Abstract
The nuclear pore complex (NPC) constitutes the sole gateway for bidirectional nucleocytoplasmic transport. Despite half a century of structural characterization, the architecture of the NPC remains unknown. Here, we present the crystal structure of a reconstituted ~400 kDa coat nucleoporin complex (CNC) from S. cerevisiae at a 7.4-Å resolution. The crystal structure revealed a curved Y-shaped architecture and the molecular details of the coat nucleoporin interactions forming the central “triskelion” of the Y. A structural comparison of the yeast CNC with an electron microscopy reconstruction of its human counterpart suggested the evolutionary conservation of the elucidated architecture. Moreover, thirty-two copies of the CNC crystal structure docked readily into a cryoelectron tomographic reconstruction of the fully-assembled human NPC, thereby accounting for ~16 MDa of its mass.
The nuclear pore complex (NPC) is composed of ~34 different proteins, termed nucleoporins (nups), that assemble in numerous copies to yield a ~120 MDa transport channel embedded in the nuclear envelope (NE) (1). To facilitate the extensive membrane curvature generated in each NE pore, NPCs require a membrane-bending coat. The NPC coat is believed to be formed by an evolutionarily conserved coat nup complex (CNC), the Nup107/160 complex in humans and the Nup84 complex in S. cerevisiae, the latter of which is composed of Nup120, Sec13, Nup145C, Seh1, Nup85, Nup84, and Nup133 (1, 2).
We reconstituted a hetero-hexameric CNC containing the yeast nups Nup120, Sec13, Nup145C, Seh1, Nup85, and the Nup84 N-terminal domain (NTD) (Fig. 1, A and B). Our reconstituted CNC did not include Nup133 because this nup is conformationally flexible and loosely associated (2–4). Because the initial crystals of this reconstituted CNC diffracted poorly, we generated a series of conformation-specific, high-affinity synthetic antibodies (sABs) and tested them as crystallization chaperones (5). This approach yielded crystals of the CNC in complex with sAB-57, which allowed us to solve the structure to 7.4 Å by molecular replacement using high-resolution crystal structures of CNC components and the sAB scaffold (fig. S1 and S2) (6–10). The inclusion of a second sAB (sAB-87) produced another crystal form, for which we collected anomalous X-ray diffraction data of Seleno-L-methionine and heavy metal-labeled crystals to confirm the placement of the CNC components (fig. S1, S2 and S3). Because the coat nups in both CNC•sAB complexes adopted the same arrangement, we focused our analysis on the better ordered CNC•sAB-57 structure (fig. S4, S5, and S6).
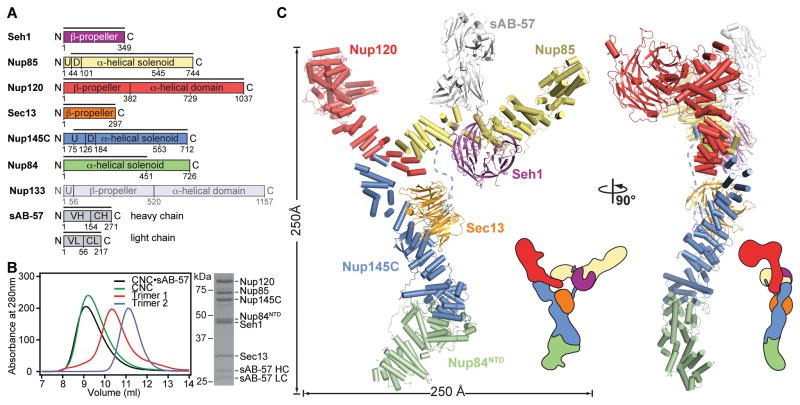
Fig. 1. Overall architecture of the CNC.
(A) Domain structures of the yeast coat nups and sAB-57. Black lines indicate the crystallized fragments. U: unstructured, D: domain invasion motif, VH: heavy chain variable region, CH: heavy chain constant region, VL: light chain variable region, CL: light chain constant region. (B) Reconstitution of the yeast CNC•sAB-57, lacking Nup133. Elution profiles from a Superdex 200 10/300 column are shown for Nup120•Seh1•Nup85 (Trimer 1), Sec13•Nup145C•Nup84NTD (Trimer 2), CNC, and CNC•sAB-57 (left). SDS-PAGE gel of the reconstituted CNC•sAB-57 used for crystallization (right). (C) Cartoon and schematic representations of the yeast CNC•sAB-57 crystal structure viewed from two sides.
The CNC adopted a curved Y-shaped structure spanning ~250 Å in length and width, consistent with previous negative-stain electron microscopy (EM) analyses (Fig. 1C and movie S1) (2–4, 11). The Seh1•Nup85 pair and Nup120 constituted the upper arms of the Y, which were connected to the rest of the CNC through a central triskelion. Sec13•Nup145C•Nup84NTD formed the stalk at the bottom of the triskelion and would attach the tail formed by Nup84CTD and Nup133, which were absent in the structure. Both arms curved out such that the Nup120 β-propeller domain was perpendicular to the plane of the Y. Nup145C organized the CNC through four distinct interaction surfaces contacting nearly every member of the complex. sAB-57 bound at the Nup145C-Nup85 interface and formed crystal packing contacts (Fig. 2 and fig. S4).
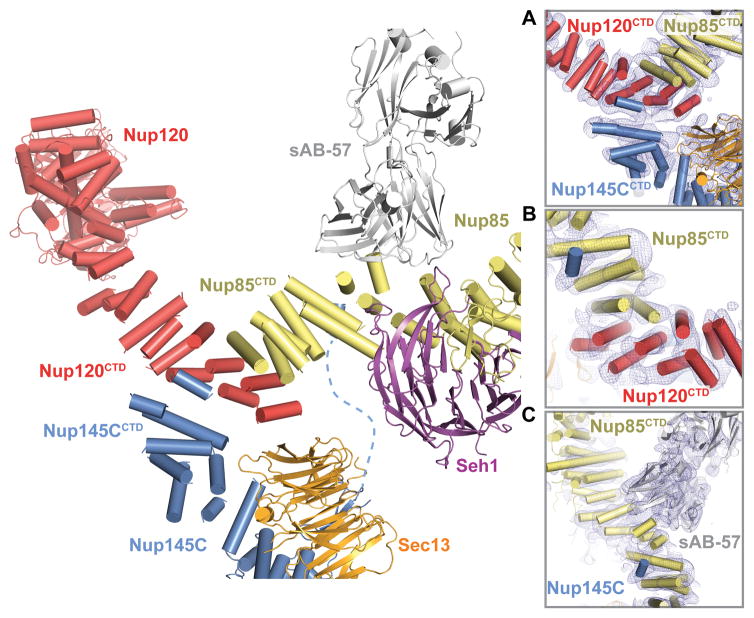
Fig. 2. Architecture of the CNC triskelion.
Cartoon representation of the triskelion formed by Nup120, Nup85 and Nup145C. Insets (A–C) depict magnified views for the interactions between (A) Nup120CTD, Nup85CTD, and Nup145CCTD (B) Nup120CTD, Nup85CTD, and N-terminal Nup145C helix; and (C) Nup145C, Nup85CTD, and sAB-57. The density modified electron density map is contoured at 1.0 σ.
The C-terminal domains (CTDs) of Nup145C (residues 553–712), Nup85 (residues 545–744), and Nup120 (residues 729–1037) converged to form the CNC triskelion. While we observed clear electron density that revealed the connectivity of the three CTDs and their interactions (Fig. 2 and fig. S2), the sequence register in the triskelion was only approximate due to the absence of side chain density. Nup120CTD was sandwiched between Nup85CTD and Nup145CCTD and no direct contacts were observed between Nup85CTD and Nup145CCTD (Fig. 2, A and B). The interactions between Nup85CTD, Nup145CCTD, and Nup120CTD were mediated predominantly by their most C-terminal helices. An additional interaction was made by an N-terminal Nup145C helix bound to a groove in the Nup85CTD surface ~60 Å away from the triskelion center, an interaction that was recognized by sAB-57 (Fig. 2C).
Consistent with our structural data, we reconstituted a stoichiometric complex between Nup120 and Nup85CTD as monitored by size-exclusion chromatography interaction experiments (fig. S7A). Furthermore Nup120 failed to interact with Sec13•Nup145C in the absence of Nup145CCTD (fig. S7, B and C). The interaction between Seh1•Nup85 and Sec13•Nup145C depended on the presence of an N-terminal Nup145C fragment (residues 75–125) (fig. S7, D and E). Further mapping identified a region of Nup145C (residues 75–109) that was sufficient for Nup85CTD binding (fig. S7F), confirming that this fragment bridged the two subcomplexes. Nup120CTD and Nup85CTD were essential for the formation of the CNC (fig. S8 and S9). These data were consistent with published CNC cross-linking data of three different species, more so than the models generated by coarse-grained analysis (3, 11, 12). Lastly, to validate their placement in the structures, we confirmed the interactions of sAB-57 and sAB-87 with Seh1•Nup85•Nup145C1-123 and Nup120NTD, respectively (fig. S10).
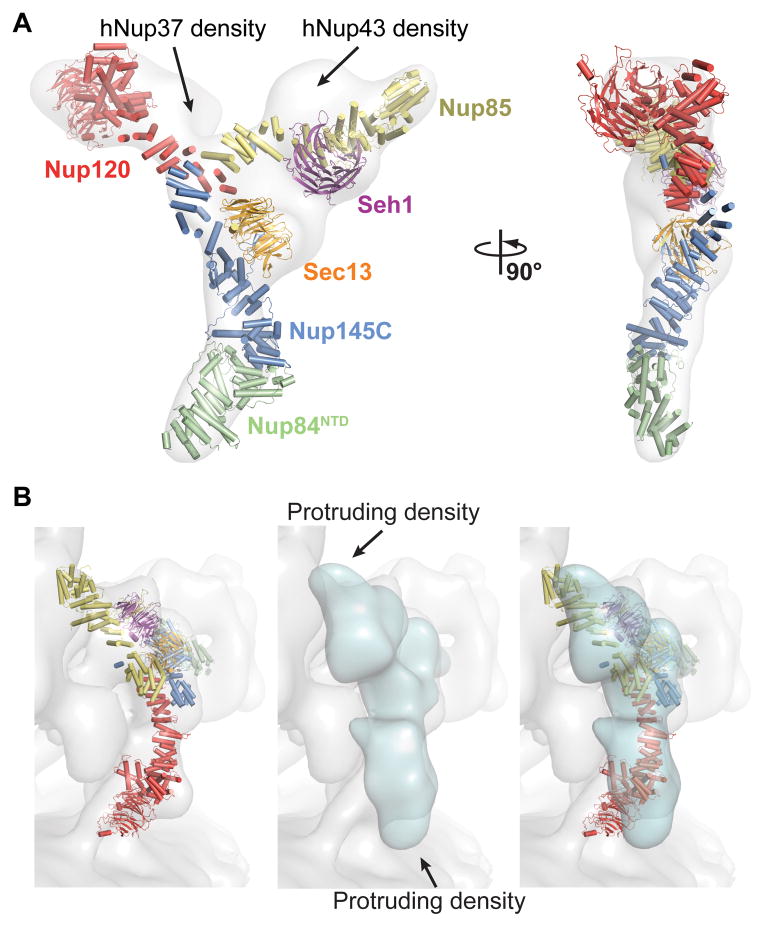
Next, we compared the CNC structure to previously determined EM reconstructions of the yeast and human CNCs (3, 4). While the EM reconstruction of the yeast CNC recapitulates its overall shape, significant deviations were apparent (fig. S11 and movie S2). No density is observed in the EM structure for the U-shaped tip of Nup85. The overall shape of the crystal structure was also consistent with the human CNC EM reconstruction, which contains the crystallized evolutionarily conserved core as well as two additional human components, Nup37 and Nup43 (Fig. 3A and movie S3). Our crystal structure did not account for additional EM density directly adjacent to the Seh1•Nup85 and Nup120 arms, which reportedly accommodate Nup43 and Nup37, respectively (Fig. 3A) (3). In the human CNC, Nup43 appears to bind to the same site as sAB-57 on the Seh1•Nup85 arm of the Y. The major difference between the CNC crystal structure and both EM reconstructions is the curvature of the arms of the Y, and thus the orientation of the Nup120 β-propeller was substantially different in the crystal structure (Fig. 3A). The flatness of both EM reconstructions suggests that these deviations may be a result of EM sample preparation. Despite this, the degree of similarity between the yeast CNC crystal structure and the human CNC EM reconstruction suggested substantial evolutionary conservation of the CNC architecture.
Fig. 3. Comparison of yeast and human CNCs.
(A) Fit of the yeast CNC crystal structure into the human CNC negative-stain EM reconstruction (gray) (3). Arrows indicate density accounted for by the additional human coat nups Nup37 or Nup43. (B) Comparison of the quality of fit for the yeast CNC crystal structure and human CNC EM reconstruction (cyan) into the intact human NPC cryoelectron tomographic reconstruction (gray) (3). Arrows indicate regions where the human CNC EM reconstruction protrudes from the cryoelectron tomographic reconstruction.
The higher-order arrangement of CNCs in a fully assembled NPC has been debated, with several models proposed based on various structural, biophysical, or computational approaches (3, 7, 13, 14). Given the evolutionary conservation of the CNC architecture, we tested whether our crystal structure could be docked into the ~32-Å resolution tomographic reconstruction of an intact human NPC (3). Indeed, an unbiased 6-dimensional search combined with a cross-correlation analysis confidently docked 32 copies of the CNC crystal structure in the tomographic reconstruction, yielding a model for the NPC coat (Figs. 3B, 4A and S12). These results agreed with the stoichiometry and approximate localization previously proposed based on crosslinking mass spectrometry and the docking of the human CNC EM reconstruction (3). However, the crystal structure fit the tomographic reconstruction substantially better than the human CNC EM reconstruction (Fig. 3B).
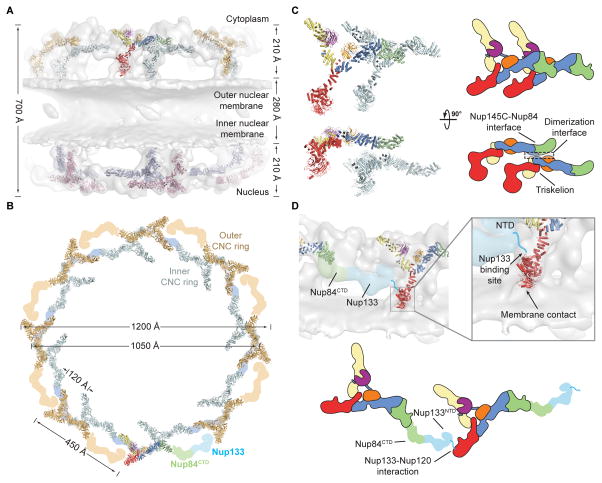
Fig. 4. Architecture of the NPC coat.
(A) 32 copies of the yeast CNC, shown in cartoon representation with a representative subunit colored as in Fig. 1, docked into the cryoelectron tomographic reconstruction of the intact human NPC (3), shown as a gray surface. The outer and inner cytoplasmic and nuclear CNC rings are highlighted in orange, cyan, pink, and blue, respectively. (B) Cartoon representations of 16 yeast CNC copies from the cytoplasmic side of the NPC coat. Schematics indicating the positions assigned to Nup84CTD and Nup133, which were not crystallized, are shown. (C) Interface between the inner and outer CNC rings. Two views of the yeast CNC and its mate from the inner ring are shown. (D) Orientation of the Nup120 β-propeller relative to neighboring coat nups and the membrane. Portions of two CNCs from the cytoplasmic outer ring are shown in cartoon representation. Green and cyan shading indicates the positioning of Nup84CTD and Nup133, respectively. The cyan line represents the N-terminal unstructured segment of Nup133 that binds to Nup120 (9). A schematic representation of the ring-forming Nup120-Nup133 interaction is shown below.
The NPC coat was formed by 32 copies of the CNC arranged in four eight-membered rings (Fig. 4, A and B and movie S4). The eight CNCs in each ring were oriented horizontally with their long axis positioned parallel to the surface of the NE in a head-to-tail fashion. On each side of the NE a pair of inner and outer CNC rings emerged up to ~210 Å (Fig. 4A). These rings were separated by a ~280 Å gap, yielding a total height of ~700 Å. The diameters of the outer CNC rings were slightly larger than those of the inner CNC rings, spanning ~1,200 Å and ~1,050 Å, respectively (Fig. 4B). While the CNCs in both rings were arranged with the same directionality, each CNC from the outer ring was offset from its mate in the inner ring by ~120 Å in a clockwise direction (Fig. 4B). Moreover, the tandem CNC rings on the nuclear and cytoplasmic side of the NE possessed the same handedness and were related by two-fold rotational symmetry (Fig. 4A and S12D).
The unambiguous placement and orientation of the coat nups and their conserved surfaces allowed for an investigation into the details of their interactions in the assembled NPC coat. Each CNC was situated on top of the NE and was oriented such that the plane of the Y was nearly perpendicular to the membrane, with the Nup120 and Seh1•Nup85 arms pointed at or away from the membrane, respectively (Fig. 4A). Only two interfaces appeared to be responsible for oligomerization of individual CNCs into the NPC coat. The inner and outer rings only interacted where the top of the triskelion of each inner ring CNC met the bottom of the Nup84-Nup145C interface of each outer ring CNC (Fig. 4C). Nup120 was oriented such that its Nup133 binding site was directly adjacent to the density assigned to the N-terminus of Nup133, consistent with our previous findings that this interaction is responsible for CNC ring formation and critical for NPC assembly (Fig. 4D) (9). This Nup120 orientation also pointed the apex of its β-propeller directly towards the NE, which was the only membrane contact that we observed in our model (Fig. 4D). This region of the Nup120 β-propeller domain also contains a conserved surface patch on its side (9) that may serve as a NE anchor point for the entire NPC coat, either through a direct interaction with the membrane or via a membrane-anchored nup, as previously reported (15).
The NPC coat architecture is dissimilar to other structurally characterized membrane coats. Whereas the latter are generated by homotypic vertex elements (16, 17), the NPC coat is formed by heterotypic interactions of its asymmetric CNC protomers. Given the location of the CNC rings above and below the NE, other nups likely play a role in generating the complex curvature of the NE pores. While the placement of the CNCs in the NPC coat did not directly address the organization of the central transport channel (fig. S13), it accounted for ~16 MDa of the total mass of the NPC, bridged the resolution gap between low-resolution EM analyses and high-resolution crystallographic studies, and suggested the evolutionarily conservation of its architecture.
Supplementary Material
Acknowledgments
We thank C.J. Bley, W.M. Clemons, A.M. Davenport, O. Dreesen, A. Patke, D.C. Rees, S.O. Shan, P. Stravropoulos, and K. Thierbach for critical reading of the manuscript, K. Kato and K. Kato for their contributions at the initial stages of this project, L.N. Collins for technical support, D. King for mass spectrometry analysis, E. Hurt and P. Loppnau for material, S. Koide for providing the phage display library, S. Gräslund for the pSFV4 vector, P. Afonine for advice regarding structure refinement in PHENIX, and J. Kaiser and the scientific staff of SSRL Beamline 12-2 and the APS Beamline GM/CA-CAT for their support with X-ray diffraction measurements. We acknowledge the Gordon and Betty Moore Foundation, the Beckman Institute, and the Sanofi-Aventis Bioengineering Research Program for their support of the Molecular Observatory at the California Institute of Technology. The operations at the SSRL and APS are supported by the Department of Energy and the National Institutes of Health. T.S. was supported by a Postdoctoral Fellowship of the Deutsche Forschungsgemeinschaft. D.H.L. was supported by a National Institutes of Health Research Service Award (5 T32 GM07616). A.A.K. was supported by National Institutes of Health Awards (U01 GM094588, U54 GM087519) and the Chicago Biomedical Consortium. A.H. was supported by Caltech startup funds, the Albert Wyrick V Scholar Award of the V Foundation for Cancer Research, the 54th Mallinckrodt Scholar Award of the Edward Mallinckrodt, Jr. Foundation, and a Kimmel Scholar Award of the Sidney Kimmel Foundation for Cancer Research. The coordinates and structure factors have been deposited with the Protein Data Bank with accession codes 4XMM and 4XMN. The authors declare no financial conflicts of interest.
Footnotes
Materials and Methods
REFERENCES AND NOTES
- 1.Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 2.Lutzmann M, Kunze R, Buerer A, Aebi U, Hurt E. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 2002;21:387–397. doi: 10.1093/emboj/21.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bui KH, et al. Integrated structural analysis of the human nuclear pore complex scaffold. Cell. 2013;155:1233–1243. doi: 10.1016/j.cell.2013.10.055. [DOI] [PubMed] [Google Scholar]
- 4.Kampmann M, Blobel G. Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat Struct Mol Biol. 2009;16:782–788. doi: 10.1038/nsmb.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paduch M, et al. Generating conformation-specific synthetic antibodies to trap proteins in selected functional states. Methods. 2013;60:3–14. doi: 10.1016/j.ymeth.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Debler EW, et al. A fence-like coat for the nuclear pore membrane. Mol Cell. 2008;32:815–826. doi: 10.1016/j.molcel.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Hsia KC, Stavropoulos P, Blobel G, Hoelz A. Architecture of a coat for the nuclear pore membrane. Cell. 2007;131:1313–1326. doi: 10.1016/j.cell.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagy V, et al. Structure of a trimeric nucleoporin complex reveals alternate oligomerization states. Proc Natl Acad Sci USA. 2009;106:17693–17698. doi: 10.1073/pnas.0909373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seo HS, et al. Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex. Proc Natl Acad Sci USA. 2009;106:14281–14286. doi: 10.1073/pnas.0907453106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizk SS, et al. Allosteric control of ligand-binding affinity using engineered conformation-specific effector proteins. Nat Struct Mol Biol. 2011;18:437–442. doi: 10.1038/nsmb.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thierbach K, et al. Protein interfaces of the conserved Nup84 complex from Chaetomium thermophilum shown by crosslinking mass spectrometry and electron microscopy. Structure. 2013;21:1672–1682. doi: 10.1016/j.str.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Shi Y, et al. Structural Characterization by Cross-linking Reveals the Detailed Architecture of a Coatomer-related Heptameric Module from the Nuclear Pore Complex. Mol Cell Proteomics. 2014;13:2927–2943. doi: 10.1074/mcp.M114.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alber F, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 14.Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 2008;322:1369–1373. doi: 10.1126/science.1165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchell JM, Mansfeld J, Capitanio J, Kutay U, Wozniak RW. Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane. J Cell Biol. 2010;191:505–521. doi: 10.1083/jcb.201007098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison SC, Kirchhausen T. Structural biology: Conservation in vesicle coats. Nature. 2010;466:1048–1049. doi: 10.1038/4661048a. [DOI] [PubMed] [Google Scholar]
- 17.Debler EW, Hsia KC, Nagy V, Seo HS, Hoelz A. Characterization of the membrane-coating Nup84 complex: paradigm for the nuclear pore complex structure. Nucleus. 2010;1:150–157. doi: 10.4161/nucl.1.2.11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoelz A, Nairn AC, Kuriyan J. Crystal structure of a tetradecameric assembly of the association domain of Ca2+/calmodulin-dependent kinase II. Mol Cell. 2003;11:1241–1251. doi: 10.1016/s1097-2765(03)00171-0. [DOI] [PubMed] [Google Scholar]
- 19.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joosten RP, Long F, Murshudov GN, Perrakis A. The PDB_REDO server for macromolecular structure model optimization. IUCrJ. 2014;1:213–220. doi: 10.1107/S2052252514009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 28.Kleywegt GJ, Jones TA. Template convolution to enhance or detect structural features in macromolecular electron-density maps. Acta Crystallogr D Biol Crystallogr. 1997;53:179–185. doi: 10.1107/S0907444996012279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.