Abstract
Background
The forkhead box F2 gene (FOXF2) located in chromosome 6p25.3 has been shown to play a crucial role in palatal development in mouse and rat models. To date, no evidence of linkage or association has been reported for this gene in humans with oral clefts.
Methods
Allelic transmission disequilibrium tests were used to robustly assess evidence of linkage and association with nonsyndromic cleft lip with or without cleft palate (NSCL/P) for 9 SNPs in and around FOXF2 in both Asian and European trios using PLINK.
Results
Statistically significant evidence of linkage and association was shown for two SNPs (rs1711968, and rs732835) in 216 Asian trios where the empiric P values with permutation tests were 0.0016 and 0.005, respectively. The corresponding estimated odds ratios for carrying the minor allele at these SNPs were 2.05 (95%CI=1.41, 2.98) and 1.77 (95%CI=1.26, 2.49), respectively.
Conclusions
Our results provided statistical evidence of linkage and association between FOXF2 and NSCL/P.
Keywords: FBAT, FOXF2, nonsyndromic cleft lip with or without cleft palate, PLINK, SNP, transmission disequilibrium test
INTRODUCTION
Oral clefts (which include cleft lip, cleft lip and palate, and cleft palate) represent a collection of common birth defects that severely influence quality of life of the affected and their families (Wehby et al., 2010). There is strong evidence for some genetic etiology for oral clefts from many family, twin and population studies (Marazita, 2012), and a large number of candidate genes have been suggested as important in the etiology of these complex and heterogeneous malformations (Jugessur et al., 2009; Dixon et al., 2011). Genome-wide association studies (GWAS) have confirmed at least a dozen genes playing some role in the etiology of oral clefts in recent years (Birnbaum et al., 2009; Beaty et al., 2010; Mangold et al., 2010; Grant et al., 2009), although specific causal variants and biological mechanisms remain unclear.
The human forkhead box F2 gene (FOXF2), located in chromosome 6p25.3, has been shown to play a crucial role in palatal development in mouse models (Wang et al., 2003). However, no evidence of linkage or association has been reported for this gene in humans including linkage studies of multiplex NSCL/P families recruited from various populations (Marazita et al., 2004 &2009), two candidate gene studies of population-based Scandinavia NSCL/P case-parent trios (Jugessur et al., 2009&2010) and several GWAS (Birnbaum et al., 2009; Beaty et al., 2010; Mangold et al., 2010; Grant et al., 2009).
To clarify the potential role that FOXF2 may play in the etiology of this common and complex disorder in humans, we performed allelic transmission disequilibrium tests (TDT) for NSCL/P with markers in and near FOXF2 gene using both Asian and European case-parent trios. Our results provide statistical evidence of association between markers in FOXF2 and NSCL/P in Asian trios, and provide new insight to the etiology of this birth defect.
MATERIALS AND METHODS
Ethics statement
Institutional review boards (IRB) or ethical committees from the following institutions reviewed and approved the study: KK Women’s and Children’s Hospital in Singapore, National University of Singapore, Chang Gung Memorial Hospital in Taiwan, Yonsei University in South Korea, and Johns Hopkins University in the US. Written informed consent was obtained from adult participants (including biological parents of all probands and probands old enough to give their own consent/assent) and parents or guardians of the minor participants.
Sample description
This study includes 216 Asian NSCL/P probands and their biological parents, recruited from cleft lip and palate treatment centers in Taiwan, Singapore, Korea, and 75 European NSCL/P trios, recruited from Maryland (US) and Singapore (n=1) (Table 1). All probands received clinical genetic assessment by a health professional to check for other birth defects or developmental delays and were diagnosed as having a non-syndromic, isolated oral cleft.
Table 1.
Race and gender for 297 NSCL/P trios
| Race&Site | NSCL/P
|
||
|---|---|---|---|
| Male | Female | Total | |
| Asian | |||
| Korea | 22 | 18 | 40 |
| Singapore | 21 | 9 | 30 |
| Taiwan | 95 | 51 | 146 |
| Subtotal | 138 | 78 | 216 |
|
|
|||
| European | |||
| Maryland | 43 | 30 | 73 |
| Singapore | 1 | 1 | 2 |
| Subtotal | 44 | 31 | 75 |
|
|
|||
| AA | |||
| Maryland | 1 | 2 | 3 |
|
|
|||
| Other | |||
| Singapore | 2 | 1 | 3 |
|
|
|||
| Total | 185 | 112 | 297 |
AA: African American
SNP selection, DNA, and genotyping
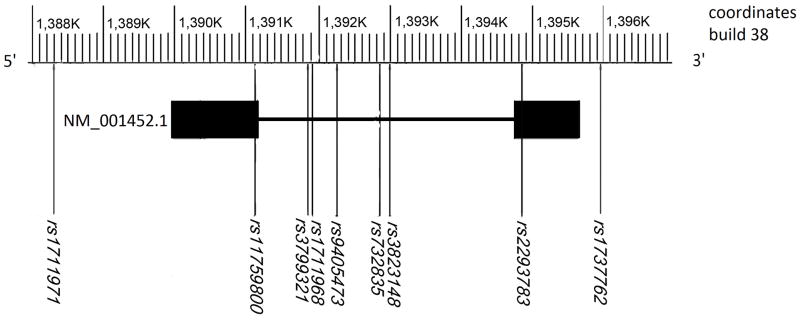
Nine SNPs spanning 7 691 base pairs (bps) in physical distance were genotyped in and near FOXF2 for the current study. SNPs were chosen with a goal of identifying an average of one SNP per 5 kilo bps of physical distance. The physical distance between adjacent genotyped SNPs ranged from 56 to 2909 bps. The target region, represented by the 9 genotyped SNPs, covered the whole gene from its 5′end (rs1711971) to its 3′ end (rs1737762) involving all the two exons (exon 1: rs11759800; exon 2:rs2293783) and the only intron (rs3799321, rs1711968, rs9405473, rs732835, and rs3823148) basing on variant NM_1400521 (Fig. 1, http://www.ncbi.nlm.nih.gov/gene/2295). SNPs with “SNP scores” >0.6, high validation levels in dbSNP, and high heterozygosity levels were given priority in selection. Three (rs1711968, rs732835, and rs2293783) of the four SNPs released by the HapMap project (HapMap Data Rel 24/phaseII Nov08, on NCBI B36 assembly, dbSNP b126) were included in the marker panel for the current study. Among these 9 SNPs, rs1711971 was the only SNP genotyped for both the current study and the published GWAS panel (Birnbaum et al., 2009; Beaty et al., 2010; Mangold et al., 2010; Grant et al., 2009).
Fig. 1.
Schematic genomic structure of FOXF2 and coordinates of 9 SNPs genotyped in this study and SNPs representing those genotyped using Illumina 610-Chips for NSCL/P GWAS. Variant NM_001452.1 of the human FOXF2 gene is shown in the figure with 2 exons aligned from left to right. Filled black boxes represent exons. Scale on top of the figure shows position of the gene and coordinates for the 9 SNPs (black) genotyped in this study and some of the 32 SNPs (gray) genotyped using Illumina 610-Chips in NSCL/P GWAS basing on GRCh build 38. There are 5 SNPs (rs1737753, rs10498654, rs1711969, rs1878475 and rs1711970) genotyped between rs12524544 and rs1711971 at the 5′ of the gene, and there are another 24 SNPs (rs4959557, rs1711973, rs746095, rs731394, rs9501718, rs9328051, rs11753773, rs9378623, rs1711959, rs1711960, rs9392288, rs1555110, rs1737786, rs9502924, rs11242687, rs9405475, rs2493158, rs9502928, rs1922932, rs10458124, rs7774941, rs17202895, rs9328053, rs932410) genotyped between rs6596817 and rs2816251 at the 3′ of the gene in the NSCL/P GWAS panel.
Genomic DNA was extracted from peripheral blood using protein precipitation method and stored at −20°C. Aliquots (4μg) of each genomic DNA sample were dispensed into a bar-coded 96-well microtiter plate at a concentration of 100ng/μl and genotyped by Illumina’s GoldenGate chemistry (Oliphant et al., 2002) at the Genetic Resources Core Facility (GRCF) of Johns Hopkins University. Two duplicates and four controls from the Centre d’Etude du Polymorphisme Humain (CEPH) collection were included in each plate to evaluate genotyping consistency within and between plates.
Statistical analysis
(1) SNP screening and preliminary analysis
We first screened SNPs for minor allele frequency (MAF) and Hardy-Weinberg equilibrium (HWE) using parents’ genotypes, for Asian and European trios, respectively. SNPs with MAF>1% and showing adherence to HWE (P>0.01) were eligible for association analysis. This screening procedure resulted in 6 and 5 eligible SNPs in Asian and European NSCL/P trios, respectively. Further, one additional SNP was excluded due to very low number of informative families in European trios. As a result, six SNPs in Asian and four SNPs in European trios were finally used in association analysis. SNP screening analysis and pairwise linkage disequilibrium (LD, measured as r2) estimation for eligible SNPs were all carried out using Haploview (v4.2, http://www.broadinstitute.org/haploview/haploview) (Barrett et al., 2005).
In addition, data from the 1000 genome project (Genomes Project Consortium, 2010) for Asian samples (Han Chinese in Beijing, CHB; Han Chinese South, CHS; Japanese individuals, JPT; as well as the combined Asian sample, Asian), and the CEPH collection were used to estimate LD patterns among SNPs genotyped for the current study and those genotyped in the GWAS panel using Haploview.
(2) Association analysis
We performed an allelic TDT analysis, originally proposed by Spielman et al., (1993) on each eligible marker using PLINK (v1.07; http://pngu.mgh.harvard.edu/purcell/plink/) (Purcell et al., 2007) and used Family Based Association Test (FBAT; http://www.biostat.harvard.edu/~fbat/fbat.htm) to calculate the number of informative families (Laird et al., 2006). In addition to the Bonferroni correction to each P-value generated from allelic TDT analysis, an overall empiric P value for each SNP was also generated from 10,000 permutations in Haploview to correct for multiple comparisons.
RESULTS
Preliminary analysis
Three and four monomorphic SNPs among the 9 genotyped SNPs in or near FOXF2 for the current study were dropped from association analysis in the Asian NSCL/P trios (rs1711971, rs1737762 and rs11759800) and in the European NSCL/P trios (rs11759800, rs3799321, rs9405473 and rs3823148), respectively. The number of informative European trios (n=3) for rs1711971 was too low for meaningful association analysis, and was excluded as well. The remaining 6 SNPs in Asian parents and 4 SNPs in European parents were all compatible with HWE (at p>0.01). Genotyping call rates in Asian and European founders were no lower than 98.3% and 97.8%, respectively (Table 2).
Table 2.
MAF for SNPs in and near FOXF2 gene in Asian and European NSCL/P trios
| SNPs | Position build 38 | Asian
|
European
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| HW pval | % Geno | MAF (%) | Minor allele | HW pval | % Geno | MAF (%) | Minor allele | ||
| rs1711971 | 1388187 | 1.00 | 100.0 | 0.0 | T | 1.00 | 99.3 | 2.6 | T |
| rs11759800 | 1391096 | 1.00 | 100.0 | 0.0 | G | 1.00 | 100.0 | 0.0 | G |
| rs3799321 | 1391817 | 0.99 | 98.3 | 4.1 | C | 1.00 | 74.6 | 0.0 | C |
| rs1711968 | 1391873 | 0.12 | 99.8 | 22.4 | C | 0.24 | 98.6 | 45.6 | C |
| rs9405473 | 1392224 | 1.00 | 100.0 | 2.9 | T | 1.00 | 100.0 | 0.0 | T |
| rs732835 | 1392765 | 0.40 | 99.8 | 26.0 | C | 0.05 | 97.8 | 20.7 | C |
| rs3823148 | 1392958 | 1.00 | 99.8 | 2.6 | G | 1.00 | 99.3 | 0.0 | G |
| rs2293783 | 1394808 | 0.62 | 99.8 | 23.8 | C | 0.36 | 100.0 | 22.1 | C |
| rs1737762 | 1395878 | 1.00 | 99.3 | 0.0 | A | 1.00 | 97.8 | 25.2 | A |
Among the Asian founders, SNPs rs1711968, rs9405473 and rs732835 formed the only linkage disequilibrium (LD) block where rs1711968 and rs732835 were in strong LD with r2= 0.72. Among European founders, the only LD block was formed by SNPs rs732835, rs2293783 and rs1737762 where rs1711968 was independent and r2 between rs1711968 and rs732835 was 0.31 (Fig. 2).
Fig. 2.
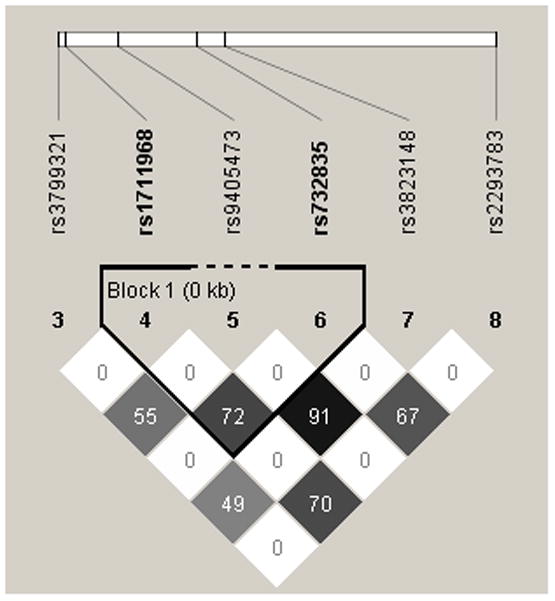
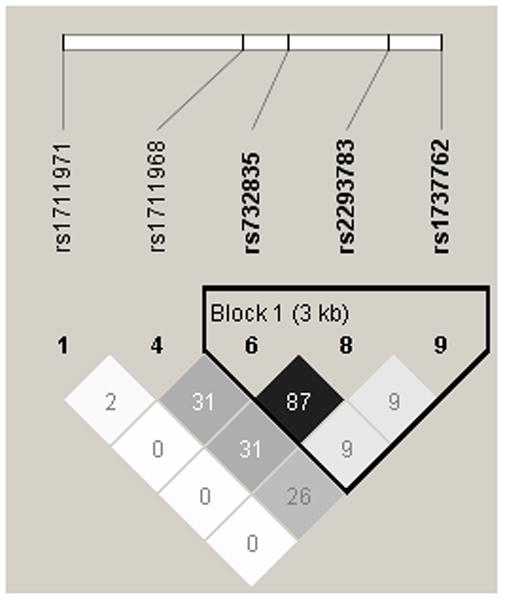
The LD patterns in founders for Asian (left) and European (right) NSCL/P probands. Numbers shown on the figure represent r2 between SNPs.
No evidence of strong LD was shown for SNPs eligible for the current study with SNPs included in the GWAS panel (r2 ranged from 0 to 34) using data from 1000 genomes for CHB, CHS, JPT, and the combined Asian group (Fig. S1).
Association analysis
Statistically significant linkage and association was shown for three SNPs (rs1711968, rs732835 and rs2293783) in the NSCL/P Asian trios by allelic TDT analysis for each marker individually using PLINK. The corresponding Bonferroni corrected P-values were 0.00072, 0.0043 and 0.032. The overall empiric P values for these three SNPs over 10,000 permutations (using Haploview) were 0.0016, 0.005 and 0.054, respectively. The corresponding estimated odds ratios (OR) for carrying the minor allele for rs1711968 and rs732835 were 2.05 (1.41, 2.98) and 1.77 (1.26, 2.49), respectively (Table 3). Significance held for rs1711968 and rs732835 for Taiwan trios (Table S1).
Table 3.
TDT analysis for SNPs in and near FOXF2 gene in 216 Asian and 75 European NSCL/P trios
| SNPs | Position build 38 | Minor allele | MAF (%) | FAM | T | NT | OR (95%CI) | P Value | Bonferroni P Value | Empiric P Value |
|---|---|---|---|---|---|---|---|---|---|---|
| Asian | ||||||||||
| rs3799321 | 1391817 | C | 4.1 | 32 | 18 | 16 | 1.13 (0.57, 2.21) | 0.73 | 1 | 1 |
| rs1711968 | 1391873 | C | 22.4 | 101 | 84 | 41 | 2.05 (1.41, 2.98) | 0.00012 | 0.00072 | 0.0016 |
| rs9405473 | 1392224 | T | 2.9 | 21 | 14 | 8 | 1.75 (0.73, 4.17) | 0.2 | 1 | 0.85 |
| rs732835 | 1392765 | C | 26.0 | 119 | 92 | 52 | 1.77 (1.26, 2.49) | 0.00086 | 0.0043 | 0.005 |
| rs3823148 | 1392958 | G | 2.6 | 19 | 13 | 7 | 1.86 (0.74, 4.66) | 0.18 | 1 | 0.75 |
| rs2293783 | 1394808 | C | 23.8 | 114 | 85 | 53 | 1.60 (1.14, 2.26) | 0.00645 | 0.032 | 0.054 |
|
| ||||||||||
| European | ||||||||||
| rs1711968 | 1391873 | C | 45.6 | 32 | 17 | 22 | 0.77 (0.41, 1.46) | 0.42 | 1 | 0.98 |
| rs732835 | 1392765 | C | 20.7 | 25 | 9 | 20 | 0.45 (0.20, 0.99) | 0.041 | 0.16 | 0.063 |
| rs2293783 | 1394808 | C | 22.1 | 27 | 10 | 23 | 0.43 (0.21, 0.91) | 0.024 | 0.1 | 0.093 |
| rs1737762 | 1395878 | A | 25.2 | 27 | 20 | 14 | 1.43 (0.72, 2.83) | 0.3 | 1 | 0.95 |
FAM: number of informative families, T: number of transmitted alleles, NT: number of un-transmitted alleles; Empiric P Value: P values for 10,000 permutations in Haploview
Among the European trios, nominally significant evidence in the allelic TDT analysis was seen for two adjacent SNPs (rs732835 and rs2293783) which were in strong LD with one another (r 2 = 0.87). However, after Bonferroni correction they were no longer significant (Table 3).
DISCUSSION
The FOXF2 gene is one of the important candidate genes for oral clefts. It has 1 intron and 2 exons and encodes a transcription factor protein which belongs to a large family of evolutionarily conserved DNA-binding proteins (http://genome.ucsc.edu/cgi-bin) (Clark et al., 1993). It was found to be expressed in developing palate, tongue, oral cavity as well as other organs by in situ hybridization in mouse (Wang et al., 2003; Aitola et al., 2000) and rat (Aitola et al., 2000). The embryonic expression levels in FOXF2-deficient homozygous mutant mouse were the same in the wild-type mouse for several other candidate genes for oral clefts including MSX1, MSX2, TBX1, FOXC2 (MFH1), and FOXF1. All the 34 examined FOXF2-deficient mice were born with cleft palate without any combination of other craniofacial malformations (Wang et al., 2003). In addition, heterozygous DNA sequence variations in FOXF2 were identified in 2 of the 18 patients with both disorder of sex development and cleft palate but not in any of the 20 normal controls (Jochumsen et al., 2008).
In the current analysis, statistically significant evidence of linkage and association was shown for NSCL/P with two adjacent intronic SNPs (rs1711968 and rs732835) in FOXF2 in Asian case-parent trios after correction for multiple comparison. These statistical results are consistent with the findings in animal models and provide further support for FOXF2 as a potential susceptibility gene for NSCL/P in humans. The failure to replicate these significant findings in our European trios may be explained by etiologic heterogeneity, the small sample size available for analysis or missing of important SNPs in the marker panel.
In analyses of Scandinavian nationwide population-based NSCL/P and NSCP case-parent trios, none of the three tested SNPs (rs1711970, rs732835, rs2293783) in or near FOXF2 in that study showed evidence of linkage or association with either NSCL/P or NSCP (Jugessur et al., 2009&2010). However, our significant findings for rs732835 in Asians do not necessarily conflict with this Scandinavian study because different population origins may well explain the inconsistency.
Significant linkage or association identified with rs1711968 for Asian NSCL/P in the current study does not conflict with the published NSCL/P GWAS either (Birnbaum et al., 2009; Beaty et al., 2010; Mangold et al., 2010; Grant et al., 2009). In one of these GWAS for NSCL/P, rs1711971 is the only SNP tested in FOXF2 (Grant et al., 2009). For the other 3 GWAS for NSCL/P, rs1711971 is the only SNP in common (Birnbaum et al., 2009; Beaty et al., 2010; Mangold et al., 2010) in FOXF2 with those in the marker panel for the current study. None of the other 32 SNPs genotyped in the GWAS panel for FOXF2 fell into the 7 691 base pair region where the 9 SNPs tested in this study are located. Also, because neither of the statistically significant SNPs (rs1711968 and rs732835) identified in the current study has been included in the previous NSCL/P GWAS, and they are unlikely in tight LD with SNPs genotyped in the GWAS panel, our study can be seen as an independent analysis although 58.3% (126 trios) of our Asian sample went into one of these GWAS where only one SNP (rs11753773, P=0.01) reached nominal significance of linkage or association with NSCL/P (Beaty et al., 2010). Because rs1711968 is a common SNP and located in the intronic region of the gene, the statistically significant association seen between NSCL/P and rs1711968 in our Asian trios could possibly be explained by LD with an unobserved causal variant.
This study reported the first significant linkage and association between markers in FOXF2 and NSCL/P in humans, building upon previous experimental animal studies. Further exploration is warranted to replicate these findings in other populations.
Supplementary Material
Fig. S1 The LD patterns for CHB, CHS, JPT, combined Asian group, and CEPH using data from1000 genome project involving SNPs in FOXF2 genotyped for the current study and for the NSCL/P GWAS using Illumina 610-Chips. Numbers shown on the figure represent r2 between SNPs.
Acknowledgments
This research was supported by grants from The National Natural Science Foundation of China (No. 81273164, HW), the National Institute of Dental & Cranial Facial Research (No. R21-DE-013707 and R01-DE-014581, TH Beaty), and Fogarty Institution (No. D43-TW006176, EW Jabs). The funders had no role in study design, data collection and data analysis, decision for publication, or preparation of the manuscript. We thank all participants who donated samples for this multi-center study of oral clefts, as well as staff at each participating site and institution.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Conceived and designed the study: HW QC EWJ AFS THB. Performed the experiments: SSC AFS. Analyzed the data: HW QC JBH HS TZ MP BZ. Wrote the paper: LB QC HW TZ EWJ AFS THB JBH MP. Subject recruitment and/or diagnosis: Y-HWC SSC VY EWJ AFS THB JBH.
Contributor Information
Dr. Lingxue Bu, Department of Oral and Maxillofacial Surgery, Affiliated Hospital of Qingdao University, Qingdao, China
Ms. Qianqian Chen, Department of Epidemiology and Biostatistics, School of Public Health, Peking University, Beijing, China. National Center for Public Health Surveillance and Information Services, Chinese Center for Disease Control and Prevention, Beijing, China
Dr. Hong Wang, Department of Epidemiology and Biostatistics, School of Public Health, Peking University; Ministry of Health Key Laboratory of Reproductive Health, Beijing, China
Mr. Tianxiao Zhang, Division of Biology and Biomedical Sciences, Washington University, St. Louis, Missouri, USA
Ms. Jacqueline B. Hetmanski, Department of Epidemiology, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland, USA
Dr. Holger Schwender, Mathematical Institute, Heinrich Heine University Duesseldorf, Duesseldorf, Germany
Ms. Margaret Parker, Department of Epidemiology, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland, USA
Dr. Yah-Huei Wu Chou, Department of Medical Research, Chang Gung Memorial Hospital, Taipei, Taiwan
Dr. Vincent Yeow, Department of Plastic Surgery, K K Women’s and Children’s Hospital, Singapore
Dr. Samuel S. Chong, Department of Pediatrics, Yong Loo Lin School of Medicine, National University of Singapore, Singapore; Khoo Teck Puat – National University Children’s Medical Institute, National University Health System, Singapore
Ms. Bo Zhang, Department of Biomedical Engineering, Xi’an JiaoTong University, Xi’an, China
Dr. Ethylin Wang Jabs, Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York City, New York, USA. Department of Pediatrics, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA
Dr. Alan F. Scott, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA
Dr. Terri H. Beaty, Department of Epidemiology, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland, USA
References
- Aitola M, Carlsson P, Mahlapuu M, et al. Forkhead transcription factor FoxF2 is expressed in mesodermal tissues involved in epithelio-mesenchymal interactions. Dev Dyn. 2000;218:136–149. doi: 10.1002/(SICI)1097-0177(200005)218:1<136::AID-DVDY12>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Beaty TH, Murray JC, Marazita ML, et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet. 2010;42:525–529. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum S, Ludwig KU, Reutter H, et al. Key susceptibility locus for nonsyndrominc cleft lip with or without cleft palate on chromosome 8q24. Nat Genet. 2009;41:473–477. doi: 10.1038/ng.333. [DOI] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, et al. Co-crystal structure of the HNF-3/forkhead DNA-recognition motif resembles histone H5. Nature. 1993;364(6436):412–20. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Dixon MJ, Marazita ML, Beaty TH, et al. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 2011;12:167–178. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SF, Wang K, Zhang H, et al. A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24. J Pediatr. 2009;155:909–913. doi: 10.1016/j.jpeds.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochumsen U, Werner R, Miura N, et al. Mutation analysis of FOXF2 in patients with disorders of sex development (DSD) in combination with cleft palate. Sex Dev. 2008;2(6):302–308. doi: 10.1159/000195679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugessur A, Farlie PG, Kilpatrick N. The genetics of isolated orofacial clefts: from genotypes to subphenotypes. Oral Dis. 2009;15:437–453. doi: 10.1111/j.1601-0825.2009.01577.x. [DOI] [PubMed] [Google Scholar]
- Jugessur A, Shi M, Gjessing HK, et al. Genetic determinants of facial clefting:analysis of 357 candidate genes using two national cleft studies from Scandinavia. PLoS ONE. 2009;4:e5385. doi: 10.1371/journal.pone.0005385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugessur A, Shi M, Gjessing HK, et al. Maternal genes and facial clefts in offspring: a comprehensive search for genetic associations in two population-based cleft studies from Scandinavia. PLoS ONE. 2010;5:e11493. doi: 10.1371/journal.pone.0011493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird NM, Lange C. Family-based designs in the age of large-scale gene-association studies. Nat Rev Genet. 2006;7:385–394. doi: 10.1038/nrg1839. [DOI] [PubMed] [Google Scholar]
- Mangold E, Ludwig KU, Birnbaum S, et al. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat Genet. 2010;42:24–26. doi: 10.1038/ng.506. [DOI] [PubMed] [Google Scholar]
- Marazita ML, Murray JC, Lidral AC, et al. Meta-analysis of 13 genome scans reveals multiple cleft lip/palate genes with novel loci on 9q21 and 2q32-35. Am J Hum Genet. 2004;75:161–173. doi: 10.1086/422475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita ML, Lidral AC, Murray JC, et al. Genome scan, fine-mapping, and candidate gene analysis of non-syndromic cleft lip with or without cleft palate reveals genotype-specific differences in linkage and association results. Hum Hered. 2009;68:151–170. doi: 10.1159/000224636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita ML. The evolution of human genetic studies of cleft lip and cleft palate. Annu Rev Genomics Hum Genet. 2012;13:263–283. doi: 10.1146/annurev-genom-090711-163729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant A, Barker DL, Stuenlpnagel JR, et al. BeadArray™ Technology: Enabling an accurate, cost efficient approach to high-throughput genotyping. Biotechniques. 2002;32:S56–S61. [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewenst WJ. Transmission test for linkage disequilibrium: The insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- Wang T, Tamakoshi T, Uezato T, et al. Forkhead transcription factor Foxf2 (LUN)-deficient mice exhibit abnormal development of secondary palate. Dev Biol. 2003;259(1):83–94. doi: 10.1016/s0012-1606(03)00176-3. [DOI] [PubMed] [Google Scholar]
- Wehby GL, Cassell CH. The impact of orofacial clefts on quality of life and healthcare use and costs. Oral Dis. 2010;16:3–10. doi: 10.1111/j.1601-0825.2009.01588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 The LD patterns for CHB, CHS, JPT, combined Asian group, and CEPH using data from1000 genome project involving SNPs in FOXF2 genotyped for the current study and for the NSCL/P GWAS using Illumina 610-Chips. Numbers shown on the figure represent r2 between SNPs.