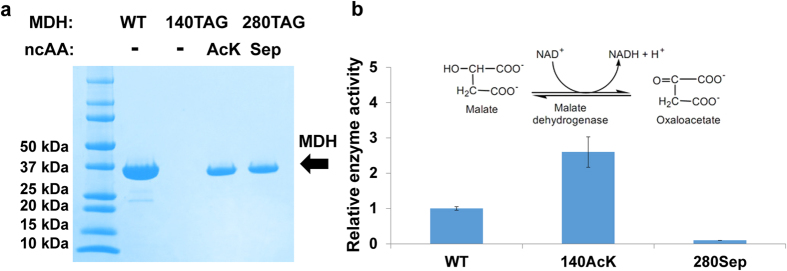
As for the target protein for validation, we chose a native Salmonella protein, malate dehydrogenase (MDH), an important metabolic enzyme in the TCA cycle43. Previous studies have shown that the acetylation of lysine residues in mammalian MDHs increases their enzyme activities, and is involved in the cross-talk between adipogenesis and the intracellular energy level44,45. It has also been shown that bacterial MDHs are subject to both lysine acetylation and serine phosphorylation38,46. We selected residue Lys140 for AcK incorporation based on the proteomic studies38, and residue Ser280 for Sep incorporation, as its corresponding position in E. coli MDH (95% sequence identity) was shown to be phosphorylated46. We mutated these two positions to amber stop codons separately, and overexpressed the MDH from pBAD plasmids (Fig. 4a). Enzyme assays of the purified MDH proteins showed that acetylation of lysine140 increased enzyme activity, while the phosphorylation of Ser280 inactivated the enzyme (Fig. 4b), indicating that Salmonella cells could use different modifications to activate or inactivate the MDH enzyme to control the flux in the TAC cycle. The AcK and Sep incorporation in MDH was confirmed by LC-MS/MS analyses (Supplementary Fig. S5).
Figure 4. Incorporation of acetyl-lysine (AcK) and phosphoserine (Sep) in Salmonella malate dehydrogenase (MDH).
(a) The SDS-PAGE gel of purified MDH and its modified variants. From left to right, lane 1 is the protein marker; lane 2 is wild-type full-length MDH; lane 3 is MDH with 140 position mutated to a UAG stop codon and without any noncanonical amino acid supplied in the growth media; lane 4 is MDH with 140 position mutated to a UAG stop codon and with 5 mM AcK supplied in the growth media; lane 5 is MDH with 280 position mutated to a UAG stop codon and with 2 mM Sep supplied in the growth media. (b) The enzyme activities of the MDH enzyme and its modified variants. The mean values and standard deviations were calculated from three replicates. The enzyme activity of wild-type MDH was set as 1, and the reaction catalyzed by MDH is demonstrated as inset.
Discussion
In this work, we used mjTyrRS to incorporate pAzF and Bpa into two different native Salmonella proteins. In other organisms, the mjTyrRS has been engineered to incorporate many other tyrosine or phenylalanine analogs2 that enable a range of biochemical investigations1,2,9,10,14: (1) Acetyl-phenylalanine (ketone group), allyl-tyrosine (alkene group), and propargyloxy-phenylalanine (alkyne group) can be modified by selective chemical reactions such as oxime condensation and click chemistry to allow site-specific protein labeling; (2) Cyano-phenylalanine, amino-phenylalanine, iodo-phenylalanine, and fluoro-phenylalanine can be used as heavy atoms for X-ray structure determination and probes for IR and NMR; (3) Photo-reactive amino acids such as bipyridyl-alanine, benzoyl-phenylalanine, and nitrobenzyl-tyrosine can be used as switches to control biological processes with light; (4) Varied post-translation modifications such nitro-tyrosine and sulfo-tyrosine can be used to study functions of protein modifications. Since we have successfully introduced the mjTyrRS systems for pAzF and Bpa incorporation in this study, the ncAAs mentioned above might also be utilized in Salmonella for a wide range of biochemical and biophysical investigations.
We also used the PylRS system to install lysine acetylation into the MDH protein of Salmonella. This system has been widely used in both prokaryotic and eukaryotic organisms, and has been evolved for not only lysine derivatives but also phenylalanine or even tryptophan analogs16,17. Other lysine modifications such as methylation and ubiquitylation have also be incorporated or generated by the PylRS systems to form complete lysine modification profiles47,48. A previous study also showed that the PylRS system can incorporate pyrrolysine analogs as crosslinkers in Salmonella49. Thus, the mjTyrRS and PylRS systems, open up a wide range of ncAA candidates with different sizes and properties for a variety of research objectives.
Proteomic studies indicate many proteins have both acetylation and phosphorylation modifications simultaneously. We have demonstrated the facile incorporation of lysine acetylation and serine phosphorylation site-specifically into proteins, individually. Moreover, PylRS and its variants have low selectivity toward the tRNA anticodon, and can be used for incorporating ncAAs towards different codons such as the opal stop codon (TGA), quadruple codons, and even sense codons17. Thus, combining two mutually orthogonal PylRS and SepRS systems with different stop codons or quadruplet codons, could allow the production of simultaneously acetylated and phosphorylated proteins, providing a powerful tool to study the crosstalk between protein acetylation and phosphorylation.
Our established ncAA incorporation systems could facilitate a number of studies on Salmonella, such as tracking proteins, mapping protein-protein interaction networks, and characterizing protein post-translational modifications. Together with this work, genetic code expansion has been successfully applied in several pathogens including Salmonella, Shigella, and Mycobacterium49,50, suggesting that it is promising to extend this strategy to other pathogens for broader applications. Furthermore, all the ncAAs and chemicals mentioned in this work are commercially available, so our systems could benefit many biological laboratories without the need for in house organic synthesis.
Methods
General molecular biology
The amino acids in this study were purchased from Sigma-Aldrich or ChemImpex. E. coli TOP10 cells (Life Technologies) were used for general cloning. Salmonella enterica Serovar Typhimurium LT2 was used as the representative Salmonella strain. Plasmids: The genes of AARSs and tRNAs were cloned by PCR from laboratory inventory and inserted into the pTech plasmid. The genes of target proteins with C-terminal His6-tag were cloned into the pBAD plasmid with the arabinose promoter, or a modified pET plasmid with the lac promoter. All the cloning experiments were performed by the Gibson Assembly kit (New England Biolabs). The mutations of target genes were made by the QuikChange II mutagenesis kit (Agilent Life Sciences). The SDS-PAGE gel is 4–20% gradient gel purchased from Bio-Rad. The chromosomal mutation PduA N67TAG was constructed as described previously51. DNA oligos used to construct the mutation were ordered from Integrated DNA Technologies (Coralville, IA). The mutation was confirmed by DNA sequencing. The information of plasmids and primers was listed in Supplementary Tables S2 and S3, respectively. The primary sequences of proteins were listed in the Supplementary information, and the locations of the ncAA substitution and the active sites of enzymes were marked as well.
sfGFP readthrough assay
The strains harboring the genes of sfGFP as well as AARSs and tRNAs were inoculated into 2 ml LB medium. The overnight culture was diluted with fresh LB medium to an absorbance of 0.2 at 600 nm, supplemented with ncAAs. 1 mM arabinose or 1 mM IPTG was added to induce the expression of sfGFP. 200 μL culture of each strain was transferred to a well in the 96 well plate. Cells were shaken for 24 hours at 37 °C, with monitoring of fluorescence intensity (excitation 485 nm, emission 528 nm, bandwidths 20 nm) and optical cell density by the microplate-reader.
Protein expression and purification
The genes of target proteins were cloned into the pBAD vector with a C-terminal His6-tag, and transformed into LT2 ΔaraBAD cells together with the plasmids harboring genes of AARSs and tRNAs for expression. The expression strain was grown on 1 L of LB medium at 37 °C to an absorbance of 0.6–0.8 at 600 nm, and protein expression was induced by the addition of 1 mM arabinose and ncAAs (1 mM for pAzF, Bpa; 2 mM for Sep; and 5 mM for AcK). Cells were incubated at 30 °C for an additional 8 hours, and harvested by centrifugation. The cell paste was suspended in 15 ml of 50 mM Tris (pH 7.5), 300 mM NaCl, 20 mM imidazole, and broken by sonication. The crude extract was centrifuged at 20,000 × g for 30 min at 4 °C. The soluble fraction was loaded onto a column containing 2 ml of Ni-NTA resin (Qiagen). The column was then washed with 50 ml of 50 mM Tris (pH 7.5), 300 mM NaCl, 50 mM imidazole, and eluted with 2 ml of 50 mM Tris (pH 7.5), 300 mM NaCl, 200 mM imidazole.
Mass spectroscopy analysis
The samples was loaded onto the SDS-PAGE gel. The bands with the corresponding molecular weight of target proteins were cut and sent for MS analysis. The proteins were trypsin digested by a standard in-gel digestion protocol, and analyzed by LC-MS/MS on an LTQ Orbitrap XL (Thermo Scientific) equipped with a nanoACQUITY UPLC system (Waters). A Symmetry C18 trap column (180 μm × 20 mm; Waters) and a nanoACQUITY UPLC column (1.7 μm, 100 μm × 250 mm, 35 °C) were used for peptide separation. Trapping was done at 15 μL min−1, 99% buffer A (0.1% formic acid) for 1 min. Peptide separation was performed at 300 nL min−1 with buffer A and buffer B (CH3CN containing 0.1% formic acid). The linear gradient was from 5% to 50% buffer B at 50 min, to 85% buffer B at 51 min. MS data were acquired in the Orbitrap with one microscan, and a maximum inject time of 900 ms followed by data-dependent MS/MS acquisitions in the ion trap (through collision induced dissociation, CID). The Mascot search algorithm was used to determine the amino acid composition at specific positions (Matrix Science, Boston, MA).
Protein labeling
The fluorescent labeling reagent was Click-IT® Alexa Fluor® 488 DIBO Alkyne purchased from Invitrogen. The labeling experiment was performed by following the protocol from the manufacturer. 100 μg purified PduA protein or 1 mg purified Salmonella microcompartment was used for labeling. The labeled proteins were separated by the SDS-PAGE gel and imaged by the ChemiDocTM MP System from Bio-Rad.
Photocrosslinking
The reactions were performed in a 96-well plate by using 100 μl of proteins with the concentration of 1 mg/ml. Samples were irradiated at 365 nm by using the UV crosslinker CL-1000L (Denville Scientific). Then, samples were removed from the wells and analyzed by the SDS-PAGE gel.
MDH activity assay
The assays were performed by following the instruction of the EnzyChromTM Malate Dehydrogenase Assay Kit (EMDH-100) from BioAssay Systems. This non-radioactive, colorimetric assay is based on the reduction of the tetrazolium salt MTT in a NADH-coupled enzymatic reaction to a reduced form of MTT which exhibits an absorption maximum at 565 nm. The increase in absorbance at 565 nm is proportional to the enzyme activity. 100 μg purified MDH and its variants were used in the assay.
Additional Information
How to cite this article: Gan, Q. et al. Expanding the genetic code of Salmonella with non-canonical amino acids. Sci. Rep. 6, 39920; doi: 10.1038/srep39920 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Professor Dieter Söll (Yale University, USA) for generous supports and suggestions. This work was financially supported by the start-up fund of University of Arkansas, the grants from the National Institutes of Allergy and Infectious Diseases (AI119813 and AI081146), and the grant from the National Institute of General Medical Sciences (GM22854).
Footnotes
Author Contributions C.F. directed the studies, designed the experiments, and interpreted data. Q.G. and C.F. performed the experiments. B.P.L. and T.A.B. constructed the Salmonella strains. C.F. and T.A.B. wrote the manuscript.
References
- O’Donoghue P., Ling J., Wang Y. S. & Soll D. Upgrading protein synthesis for synthetic biology. Nat Chem Biol 9, 594–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. C. & Schultz P. G. Adding new chemistries to the genetic code. Annu Rev Biochem 79, 413–44 (2010). [DOI] [PubMed] [Google Scholar]
- Hoesl M. G. & Budisa N. Recent advances in genetic code engineering in Escherichia coli. Curr Opin Biotechnol 23, 751–7 (2012). [DOI] [PubMed] [Google Scholar]
- Antonczak A. K., Morris J. & Tippmann E. M. Advances in the mechanism and understanding of site-selective noncanonical amino acid incorporation. Curr Opin Struct Biol 21, 481–7 (2011). [DOI] [PubMed] [Google Scholar]
- Terasaka N., Iwane Y., Geiermann A. S., Goto Y. & Suga H. Recent developments of engineered translational machineries for the incorporation of non-canonical amino acids into polypeptides. Int J Mol Sci 16, 6513–31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin J. W. Expanding and reprogramming the genetic code of cells and animals. Annu Rev Biochem 83, 379–408 (2014). [DOI] [PubMed] [Google Scholar]
- Lemke E. A. The exploding genetic code. Chembiochem 15, 1691–4 (2014). [DOI] [PubMed] [Google Scholar]
- Xie J. & Schultz P. G. A chemical toolkit for proteins–an expanded genetic code. Nat Rev Mol Cell Biol 7, 775–82 (2006). [DOI] [PubMed] [Google Scholar]
- Neumann H. Rewiring translation - Genetic code expansion and its applications. FEBS Lett 586, 2057–64 (2012). [DOI] [PubMed] [Google Scholar]
- Davis L. & Chin J. W. Designer proteins: applications of genetic code expansion in cell biology. Nat Rev Mol Cell Biol 13, 168–82 (2012). [DOI] [PubMed] [Google Scholar]
- Yanagisawa T., Umehara T., Sakamoto K. & Yokoyama S. Expanded genetic code technologies for incorporating modified lysine at multiple sites. Chembiochem 15, 2181–7 (2014). [DOI] [PubMed] [Google Scholar]
- Nikic I. & Lemke E. A. Genetic code expansion enabled site-specific dual-color protein labeling: superresolution microscopy and beyond. Curr Opin Chem Biol 28, 164–73 (2015). [DOI] [PubMed] [Google Scholar]
- Li X. & Liu C. C. Biological applications of expanded genetic codes. Chembiochem 15, 2335–41 (2014). [DOI] [PubMed] [Google Scholar]
- Wang Q., Parrish A. R. & Wang L. Expanding the genetic code for biological studies. Chem Biol 16, 323–36 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Brock A., Herberich B. & Schultz P. G. Expanding the genetic code of Escherichia coli. Science 292, 498–500 (2001). [DOI] [PubMed] [Google Scholar]
- Fekner T. & Chan M. K. The pyrrolysine translational machinery as a genetic-code expansion tool. Curr Opin Chem Biol 15, 387–91 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan W., Tharp J. M. & Liu W. R. Pyrrolysyl-tRNA synthetase: an ordinary enzyme but an outstanding genetic code expansion tool. Biochim Biophys Acta 1844, 1059–70 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. S. et al. Expanding the genetic code of Escherichia coli with phosphoserine. Science 333, 1151–4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogerson D. T. et al. Efficient genetic encoding of phosphoserine and its nonhydrolyzable analog. Nat Chem Biol 11, 496–503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedelacq J. D., Cabantous S., Tran T., Terwilliger T. C. & Waldo G. S. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24, 79–88 (2006). [DOI] [PubMed] [Google Scholar]
- Amiram M. et al. Evolution of translation machinery in recoded bacteria enables multi-site incorporation of nonstandard amino acids. Nat Biotechnol 33, 1272–1279 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C., Xiong H., Reynolds N. M. & Soll D. Rationally evolving tRNAPyl for efficient incorporation of noncanonical amino acids. Nucleic Acids Res 43, e156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. et al. A facile strategy for selective incorporation of phosphoserine into histones. Angew Chem Int Ed Engl 52, 5771–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C., Ho J. M., Chirathivat N., Soll D. & Wang Y. S. Exploring the substrate range of wild-type aminoacyl-tRNA synthetases. Chembiochem 15, 1805–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D. D. et al. An evolved aminoacyl-tRNA synthetase with atypical polysubstrate specificity. Biochemistry 50, 1894–900 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien R. Y. The green fluorescent protein. Annu Rev Biochem 67, 509–44 (1998). [DOI] [PubMed] [Google Scholar]
- Bobik T. A., Lehman B. P. & Yeates T. O. Bacterial microcompartments: widespread prokaryotic organelles for isolation and optimization of metabolic pathways. Mol Microbiol 98, 193–207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley C. S. et al. Structural insight into the mechanisms of transport across the Salmonella enterica Pdu microcompartment shell. J Biol Chem 285, 37838–46 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb H. C., Finn M. G. & Sharpless K. B. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew Chem Int Ed Engl 40, 2004–2021 (2001). [DOI] [PubMed] [Google Scholar]
- Fan C. et al. Short N-terminal sequences package proteins into bacterial microcompartments. Proc Natl Acad Sci USA 107, 7509–14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham N. D., Parker R. B. & Kohler J. J. Photocrosslinking approaches to interactome mapping. Curr Opin Chem Biol 17, 90–101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Ju T., Niu W. & Guo J. Fine-tuning interaction between aminoacyl-tRNA synthetase and tRNA for efficient synthesis of proteins containing unnatural amino acids. ACS Synth Biol 4, 207–12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin J. W., Martin A. B., King D. S., Wang L. & Schultz P. G. Addition of a photocrosslinking amino acid to the genetic code of Escherichiacoli. Proc Natl Acad Sci USA 99, 11020–4 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues L., Holden D. W. & Mota L. J. The Salmonella effector SteA contributes to the control of membrane dynamics of Salmonella-containing vacuoles. Infect Immun 82, 2923–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenal-Munoz E., Gutierrez G. & Ramos-Morales F. Global impact of Salmonella type III secretion effector SteA on host cells. Biochem Biophys Res Commun 449, 419–24 (2014). [DOI] [PubMed] [Google Scholar]
- Saxholm H. J., Pestana A., O’Connor L., Sattler C. A. & Pitot H. C. Protein acetylation. Mol Cell Biochem 46, 129–53 (1982). [DOI] [PubMed] [Google Scholar]
- Rubin C. S. & Rosen O. M. Protein phosphorylation. Annu Rev Biochem 44, 831–87 (1975). [DOI] [PubMed] [Google Scholar]
- Wang Q. et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327, 1004–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starai V. J., Celic I., Cole R. N., Boeke J. D. & Escalante-Semerena J. C. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298, 2390–2 (2002). [DOI] [PubMed] [Google Scholar]
- Starai V. J. & Escalante-Semerena J. C. Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J Mol Biol 340, 1005–12 (2004). [DOI] [PubMed] [Google Scholar]
- Macek B. & Mijakovic I. Site-specific analysis of bacterial phosphoproteomes. Proteomics 11, 3002–11 (2011). [DOI] [PubMed] [Google Scholar]
- Bernal V. et al. Regulation of bacterial physiology by lysine acetylation of proteins. N Biotechnol 31, 586–95 (2014). [DOI] [PubMed] [Google Scholar]
- Musrati R. A., Kollarova M., Mernik N. & Mikulasova D. Malate dehydrogenase: distribution, function and properties. Gen Physiol Biophys 17, 193–210 (1998). [PubMed] [Google Scholar]
- Kim E. Y. et al. Acetylation of malate dehydrogenase 1 promotes adipogenic differentiation via activating its enzymatic activity. J Lipid Res 53, 1864–76 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. Y., Han B. S., Kim W. K., Lee S. C. & Bae K. H. Acceleration of adipogenic differentiation via acetylation of malate dehydrogenase 2. Biochem Biophys Res Commun 441, 77–82 (2013). [DOI] [PubMed] [Google Scholar]
- Macek B. et al. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol Cell Proteomics 7, 299–307 (2008). [DOI] [PubMed] [Google Scholar]
- Nguyen D. P., Garcia Alai M. M., Kapadnis P. B., Neumann H. & Chin J. W. Genetically encoding N(epsilon)-methyl-L-lysine in recombinant histones. J Am Chem Soc 131, 14194–5 (2009). [DOI] [PubMed] [Google Scholar]
- Li X., Fekner T., Ottesen J. J. & Chan M. K. A pyrrolysine analogue for site-specific protein ubiquitination. Angew Chem Int Ed Engl 48, 9184–7 (2009). [DOI] [PubMed] [Google Scholar]
- Lin S. et al. Site-specific incorporation of photo-cross-linker and bioorthogonal amino acids into enteric bacterial pathogens. J Am Chem Soc 133, 20581–7 (2011). [DOI] [PubMed] [Google Scholar]
- Wang F., Robbins S., Guo J., Shen W. & Schultz P. G. Genetic incorporation of unnatural amino acids into proteins in Mycobacterium tuberculosis. PLoS One 5, e9354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S. et al. Alanine scanning mutagenesis identifies an asparagine-arginine-lysine triad essential to assembly of the shell of the Pdu microcompartment. J Mol Biol 426, 2328–45 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.