Abstract
Since the introduction of penicillin into the clinic in 1942, antibiotics have saved the lives of millions of people around the world. While penicillin and other traditional broad spectrum antibiotics were effective as monotherapies, the inexorable spread of antibiotic resistance has made alternative therapeutic approaches necessary. Compound combinations are increasingly seen as attractive options. Such combinations may include: lethal compounds; synthetically lethal compounds; or administering a lethal compound with a nonlethal compound that targets a virulence factor or a resistance factor. Regardless of the therapeutic strategy, high throughput screening is a key approach to discover potential leads. Unfortunately, the discovery of biologically active compounds that inhibit a desired pathway can be a very slow process, and an inordinate amount of time is often spent following up on compounds that do not have the desired biological activity. Here we describe a pathway-directed high throughput screening paradigm that combines the advantages of target-based and whole cell screens while minimizing the disadvantages. By exploiting this paradigm, it is possible to rapidly identify biologically active compounds that inhibit a pathway of interest. We describe some previous successful applications of this paradigm and report the discovery of a new class of D-alanylation inhibitors that may be useful as components of compound combinations to treat methicillin-resistant Staphylococcus aureus (MRSA).
Keywords: Teichoic acid, High throughput screen, Transposon sequencing, D-alanylation, Pathway-directed drug discovery
Graphical abstract
1. Introduction
The incidence of antibiotic-resistant infections is rising worldwide and these infections are increasingly difficult to treat. In the USA alone, antibiotic-resistant bacteria cause at least two million infections and 23,000 deaths annually, and nearly half of those deaths are due to MRSA.1 The burden of drug resistant infections on healthcare systems is extremely costly and despite the effort of many academic and industrial teams, antibiotic discovery has not kept pace with the rise in antibiotic resistance. The paucity of new antibiotics has been the subject of much debate and scrutiny over the years, with the lack of success in bringing compounds to market attributed to: poor quality compounds in screening libraries; poor financial incentives; unreasonable regulatory barriers; and the changing landscape of resistant microorganisms.2 It is clear that solutions to the antibiotic resistance crisis must come from multiple sources and directions at once. In this paper we address one aspect of the problem: improving the efficiency of bioactive compound discovery.
For the past two decades, high throughput screening has served as the most common approach to identify antibacterial compounds for further development, whether for use alone or in combination with other compounds.3,4 High throughput screening approaches have generally been classified into two categories: target-based screens, in which an enzyme is screened in vitro for direct binding and inhibition, and whole cell screens, in which growth inhibition is the usual readout (Table 1). In a much-discussed paper from 2007, Pompliano and coworkers described the results of 67 high throughput screening campaigns carried out over a period of seven years at GlaxoSmithKline against a wide range of antibacterial targets.5 Only 16 of those screens, each involving approximately 250,000–500,000 compounds, resulted in hits, defined as chemically tractable, low-micromolar inhibitors of a given target, and only five of those hits progressed to leads, defined as compounds with biological activity and some evidence for target engagement. As the paper made abundantly clear, target-based screening is problematic because the likelihood that a hit can be developed into a useful lead is low. While improving the quality of compounds in a library may partially address this problem, the screening process is inherently inefficient. Additionally, target-based screens can only be applied to well-behaved targets, which excludes most membrane proteins and overlooks the possibility that the best-behaved targets for an in vitro screen may not be the most druggable targets in a given pathway. Whole cell screens have a major advantage over target-based screens because biological activity is guaranteed and bacterial growth/inhibition assays are simple to implement. However, target identification is more difficult and it can also be difficult to prioritize hits for follow-up. Because nuisance compounds with non-specific activities may represent a large fraction of the hits and can be difficult to recognize, considerable time and effort may be spent sorting through the hits to identify the more promising compounds.
Table 1.
Pros and cons of different screening approaches. Pathway-directed whole cell screens attempt to merge the advantages of target-based and whole cell screens while circumventing major disadvantages.
| Target-based | Cell-based | Pathway-directed whole cell | |
|---|---|---|---|
| Pros |
|
|
|
| Cons |
|
|
|
To improve the efficiency of high throughput screening for discovery of biologically active compounds, the field has turned to screening strategies that combine the advantages of target-based and whole cell screens while minimizing the disadvantages. There are different ways to accomplish this. One way is through target depletion. For example, Merck developed an antisense platform to reduce expression of 245 essential genes in Staphylococcus aureus.6 Antisense strains were pooled based on growth rates and then the pools were screened against compound libraries to identify agents that resulted in depletion of particular antisense strains from the pools. The pathway targeted by a given compound could be deduced from the strains that were most sensitive to it. This strategy not only guarantees the discovery of biologically active compounds, but increases the likelihood that hits will have a desirable mechanism of action.7 We developed an alternative approach to accomplish the same goal, which involves screening a chemical library against a wildtype and a mutant bacterial strain to identify compounds that differentially affect growth of one of the strains.8,9 This approach can be used to discover compounds that inhibit essential targets as well as compounds that inhibit non-essential targets involved in antibiotic resistance or virulence. Below we describe the application of this approach to discover compounds that inhibit cell envelope targets in Staphylococcus aureus. Using the same wildtype/mutant strain pair, we have identified multiple biologically active scaffolds for each of two different targets. In a testament to the efficiency of the approach, we report here the discovery of a new class of teichoic acid D-alanylation inhibitors based on following up only two hits from a screen of 230,000 small molecules.
2. Teichoic acids in Staphylococcus aureus as antibacterial targets
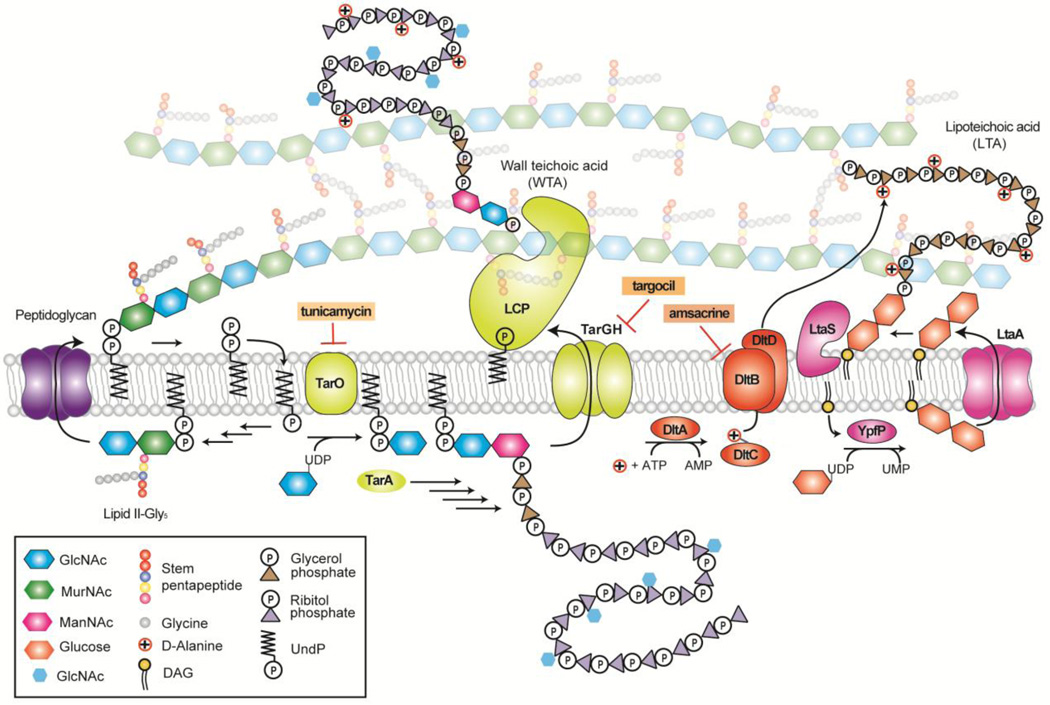
Teichoic acids are anionic polymers that are major constituents of the S. aureus cell envelope.10–13 There are two types: lipoteichoic acids, which are embedded in the cell membrane, and wall teichoic acids, which are covalently attached to peptidoglycan (Fig 1). Both types of teichoic acids play important roles in cell growth and division and are required for survival in a host, making them targets for antibacterials.14 Lipoteichoic acids are composed of a poly(glycerol phosphate) chain attached to a diglucosyl-diacylglycerol anchor.15,16 LTAs continue to be produced when synthesis or export of diglucosyl-DAG is prevented, but strain growth is compromised and polymer length is altered.17 Wall teichoic acids are composed of a disaccharide sugar linked through the reducing end to PG and through the non-reducing end to a poly(ribitol-phosphate) chain.16,18,19 Both lipo-and wall teichoic acids are functionalized with D-alanine esters; wall teichoic acids are also heavily glycosylated with N-acetyl-D-glucosamine. 10,20,21 D-alanine ester levels are regulated by at least one multicomponent sensory system, the GraRS/VraFG system, and increase under various stress conditions.22–24 The D-alanine esters on lipoteichoic acids are installed by the four protein Dlt pathway (DltABCD) and are then transferred to wall teichoic acids in a process that remains unknown.25 Strains in which Dlt pathway genes have been removed are highly susceptible to host immune defenses and are also sensitive to cationic antibiotics such as aminoglycosides.26–29 Therefore, compounds that inhibit teichoic acid D-alanylation may be useful as potentiators of aminoglycosides, which have dose-limiting toxicities, and may also attenuate S. aureus virulence.
Figure 1.
Schematic of important cell envelope biosynthetic pathways in Staphylococcus aureus. The S. aureus cell wall is composed of thick layers of peptidoglycan containing covalently bound wall teichoic acids (WTA). S. aureus also contains membrane-bound lipoteichoic acids (LTA). LTA and WTA are modified with D-alanine esters installed by DltABCD. In the schematic, selected WTA enzymes are shown in yellow, Dlt pathway enzymes are shown in orange, and LTA pathway enzymes are shown in pink. The targets of selected inhibitors mentioned in the text are indicated. Figure adapted from Rajagopal and Walker (2016)30
3. Exploiting suppression of growth inhibitory activity to target wall teichoic acid biosynthesis
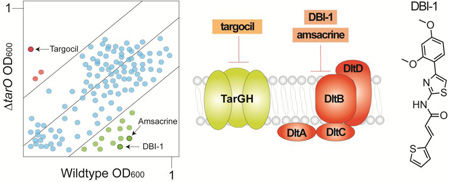
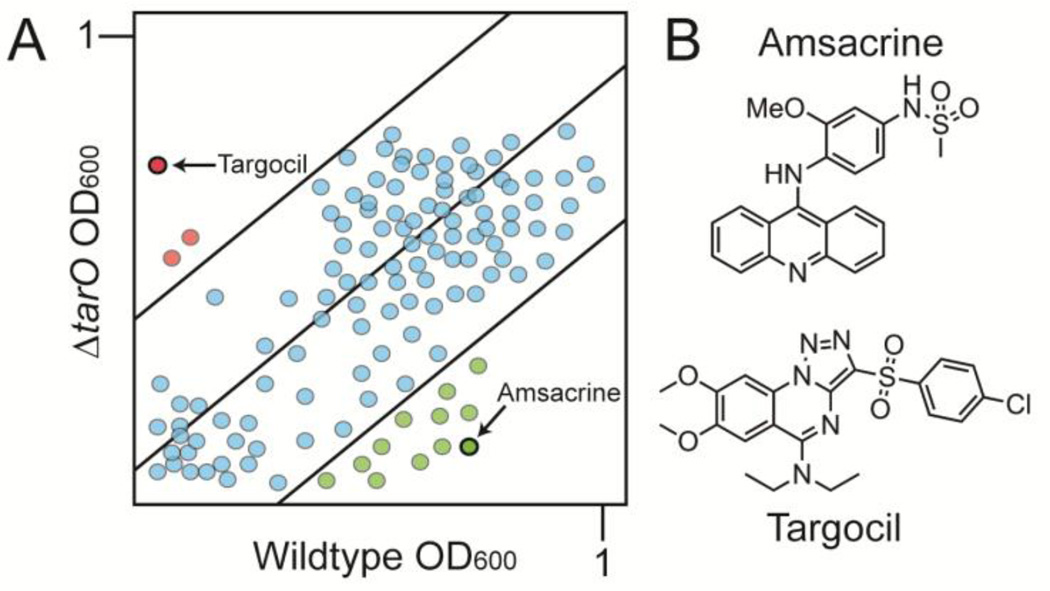
Although WTAs are not essential for survival in vitro, genes that act late in the pathway cannot be deleted unless flux into the pathway is prevented.31,32 This behavior is due to the fact that blocking a late step in WTA biosynthesis depletes Lipid II, the peptidoglycan precursor, which is synthesized on the same undecaprenyl phosphate carrier lipid as the WTA precursor.33,34 Therefore, it is possible to identify compounds that inhibit a late step in WTA biosynthesis by monitoring growth of a wildtype S. aureus strain and a ΔtarO mutant in which the first gene in the pathway has been deleted. From a screen of ~55,000 compounds, we identified three compounds that inhibited growth of the wildtype strain but not the mutant (Fig 2a, red hits).8 We raised resistant mutants and performed targeted sequencing of genes in the WTA pathway based on the expectation that the screen was pathway specific. Only two types of mutations were found: null mutations in tarO or tarA, the first two genes in the WTA pathway, and missense mutations in tarG, which encodes the transmembrane component of the two component ABC transporter that exports WTA precursors from the cytoplasmic surface to the extracellular surface of the membrane.8 Replacing wildtype tarG with the mutant alleles conferred resistance to the compound, establishing TarG as the target. Compound potency was improved ten-fold through medicinal chemistry to produce targocil, which has been used as a probe in a number of studies.35–39 Other compounds that target TarGH have been identified by Merck using a similar approach and three compounds, including targocil, have shown some efficacy in combination with a beta lactam in a MRSA infection model.40 Although none of the compounds identified has appropriate pharmacokinetic properties for clinical use, the success of this pathway-directed whole cell screen demonstrated that suppression of compound lethality is a useful screening phenotype. As long as a genetic suppressor of a lethal block in a pathway can be identified, a similar screening approach can be adapted to identify inhibitors of other essential targets.
Figure 2.
Biologically active compounds with activity against preselected pathways can be identified by screening for compounds that differentially inhibit growth of different bacterial strains. (A) Schematic of a plot depicting growth of library compounds against wildtype S. aureus and ΔtarO. Hit compounds are depicted in green or red depending on whether they are lethal to the the ΔtarO or wildtype strain, respectfully. The former are possible late stage WTA inhibitors and the latter inhibit a pathway that becomes essential when WTA biosynthesis is prevented. (B) Structures of two compounds previously identified using the pathway-directed whole cell screening approach depicted in 2A. Amsacrine inhibits DltB, which is required to install D-alanine esters on LTA (see Figure 1). Targocil inhibits TarG, which transports WTA precursors to the cell surface for attachment to peptidoglycan.
It is important to note that suppression-based screens are not limited to essential targets. In a recent successful inversion of a pathway-directed screening approach to identify WTA inhibitors, Roemer and coworkers at Merck discovered compounds that suppressed growth inhibition caused by a TarG inhibitor.41 They identified a promising TarO inhibitor, which is potentially useful for two reasons. First, S. aureus that does not produce WTAs cannot survive in a host; second, MRSA strains lacking WTAs are sensitive to beta lactams.20,40,42,43 MRSA strains develop beta lactam resistance through the acquisition of the mecA gene, which encodes an intrinsically resistant transpeptidase.44–46 It is thought that WTAs act as scaffolds for peptidoglycan biosynthetic machinery required for properly coordinated function of PBP2a.42,47 A TarO inhibitor may be useful in combination with existing beta lactams to recover the MRSA market.
4. Exploiting synthetic lethality in pathway-directed screening: identifying a DltB inhibitor
While traditional antibiotics target essential pathways, including nucleic acid synthesis, protein synthesis, and cell wall synthesis, non-essential pathways are receiving increasing attention.48,49 Some pathways that are nonessential in vitro are thought to be possible antibacterial targets because they are required for infection or survival in a host; others are known to affect virulence properties or pathogen susceptibility to traditional antibiotics.50 While motivations for pursuing a non-essential target vary, the discovery effort is not any easier. If anything, it is more difficult to identify biologically active inhibitors of non-essential targets because inhibition does not usually lead to a clear phenotype, at least in a wild type background. One approach that has been used to identify anti-virulence agents and immunomodulatory compounds involves screening in animal models (e.g., Caenorhabditis elegans) for compounds that rescue the host from a bacterial infection.51,52 This approach has a crucial advantage in that hits are not only biologically active against an organism of interest, they are also biologically active in the context of a host. Key disadvantages include the fact that throughput is only moderate and follow-up can be difficult. It can be challenging to determine whether biological activity is due to inhibition of a bacterial target or a result of immunomodulation. Hence, although the first commercially used class of antibiotics, the sulfa drugs, was fortuitously discovered by screening compounds in mice, in vivo screening remains challenging.53
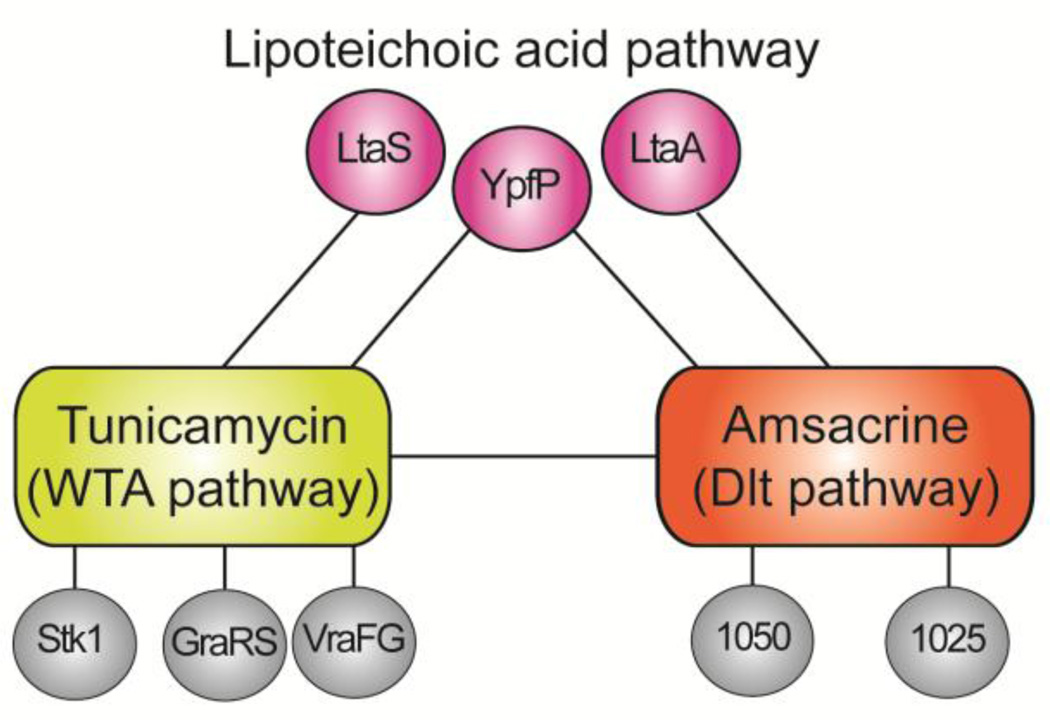
We decided to approach the discovery of biologically active compounds for non-essential cell envelope targets in Staphylococcus aureus using a synthetic lethal screening approach. The approach was grounded in the discovery that the wall teichoic acid pathway is at the center of a dense network of synthetic lethal relationships.54 Pathways connected through synthetic lethality to the WTA pathway include the D-alanylation pathway, the LTA pathway, and the GraRS/VraFG stress response pathway that confers protection to several classes of antibiotics (Fig. 3).55,56 We identified these connections by growing a pooled S. aureus transposon mutant library in the presence and absence of the natural product tunicamycin, a potent and highly selective TarO inhibitor.42,54,57,58 By sequencing the transposon insertion sites, we were able to identify the genes that became essential when the WTA pathway was inhibited because reads mapping to these genes were depleted in tunicamycin-treated samples. After discovering that some of the pathways synthetically lethal with depletion of WTAs included other proposed targets, we realized it should be possible to identify biologically active inhibitors of some of these targets using a pathway-directed whole cell screen.
Figure 3.
Schematic showing selected synthetic lethal interactions between three cell envelope pathways in Staphylococcus aureus. Synthetic lethal interactions were identified by probing a high density transposon mutant library with tunicamycin, which inhibits TarO and prevents WTA synthesis, and amsacrine, which inhibits DltB and prevents D-alanylation of lipoteichoic acids. Inhibiting D-alanylation is lethal to mutants that make abnormal LTA due to deletion of ypfP or ltaA, but not to mutants that make no LTA (ΔltaS strains). Information on synthetic lethal interactions between the Dlt pathway and other pathways enabled design of a strain panel diagnostic for Dlt pathway inhibitors (see Figure 4).
We returned to the same differential growth screen that led to the identification of targocil, but this time with a focus on compounds that killed the ΔtarO strain rather than wildtype (Fig 2b, green dots). In a pilot screen of ~28,000 compounds, we identified twenty possible hits, of which five were confirmed in a cherry pick. The best of these, amsacrine, inhibited growth of ΔtarO and ΔtarA strains at a concentration of five µg/mL.57 We grew our previously prepared transposon mutant library in the presence and absence of amsacrine to identify other susceptible mutants on which to select resistant colonies. Reads mapping to only a handful of genes were depleted from the library. These genes included ypfP (ugtP) and ltaA, encoding components of the lipoteichoic acid biosynthetic pathway, and SAOUHSC_01025 and SAOUHSC_01050, encoding polytopic membrane proteins of unknown function (Fig. 3). These results increased our confidence that the target of amsacrine was in the previously defined WTA synthetic lethal network. To identify the target, we raised resistant mutants in a background containing an inactivating transposon insertion in SAOUHSC_01050 (hereafter designated as tn::1050) and sequenced seven mutants from independent cultures that contained putative target mutations. The resistance mutations mapped to DltB, a polytopic membrane protein that is a core component of the D-alanylation machinery. Genetic and biochemical studies confirmed DltB as the target of amsacrine.57
5. Exploiting synthetic lethality in pathway-directed screening: rapid identification of a new D-alanylation inhibitor scaffold
The discovery of amsacrine provided proof of concept for our strategy to identify biologically active inhibitors of non-essential targets by exploiting synthetic lethality. Gratifyingly, DltB was in the WTA synthetic lethal network and it was a target of interest due to its role in virulence and resistance to cationic antimicrobial peptides and aminoglycosides.10,27,57 Amsacrine has already proven useful as a probe to map synthetic interactions with the Dlt pathway, but it cannot be used to validate the Dlt pathway as a pharmacological target in animals because it acts as a eukaryotic topoisomerase poison.59 Poor pharmacological properties and/or unacceptable safety profiles are commonly encountered barriers to development. For this reason, it is desirable, if not imperative, to have multiple scaffolds against a particular target in order to move forward if animal studies are a goal. The ability to identify new scaffolds rapidly is crucial. Because the pilot screen of 28,000 compounds had been successful and the screening approach offered the possibility of identifying inhibitors to several different targets, we screened an additional 230,000 compounds in duplicate at a final concentration of ~15 micromolar and identified 200 synthetic lethal ‘hits.’ The next challenge was to sort the compounds into possible Dlt pathway inhibitors and other inhibitors.
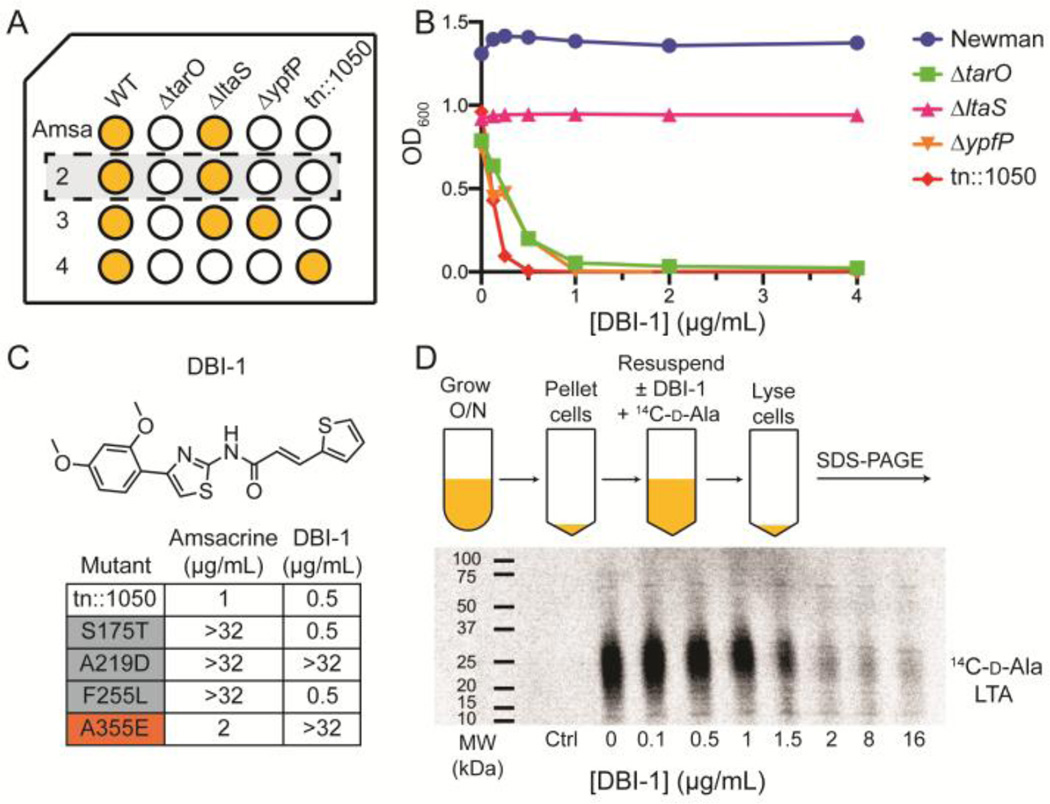
Based on the results obtained through analyzing amsacrine, we organized our cherry pick stage of hit validation to identify putative Dlt pathway inhibitors. Screening at Harvard Medical School is typically carried out at a single concentration, with cherry pick validation performed using only the screening strain(s) in a dose-response format. By testing three additional strains, tn::1050, ΔypfP, and ΔltaS, along with the wildtype and ΔtarO strains, we expected to be able to recognize Dlt pathway inhibitors based on the fact that they would not affect growth of the wildtype of ΔltaS strains, but would inhibit growth of the other three strains (Fig 4a).54,57,60 The ΔltaS strain completely lacks LTAs and tends to be more susceptible than the other strains to a wide range of compounds, but it is not susceptible to Dlt pathway inhibition, which is perhaps not surprising given that there is no LTA to attach them to. We screened the 200 ‘hits’ identified in the primary screen against our five test strains and three compounds representing two different scaffolds emerged as possible Dlt pathway inhibitors. Two compounds, one from each scaffold, were purchased for follow-up, but one of the scaffolds proved toxic to wildtype S. aureus at higher concentrations, suggesting a secondary, lethal target. Therefore, we focused on the other scaffold, DBI-1, which displayed the same susceptibility profile as amsacrine (Fig. 4b, Supplemental Fig. 1).
Figure 4.
A new DltB inhibitor was rapidly identified by testing synthetic lethal hits from a high throughput screen against a strain panel diagnostic for Dlt pathway inhibitors. (A) Schematic of a plate showing possible outcomes of treatment of five different strains against test compounds. Amsacrine, a validated DltB inhibitor, was used as a positive control. It inhibits growth of the ΔtarO, ΔypfP and tn::1050 strains, but not of the wildtype or ΔltaS strains. 200 synthetic lethal hits from a 230,000 compound screen were tested against these five strains and three possible Dlt pathway inhibitors were identified. (B) Plots showing growth as a function of inhibitor concentration for DBI-1, one of the three compounds identified as a Dlt pathway inhibitor, against the test strains. (C) Structure of DBI-1 and minimum inhibitory concentrations against a panel of mutants selected by plating amsacrine (gray boxes) or DBI-1 (red box) on a strain containing an inactivating transposon insertion in SAOUHSC_01050 (tn::1050). The A355E mutant was identified in independent selections on DBI-1. (D) PAGE autoradiograph showing 14C-D-Ala LTA after treatment of cells with increasing concentrations of DBI-1.
To identify the target of the compound, we raised resistant mutants in the susceptible tn::1050 background (Fig. 4c). Two of four colonies selected in independent experiments and identified as possible target mutants displayed a stable resistance phenotype; both of these were found to contain the same mutation in dltB, a C→A transversion that resulted in substitution of glutamate for alanine at amino acid 355. We also tested the compound against amsacrine-resistant mutants containing other amino acid substitutions at three different sites in DltB. One of these mutants was cross-resistant to DBI-1. Finally, we tested DBI-1 in a cell-based assay for inhibition of Dalanylation and found that it blocks incorporation of radiolabeled D-ala into lipoteichoic acid (Fig. 4d). The concentration at which inhibition is observed is similar to the concentration that inhibits growth of susceptible strains. DBI-1 is more potent than amsacrine against ΔtarO and other susceptible strains (1 µg/mL versus 5 µg/mL). It is not yet clear whether this class of compounds can be developed for administration in animals, but our ability to correctly identify a new Dlt pathway inhibitor during the cherry-pick stage of hit validation demonstrates how efficient pathway-directed whole cell screening can be for discovering new scaffolds rapidly.
6. Summary and conclusions
High throughput screening has become the primary way in which new antibacterial compounds and chemical probes are discovered.61,62 Historically, high throughput assays have been divided into two types: targetbased and whole cell. Although target-based assays can work, they are likely best suited for targets that have already been pharmacologically validated through previous discovery of a bioactive natural product or small molecule. For other targets, translating in vitro binding to biological activity may be particularly difficult, and the problem may not only be the properties of the compound, but the target itself. As noted, some targets in a given pathway appear more druggable than others. Relevant to the pathways described here, several late stage wall teichoic acid inhibitors have been identified using cell-based screening approaches, and every one identified so far inhibits TarG, the transmembrane component of the ABC transporter that exports WTA precursors to the cell surface. TarG would likely not have been chosen as a target for an in vitro screen because it is a polytopic membrane protein for which no biochemical assay exists. What makes TarG a more “druggable” target than other proteins in the WTA pathway is unclear, but its membrane location may make it more accessible to compounds than intracellular targets are. It is also possible that TarGH is more sensitive to inhibition because WTA precursor export may be the rate-limiting step in the pathway. In any event, one lesson learned from screening for WTA inhibitors is that success is more likely if a pathway -- rather than a specific target within a pathway -- is targeted. The challenge is to design whole cell screens so that hits are strongly biased towards one, or at most a few, pathways of interest.
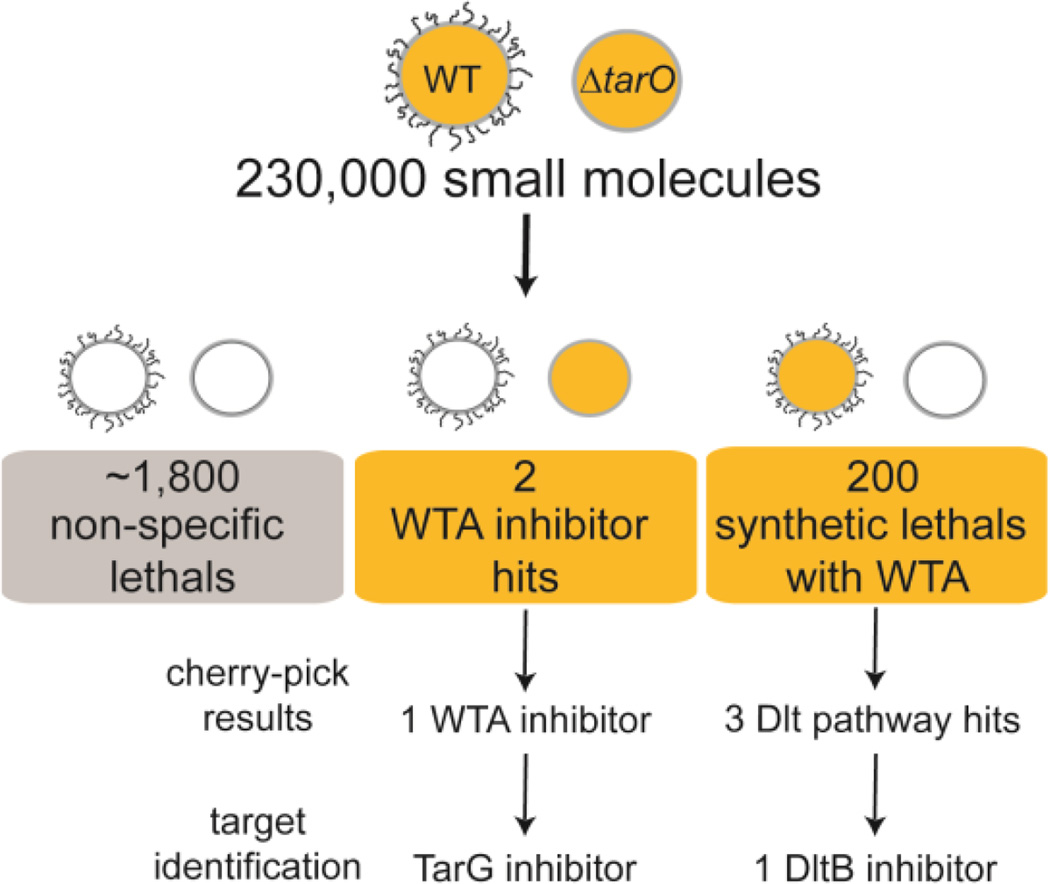
Here we have described two ways in which whole cell screens based on growth inhibition can be designed to identify compounds that inhibit predefined pathways. One is by exploiting suppression of bioactivity and the other is by exploiting synthetic lethality. Both strategies involve screening a wildtype and a mutant strain for growth inhibition. Time spent sorting through bioactive compounds for those with desired mechanisms is minimized by excluding all compounds that inhibit growth of both strains (Figure 5). This is a crucially important advantage of pathway-directed whole cell screens. For a ΔtarO versus wildtype growth inhibition screen, the outcome matrix results in four classes of compounds: 1) non-actives; 2) bioactive compounds having undesired mechanisms or nonspecific toxicity; 3) bioactive compounds that are possible late stage wall teichoic acid inhibitors; and 4) bioactive compounds that inhibit a pathway connected through synthetic lethality to wall teichoic acids. In the 230,000 compound screen reported here, we identified approximately 2,000 compounds with biological activity due to all mechanisms, but only two primary hits were identified as new possible WTA inhibitors; another 200 were identified as inhibitors of pathways connected to WTAs. One of the possible WTA inhibitors has been confirmed and will be described elsewhere. Of the 200 synthetic lethal hits, three were identified as possible Dlt pathway inhibitors through cherry pick testing against a diagnostic panel of strains. One of these has been identified as a DltB inhibitor based on the following lines of evidence: 1) it has the expected growth inhibition profile against the diagnostic panel of strains; 2) we identified two mutants from independent cultures that contained putative target mutations and both were found to have the same mutation in dltB; 3) the compound was cross-resistant to an allele of dltB that was previously found in a selection with the validated DltB inhibitor, amsacrine; and 4) the compound was shown to inhibit D-alanylation of lipoteichoic acids in a cell-based biochemical assay. This new inhibitor scaffold contains a cinnamide substituent, placing it in a large family of compounds containing cinnamic acid derivatives. Cinnamic acid derivatives are produced by many different plants, and various natural products as well as synthetic analogs containing this feature have anticancer,63 antifungal,64 antimalarial,65 and antimicrobial activities.66 DBI-1, however, had no effect on viability of Vero cells (Supplemental Table 2) and did not have antimicrobial activity against wildtype S. aureus. Furthermore, limited SAR studies we have carried out show that the cinnamide moiety is not essential for activity against the tn::1050 strain, which is susceptible to D-alanylation inhibitors (Supplemental Table 3). In contrast, the methoxy-substituted benzene ring and the thiazole are critical for activity. Further investigation of this new DltB inhibitor scaffold is underway.
Figure 5.
Summary of results from a pathway-directed screen of 230,000 small molecules. Approximately 2,000 compounds had some biological activity. The majority of these inhibited growth of both wildtype and ΔtarO S. aureus and were not considered further. We identified two possible WTA inhibitors, of which one has been confirmed as a new TarG inhibitor and will be reported elsewhere. We identified 200 synthetic lethal compounds and designed a cherry-pick screening panel to sort these compounds into Dlt pathway inhibitors and other types of inhibitors. Of three possible Dlt pathway inhibitors, one has been confirmed as a new DltB inhibitor.
Can these screening results – the identification of biologically active compounds with on-target activity in a primary screen or at the cherry-pick stage of hit validation – be duplicated for other cases? We are confident that the answer is yes. Advances in genetic and genomic technologies have made it possible to rapidly characterize genetic suppressors of lethal blocks in cellular pathways as well as synthetic lethal interactions between pathways, which in turn enables the design of simple cell-based screens based on an informed understanding of possible outcomes. Reducing the time spent following up on compounds that cannot be developed as therapeutics or have no applications as biological probes is crucial to leveraging the power of high throughput screening. By integrating a sophisticated understanding of biological pathways with simple screening readouts such as differential growth, we have shown that it is possible to rapidly identify multiple biologically active compounds with cellular activity against desired targets.
7. Materials and methods
7.1 Reagents and general methods
S. aureus was grown in tryptic soy broth (TSB) or on TSB with 1.5% agar at 30°C. High-throughput screening was done at the ICCB-Longwood Screening Facility. DBI-1 and DBI-3 were purchased from Life Chemicals (catalog numbers F0715-0438 and F2553-0110 respectively). Analogs DBI-2, DBI-4, DBI-5, and DBI-6 were purchased from Life Chemicals (catalog numbers Z763933882, Z27852313, Z119631630 and Z27772254 respectively). Amsacrine was purchased from Abcam (ab142742).
7.2 High throughput chemical screening
Overnight cultures of S. aureus Newman WT and ΔtarO were grown in tryptic soy broth (TSB) at 30°C. Before dispensing into 384 well plates, cultures were diluted to an OD600 of one. 30 µL of TSB was predispensed into 384 well plates (Corning 3702) using a Matrix Wellmate plate filler. 100 nL of compound was then pin transferred into lanes 1–22 of the prefilled 384 well plates (final concentration ~15 µM). 50 µL of diluted overnight cultures (diluted 1:625) were added to the wells for a final volume of 80 µL and a final dilution of 1:1000. 10 µg/mL erythromycin final concentration was added to lane 24 as a positive control. Screening was done in duplicate for each strain. Plates were incubated at 30°C for 16–18 hrs and OD600 was measured on a PerkinElmer Envision plate reader.
7.3 Principle component analysis
PCA was done as previously described.56 An X-Y scatter plot of compound data with optical density of each of the strains on each axis. The line of best fit was yielded through the positive and negative controls as the first component and the distance of each point from that line was the second component. The Z score was calculated for each compound by dividing the distance over the standard deviation.
7.4 Cherry-pick strains and procedures
Overnight cultures of S. aureus Newman WT, Newman ΔtarO, SEJ1 ΔltaS, RN4220 ΔypfP and USA300 JE2 tn::1050 were grown in TSB at 30°C. Before dispensing into 384 well plates, cultures were diluted to an OD600 of 1 and then further diluted 1:1000. Compounds provided by the ICCB-L for the cherry pick were administered to 384 well plates (100 nL of 5 mg/mL stock) using the Hewlett Packard D300 Digital Dispenser. Amsacrine was used as a positive control. Then 80 µL of diluted cultures were added to the wells using a Matrix Wellmate plate filler. Compounds were tested in duplicate for each strain. Plates were incubated at 30°C for 16–18 hours and the OD600 was measured on a PerkinElmer Envision plate reader.
7.5 Minimum inhibitory concentration determination
Overnight cultures of wildype and mutant strains were diluted 1:100 and grown until OD600 ~1. Strains were normalized to OD600 of 1, diluted 1:1000 and 147 µL was dispensed into a 96 well plate. All compound dilution series were made in DMSO. 3 µL aliquots were made and transferred into 147 µL for final concentration 0–32 µg/mL in 150 µL. Plates were incubated overnight at 30°C for 16–18 hours and the OD600 was read on a 384SpectramaxPlus plate reader.
7.6 Raising resistant mutants
Raising resistant mutants on DBI-1 (5 µg/mL) and sorting to identify possible target mutants was performed as previously described for amsacrine.56
7.6 14C-D-Ala detection assay
14C-D-Ala incorporation into LTA was performed as described previously.56 Briefly, an overnight of S. aureus Newman was diluted 1:100 and grown at 30°C in TSB until an OD600 of 0.6. Eight 1 mL aliquots were spun down and resuspended in buffer (0.25X TSB, pH 6.0, 200 µg/mL D-cycloserine) plus eight DBI-1 concentrations (0,0.1,0.5,1,1.5,2,8,16 µg/mL DBI-1) for 30 minutes at 30°C shaking at 300 rpm. Samples were then spun down, resuspended in the same buffer containing appropriate DBI-1 concentration plus the addition of 14C-D-Ala and were incubated for another 30 minutes at 30°C shaking at 300 rpm. Samples were then spun down, resuspended in SDS loading buffer, boiled for 5 minutes at 95°C, spun down again and 20 µL of supernatant were added to a 4–20% Trisglycine gel (Bio-Rad). 14C-D-Ala was added to the first lane as a control. The gel was dried and exposed to a phosphor storage screen for 72 hours before imaging.
7.7 NMR of DBI-1
1H NMR was taken on a Varian 400 MHz instrument and recorded at ambient temperature. DBI-1 was prepared in deuterated-DMSO at a concentration of 10 mg/mL. NMR was calibrated to the solvent peak at 2.5 ppm.
Supplementary Material
Acknowledgments
The authors thank the staff at the ICCB-Longwood Screening Facility for compound screening and Christine Anderson in the CETR Discovery and Translational Services Core for toxicity testing. This work was funded by National Institutes of Health grants U19AI109764 and P01AI083214.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Sources
- 1.Centers for Disease Control and Prevention. [accessed May 21];Antibiotic Resistance Threats in the United States. 2013 http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf.
- 2.Walsh CT, Wencewicz TA. J. Antibiot. 2014;67:7. doi: 10.1038/ja.2013.49. [DOI] [PubMed] [Google Scholar]
- 3.Brown ED, Wright GD. Nature. 2016;529:336. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- 4.Brooks BD, Brooks AE. Adv. Drug Delivery Rev. 2014 [Google Scholar]
- 5.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Nat. Rev. Drug Discovery. 2007;6:29. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 6.Donald RGK, Skwish S, Forsyth RA, Anderson JW. Cell Chem. Biol. 2009 doi: 10.1016/j.chembiol.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Soisson SM, Young K, Shoop W, Kodali S. Nat. Lett. 2006 [Google Scholar]
- 8.Swoboda JG, Meredith TC, Campbell J, Brown S, Suzuki T, Bollenbach T, Malhowski AJ, Kishony R, Gilmore MS, Walker S. ACS Chem. Biol. 2009;4:875. doi: 10.1021/cb900151k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen W, Woodward R, Wang PG. ACS Chem. Biol. 2009 doi: 10.1021/cb900259w. [DOI] [PubMed] [Google Scholar]
- 10.Neuhaus FC, Baddiley J. Microbiol. Mol. Biol. Rev. 2003;67:686. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown S, Jr, J P, Walker S. Annu. Rev. Microbiol. 2013;67:313. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Percy MG, Gründling A. Annu. Rev. Microbiol. 2014 doi: 10.1146/annurev-micro-091213-112949. [DOI] [PubMed] [Google Scholar]
- 13.Schneewind O, Missiakas D. J. Bacteriol. 2014;196:1133. doi: 10.1128/JB.01155-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pasquina LW, Maria JP, Walker S. Curr. Opin. Microbiol. 2013;16:531. doi: 10.1016/j.mib.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gründling A, Schneewind O. Proc. Natl. Acad. Sci. U.S.A. 2007;104:8478. doi: 10.1073/pnas.0701821104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baddiley J. Acc. Chem. Res. 1970;3:98. [Google Scholar]
- 17.Grundling A, Schneewind O. J. Bacteriol. 2007;189:2521. doi: 10.1128/JB.01683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown S, Zhang YH, Walker S. Cell Chem. Biol. 2008;15:12. doi: 10.1016/j.chembiol.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meredith TC, Swoboda JG, Walker S. J. Bacteriol. 2008;190:3046. doi: 10.1128/JB.01880-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown S, Xia G, Luhachack LG, Campbell J, Meredith TC, Chen C, Winstel V, Gekeler C, Irazoqui JE, Peschel A, Walker S. Proc. Natl. Acad. Sci. U.S.A. 2012;109:18909. doi: 10.1073/pnas.1209126109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia G, Maier L, Sanchez-Carballo P, Li M, Otto M. J. Biol. Chem. 2010 doi: 10.1074/jbc.M109.096172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herbert S, Bera A, Nerz C, Kraus D, Peschel A. PLoS Pathogens. 2007 doi: 10.1371/journal.ppat.0030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui L, Lian JQ, Neoh H, Reyes E. Antimicrob. Agents Chemother. 2005 doi: 10.1128/AAC.49.8.3404-3413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meehl M, Herbert S, Götz F. Antimicrob. Agents Chemother. 2007 doi: 10.1128/AAC.00209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reichmann NT, Cassona CP, Grundling A. Micriobiol. 2013 doi: 10.1099/mic.0.069898-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaito C, Sekimizu K. J. Bacteriol. 2007;189:2553. doi: 10.1128/JB.01635-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins LV, Kristian SA, Weidenmaier C, Faigle M, Van Kessel KP, Van Strijp JA, Götz F, Neumeister B, Peschel A. J. Infect. Dis. 2002;186:214. doi: 10.1086/341454. [DOI] [PubMed] [Google Scholar]
- 28.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Götz F. J. Biol. Chem. 1999;274:8405. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 29.Peschel A, Vuong C, Otto M, Götz F. Antimicrob. Agents Chemother. 2000;44:2845. doi: 10.1128/aac.44.10.2845-2847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajagopal M, Walker S. Curr. Top. Micriobiol. Immunobiol. 2016 [Google Scholar]
- 31.D'Elia MA, Pereira MP, Chung YS, Zhao W, Chau A, Kenney TJ, Sulavik MC, Black TA, Brown ED. J. Bacteriol. 2006;188:4183. doi: 10.1128/JB.00197-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D'Elia MA, Henderson JA, Beveridge TJ. J. Bacteriol. 2009 doi: 10.1128/JB.00611-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiao Y, Lebar MD, Schirner K, Schaefer K, Tsukamoto H, Kahne D, Walker S. J. Am. Chem. Soc. 2014;136:14678. doi: 10.1021/ja508147s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee W, Schaefer K, Qiao Y, Srisuknimit V. J. Am Chem. Soc. 2015 [Google Scholar]
- 35.Lee K, Campbell J, Swoboda JG, Cuny GD, Walker S. Bioorg. Med. Chem. Lett. 2010;20:1767. doi: 10.1016/j.bmcl.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell J, Singh AK, Swoboda JG, Gilmore MS, Wilkinson BJ, Walker S. Antimicrob. Agents Chemother. 2012;56:1810. doi: 10.1128/AAC.05938-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schirner K, Stone LK, Walker S. ACS Chem. Biol. 2011;6:407. doi: 10.1021/cb100390w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schirner K, Eun Y-J, Dion M, Luo Y, Helmann JD, Garner EC, Walker S. Nat. Chem. Biol. 2015;11:38. doi: 10.1038/nchembio.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farha MA, Czarny TL, Myers CL. Proc. Natl. Acad. Sci. U.S.A. 2015 [Google Scholar]
- 40.Wang H, Gill CJ, Lee SH, Mann P, Zuck P, Meredith TC, Murgolo N, She X, Kales S, Liang L, Liu J, Wu J, Santa Maria J, Su J, Pan J, Hailey J, McGuinness D, Tan CM, Flattery A, Walker S, Black T, Roemer T. Cell Chem. Biol. 2013;20:272. doi: 10.1016/j.chembiol.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee SH, Wang H, Labroli M, Koseoglu S, Zuck P, Mayhood T, Gill C, Mann P, Sher X, Ha S, Yang S-WW, Mandal M, Yang C, Liang L, Tan Z, Tawa P, Hou Y, Kuvelkar R, DeVito K, Wen X, Xiao J, Batchlett M, Balibar CJ, Liu J, Xiao J, Murgolo N, Garlisi CG, Sheth PR, Flattery A, Su J, Tan C, Roemer T. Sci. Transl. Med. 2016:8. doi: 10.1126/scitranslmed.aad7364. [DOI] [PubMed] [Google Scholar]
- 42.Campbell J, Singh AK, Maria JP, Kim Y, Brown S, Swoboda JG, Mylonakis E, Wilkinson BJ, Walker S. ACS Chem. Biol. 2011;6:106. doi: 10.1021/cb100269f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atilano ML, Pereira PM, Yates J, Reed P, Veiga H, Pinho MG, Filipe SRR. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18991. doi: 10.1073/pnas.1004304107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zapun A, Contreras-Martel C, Vernet T. FEMS Microbiol. Rev. 2008;32:361. doi: 10.1111/j.1574-6976.2007.00095.x. [DOI] [PubMed] [Google Scholar]
- 45.Matsuhashi M, Song MD, Ishino F, Wachi M. J. Bacteriol. 1986 [Google Scholar]
- 46.Pinho MG, de Lencastre H, Tomasz A. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10886. doi: 10.1073/pnas.191260798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farha MA, Leung A, Sewell EW, D'Elia MA, Allison SE, Ejim L, Pereira PM, Pinho MG, Wright GD, Brown ED. ACS Chem. Biol. 2013;8:226. doi: 10.1021/cb300413m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clatworthy AE, Pierson E, Hung DT. Nat. Chem. Biol. 2007;3:541. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 49.Smith PA, Romesberg FE. Nat. Chem. Biol. 2007 doi: 10.1038/nchembio.2007.27. [DOI] [PubMed] [Google Scholar]
- 50.Rasko DA, Sperandio V. Nat. Rev. Drug Discovery. 2010 doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 51.Casadei G, Lewis K, Ausubel FM. Proc. Natl. Acad. Sci. U.S.A. 2006 [Google Scholar]
- 52.Irazoqui JE, Urbach JM, Ausubel FM. Nat. Rev. Immunol. 2010 doi: 10.1038/nri2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rubin RP. Pharmacological Reviews. 2007;59:289. doi: 10.1124/pr.107.70102. [DOI] [PubMed] [Google Scholar]
- 54.Santa Maria JP, Sadaka A, Moussa SH, Brown S, Zhang YJ, Rubin EJ, Gilmore MS, Walker S. Proc. Natl. Acad. Sci. U.S.A. 2014;111:12510. doi: 10.1073/pnas.1404099111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Falord M, Mäder U, Hiron A, Debarbouille M. PLoS One. 2011 doi: 10.1371/journal.pone.0021323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Falord M, Karimova G, Hiron A. Antimicrob. Agents Chemother. 2012 doi: 10.1128/AAC.05054-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pasquina L, Jr, J P, Wood MB, Moussa SH, Matano LM, Santiago M, Martin SES, Lee W, Meredith TC, Walker S. Nat. Chem. Biol. 2015 doi: 10.1038/nchembio.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hancock IC, Wiseman G, Baddiley J. FEBS Lett. 1976 doi: 10.1016/0014-5793(76)80657-6. [DOI] [PubMed] [Google Scholar]
- 59.Cain BF, Atwell GJ. European Journal of Cancer. 1974 doi: 10.1016/0014-2964(74)90079-6. [DOI] [PubMed] [Google Scholar]
- 60.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Grundling A. PLoS Pathogens. 2011;7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Macarron R, Banks MN, Bojanic D, Burns DJ, Cirovic DA, Garyantes T, Green DVS, Hertzberg RP, Janzen WP, Paslay JW. Nat. Rev. Drug Discovery. 2011;10:188. doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]
- 62.Castoreno AB, Eggert US. ACS Chem. Biol. 2010 [Google Scholar]
- 63.De PBM, Bedos-Beval F. Curr. Med. Chem. 2011;18:1672. doi: 10.2174/092986711795471347. [DOI] [PubMed] [Google Scholar]
- 64.Tawata S, Taira S, Kobamoto N, Zhu J. Bioscience. 1996 doi: 10.1271/bbb.60.1643. [DOI] [PubMed] [Google Scholar]
- 65.Wiesner J, Mitsch A, Wißner P, Jomaa H, Schlitzer M. Bioorg. Med. Chem. Lett. 2001;11:423. doi: 10.1016/s0960-894x(00)00684-3. [DOI] [PubMed] [Google Scholar]
- 66.Guzman JD. Molecules. 2014;19:19292. doi: 10.3390/molecules191219292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.