Abstract
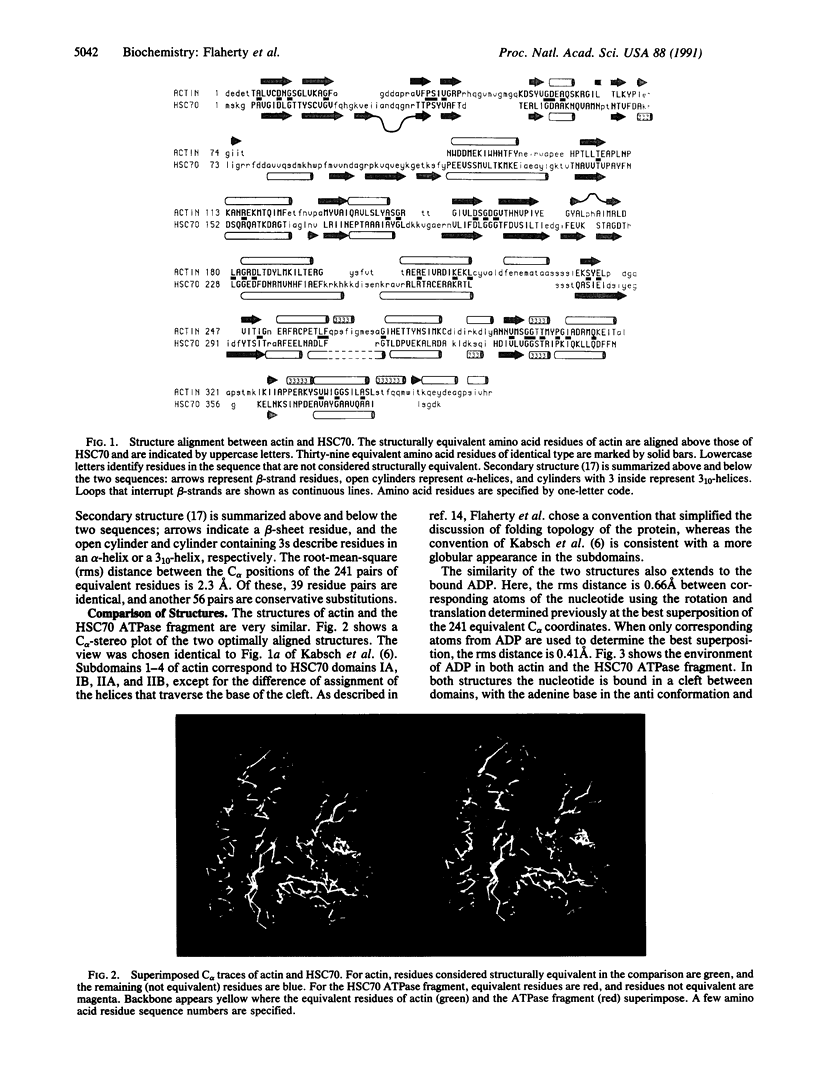
Although there is very little sequence identity between the two proteins, the structures of rabbit skeletal muscle actin (375-amino acid residues) and the 44-kDa ATPase fragment of the bovine 70-kDa heat shock cognate protein (HSC70; 386 residues) are very similar. The alpha-carbon positions of 241 pairs of amino acid residues that are structurally equivalent within the two proteins can be superimposed with a root-mean-square difference in distance of 2.3 A; of these, 39 residues are identical, and 56 are conservative substitutions. In addition, the conformations of ADP are very similar in both proteins. A local sequence "fingerprint," which may be diagnostic of the adenine nucleotide beta-phosphate-binding pocket, has been derived. The fingerprint identifies members of the glycerol kinase family as candidates likely to have a similar structure in their nucleotide-binding domains. The structural differences between the two molecules mainly occur in loop regions of actin known to be involved in interactions with other monomers in the actin filament or in the binding of myosin; the corresponding regions in heat shock proteins may have functions that are as yet undetermined. Placing the Ca2+ ATP of actin on the ATPase fragment structure suggests Asp-206 (corresponding to His-161 of actin) as a candidate proton acceptor for the ATPase reaction.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson C. M., McDonald R. C., Steitz T. A. Sequencing a protein by x-ray crystallography. I. Interpretation of yeast hexokinase B at 2.5 A resolution by model building. J Mol Biol. 1978 Jul 25;123(1):1–13. doi: 10.1016/0022-2836(78)90373-x. [DOI] [PubMed] [Google Scholar]
- Andreone T. L., Printz R. L., Pilkis S. J., Magnuson M. A., Granner D. K. The amino acid sequence of rat liver glucokinase deduced from cloned cDNA. J Biol Chem. 1989 Jan 5;264(1):363–369. [PubMed] [Google Scholar]
- Beckmann R. P., Mizzen L. E., Welch W. J. Interaction of Hsp 70 with newly synthesized proteins: implications for protein folding and assembly. Science. 1990 May 18;248(4957):850–854. doi: 10.1126/science.2188360. [DOI] [PubMed] [Google Scholar]
- Carlier M. F. Role of nucleotide hydrolysis in the dynamics of actin filaments and microtubules. Int Rev Cytol. 1989;115:139–170. doi: 10.1016/s0074-7696(08)60629-4. [DOI] [PubMed] [Google Scholar]
- Chappell T. G., Konforti B. B., Schmid S. L., Rothman J. E. The ATPase core of a clathrin uncoating protein. J Biol Chem. 1987 Jan 15;262(2):746–751. [PubMed] [Google Scholar]
- Chirico W. J., Waters M. G., Blobel G. 70K heat shock related proteins stimulate protein translocation into microsomes. Nature. 1988 Apr 28;332(6167):805–810. doi: 10.1038/332805a0. [DOI] [PubMed] [Google Scholar]
- Cupples C. G., Pearlman R. E. Isolation and characterization of the actin gene from Tetrahymena thermophila. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5160–5164. doi: 10.1073/pnas.83.14.5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca-Flaherty C., Flaherty K. M., McIntosh L. J., Bahrami B., McKay D. B. Crystals of an ATPase fragment of bovine clathrin uncoating ATPase. J Mol Biol. 1988 Apr 20;200(4):749–750. doi: 10.1016/0022-2836(88)90487-1. [DOI] [PubMed] [Google Scholar]
- DeLuca-Flaherty C., McKay D. B. Nucleotide sequence of the cDNA of a bovine 70 kilodalton heat shock cognate protein. Nucleic Acids Res. 1990 Sep 25;18(18):5569–5569. doi: 10.1093/nar/18.18.5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R. J., Koch B. D., Werner-Washburne M., Craig E. A., Schekman R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature. 1988 Apr 28;332(6167):800–805. doi: 10.1038/332800a0. [DOI] [PubMed] [Google Scholar]
- Ellis R. J., van der Vies S. M., Hemmingsen S. M. The molecular chaperone concept. Biochem Soc Symp. 1989;55:145–153. [PubMed] [Google Scholar]
- Elzinga M., Collins J. H., Kuehl W. M., Adelstein R. S. Complete amino-acid sequence of actin of rabbit skeletal muscle. Proc Natl Acad Sci U S A. 1973 Sep;70(9):2687–2691. doi: 10.1073/pnas.70.9.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber H. R., Pettigrew D. W., Remington S. J. Crystallization and preliminary X-ray studies of Escherichia coli glycerol kinase. J Mol Biol. 1989 Jun 5;207(3):637–639. doi: 10.1016/0022-2836(89)90473-7. [DOI] [PubMed] [Google Scholar]
- Flaherty K. M., DeLuca-Flaherty C., McKay D. B. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990 Aug 16;346(6285):623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- Hirono M., Endoh H., Okada N., Numata O., Watanabe Y. Tetrahymena actin. Cloning and sequencing of the Tetrahymena actin gene and identification of its gene product. J Mol Biol. 1987 Mar 20;194(2):181–192. doi: 10.1016/0022-2836(87)90367-6. [DOI] [PubMed] [Google Scholar]
- Hirono M., Tanaka R., Watanabe Y. Tetrahymena actin: copolymerization with skeletal muscle actin and interactions with muscle actin-binding proteins. J Biochem. 1990 Jan;107(1):32–36. doi: 10.1093/oxfordjournals.jbchem.a123007. [DOI] [PubMed] [Google Scholar]
- Hitchcock S. E. Actin deoxyroboncuclease I interaction. Depolymerization and nucleotide exchange. J Biol Chem. 1980 Jun 25;255(12):5668–5673. [PubMed] [Google Scholar]
- Hohn T., Hohn B., Engel A., Wurtz M., Smith P. R. Isolation and characterization of the host protein groE involved in bacteriophage lambda assembly. J Mol Biol. 1979 Apr 15;129(3):359–373. doi: 10.1016/0022-2836(79)90501-1. [DOI] [PubMed] [Google Scholar]
- Holmes K. C., Popp D., Gebhard W., Kabsch W. Atomic model of the actin filament. Nature. 1990 Sep 6;347(6288):44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Mannherz H. G., Suck D., Pai E. F., Holmes K. C. Atomic structure of the actin:DNase I complex. Nature. 1990 Sep 6;347(6288):37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Korn E. D., Carlier M. F., Pantaloni D. Actin polymerization and ATP hydrolysis. Science. 1987 Oct 30;238(4827):638–644. doi: 10.1126/science.3672117. [DOI] [PubMed] [Google Scholar]
- Lazarides E., Lindberg U. Actin is the naturally occurring inhibitor of deoxyribonuclease I. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4742–4746. doi: 10.1073/pnas.71.12.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan R. A., Whittaker M., Safer D. Molecular structure of F-actin and location of surface binding sites. Nature. 1990 Nov 15;348(6298):217–221. doi: 10.1038/348217a0. [DOI] [PubMed] [Google Scholar]
- Pelham H. R. Speculations on the functions of the major heat shock and glucose-regulated proteins. Cell. 1986 Sep 26;46(7):959–961. doi: 10.1016/0092-8674(86)90693-8. [DOI] [PubMed] [Google Scholar]
- Pettigrew D. W., Ma D. P., Conrad C. A., Johnson J. R. Escherichia coli glycerol kinase. Cloning and sequencing of the glpK gene and the primary structure of the enzyme. J Biol Chem. 1988 Jan 5;263(1):135–139. [PubMed] [Google Scholar]
- Pollard T. D. Actin. Curr Opin Cell Biol. 1990 Feb;2(1):33–40. doi: 10.1016/s0955-0674(05)80028-6. [DOI] [PubMed] [Google Scholar]
- Schlossman D. M., Schmid S. L., Braell W. A., Rothman J. E. An enzyme that removes clathrin coats: purification of an uncoating ATPase. J Cell Biol. 1984 Aug;99(2):723–733. doi: 10.1083/jcb.99.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachelek C., Stachelek J., Swan J., Botstein D., Konigsberg W. Identification, cloning and sequence determination of the genes specifying hexokinase A and B from yeast. Nucleic Acids Res. 1986 Jan 24;14(2):945–963. doi: 10.1093/nar/14.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A., Fletterick R. J., Anderson W. F., Anderson C. M. High resolution x-ray structure of yeast hexokinase, an allosteric protein exhibiting a non-symmetric arrangement of subunits. J Mol Biol. 1976 Jun 14;104(1):197–122. doi: 10.1016/0022-2836(76)90009-7. [DOI] [PubMed] [Google Scholar]
- Vandekerckhove J. Actin-binding proteins. Curr Opin Cell Biol. 1990 Feb;2(1):41–50. doi: 10.1016/s0955-0674(05)80029-8. [DOI] [PubMed] [Google Scholar]