Abstract
Horizontal gene transfer plays a major role in microbial evolution, allowing microbes to acquire new genes and phenotypes. Integrative and conjugative elements (ICEs, a.k.a. conjugative transposons) are modular mobile genetic elements integrated into a host genome and are passively propagated during chromosomal replication and cell division. Induction of ICE gene expression leads to excision, production of the conserved conjugation machinery (a type IV secretion system), and the potential to transfer DNA to appropriate recipients. ICEs typically contain cargo genes that are not usually related to the ICE life cycle and that confer phenotypes to host cells. We summarize the life cycle and discovery of ICEs, some of the regulatory mechanisms, and how the types of cargo have influenced our view of ICEs. We discuss how ICEs can acquire new cargo genes and describe challenges to the field and various perspectives on ICE biology.
Keywords: ICE, conjugation, bacteria, gene transfer, Tn916, ICEBs1
INTRODUCTION
Microbes can acquire (and donate) new genes and phenotypes rapidly by horizontal gene transfer (HGT), the transfer of DNA from one organism to another. There are three well-studied types of HGT in microbes (see sidebar, Types of Horizontal Gene Transfer Used by Bacteria): (a) transformation, the natural ability to take up exogenous DNA from the environment (reviewed in 84); (b) transduction, the transfer of DNA from one cell to another by bacteriophage (140 and references therein); and (c) conjugation, the contact-dependent, unidirectional transfer of DNA from a donor to a recipient via a conjugation (or mating) apparatus expressed in the donor [first described by Lederberg & Tatum (87)]. Transduction and conjugation are both mediated by mobile genetic elements, which can mediate their own transfer from one cell to another. Other types of mobile genetic elements, such as transposons and insertion sequences, are mobile within an organism but not necessarily between organisms (see sidebar, General Types of Mobile Genetic Elements That Can Move Within or Between Cells). A fourth type of HGT, fusion of two cells, and perhaps fusion of cells with DNA-containing vesicles, appears to be less common in prokaryotes than transformation, transduction, and conjugation.
TYPES OF HORIZONTAL GENE TRANSFER USED BY BACTERIA.
Transformation: The direct uptake of DNA from the environment and its incorporation into an organism’s genome.
Transduction: Phage-mediated transfer of DNA from one host to another. The DNA is generally genomic DNA from the original host of the phage that was packaged into the phage head instead of or in addition to phage DNA.
Conjugation: Contact-dependent, unidirectional transfer of DNA from one host to another, mediated by the mating pore of a conjugative element.
Fusion: Acquisition of DNA via fusion with a DNA-containing outer membrane vesicle or with another cell (protoplast fusion).
GENERAL TYPES OF MOBILE GENETIC ELEMENTS THAT CAN MOVE WITHIN OR BETWEEN CELLS.
Individual elements may have characteristics of more than one type of element. Many types of elements are not themselves mobile but can be mobilized by one or another of the mobile elements.
Transposable elements: Includes transposons and insertion sequences with many different subtypes. Capable of moving to different DNA sites with varying degrees of site selectivity. Some excise from the original site and insert into new site (cut and paste), whereas others use replicative mechanisms to create a copy at a new site (reviewed in 41, 132).
Phages/viruses: Mobile between cells. Viral nucleic acid contained in proteinaceous particles. Many kill the host during growth.
Conjugative plasmids: Mobile between cells using conjugation machinery. Require cell-cell contact for transfer. Replicate independently of the host chromosome.
ICEs: Mobile between cells using conjugation machinery. Able to integrate into DNA sites via site-specific recombination. Some are also mobile within cells.
This review focuses on integrative and conjugative elements (ICEs), also called conjugative transposons, which make up a large family of mobile genetic elements. There are two defining features of ICEs: (a) They are found integrated in a host genome; and (b) they encode a functional conjugation system, a type IV secretion system (described below and reviewed in 2, 39, 144), that mediates their transfer to other cells. We limit our scope to ICEs that transfer linear single-stranded DNA (ssDNA) and do not cover the ICEs of actinomycetes (AICEs), which are transferred as double-stranded DNA (dsDNA) by an FtsK-like ATPase (reviewed in 18, 62, 139) and not a type IV secretion system.
There are many excellent reviews of ICEs (e.g., 14, 123, 142, 158) and various functions associated with ICEs and other mobile elements (e.g., 14, 142). Here, we briefly summarize some of what has been previously reviewed and describe some of the key features of the ICE life cycle. We focus on aspects of ICE biology that are emergent and have not been extensively summarized.
Integrative and Conjugative Elements
ICEs and conjugative plasmids are both mobile genetic elements that carry genes that encode the machinery necessary for conjugation. ICEs are typically found integrated in the host chromosome and contain genes needed for integration and excision. They are propagated passively during chromosomal replication, segregation, and cell division. In contrast, conjugative plasmids exist as extrachromosomal elements that replicate separately from the host chromosome. However, several ICEs, and we postulate many more, are capable of autonomous plasmid-like replication (see below), which blurs the line separating these two classes of elements.
ICEs and conjugative plasmids contain genes and sites needed for processing their DNA for transfer. Most of these genes are not expressed when the ICE is integrated in the chromosome. However, under certain conditions, or perhaps spontaneously, expression of the ICE genes needed for excision and conjugation is induced, and the ICE excises from the host chromosome. Cells then have the ability to transfer the ICE (or other DNA) through the ICE-encoded conjugation machinery to an appropriate recipient.
ICEs are typically mosaic and modular, ranging from ~20 kb to >500 kb in size (see sidebar, Diversity of ICEs). They contain functional modules from different sources. Genes of similar function are typically grouped together on the element (80, 106, 111, 122, 123, 143, 157, 158).
DIVERSITY OF ICEs.
Size range: approximately 18 kb (Tn916) to more than 500 kb (ICEMlSymR7A).
Some phenotypes conferred by ICEs: antibiotic resistance(s) (Tn916, SXT, CTnDOT, and many others); heavy metal resistance (R391); carbon-source utilization (ICEclc, bph-sal, CTnScr94, Tn5276); symbiosis (ICEMlSymR7A); pathogenesis (PAPI-1); restriction-modification (ICESt3); bacteriocin synthesis (Tn5276); and biofilm formation (PAPI-1).
Because of the mosaic and modular nature of ICEs, knowledge of other elements greatly informs our views of different aspects of the ICE life cycle. Regulatory mechanisms controlling ICE gene expression can be similar to those of phages, plasmids, and host genes. Mechanisms of integration and excision from the host genome are similar to those of viruses and transposons. Processing of ICE DNA for conjugative transfer is similar to that of conjugative plasmids and analogous to rolling-circle replication of some plasmids and viruses.
The conjugation machinery encoded by ICEs, a type IV secretion system, is homologous to that encoded by conjugative plasmids, and much of what we know about the mechanism of conjugation comes from many beautiful studies of conjugative plasmids, including the F plasmid from Escherichia coli, R plasmids, and the Agrobacterium Ti plasmid (reviewed in 2, 3, 81, 156). In some cases, ICEs and plasmids use conjugation systems to mobilize nonconjugative elements to new hosts. As with some plasmids, conjugation can be regulated by cell-cell signaling. As with plasmids and phages, many ICEs carry cargo genes with functions unrelated to the ICE life cycle. These cargo genes are typically thought to provide some benefit to the host cells (see sidebar, Diversity of ICEs).
Historically, the study of cargo genes and their transfer led to the discovery of ICEs. The phenotypes conferred by some cargo genes, including antibiotic resistance and the ability to metabolize a new carbon source, are readily identifiable and greatly facilitate study of the ICE. Recently, ICEs have been identified by sequence analysis, and in many cases it is not obvious what benefit, if any, these ICEs confer to their hosts. Therefore, our current understanding of ICE-encoded phenotypes is skewed by the selective studies of ICEs that confer specific phenotypes. We speculate that many of the recently identified ICEs confer beneficial phenotypes that are outside the range of those already associated with ICEs.
The biology of ICEs can be viewed from both mechanistic and evolutionary perspectives. We believe that the most interesting aspects of ICE biology are those that are likely to be conserved and are not well understood for other elements. This includes understanding the mechanisms of transfer, the processing and fate of ICEs upon introduction into a transconjugant, and the identity and roles of host genes in ICE biology. In addition, the study of cargo genes and the phenotypes conferred provide insight into the evolutionary aspects of ICEs, their interactions with hosts and other mobile genetic elements, and their roles in enabling organisms to grow in different niches.
The ICE Life Cycle
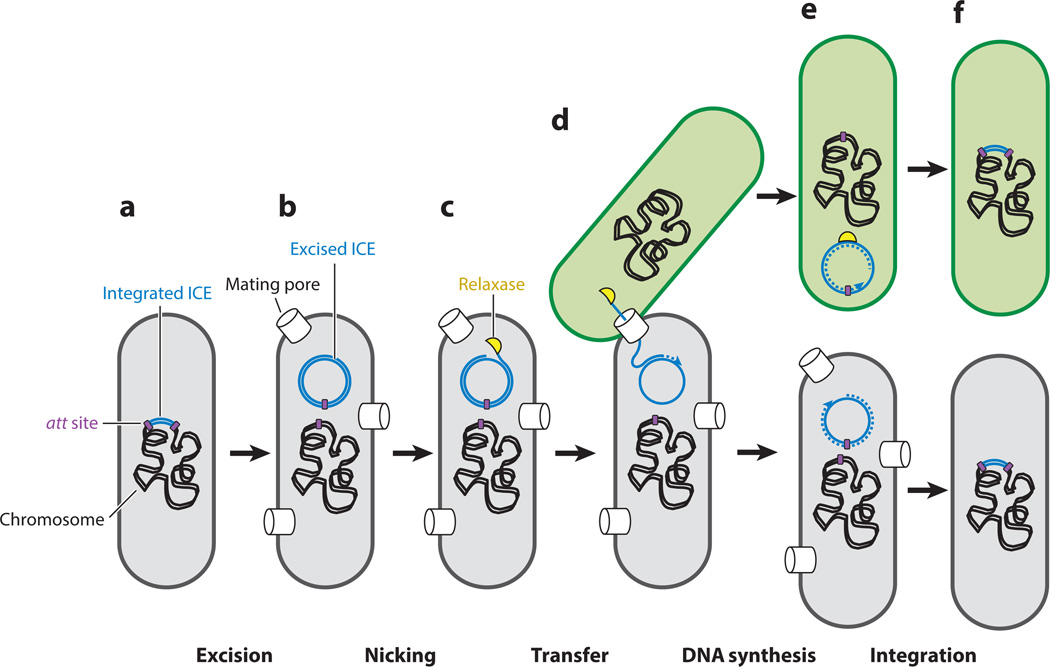
Under normal circumstances, most ICEs are integrated into the host chromosome and conjugation genes are not expressed (Figure 1a). When ICE gene expression is induced, by specific cellular conditions or perhaps stochastically, the ICE excises from the chromosome and forms a circular dsDNA molecule, essentially a plasmid (Figure 1b). Several of the ICE gene products assemble into a mating pore that is capable of transferring the ICE DNA. Other host and ICE-encoded proteins recognize the origin of transfer (oriT) and process the ICE DNA to generate a linear ssDNA-protein complex, referred to as the transfer DNA (T-DNA) (Figure 1c). The mating machinery pumps the T-DNA into the recipient (Figure 1d), where the ICE likely recircularizes, becomes double stranded (Figure 1e), and then recombines into the chromosome using an ICE-encoded recombinase (integrase) (Figure 1f). Because the known recombinases require dsDNA as a template for recombination, it is inferred that the ICE ssDNA must be converted to dsDNA. Many ICEs integrate into a specific chromosomal site, often a tRNA gene. Others are more promiscuous and can integrate into many locations. In all cases, if the ICE is to be maintained in the original donor, it must eventually integrate back into that chromosome (Figure 1f).
Figure 1.
The ICE (integrative and conjugative element) life cycle. A model of ICE conjugation is shown. The bacterium bearing the ICE (the donor) is shown in gray, and the bacterium acquiring the ICE (the recipient) is shown in green. The ICE DNA is shown in blue. (a) The ICE is found integrated into the host chromosome. Most ICE genes are not expressed, because of either repression or lack of activation. (b)When ICE gene expression is induced, the ICE excises from the host chromosome and forms a dsDNA circular plasmid. ICE-encoded proteins are produced, some of which assemble into the mating pore (cylinders spanning the donor cell envelope). (c) The ICE-encoded relaxase nicks one strand of the ICE dsDNA and covalently attaches to the 5′ end of the nicked DNA, forming the transfer DNA (T-DNA). (d) If an appropriate recipient is available, the conjugation machinery transports the T-DNA into the recipient cell. (e) In the recipient cell, the relaxase ligates the 5′ and 3′ ends of the DNA to form a covalently closed ssDNA circle. The complementary DNA strand is synthesized to generate a dsDNA circle that is the substrate for integration into the host chromosome. In the donor, the remaining DNA strand likely serves as the template for rolling-circle replication, generating a dsDNA circle that can then reintegrate into the host chromosome. Without this synthesis and reintegration, the ICE would be lost from cells in which it had excised. (f) In both the donor and recipient, the circular dsDNA ICE integrates into the host chromosome.
CARGO GENES AND THE DISCOVERY OF ICEs
Identification of ICEs That Confer Antibiotic Resistances
The discovery and earliest studies of ICEs resulted from interest in resistances to antibiotics and heavy metals, and how those resistances were spread between organisms. At the time, the spread of many of these types of resistances was known to be mediated by conjugative plasmids that harbored the resistance genes. Work with Enterococcus faecalis (53), Bacteroides species (99, 118), Haemophilus influenzae (125, 137), Streptococcus pneumoniae (129), Proteus rettgeri (109), and Clostridium species (95, 135) identified antibiotic and heavy metal resistance determinants that were transferred via conjugation. Importantly, the resistance genes were later found to be transiently or permanently located on the chromosome and not on a stable plasmid.
Conclusive evidence of a conjugative element that integrated into DNA came from studies of Tn916 in E. faecalis (then called Streptococcus faecalis). It was found that tetracycline resistance could be transferred between strains of E. faecalis via conjugation in the absence of plasmids and that the recipients in these experiments were converted to donors that could then transfer the resistance to another recipient. This work demonstrated that Tn916 encoded genes necessary for its own conjugative transfer and that it could integrate into various sites in a plasmid or the host chromosome (often in multiple copies), much like a transposon. Furthermore, Tn916 functioned in cells defective in homologous recombination, demonstrating that host recombination functions were not needed for integration of the element into other DNA (53, 59). Because of these properties, Tn916 was called a conjugative transposon.
Identification of ICEs That Confer Other Phenotypes
ICEs were also identified based on their ability to enable cells to utilize an alternative carbon source. This trait, like antibiotic resistance, provided a selective phenotype that facilitated study of the ICE. The ability of Pseudomonas sp. strain B13 to degrade chlorocatechols and use them as a carbon source was found to be transferable to other strains via conjugation of the chromosomal element ICEclc (120). Genes that allow the fermentation of sucrose were determined to be on the ICE CTnScr94 in Salmonella (72) and on the ICE Tn5276 in Lactococcus lactis (119). Genes that allow Pseudomonas putida to metabolize biphenyls and salicylate are located on the ICEbph-sal (108).
Some genomic, pathogenicity, and symbiosis islands are also ICEs. These are regions of bacterial genomes that are present or absent in otherwise closely related bacterial strains. For example, the opportunistic pathogen Pseudomonas aeruginosa has an extremely plastic genome, in large part due to pathogenicity islands (68, 98). One of these islands, PAPI-1, is an ICE (33). The genes that allow Mesorhizobium (previously called Rhizobium) species to form nodules on Lotus species are on a symbiosis island, ICEMlSymR7A (117).
The selective advantages and phenotypes conferred to the bacterial hosts by ICEs led directly to the identification of these mobile elements and have been a convenient and powerful means for identifying and tracking ICEs. However, identification of ICEs solely based on the phenotypes conferred by the cargo genes provides a limited view of ICEs. Other means for identifying ICEs provide a more complete view of these elements, their distribution in various organisms, and their potential contributions to microbial phenotypes.
Identification of ICEs Based on Conserved Features
Many ICEs, or putative ICEs, have now been identified using sequence similarities rather than the phenotypes conferred by cargo genes. Bioinformatic approaches have been used to (a) find new ICEs using the presence of conserved conjugation and DNA processing genes; (b) compare closely related ICEs to identify conserved features; and (c) survey a wide variety of genomes for conserved features indicative of conjugative elements.
Functional ICEs have been identified in diverse organisms using bioinformatic approaches. For example, ICEA of Mycoplasma agalactiae (48, 97), ICEPmu1 of Pasteurella multocida (102), and ICE-ßox (also LpPI-1) of Legionella pneumophilia (21, 52) were found by identifying variable regions that contained putative conjugation genes within the chromosomes of closely related bacteria. ICEBs1 of B. subtilis was identified both bioinformatically (25) and by analysis of a peptide signaling system (rapI-phrI) that was found to regulate conjugation (5).
Examination of genome sequences and sequencing of known ICEs has facilitated comparisons between closely related ICEs, such as those of the ICEHin1056 (80) and SXT-R391 families (8, 157). Such studies have helped to distinguish conserved genes that contribute to the ICE life cycle from cargo genes. In addition, they help to reveal the breadth of cargo genes associated with closely related ICEs and to highlight the fact that ICEs carry many of their cargo genes in defined regions (hot spots) that tolerate gene insertion without disrupting ICE function. An online tool, ICEberg (http://db-mml.sjtu.edu.cn/ICEberg), facilitates the identification and comparison of ICEs (16).
Recently, a systematic approach has been applied to the broad-scale identification of candidate conjugative elements, including ICEs, in genomic sequence databases (67). Regions that contained conserved features of conjugative elements, including genes predicted to encode conjugative relaxases, type IV coupling proteins, and ATPases of type IV secretion systems, were identified in genomic sequences. A search of more than 1,000 genomes revealed 335 putative ICEs and 180 putative conjugative plasmids, documenting that ICEs are present in most clades of bacteria and are likely more common than conjugative plasmids (67).
Approaches to Identify Cargo Genes and the Benefits of ICEs to Host Cells
Many of the putative ICEs that have been identified bioinformatically are likely to have cargo genes with functions distinct from those already associated with most well-characterized ICEs. Understanding the function of these cargo genes can reveal important information about the specific ICE, its host, and the environment in which the current and/or previous host normally resides.
A variety of approaches has been used to identify phenotypes conferred by ICEs. In some cases, a phenotype can be inferred using bioinformatic analyses based on the homology of cargo gene(s) to genes of known function and testing for that function (52, 102). High-throughput screening of multiple possible phenotypes (phenotype microarrays) has also been used to identify phenotypes conferred by ICEs (52, 56).Many ICEs carry multiple cargo genes that confer multiple, seemingly unrelated phenotypes to their hosts; thus, the identification of one phenotype conferred by an ICE does not preclude the existence of another.
It is possible that not all ICEs benefit their hosts. Some ICEs have a broad host range, and, as such, an ICE that confers a benefit to one host may not confer a benefit to another. In some cases, particular cargo genes may have mutated and become nonfunctional. Genes associated with mobile genetic elements are more likely to be pseudogenes than are chromosomal genes not associated with mobile genetic elements (92).
Generation of Diversity Among ICEs
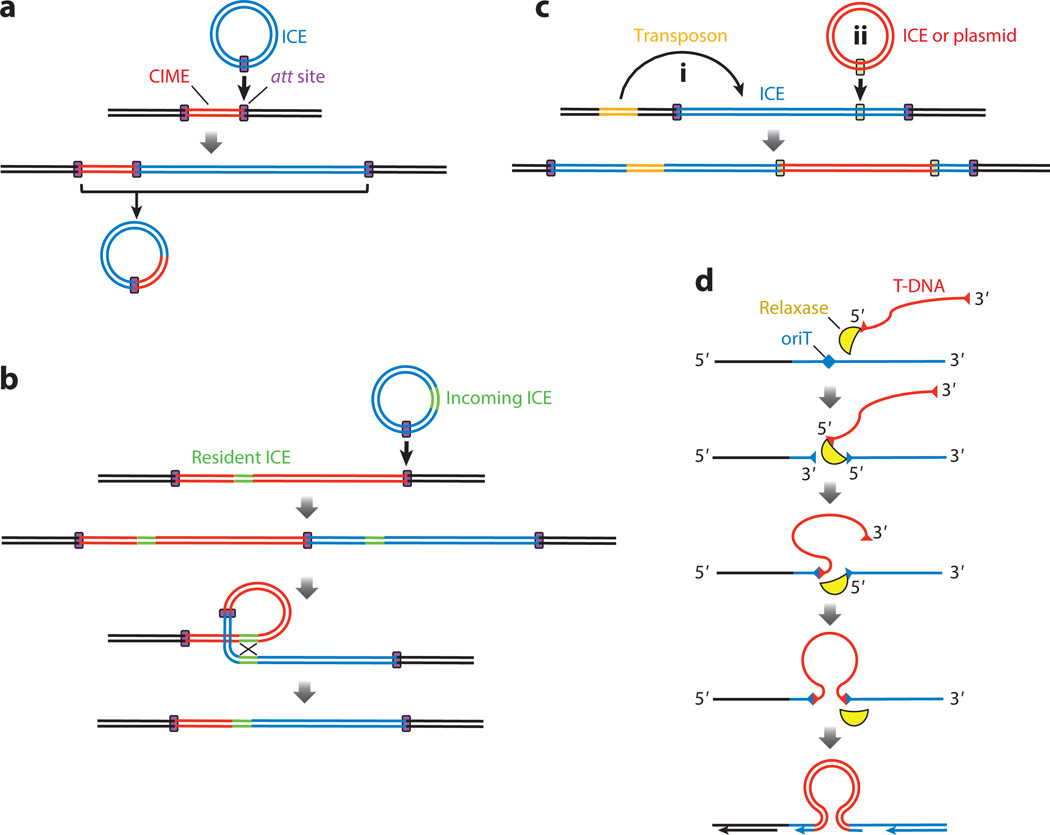
ICEs are largely modular. Genes responsible for related functions, including conjugation, recombination, and regulation, and cargo genes are often grouped together. Conjugation genes are usually the most highly conserved, but the specific gene order can vary between otherwise closely related ICEs because of the insertion of other genes (8, 65, 80, 157). Several mechanisms that generate this diversity have been described (Figure 2).
Figure 2.
Mechanisms that generate diversity between ICEs (integrative and conjugative elements). Double black lines represent the host chromosome and double red, orange, and blue lines represent mobile genetic elements as indicated. Double green lines indicate regions of homology. Rectangles behind double lines indicate att (attachment) sites or insertion sites. (a) An ICE can insert in an att site (purple boxes) next to a heterologous mobile element, such as a CIME (cis-mobilizable element), generating a construct with a total of three att sites. During excision the ICE might utilize the two att sites farthest apart, thereby generating a single dsDNA circle that contains both elements (CIME and ICE) that then serves as the substrate for conjugation. (b) An ICE can insert into an att site next to another ICE that is occupying the preferred att site, forming a tandem insertion. The tandem ICEs can recombine at regions of homology, removing the intervening sequence and forming a single, chimeric ICE. (c,d) Other mobile genetic elements can insert into an ICE. (c) Elements can transpose from another chromosomal location (Ci), or extrachromosomal elements can recombine into an ICE (Cii) by either site-specific recombination or homologous recombination. These elements then become part of the ICE and are transferred with the ICE in cis during conjugation. (d) T-DNA from an external element can recombine into a cognate oriT site on the ICE. Insertion is more efficient when the oriT is located on the lagging strand, likely indicating that the target is ssDNA, as shown. The relaxase is bound to the transferred DNA at one catalytic tyrosine residue, and nicks and binds the oriT of the resident element with a second catalytic tyrosine residue [both catalytic tyrosine residues are required for this activity (1)]. The relaxase also joins the free 3′-OH end of the chromosomal nick to the 5′ end of the T-DNA and the 3′-OH end of the T-DNA to the 5′ end of the chromosomal nick. The insertion is then replicated by the host machinery and becomes established in the chromosome.
Tandem insertion and accretion at the cognate att site
Composite ICEs can be generated by tandem insertion of an ICE alongside an existing mobile genetic element, followed by transfer of both elements. This occurs when the att site normally targeted by an ICE is already occupied by an element and the ICE inserts into one of that element’s flanking att sites (Figure 2a). Cis-mobilizable elements (CIMEs) are nonconjugative elements that occupy an att site used by an ICE (13, 23, 112). When the ICE inserts next to the CIME, the resulting integrant contains three att sites. Use of the two most distal att sites during excision and conjugation results in an element that contains both the CIME and the ICE (13, 112).
An ICE can insert next to another ICE that is occupying its normal att site (Figure 2b), forming a tandem array [e.g., ICEclc (120) and SXT (27, 71)]. These tandem ICEs may be capable of moving together to a new host. However, if the tandem ICEs carry genes with regions of sequence identity, then recombination between the ICEs can occur, removing the intervening sequence and generating a new ICE that inherits sequences from both parents. The SXT-R391 family of ICEs carries homologs of the phage lambda Red genes bet and exo that enhance RecA-dependent recombination between tandem ICEs, promoting genetic exchange within this family of ICEs (58).
Recombination into an existing ICE
ICEs can accrue other mobile genetic elements within the boundaries of their attachment sites and thereby transfer these elements during conjugation (Figure 2c). Several ICEs, including ICEHin1056 (80), SXT-R391 (17, 73), and ICEEc2 (126), carry insertion sequences and/or transposons. Other ICEs, including SXTET (73), ICEEc2 (126), and Tn5253 (6, 77), contain integrons (reviewed in 142). In some cases, a plasmid has integrated, apparently by single crossover, into an ICE, for example in the case of Tn5253 (6, 77).
Relaxase-mediated recombination
Some conjugative elements are able to insert into oriT-like sequences by a relaxase-mediated recombination event (Figure 2d). The relaxase of the conjugative plasmid R388 is able to direct recombination of plasmid T-DNA into a cognate oriT site in a recipient (1, 50). Although this activity is not conserved in all conjugative relaxases (1, 36, 50), we suggest that relaxases from some ICEs or plasmids could mediate this type of integration into a cognate oriT, either in another mobile element or in a host chromosome.
Imprecise excision
In addition to the mechanisms described above, we suspect that imprecise excision of an ICE could bring along flanking genes, analogous to the imprecise excisions that generate lambda-specialized transducing phages (reviewed in 30). Acquisition of flanking sequences could also occur if multiple sequences resembling an attachment site are present beyond the ends of an ICE, for example in the ICE in Bacillus atrophaeus (ICEBat1) (61; J. Jones, C.M. Johnson, A.D. Grossman, unpublished results). Imprecise excision might be more common with ICEs that have promiscuous integration sites (e.g., Tn916) (59) rather than a single preferred site. It might also be more prevalent with ICEs that use DDE recombinases (24) that usually have low sequence specificity (see below).
Summary of Cargo Genes and Our View of ICE Biology
ICEs were initially discovered because of interest in the genes they carry. Thus, it appeared that most ICEs (at least many of those that have been characterized) contain genes that provide obvious phenotypes and benefits to the host, including antibiotic resistances, the ability to metabolize various compounds, and the ability to colonize various hosts (symbiosis, pathogenesis). However, recent bioinformatic analyses have identified ICEs throughout the prokaryotic domain (67), and there are at least a few functional ICEs that provide no obvious (known) benefit to the host. We postulate that there are many ICEs with cargo genes that confer previously uncharacterized phenotypes to the host cells, and that these phenotypes are likely to be advantageous under conditions normally experienced by the host. These phenotypes may be difficult to identify initially but could contribute to interactions between ICEs and the host or between ICEs and other mobile genetic elements of a cognate host. Understanding some of these phenotypes is likely to provide further insight into the continuing evolution of ICEs and microbes as well as how cells inhabit particular niches.
MECHANISMS OF ICE FUNCTION
As described above, the two defining features of ICEs are that they integrate into the host genome and that they encode a functional conjugation system that mediates their intercellular transfer. To integrate into the host chromosome, ICEs employ integrases that are homologous to those encoded by phage and some genomic islands. To mediate conjugation, most ICEs encode a type IV secretion system and DNA processing proteins that are homologous to those used by conjugative plasmids. In addition, like certain plasmids, some ICEs undergo autonomous rolling-circle replication. This is likely to be a critical property of many ICEs that facilitates maintenance of an ICE in a population of cells (89).
Integration and Excision
ICEs integrate into and excise from DNA using an ICE-encoded recombinase. The recombinase, often referred to as an integrase, is needed for both integration and excision. The recombinase is often homologous to phage integrases and, like temperate phages, many ICEs insert at a specific attachment site in the bacterial chromosome (attB). For many ICEs, attB is often in a tRNA gene. The attachment site on an ICE is called attP (based on the phage nomenclature) or attICE. Many ICE integrases are tyrosine recombinases, but some ICEs use serine or DDE recombinases (for example, see 24, 48, 66, 148). Tyrosine and serine recombinases catalyze slightly different biochemical reactions, but both have the same functional consequence for the ICE. These enzymes mediate site-specific recombination between dsDNA molecules at short stretches of similar sequences (reviewed in 63, 115). When these sequences are on a circular ICE and the chromosomal target, recombination results in insertion of the ICE flanked by two att sites (attL and attR, on the left and right sides, respectively). When recombination is between two att sites flanking the ICE, the result is excision of the ICE and reestablishment of the unoccupied chromosomal attB site. Frequently, in addition to the integrase, an ICE-encoded recombination directionality factor (RDF), sometimes called Xis, is required for excision. This factor influences the direction of recombination mediated by the recombinase (integrase) to favor excision (reviewed in 70).
There is a great deal of variation in the specificity of ICEs for a particular integration site. Many ICEs target a single att site in the host chromosome that is similar to the att site in the ICE. If the normal att site is unavailable, some ICEs target alternate sites at lower efficiency (26, 88, 101). Other ICEs have lower specificity for a specific att site. For example, CTnDOT inserts at multiple sites that match a consensus sequence (11, 38), and Tn916 insertion is not site-specific in most organisms (see references in 123).
Recently, ICEs have been identified that integrate into the host chromosome using DDE recombinases. These recombinases are typically associated with transposons, insertion sequences, and phages. DDE recombinases employ a variety of recombination mechanisms and frequently do not target a specific site for integration (reviewed in 69). The ICEs TnGBS1, TnGBS2, and ICEA all encode DDE recombinases (24, 66). None of these ICEs integrate at a specific target site; however, TnGBS2 inserts upstream of promoter sequences and has a preferential insertion site that it uses more frequently (24, 48).
Overview of Conjugation and the Type IV Secretion System
Like conjugative plasmids, ICEs encode conjugation machinery, a type IV secretion system, for the transfer of DNA to another cell. The mechanism by which type IV secretion systems transfer DNA has been extensively studied in Gram-negative bacteria (reviewed in 2, 29, 39).Homologous type IV secretion systems of conjugative elements have been identified inmost bacterial phyla (67), indicating that the general mechanism of conjugation is likely to be widely conserved. Some of the details of the transfer mechanism are likely to vary, as the number and composition of proteins within conjugation systems varies widely. There are also different challenges in transferring across the larger cell wall in Gram-positive bacteria versus the challenges found in transferring across both the inner and outer membranes of Gram-negative bacteria, although it is noteworthy that the minimal conjugation system of Tn916 successfully negotiates both. Extensive descriptions of type IV secretion systems have appeared elsewhere (reviewed in 15, 29, 39, 144, 147).
DNA Processing by a Relaxase
Plasmids and excised ICEs exist as covalently closed circles of dsDNA and must be processed prior to conjugation. The dsDNA circle is converted to linear ssDNA covalently bound to a relaxase protein, which serves as the substrate for conjugative type IV secretion systems. The proteins that perform these functions are homologous for ICEs and plasmids.
During conjugation, an ICE-encoded relaxase recognizes its cognate oriT. The relaxase then nicks one strand of the ICE DNA and is covalently attached to the 5′ terminus to form the TDNA. Host and ICE factors both assist in unwinding the two DNA strands, and an ICE-encoded type IV coupling protein (an ATPase) engages the T-DNA with the type IV secretion system. The type IV secretion system then enables translocation of the T-DNA into the recipient cell, where the DNA is recircularized by the relaxase. The ssDNA is presumably converted to dsDNA, which is eventually integrated into the host chromosome by site-specific recombination. This conversion of circular ssDNA to dsDNA is thought to be the same as second-strand synthesis of rolling-circle replication employed by some plasmids and phages (reviewed in 83).Many conjugative plasmids use separate origins for replication and transfer and distinct proteins for processing each origin (78). In contrast, some ICEs use the same origin and DNA processing proteins both for conjugation and to replicate using a rolling-circle replication mechanism (see below).
Autonomous Replication of Some ICEs
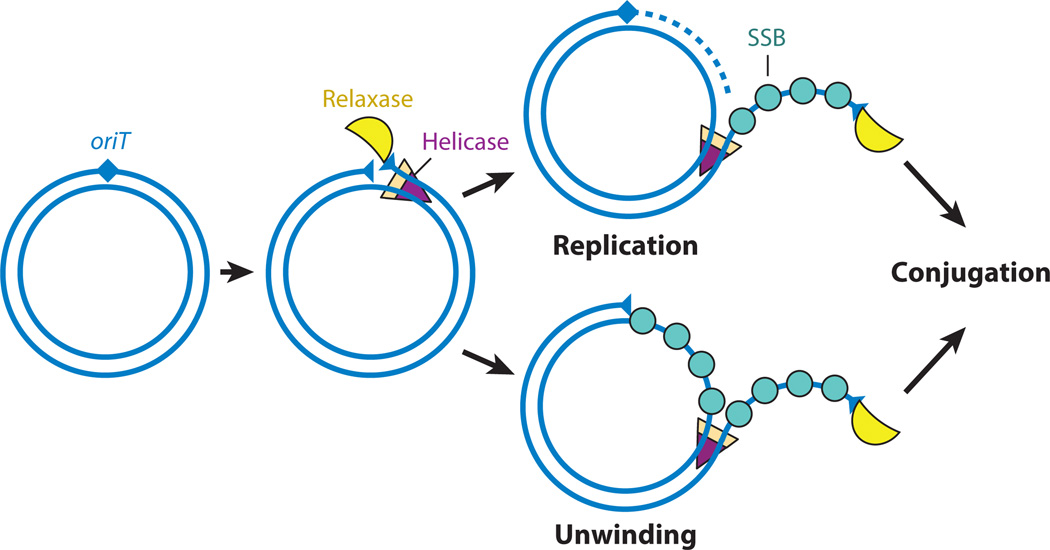
Studies with ICEBs1 provide the most direct evidence that an ICE can replicate autonomously. When ICEBs1 is induced, it excises from the chromosome, and the copy number of all ICE genes, but not adjacent chromosomal genes, increases (89). Replication is unidirectional, starts at oriT, and requires the conjugative relaxase NicK, the ICE-encoded helicase processivity factor HelP, the chromosomally encoded DNA translocase and helicase PcrA, the catalytic subunit of DNA polymerase PolC, and the β-clamp (DnaN) (89, 141). Replacement synthesis of the unwound (leading) strand is presumably primed from the 3′-OH terminus created by the nicking event. On the basis of analogies to other systems, the leading strand is presumed to be recircularized by NicK (reviewed in 83). Synthesis of the complementary, or lagging, strand is primed by a single-strand origin (sso) (160). The nicking and unwinding of DNA for rolling-circle replication are very similar to that needed to transfer ICE ssDNA during conjugation (Figure 3) (reviewed in 37).
Figure 3.
Conjugation resembles rolling-circle replication. Both processes require similar initial steps that generate a substrate that may be suitable for transfer during conjugation. The ICE (integrative and conjugative element) is shown as double blue lines. In both processes, a relaxase recognizes and nicks a cognate ori, binding to the free 5′ end. Helicase activity (purple triangle) and a single-strand binding protein (turquoise circle) are required to unwind the single-stranded DNA. Replacement synthesis of the unwound strand can occur but is not required for unwinding. If replacement synthesis occurs, a second nicking event at the reconstituted ori is likely to be required to generate a free 3′-OH group for recircularization of the unwound strand.
ICEMlSymR7A of Mesorhizobium loti R7A also replicates using its conjugative relaxase. When ICEMlSymR7A is induced by overproduction of the ICE-encoded regulator TraR, the element excises and is maintained in a host population at an average copy number of 1.5. If the conjugative relaxase is deleted, the element is lost from the population (116, 117).
Some of the earliest evidence for autonomous replication of an ICE comes from Haemophilus influenzae. ICEHin1056 and related elements integrate into a host tRNALeu gene (47). These elements were originally described as a family of antibiotic resistance vectors that were integrated into the chromosome of the donor but could be recovered as plasmids from outgrown transconjugants (46, 137). This implies that these elements are maintained as replicative plasmids in the recipient for some generations before integrating into the chromosome.
There are additional elements in which replication can be inferred because the copy number of the circular form of the ICE is greater than the copy number of the chromosomal attachment site. The ICE RD2 of Group A Streptococcus exists as both an integrated and a plasmid form in stationary-phase cultures. Treating the host with the DNA damaging agent mitomycin C increases the copy number of the plasmid form (133). ICESt3 of Streptococcus thermophilus also excises from the host chromosome and exists as a multicopy plasmid following induction with mitomycin C (31). Additionally, SXT of V. cholerae was recently found to undergo autonomous replication (32).
There are also ICEs that replicate via dedicated replication systems, independent of the conjugative relaxase. TnGBS1 and TnGBS2 of Streptococcus agalactiae encode a replication initiation protein and a conjugative relaxase. Loss of the replication initiation protein reduces copy number, whereas loss of the conjugative relaxase does not (24, 65). Additionally, after conjugation, circular forms of TnGBS1 and TnGBS2 can be isolated from outgrown transconjugants. In the majority of cases, the attachment site in the ICE matches that of the donor and does not match a potential integration site in the recipient, indicating that the circular forms result from replication rather than integration and subsequent excision in the recipient (65).
ICEs Can Mobilize Nonconjugative Elements
Nonconjugative mobile genetic elements can use the conjugation machinery encoded by an ICE or a conjugative plasmid to transfer to new hosts, a phenomenon known as mobilization. Mobilizable elements can exist as freely replicating plasmids or as chromosomally integrated genomic islands (mobilizable genomic islands) that excise prior to mobilization.
ICEs can mobilize plasmids and genomic islands. CTnDOT-ERL (128, 146), Tn916 (107), SXT (74), and ICEBs1 (91) have all been shown tomobilize plasmids. Additionally, the CTnDOT-ERL family of ICEs can mobilize genomic islands known as nonreplicating Bacteroides units (NBUs) (130, 136), and the SXT-R391 family of ICEs has recently been shown to mobilize genomic islands of Vibrio species and related organisms (42–44). In both cases, these genomic islands encode their own integrase, but excision is regulated by the mobilizing ICE and transfer depends on the ICE conjugation machinery. Once in a new host, these mobilizable genomic islands are capable of independently integrating into the chromosome.
Plasmid Mobilization by ICEBs1 Blurs the Distinction Between Conjugative and Replicative Relaxases
Relaxases are frequently classified as conjugative or replicative, depending on whether they are required for conjugation or replication. Sequence analysis has revealed that these relaxases are related but appear to be in distinct families (64, 78). However, this distinction has become blurred, as evidenced by the ability of some relaxases from plasmids and ICEs in Firmicutes to function in both replication and conjugation. For example, the replicative relaxase of the staphylococcal plasmid pC194 and the conjugative relaxase of ICEBs1 are both bifunctional and can act as replicative and conjugative relaxases (89, 90). ICEBs1 and Tn916 are both capable of mobilizing pC194 (91, 107). In the case of ICEBs1, this requires the pC194 replication protein, which does not resemble a conjugative relaxase (91, 134). The sequences of the conjugative relaxases of ICEBs1 (NicK) and Tn916 (Orf20) more closely resemble replicative relaxases than other conjugative relaxases (57, 90).The conjugative relaxase of ICEBs1 supports replication of the extrachromosomal form of this ICE, as previously discussed (89). These examples blur the distinction between conjugative and replicative relaxases and raise the possibility that replicative relaxases may also serve as conjugative relaxases when recognized by the correct type IV secretion system. These findings also blur the distinction between ICEs and rolling-circle replicating plasmids.
Lagging-Strand Synthesis
During conjugation, ssDNA is transferred into the recipient. Once there, the ICE must either function as ssDNA or convert to dsDNA. ICEBs1 of Bacillus subtilis possesses an sso, located on the transferred strand, that enables its conversion from ssDNA to dsDNA (160). ssos are DNA sequences that form a particular secondary structure when single stranded. This structure mimics other DNA elements to recruit host factors that synthesize a short RNA to prime DNA synthesis. The sso of ICEBs1 is homologous to the ssos of plasmids of Gram-positive bacteria that replicate by a rolling-circle mechanism. These ssos fold into a structure recognized as a promoter by RNA polymerase, which then synthesizes a leader RNA that is used to prime DNA synthesis (reviewed in 83).Other ssos use the primase DnaG for synthesis of the RNA (85, 154). Once a primer is generated at the sso, the host replication machinery is recruited to complete synthesis of the second strand.
Plasmids are known to prime complementary strand synthesis after conjugation. The rolling-circle replication plasmid pMV158 is mobilized by conjugative elements and contains two ssos that function in different recipient species (93). In addition, several plasmids in Gram-negative bacteria encode primase proteins that directly polymerize RNA synthesis on the T-DNA, either through recognition of a cognate oriV (76) or via a general mechanism that functions on ssDNA (155).
Summary of ICE Replication
We postulate that many (perhaps most or all) ICEs undergo autonomous rolling-circle replication. This view is based on the conserved nature of ICE-encoded relaxases, the known roles of some of these relaxases in conjugation and replication, and the role of replication in the stability of ICEs in a population of cells. We suspect that it has been difficult to detect autonomous replication of most ICEs due to the low frequencies of induction and excision.
REGULATION OF ICE ACTIVATION
Normally, ICEs are maintained as quiescent elements in the host chromosome. The excision and conjugation genes are not expressed, often because of active repression of transcription. Constitutive expression of conjugation genes has been found to be detrimental to the host and to the maintenance of the ICE, providing selective pressure for repression of these genes. Under certain conditions, the ICE can become induced, resulting in de-repression (or activation) of expression of ICE genes, which leads to excision of the ICE and the potential for conjugation. Even under inducing conditions, most ICEs appear to excise from the chromosome of a relatively small subpopulation of cells (96, 103). The signals that induce ICE gene expression and the mechanisms of repression are not universally conserved, but there are some common themes.
Pressures Against ICE Activation
Expression of conjugation genes is likely to be maladaptive under most circumstances for both the host and the ICE. In general, expression of the genes is likely to create a metabolic burden on the host, diverting cellular resources away from essential processes. It has been shown that constitutive activation of SXT—by either deletion of the CI-like repressor SetR or overexpression of the transcriptional activators SetC and SetD—is deleterious to the host (9, 10). This, however, may be due to loss of the element and subsequent activation of a toxin-antitoxin system carried by SXT (159). For its part, an excised ICE can no longer rely on chromosomal replication to ensure vertical inheritance. Even ICEs that replicate are lost from a population if they are constitutively activated (5, 117). In addition to these general considerations, ICE activation can be deleterious to a host for specific reasons. In the case of Gram-negative conjugation systems, expression of a mating pilus may make the host susceptible to male-specific phages. In other cases, induction of the ICE results in host death. Activation of ICEclc causes a portion of the cells with the element to differentiate into potential donors that have reduced growth kinetics and eventually lyse (121). In the absence of its preferred attachment site, ICEBs1 integrates into secondary sites within the host chromosome. If the ICE cannot efficiently excise from these secondary sites, induction of the ICE kills the host in a manner dependent on the activity of the relaxase (101).
ICEs Are Induced by a Variety of Signals
ICEs are induced in response to a wide variety of signals. The inducing signals vary for each ICE, but there are common stimuli known to activate multiple ICEs. These include induction of the cellular response to DNA damage (the recA-dependent SOS response; see below), secreted signaling molecules from potential recipients, the growth phase of the host, and mechanisms tied to selective advantages conferred by the cargo genes in an ICE. Some ICEs are known to respond to more than one cue.
Some of the signals that induce ICEs activate other mobile genetic elements. The SOS response to DNA damage induces many lysogenic phages to enter the lytic cycle (45, 110).Cell-cell signaling regulates conjugation of some plasmids (51, 114).
Induction during the SOS response
DNA damaging agents cause induction of the recAdependent SOS response in host cells and also induce several ICEs (5, 10, 12, 19, 133). During the SOS response, DNA damage generates ssDNA. This is bound by and activates RecA, which causes auto-cleavage of the LexA repressor, the phage lambda repressor CI, and related repressors (reviewed in 28, 45, 110).
SXT, ICEBs1, and ICESt3 all contain genes homologous to phage repressors and these ICEs are induced by the SOS response (4, 5, 10, 12, 26). This induction is known to be RecA-dependent for SXT and ICEBs1. Whereas canonical CI-like repressors (LexA,CI) mediate their own cleavage, the ICEBs1 repressor, ImmR, is cleaved by a separate protease, ImmA (19). immR and immA are linked in ICEBs1 and are immediately upstream of int, the gene for the site-specific recombinase. In addition to ICEBs1, this gene arrangement and protease-mediated cleavage of a repressor appears to be a common property of many phages (19).
Induction of ICEs by the SOS response likely indicates that the host is facing a potentially lethal challenge and that the ICE must rely on horizontal rather than vertical transmission in order to propagate. Alternately, induction via the SOS response may be a way of maintaining a low level of horizontal transmission without incurring an undue metabolic burden on the population as a whole. The SOS response is generally activated in a small subset of growing cells (82, 113), usually in response to replication fork stress (40, 113). Induction of ICE within these cells would ensure that a small portion of an ICE-bearing population is primed to act as donor cells at any given time.
Control by cell-cell signaling
Some ICEs are activated in response to cell-cell signaling (quorum sensing), becoming induced at high population densities. The use of quorum-sensing pathways likely signals to ICE-containing cells the presence of potential mating partners. In some cases, multiple signals are used to indicate the presence of potential partners and whether or not these cells already contain a copy of the cognate ICE, allowing an additional level of control such that conjugation is induced only when it is likely to result in horizontal transmission to a naïve recipient rather than to a cell with an established ICE.
The Mesorhizobium loti symbiosis island ICEMlsymR7A is induced by quorum signals. It specifies the production of at least one and possibly two acyl homoserine lactones (AHLs), as well as a transcription factor, TraR, that drives expression of genes needed for conjugation (116, 117). TraR is activated by several different AHLs, likely enabling this element to respond to a variety of potential recipients.
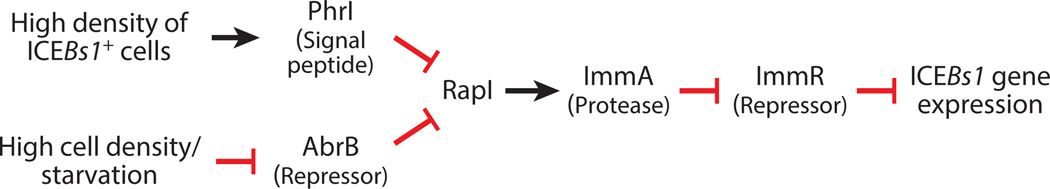
ICEBs1 of B. subtilis is controlled by cell-cell signaling in two ways (Figure 4). First, secreted peptides produced and sensed by B. subtilis strains indicate the presence of neighboring cells and potential mating partners. These peptides stimulate transcription of rapI, leading to activation (derepression) of ICEBs1. RapI activates ICEBs1 gene expression by stimulating the ImmA-mediated proteolysis of the ICEBs1 repressor ImmR (4, 5, 19). Second, ICEBs1-containing cells produce an additional secreted peptide that inhibits activation of ICEBs1 gene expression. This inhibition occurs when the peptide PhrI is imported and inhibits the activity of RapI. The ICEBs1-encoded peptide PhrI signals that the surrounding cells already have a copy of ICEBs1 and serves to limit activation and potential loss of the element. In addition to this peptide-mediated mechanism of limiting activation of ICEBs1, there are other mechanisms for limiting acquisition of ICEBs1 by cells that already have a copy (e.g., 4). The study of quorum sensing and RapI-PhrI led to the initial experimental discovery of ICEBs1 (5).
Figure 4.
Cell-cell signaling induces ICEBs1. The pathway by which cell-cell signaling regulates ICEBs1 gene expression is shown. Arrows indicate positive regulatory effects. Lines with cross bars indicate negative regulatory effects. Proteins and peptides are shown with a brief explanation of their role or activity. The PhrI signaling peptide is likely either a pentapeptide (5) or hexapeptide (104) with sequence (A)DRVGA.
Induction during stationary phase
Several ICEs are induced during the stationary phase. In ICEclc, expression of genes that regulate conjugation is driven by the host-encoded stationary phase sigma factor RpoS (105). ICEMlSymR7A preferentially excises during the stationary phase, likely in response to the growth phase of the host in a manner independent of cell-cell signaling. Deletion of TraR, the ICE-encoded transcription factor that responds to acyl-homoserine lactones, has a negligible effect on ICE excision, although conjugation is reduced (116). ICESt3 is also activated during the stationary phase, as indicated by an increase in excision from the chromosome and expression of conjugation genes (31). Induction of an ICE during the stationary phase does not necessarily lead to conjugation but could indicate that the ICE is primed to transfer once nutrients become available, as with ICEclc. The stationary phase may also serve as an indicator that the host is poorly adapted to grow under current conditions. In this case, it might be beneficial for the ICE to move to a different host rather than rely on vertical inheritance in the starving-stressed host.
Phenotype-dependent induction
In several instances, the induction of an ICE is tied to a selective advantage that the ICE provides to the host. Tn916 was originally identified because it confers tetracycline resistance to its hosts (53). Tetracycline induces conjugation of Tn916 and other related elements (49, 131). This induction is initially regulated at the level of transcriptional attenuation. Under noninducing conditions, most of the transcripts that initiate at the promoter for tetM (Ptet) in Tn916 terminate at a factor-independent terminator upstream of tetM (the gene for tetracycline resistance) and other genes. After exposure to tetracycline, transcription extends past this terminator into tetM and downstream regulatory genes. Full expression of the conjugation genes occurs when Tn916 circularizes upon excision from the chromosome (Figure 5; 34, 137).
Figure 5.
Excision of Tn916 allows expression of conjugation genes. Both linear and circular maps of Tn916 are shown. Genes are shown as arrows on the map. Some known promoters are shown as bent arrows. While the element is in the chromosome, there is a low level of transcription (red dashed arrows) of genes in the regulatory region (white arrows), including xis and int. The genes upstream of tetM, including the relaxase and conjugation genes, are not expressed. Excision and circularization of Tn916 make orf20 (relaxase) and the conjugation operon contiguous and codirectional with the regulatory region, allowing all these genes to be transcribed. The dashed red arrows are used to illustrate this phenomenon and do not depict the known variety or relative abundance of transcripts produced by Tn916.
Tetracycline also induces conjugation of the CTnDOT-ERL family of ICEs from Bacteroides sp., which confers tetracycline resistance to their hosts. In this case, induction is regulated at the level of translation initiation rather than transcriptional attenuation. Transcription of tetQ is constitutive; however, the ribosome binding site is inaccessible because of the secondary structure of the mRNA. Tetracycline causes ribosomes to stall during synthesis of a leader peptide, which is also encoded by the tetQ mRNA. The paused ribosomes change them RNA secondary structure so that the tetQ ribosome binding site is accessible, allowing translation of TetQ and the downstream regulators RteA and RteB (149, 150). This initiates a regulatory cascade that leads to excision and conjugation of CTnDOT (reviewed in 151).
Other ICE-encoded phenotypes are also related to activation signals. ICEclc enables Pseudomonas spp. to metabolize chlorocatechols, permitting some species to use 3-chlorobenzoate as their sole carbon source. Growth of ICEclc donors on 3-chlorobenzoate enhances expression of conjugation genes and conjugation of ICEclc (103, 127). The advantage of linking conjugation to an adaptive phenotype conferred by the ICE is twofold. Any bacterium that acquires the ICE and can benefit from the phenotype immediately gains a selective advantage over its peers, promoting vertical transmission of the ICE. Additionally, any metabolic burden on the donor due to the expression of conjugation genes may be offset by the ability to exploit a distinct niche made available by the ICE.
Induction upon entry into a new host
Some ICEs are active immediately upon entering a recipient cell, analogous to zygotic induction of some phages. When ICEBs1 is introduced into a new recipient, it is able to spread rapidly to other cells in a manner that requires the conjugation machinery (7). This activity does not require rapI, which is needed for activation of ICEBs1 in response to cell density. Similarly, if Tn916 is delivered to a cell on a conjugative plasmid, it frequently transposes to a new location before becoming quiescent (59, 60). The initial burst of activity seen in these ICEs is likely due to a lack of repression. When the ICE enters a recipient cell, the ICE-encoded regulators that normally repress gene activity are absent. It is only after the ICE has expressed these regulatory genes that repression of ICE functions is achieved.
CHALLENGES TO THE FIELD
Although there has been a tremendous increase in our knowledge of ICE biology since the description of Tn916 (53), there is still a tremendous amount to learn. One area of intense interest is the nature of the mating machinery and how a protein attached to ssDNA is transferred out of the donor cell and into the recipient. Many components of the type IV secretion system encoded by ICEs and conjugative plasmids are conserved across the bacterial domain. Recent biochemical and structural advances (e.g., 34, 54, 55, 94) have improved our understanding of this macromolecular machine, but a thorough understanding of its structure and function remains elusive.
It is not known whether there is a specific cue that signals the mating machinery to export the T-DNA. There is reason to believe that such a cue exists and that conjugation systems do not pump DNA into the environment when no recipient is present. Studies of conjugative plasmid R1 indicate that the mating channel is gated and that communication occurs between the inside and outside. Entry of the R17 phage via the conjugation channel requires that a T-DNA and type IV coupling protein be docked at the inner opening of the channel (86).
It is becoming evident that there is not a clear distinction between conjugative and replicative relaxases in Firmicutes. The distinction between conjugative plasmids and ICEs is also becoming blurred, as several ICEs appear to undergo plasmid-like rolling-circle replication. Further studies are needed to identify and characterize interactions between relaxases and conjugation systems of Gram-positive bacteria. In particular, investigation into the role of type IV coupling proteins in determining the range of relaxases that can be recognized might prove fruitful (153).
Host genes, both essential and nonessential, are likely to be involved in every step of the ICE life cycle. Some of these gene products, such as integration host factors (100) and components of the host replication machinery (89, 141), have been identified and their roles are understood to varying degrees.However, the contribution of other host genes remains to be elucidated (79), and it is likely that other unidentified genes are also involved in conjugation.
Similarly, the relationship between the host range of an ICE and the genetic content of the permissive (or nonpermissive) hosts is not well understood. Some ICEs, such as Tn916, have a very broad host range, whereas others are more restricted. Host range could be limited by incompatibility between a given type IV secretion (conjugation) system and a particular recipient’s cell envelope. Host range could also be restricted by cytoplasmic factors, for example restriction-modification or CRISPR systems, an inability of a specific ICE to replicate or integrate in some hosts, or incompatibility with other resident mobile genetic elements.
A mechanistic understanding of what happens to an ICE once it enters a recipient has not been thoroughly developed. It is generally assumed that the ICE must generate a complementary strand to the T-DNA prior to integrating into the chromosome. For ICEs that replicate prior to integration, this is certainly the case, and this is also likely for those that encode a mechanism for generating a complementary strand. However, it is not clear that such a requirement exists for all ICEs. Some integrases of the tyrosine-recombinase family can insert ssDNA elements into the chromosome, provided the DNA forms a double-stranded structure at the attachment site (20, 145). The integrase itself might be expressed from a single-stranded promoter on the TDNA or transferred from the donor to the recipient, as is thought to be the case for Tn916 (22). Alternately, the relaxase could directly mediate integration of the covalently bound T-DNA into the chromosome, although this would require that the chromosome already harbor a cognate oriT (50).
ICEs have tremendous potential to be developed as tools for genetic engineering. Conjugation can be used to deliver DNA to organisms from all domains of life, provided there is a match between the conjugation system and the recipient. ICEs have the added benefit over plasmids of being able to insert into a host chromosome, provided the integration system functions in the recipient. This avoids the need for replication of and selection for the element to ensure maintenance and the inherent variability in the copy number of most plasmids. In the past, conjugative delivery of Tn916 has been used to mutagenize a variety of bacteria, including Gram positives (152), Gram negatives (75), and the wall-less Mollicutes (124), and to mobilize other elements. In the future, ICEs could allow the delivery of specific genes or metabolic pathways to an incredibly diverse array of organisms.
Acknowledgments
We thank Dr. Catherine Lee, Dr. Mohan Viswanathan, Laurel Wright, and Monica Avello for suggestions on this review and Laurel Wright and Joshua Jones for sharing unpublished results. Work on HGT in the lab of A.D.G. is supported in part by grant GM050895 from the National Institute of General Medical Sciences of the National Institutes of Health.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Agundez L, Gonzalez-Prieto C, Machen C, Llosa M. Site-specific integration of foreign DNA into minimal bacterial and human target sequences mediated by a conjugative relaxase. PLOS ONE. 2012;7:e31047. doi: 10.1371/journal.pone.0031047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arutyunov D, Frost LS. F conjugation: back to the beginning. Plasmid. 2013;70:18–32. doi: 10.1016/j.plasmid.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 4.Auchtung JM, Lee CA, Garrison KL, Grossman AD. Identification and characterization of the immunity repressor (ImmR) that controls the mobile genetic element ICE Bs1 of Bacillus subtilis . Mol. Microbiol. 2007;64:1515–1528. doi: 10.1111/j.1365-2958.2007.05748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auchtung JM, Lee CA, Monson RE, Lehman AP, Grossman AD. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. PNAS. 2005;102:12554–12559. doi: 10.1073/pnas.0505835102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayoubi P, Kilic AO, Vijayakumar MN. Tn 5253 the Pneumococcal omega ( cat tet ) BM6001 element, is a composite structure of two conjugative transposons, Tn 5251 and Tn 5252 . J. Bacteriol. 1991;173:1617–1622. doi: 10.1128/jb.173.5.1617-1622.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babic A, Berkmen MB, Lee CA, Grossman AD. Efficient gene transfer in bacterial cell chains. mBio. 2011;2:00027–00011. doi: 10.1128/mBio.00027-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beaber JW, Burrus V, Hochhut B, Waldor MK. Comparison of SXT and R391, two conjugative integrating elements: definition of a genetic backbone for the mobilization of resistance determinants. Cell Mol. Life Sci. 2002;59:2065–2070. doi: 10.1007/s000180200006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beaber JW, Hochhut B, Waldor MK. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae . J. Bacteriol. 2002;184:4259–4269. doi: 10.1128/JB.184.15.4259-4269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427:72–74. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- 11.Bedzyk LA, Shoemaker NB, Young KE, Salyers AA. Insertion and excision of Bacteroides conjugative chromosomal elements. J. Bacteriol. 1992;174:166–172. doi: 10.1128/jb.174.1.166-172.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellanger X, Morel C, Decaris B, Guedon G. Derepression of excision of integrative and potentially conjugative elements from Streptococcus thermophilus by DNA damage response: implication of a cI-related repressor. J. Bacteriol. 2007;189:1478–1481. doi: 10.1128/JB.01125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bellanger X, Morel C, Gonot F, Puymege A, Decaris B, Guedon G. Site-specific accretion of an integrative conjugative element together with a related genomic island leads to cis mobilization and gene capture. Mol. Microbiol. 2011;81:912–925. doi: 10.1111/j.1365-2958.2011.07737.x. [DOI] [PubMed] [Google Scholar]
- 14.Bellanger X, Payot S, Leblond-Bourget N, Guedon G. Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol. Rev. 2014;38:720–760. doi: 10.1111/1574-6976.12058. [DOI] [PubMed] [Google Scholar]
- 15.Bhatty M, Laverde Gomez JA, Christie PJ. The expanding bacterial type IV secretion lexicon. Res. Microbiol. 2013;164:620–639. doi: 10.1016/j.resmic.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bi D, Xu Z, Harrison EM, Tai C, Wei Y, et al. ICEberg: a web-based resource for integrative and conjugative elements found in Bacteria. Nucleic Acids Res. 2012;40:D621–D626. doi: 10.1093/nar/gkr846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boltner D, MacMahon C, Pembroke JT, Strike P, Osborn AM. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J. Bacteriol. 2002;184:5158–5169. doi: 10.1128/JB.184.18.5158-5169.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bordeleau E, Ghinet MG, Burrus V. Diversity of integrating conjugative elements in Actinobacteria: coexistence of two mechanistically different DNA-translocation systems. Mob. Genet. Elem. 2012;2:119–124. doi: 10.4161/mge.20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bose B, Auchtung JM, Lee CA, Grossman AD. A conserved anti-repressor controls horizontal gene transfer by proteolysis. Mol. Microbiol. 2008;70:570–582. doi: 10.1111/j.1365-2958.2008.06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouvier M, Demarre G, Mazel D. Integron cassette insertion: a recombination process involving a folded single strand substrate. EMBO J. 2005;24:4356–4367. doi: 10.1038/sj.emboj.7600898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brassinga AKC, Hiltz MF, Sisson GR, Morash MG, Hill N, et al. A 65-kilobase pathogenicity island is unique to Philadelphia-1 strains of Legionella pneumophila . J. Bacteriol. 2003;185:4630–4637. doi: 10.1128/JB.185.15.4630-4637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bringel F, Van Alstine GL, Scott JR. Conjugative transposition of Tn 916: the transposon int gene is required only in the donor. J. Bacteriol. 1992;174:4036–4041. doi: 10.1128/jb.174.12.4036-4041.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brochet M, Couve E, Glaser P, Guedon G, Payot S. Integrative conjugative elements and related elements are major contributors to the genome diversity of Streptococcus agalactiae . J. Bacteriol. 2008;190:6913–6917. doi: 10.1128/JB.00824-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brochet M, Da Cunha V, Couve E, Rusniok C, Trieu-Cuot P, Glaser P. Atypical association of DDEtransposition with conjugation specifies a new family of mobile elements. Mol. Microbiol. 2009;71:948–959. doi: 10.1111/j.1365-2958.2008.06579.x. [DOI] [PubMed] [Google Scholar]
- 25.Burrus V, Pavlovic G, Decaris B, Guedon G. The ICE St1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid. 2002;48:77–97. doi: 10.1016/s0147-619x(02)00102-6. [DOI] [PubMed] [Google Scholar]
- 26.Burrus V, Waldor MK. Control of SXT integration and excision. J. Bacteriol. 2003;185:5045–5054. doi: 10.1128/JB.185.17.5045-5054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burrus V, Waldor MK. Formation of SXT tandem arrays and SXT-R391 hybrids. J. Bacteriol. 2004;186:2636–2645. doi: 10.1128/JB.186.9.2636-2645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butala M, Zgur-Bertok D, Busby SJW. The bacterial LexA transcriptional repressor. Cell. Mol. Life Sci. 2009;66:82–93. doi: 10.1007/s00018-008-8378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cabezon E, Ripoll-Rozada J, Pena A, de la Cruz F, Arechaga I. Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 2014;39:81–95. doi: 10.1111/1574-6976.12085. [DOI] [PubMed] [Google Scholar]
- 30.Campbell A. Phage integration and chromosome structure. A personal history. Annu. Rev. Genet. 2007;41:1–11. doi: 10.1146/annurev.genet.41.110306.130240. [DOI] [PubMed] [Google Scholar]
- 31.Carraro N, Libante V, Morel C, Decaris B, Charron-Bourgoin F, et al. Differential regulation of two closely related integrative and conjugative elements from Streptococcus thermophilus . BMC Microbiol. 2011;11:238. doi: 10.1186/1471-2180-11-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carraro N, Poulin D, Burrus V. Replication and active partition of integrative and conjugative elements (ICEs) of the SXT/R391 family: the line between ICEs and conjugative plasmids is getting thinner. PLoS Genet. 2015;11:e1005298. doi: 10.1371/journal.pgen.1005298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carter MQ, Chen J, Lory S. The Pseudomonas aeruginosa pathogenicity island PAPI-1 is transferred via a novel type IV pilus. J. Bacteriol. 2010;192:3249–3258. doi: 10.1128/JB.00041-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cascales E, Christie PJ. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004;304:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Celli J, Trieu-Cuot P. Circularization of Tn 916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol. Microbiol. 1998;28:103–117. doi: 10.1046/j.1365-2958.1998.00778.x. [DOI] [PubMed] [Google Scholar]
- 36.Cesar CE, Machon C, de la Cruz F, Llosa M. A new domain of conjugative relaxase TrwC responsible for efficient oriT -specific recombination on minimal target sequences. Mol. Microbiol. 2006;62:984–996. doi: 10.1111/j.1365-2958.2006.05437.x. [DOI] [PubMed] [Google Scholar]
- 37.Chandler M, de la Cruz F, Dyda F, Hickman AB, Moncalian G, Ton-Hoang B. Breaking and joining single-stranded DNA: the HUH endonuclease superfamily. Nat. Rev. Microbiol. 2013;11:525–538. doi: 10.1038/nrmicro3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng Q, Paszkiet BJ, Shoemaker NB, Gardner JF, Salyers AA. Integration and excision of a Bacteroides conjugative transposon, CTnDOT. J. Bacteriol. 2000;182:4035–4043. doi: 10.1128/jb.182.14.4035-4043.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christie PJ, Whitaker N, Gonzalez-Rivera C. Mechanism and structure of the bacterial type IV secretion systems. Biochim. Biophys. Acta. 2014;1843:1578–1591. doi: 10.1016/j.bbamcr.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 41.Curcio MJ, Derbyshire KM. The outs and ins of transposition: from Mu to Kangaroo. Nat. Rev. Mol. Cell Biol. 2003;4:865–877. doi: 10.1038/nrm1241. [DOI] [PubMed] [Google Scholar]
- 42.Daccord A, Ceccarelli D, Burrus V. Integrating conjugative elements of the SXT/R391 family trigger the excision and drive the mobilization of a new class of Vibrio genomic islands. Mol. Microbiol. 2010;78:576–588. doi: 10.1111/j.1365-2958.2010.07364.x. [DOI] [PubMed] [Google Scholar]
- 43.Daccord A, Ceccarelli D, Rodrigue S, Burrus V. Comparative analysis of mobilizable genomic islands. J. Bacteriol. 2013;195:606–614. doi: 10.1128/JB.01985-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daccord A, Mursell M, Poulin-Laprade D, Burrus V. Dynamics of the SetCD-regulated integration and excision of genomic islands mobilized by integrating conjugative elements of the SXT/R391 family. J. Bacteriol. 2012;194:5794–5802. doi: 10.1128/JB.01093-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.d’Ari R. The SOS system. Biochimie. 1985;67:343–347. doi: 10.1016/s0300-9084(85)80077-8. [DOI] [PubMed] [Google Scholar]
- 46.Dimopoulou ID, Jordens JZ, Legakis NJ, Crook DW. A molecular analysis of Greek and UK Haemophilus influenzae conjugative resistance plasmids. J. Antimicrob. Chemother. 1997;39:303–307. doi: 10.1093/jac/39.3.303. [DOI] [PubMed] [Google Scholar]
- 47.Dimopoulou ID, Russell JE, Mohd-Zain Z, Herbert R, Crook DW. Site-specific recombination with the chromosomal tRNALeugene by the large conjugative Haemophilus resistance plasmid. Antimicrob. Agents Chemother. 2002;46:1602–1603. doi: 10.1128/AAC.46.5.1602-1603.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dordet Frisoni E, Marenda MS, Sagne E, Nouvel LX, Guerillot R, et al. ICEA of Mycoplasma agalactiae: a new family of self-transmissible integrative elements that confers conjugative properties to the recipient strain. Mol. Microbiol. 2013;89:1226–1239. doi: 10.1111/mmi.12341. [DOI] [PubMed] [Google Scholar]
- 49.Doucet-Populaire F, Trieu-Cuot P, Dosbaa I, Andremont A, Courvalin P. Inducible transfer of conjugative transposon Tn 1545 from Enterococcus faecalis to Listeria monocytogenes in the digestive tracts of gnotobiotic mice. Antimicrob. Agents Chemother. 1991;35:185–187. doi: 10.1128/aac.35.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Draper O, Cesar CE, Machon C, de la Cruz F, Llosa M. Site-specific recombinase and integrase activities of a conjugative relaxase in recipient cells. PNAS. 2005;102:16385–16390. doi: 10.1073/pnas.0506081102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dunny GM, Brown BL, Clewell DB. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. PNAS. 1978;75:3479–3483. doi: 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flynn KJ, Swanson MS. Integrative conjugative element ICE-ßox confers oxidative stress resistance to Legionella pneumophila in vitro and in macrophages. mBio. 2014;5:e01091-14. doi: 10.1128/mBio.01091-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franke AE, Clewell DB. Evidence for a chromosome-borne resistance transposon (Tn 916 ) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 1981;145:494–502. doi: 10.1128/jb.145.1.494-502.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fronzes R, Christie PJ, Waksman G. The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 2009;7:703–714. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fronzes R, Schafer E, Wang L, Saibil HR, Orlova EV, Waksman G. Structure of a type IV secretion system core complex. Science. 2009;323:266–268. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaillard M, Pernet N, Vogne C, Hagenbuchle O, van der Meer JR. Host and invader impact of transfer of the clc genomic island into Pseudomonas aeruginosa PAO1. PNAS. 2008;105:7058–7063. doi: 10.1073/pnas.0801269105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garcillan-Barcia MP, Francia MV, de la Cruz F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 2009;33:657–687. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- 58.Garriss G, Waldor MK, Burrus V. Mobile antibiotic resistance encoding elements promote their own diversity. PLOS Genet. 2009;5:e1000775. doi: 10.1371/journal.pgen.1000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gawron-Burke C, Clewell DB. A transposon in Streptococcus faecalis with fertility properties. Nature. 1982;300:281–284. doi: 10.1038/300281a0. [DOI] [PubMed] [Google Scholar]
- 60.Gawron-Burke C, Clewell DB. Regeneration of insertionally inactivated streptococcal DNA fragments after excision of transposon Tn 916 in Escherichia coli: strategy for targeting and cloning of genes from Gram-positive bacteria. J. Bacteriol. 1984;159:214–221. doi: 10.1128/jb.159.1.214-221.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gibbons HS, Broomall SM, McNew LA, Daligault H, Chapman C, et al. Genomic signatures of strain selection and enhancement in Bacillus atrophaeus var. globigii a historical biowarfare simulant. PLOS ONE. 2011;6:e17836. doi: 10.1371/journal.pone.0017836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goessweiner-Mohr N, Arends K, Keller W, Grohmann E. Conjugative type IV secretion systems in Gram-positive bacteria. Plasmid. 2013;70:289–302. doi: 10.1016/j.plasmid.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grindley NDF, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 64.Guasch A, Lucas M, Moncalian G, Cabezas M, Perez-Luque R, et al. Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC. Nat. Struct. Biol. 2003;10:1002–1010. doi: 10.1038/nsb1017. [DOI] [PubMed] [Google Scholar]
- 65.Guerillot R, Da Cunha V, Sauvage E, Bouchier C, Glaser P. Modular evolution of TnGBSs, a new family of integrative and conjugative elements associating insertion sequence transposition, plasmid replication, and conjugation for their spreading. J. Bacteriol. 2013;195:1979–1990. doi: 10.1128/JB.01745-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guerillot R, Siguier P, Gourbeyre E, Chandler M, Glaser P. The diversity of prokaryotic DDE transposases of the mutator superfamily, insertion specificity, and association with conjugation machineries. Genome Biol. Evol. 2014;6:260–272. doi: 10.1093/gbe/evu010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guglielmini J, Quintais L, Garcillan-Barcia MP, de la Cruz F, Rocha EP. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLOS Genet. 2011;7:e1002222. doi: 10.1371/journal.pgen.1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He J, Baldini RL, Deziel E, Saucier M, Zhang Q, et al. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. PNAS. 2004;101:2530–2535. doi: 10.1073/pnas.0304622101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hickman AB, Chandler M, Dyda F. Integrating prokaryotes and eukaryotes: DNA transposases in light of structure. Crit. Rev. Biochem. Mol. Biol. 2010;45:50–69. doi: 10.3109/10409230903505596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hirano N, Muroi T, Takahashi H, Haruki M. Site-specific recombinases as tools for heterologous gene integration. Appl. Microbiol. Biotechnol. 2011;92:227–239. doi: 10.1007/s00253-011-3519-5. [DOI] [PubMed] [Google Scholar]
- 71.Hochhut B, Beaber JW, Woodgate R, Waldor MK. Formation of chromosomal tandem arrays of the SXT element and R391, two conjugative chromosomally integrating elements that share an attachment site. J. Bacteriol. 2001;183:1124–1132. doi: 10.1128/JB.183.4.1124-1132.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hochhut B, Jahreis K, Lengeler JW, Schmid K. CTn scr94 a conjugative transposon found in enterobacteria. J. Bacteriol. 1997;179:2097–2102. doi: 10.1128/jb.179.7.2097-2102.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hochhut B, Lotfi Y, Mazel D, Faruque SM, Woodgate R, Waldor MK. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 2001;45:2991–3000. doi: 10.1128/AAC.45.11.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hochhut B, Marrero J, Waldor MK. Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a constin found in Vibrio cholerae O139. J. Bacteriol. 2000;182:2043–2047. doi: 10.1128/jb.182.7.2043-2047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Holland J, Towner KJ, Williams P. Tn 916 insertion mutagenesis in Escherichia coli and Haemophilus influenzae type b following conjugative transfer. J. Gen. Microbiol. 1992;138:509–515. doi: 10.1099/00221287-138-3-509. [DOI] [PubMed] [Google Scholar]
- 76.Honda Y, Sakai H, Komano T, Bagdasarian M. RepB′ is required in trans for the two single-strand DNA initiation signals in oriV of plasmid RSF1010. Gene. 1989;80:155–159. doi: 10.1016/0378-1119(89)90261-8. [DOI] [PubMed] [Google Scholar]
- 77.Iannelli F, Santoro F, Oggioni MR, Pozzi G. Nucleotide sequence analysis of integrative conjugative element Tn 5253 of Streptococcus pneumoniae . Antimicrob. Agents Chemother. 2014;58:1235–1239. doi: 10.1128/AAC.01764-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ilyina TV, Koonin EV. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Johnson CM, Grossman AD. Identification of host genes that affect acquisition of an integrative and conjugative element in Bacillus subtilis . Mol. Microbiol. 2014;93:1284–1301. doi: 10.1111/mmi.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Juhas M, Power PM, Harding RM, Ferguson DJP, Dimopoulou ID, et al. Sequence and functional analyses of Haemophilus spp. genomic islands. Genome Biol. 2007;8:R237. doi: 10.1186/gb-2007-8-11-r237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kado CI. Historical account on gaining insights on the mechanism of crown gall tumorigenesis induced by Agrobacterium tumefaciens . Front. Microbiol. 2014;5:340. doi: 10.3389/fmicb.2014.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kamensek S, Podlesek Z, Gillor O, Zgur-Bertok D. Genes regulated by the Escherichia coli SOS repressor LexA exhibit heterogeneous expression. BMC Microbiol. 2010;10:283. doi: 10.1186/1471-2180-10-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Khan SA. Plasmid rolling-circle replication: highlights of two decades of research. Plasmid. 2005;53:126–136. doi: 10.1016/j.plasmid.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 84.Krüger N-J, Stingl K. Two steps away from novelty: principles of bacterial DNA uptake. Mol. Microbiol. 2011;80:860–867. doi: 10.1111/j.1365-2958.2011.07647.x. [DOI] [PubMed] [Google Scholar]
- 85.Lambert PF, Waring DA, Wells RD, Reznikoff WS. DNA requirements at the bacteriophage G4 origin of complementary-strand DNA synthesis. J. Virol. 1986;58:450–458. doi: 10.1128/jvi.58.2.450-458.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lang S, Kirchberger PC, Gruber CJ, Redzej A, Raffl S, et al. An activation domain of plasmid R1 TraI protein delineates stages of gene transfer initiation. Mol. Microbiol. 2011;82:1071–1085. doi: 10.1111/j.1365-2958.2011.07872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lederberg J, Tatum EL. Gene recombination in Escherichia coli . Nature. 1946;158:558. doi: 10.1038/158558a0. [DOI] [PubMed] [Google Scholar]
- 88.Lee CA, Auchtung JM, Monson RE, Grossman AD. Identification and characterization of int (integrase), xis (excisionase) and chromosomal attachment sites of the integrative and conjugative element ICE Bs1 of Bacillus subtilis . Mol. Microbiol. 2007;66:1356–1369. doi: 10.1111/j.1365-2958.2007.06000.x. [DOI] [PubMed] [Google Scholar]
- 89.Lee CA, Babic A, Grossman AD. Autonomous plasmid-like replication of a conjugative transposon. Mol. Microbiol. 2010;75:268–279. doi: 10.1111/j.1365-2958.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee CA, Grossman AD. Identification of the origin of transfer ( oriT ) and DNA relaxase required for conjugation of the integrative and conjugative element ICE Bs1 of Bacillus subtilis . J. Bacteriol. 2007;189:7254–7261. doi: 10.1128/JB.00932-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee CA, Thomas J, Grossman AD. The Bacillus subtilis conjugative transposon ICE Bs1 mobilizes plasmids lacking dedicated mobilization functions. J. Bacteriol. 2012;194:3165–3172. doi: 10.1128/JB.00301-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu Y, Harrison PM, Kunin V, Gerstein M. Comprehensive analysis of pseudogenes in prokaryotes: widespread gene decay and failure of putative horizontally transferred genes. Genome Biol. 2004;5:R64. doi: 10.1186/gb-2004-5-9-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lorenzo-Diaz F, Espinosa M. Lagging-strand DNA replication origins are required for conjugal transfer of the promiscuous plasmid pMV158. J. Bacteriol. 2009;191:720–727. doi: 10.1128/JB.01257-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Low HH, Gubellini F, Rivera-Calzada A, Braun N, Connery S, et al. Structure of a type IV secretion system. Nature. 2014;508:550–553. doi: 10.1038/nature13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Magot M. Transfer of antibiotic resistances from Clostridium innocuum to Clostridium perfringens in the absence of detectable plasmid DNA. FEMS Microbiol. Lett. 1983;18:149–151. [Google Scholar]
- 96.Manganelli R, Romano L, Ricci S, Zazzi M, Pozzi G. Dosage of Tn 916 circular intermediates in Enterococcus faecalis . Plasmid. 1995;34:48–57. doi: 10.1006/plas.1995.1032. [DOI] [PubMed] [Google Scholar]
- 97.Marenda M, Barbe V, Gourgues G, Mangenot S, Sagne E, Citti C. A new integrative conjugative element occurs in Mycoplasma agalactiae as chromosomal and free circular forms. J. Bacteriol. 2006;188:4137–4141. doi: 10.1128/JB.00114-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mathee K, Narasimhan G, Valdes C, Qiu X, Matewish JM, et al. Dynamics of Pseudomonas aeruginosa genome evolution. PNAS. 2008;105:3100–3105. doi: 10.1073/pnas.0711982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mays TD, Smith CJ, Welch RA, Delfini C, Macrina FL. Novel antibiotic resistance transfer in Bacteroides . Antimicrob. Agents Chemother. 1982;21:110–118. doi: 10.1128/aac.21.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McLeod SM, Burrus V, Waldor MK. Requirement for Vibrio cholerae integration host factor in conjugative DNA transfer. J. Bacteriol. 2006;188:5704–5711. doi: 10.1128/JB.00564-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Menard KL, Grossman AD. Selective pressures to maintain attachment site specificity of integrative and conjugative elements. PLOS Genet. 2013;9:e1003623. doi: 10.1371/journal.pgen.1003623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Michael GB, Kadlec K, Sweeney MT, Brzuszkiewicz E, Liesegang H, et al. ICE Pmu1 an integrative conjugative element (ICE) of Pasteurella multocida: structure and transfer. J. Antimicrob. Chemother. 2012;67:91–100. doi: 10.1093/jac/dkr411. [DOI] [PubMed] [Google Scholar]
- 103.Minoia M, Gaillard M, Reinhard F, Stojanov M, Sentchilo V, van der Meer JR. Stochasticity and bistability in horizontal transfer control of a genomic island in Pseudomonas . PNAS. 2008;105:20792–20797. doi: 10.1073/pnas.0806164106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mirouze N, Parashar V, Baker MD, Dubnau DA, Neiditch MB. An atypical Phr peptide regulates the developmental switch protein RapH. J. Bacteriol. 2011;193:6197–6206. doi: 10.1128/JB.05860-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Miyazaki R, Minoia M, Pradervand N, Sulser S, Reinhard F, van der Meer JR. Cellular variability of RpoS expression underlies subpopulation activation of an integrative and conjugative element. PLOS Genet. 2012;8:E1002818. doi: 10.1371/journal.pgen.1002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mohd-Zain Z, Turner SL, Cerdeno-Tarraga AM, Lilley AK, Inzana TJ, et al. Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J. Bacteriol. 2004;186:8114–8122. doi: 10.1128/JB.186.23.8114-8122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Naglich JG, Andrews RE., Jr Tn 916 -dependent conjugal transfer of pC194 and pUB110 from Bacillus subtilis into Bacillus thuringiensis subsp. israelensis . Plasmid. 1988;20:113–126. doi: 10.1016/0147-619x(88)90014-5. [DOI] [PubMed] [Google Scholar]
- 108.Nishi A, Tominaga K, Furukawa K. A 90-kilobase conjugative chromosomal element coding for biphenyl and salicylate catabolism in Pseudomonas putida KF715. J. Bacteriol. 2000;182:1949–1955. doi: 10.1128/jb.182.7.1949-1955.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nugent ME. A conjugative “plasmid” lacking autonomous replication. J. Gen. Microbiol. 1981;126:305–310. doi: 10.1099/00221287-126-2-305. [DOI] [PubMed] [Google Scholar]
- 110.Ogawa H, Ogawa T. Regulation in repressor inactivation by RecA protein. Adv. Biophys. 1990;26:33–49. doi: 10.1016/0065-227x(90)90006-f. [DOI] [PubMed] [Google Scholar]
- 111.Osborn AM, Boltner D. When phage, plasmids, and transposons collide: genomic islands, and conjugative- and mobilizable-transposons as a mosaic continuum. Plasmid. 2002;48:202–212. doi: 10.1016/s0147-619x(02)00117-8. [DOI] [PubMed] [Google Scholar]
- 112.Pavlovic G, Burrus V, Gintz B, Decaris B, Guedon G. Evolution of genomic islands by deletion and tandem accretion by site-specific recombination: ICE St1 -related elements from Streptococcus thermophilus . Microbiology. 2004;150:759–774. doi: 10.1099/mic.0.26883-0. [DOI] [PubMed] [Google Scholar]
- 113.Pennington JM, Rosenberg SM. Spontaneous DNA breakage in single living Escherichia coli cells. Nat. Genet. 2007;39:797–802. doi: 10.1038/ng2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Piper KR, Beck von Bodman S, Farrand SK. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature. 1993;362:448–450. doi: 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- 115.Rajeev L, Malanowska K, Gardner JF. Challenging a paradigm: the role of DNA homology in tyrosine recombinase reactions. Microbiol. Mol. Biol. Rev. 2009;73:300–309. doi: 10.1128/MMBR.00038-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ramsay JP, Sullivan JT, Jambari N, Ortori CA, Heeb S, et al. A LuxRI-family regulatory system controls excision and transfer of the Mesorhizobium loti strain R7A symbiosis island by activating expression of two conserved hypothetical genes. Mol. Microbiol. 2009;73:1141–1155. doi: 10.1111/j.1365-2958.2009.06843.x. [DOI] [PubMed] [Google Scholar]
- 117.Ramsay JP, Sullivan JT, Stuart GS, Lamont IL, Ronson CW. Excision and transfer of the Mesorhizobium loti R7A symbiosis island requires an integrase IntS, a novel recombination directionality factor RdfS, and a putative relaxase RlxS. Mol. Microbiol. 2006;62:723–734. doi: 10.1111/j.1365-2958.2006.05396.x. [DOI] [PubMed] [Google Scholar]
- 118.Rashtchian A, Dubes GR, Booth SJ. Tetracycline-inducible transfer of tetracycline resistance in Bacteroides fragilis in the absence of detectable plasmid DNA. J. Bacteriol. 1982;150:141–147. doi: 10.1128/jb.150.1.141-147.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rauch PJ, De Vos WM. Characterization of the novel nisin-sucrose conjugative transposon Tn 5276 and its insertion in Lactococcus lactis . J. Bacteriol. 1992;174:1280–1287. doi: 10.1128/jb.174.4.1280-1287.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ravatn R, Studer S, Springael D, Zehnder AJ, van der Meer JR. Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. strain B13. J. Bacteriol. 1998;180:4360–4369. doi: 10.1128/jb.180.17.4360-4369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Reinhard F, Miyazaki R, Pradervand N, van der Meer JR. Cell differentiation to “mating bodies” induced by an integrating and conjugative element in free-living bacteria. Curr. Biol. 2013;23:255–259. doi: 10.1016/j.cub.2012.12.025. [DOI] [PubMed] [Google Scholar]
- 122.Roberts AP, Johanesen PA, Lyras D, Mullany P, Rood JI. Comparison of Tn 5397 from Clostridium difficile Tn 916 from Enterococcus faecalis and the CW459 tet ( M ) element from Clostridium perfringens shows that they have similar conjugation regions but different insertion and excision modules. Microbiology. 2001;147:1243–1251. doi: 10.1099/00221287-147-5-1243. [DOI] [PubMed] [Google Scholar]
- 123.Roberts AP, Mullany P. A modular master on the move: the Tn 916 family of mobile genetic elements. Trends Microbiol. 2009;17:251–258. doi: 10.1016/j.tim.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 124.Roberts MC, Kenny GE. Conjugal transfer of transposon Tn 916 from Streptococcus faecalis to Mycoplasma hominis . J. Bacteriol. 1987;169:3836–3839. doi: 10.1128/jb.169.8.3836-3839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Roberts MC, Smith AL. Molecular characterization of “plasmid-free” antibiotic-resistant Haemophilus influenzae . J. Bacteriol. 1980;144:476–479. doi: 10.1128/jb.144.1.476-479.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Roche D, Flechard M, Lallier N, Reperant M, Bree A, et al. ICE Ec2 a new integrative and conjugative element belonging to the pKLC102/PAGI-2 family, identified in Escherichia coli strain BEN374. J. Bacteriol. 2010;192:5026–5036. doi: 10.1128/JB.00609-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sentchilo V, Ravatn R, Werlen C, Zehnder AJB, van derMeer JR. Unusual integrase gene expression on the clc genomic island in Pseudomonas sp. strain B13. J. Bacteriol. 2003;185:4530–4538. doi: 10.1128/JB.185.15.4530-4538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shoemaker NB, Getty C, Guthrie EP, Salyers AA. Regions in Bacteroides plasmids pBFTM10 and pB8-51 that allow Escherichia coli–Bacteroides shuttle vectors to be mobilized by IncP plasmids and by a conjugative Bacteroides tetracycline resistance element. J. Bacteriol. 1986;166:959–965. doi: 10.1128/jb.166.3.959-965.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shoemaker NB, Smith MD, Guild WR. DNase-resistant transfer of chromosomal cat and tet insertions by filter mating in Pneumococcus . Plasmid. 1980;3:80–87. doi: 10.1016/s0147-619x(80)90036-0. [DOI] [PubMed] [Google Scholar]
- 130.Shoemaker NB, Wang GR, Stevens AM, Salyers AA. Excision, transfer, and integration ofNBU1, a mobilizable site-selective insertion element. J. Bacteriol. 1993;175:6578–6587. doi: 10.1128/jb.175.20.6578-6587.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Showsh SA, Andrews RE., Jr Tetracycline enhances Tn 916 -mediated conjugal transfer. Plasmid. 1992;28:213–224. doi: 10.1016/0147-619x(92)90053-d. [DOI] [PubMed] [Google Scholar]
- 132.Siguier P, Gourbeyre E, Chandler M. Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol. Rev. 2014;38:865–891. doi: 10.1111/1574-6976.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sitkiewicz I, Green NM, Guo N, Mereghetti L, Musser JM. Lateral gene transfer of streptococcal ICE element RD2 (region of difference 2) encoding secreted proteins. BMC Microbiol. 2011;11:65. doi: 10.1186/1471-2180-11-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EP, de la Cruz F. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 2010;74:434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Smith CJ, Markowitz SM, Macrina FL. Transferable tetracycline resistance in Clostridium difficile . Antimicrob. Agents Chemother. 1981;19:997–1003. doi: 10.1128/aac.19.6.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stevens AM, Sanders JM, Shoemaker NB, Salyers AA. Genes involved in production of plasmidlike forms by a Bacteroides conjugal chromosomal element share amino acid homology with two-component regulatory systems. J. Bacteriol. 1992;174:2935–2942. doi: 10.1128/jb.174.9.2935-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Stuy JH. Chromosomally integrated conjugative plasmids are common in antibiotic-resistant Haemophilus influenzae . J. Bacteriol. 1980;142:925–930. doi: 10.1128/jb.142.3.925-930.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Su YA, He P, Clewell DB. Characterization of the tet ( M ) determinant of Tn 916: evidence for regulation by transcription attenuation. Antimicrob. Agents Chemother. 1992;36:769–778. doi: 10.1128/aac.36.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.te Poele EM, Bolhuis H, Dijkhuizen L. Actinomycete integrative and conjugative elements. Antonie Van Leeuwenhoek. 2008;94:127–143. doi: 10.1007/s10482-008-9255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Thierauf A, Perez G, Maloy Stanley. Generalized transduction. Methods Mol. Biol. 2009;501:267–286. doi: 10.1007/978-1-60327-164-6_23. [DOI] [PubMed] [Google Scholar]
- 141.Thomas J, Lee CA, Grossman AD. A conserved helicase processivity factor is needed for conjugation and replication of an integrative and conjugative element. PLOS Genet. 2013;9:e103198. doi: 10.1371/journal.pgen.1003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Toleman MA, Walsh TR. Combinatorial events of insertion sequences and ICE in Gram-negative bacteria. FEMS Microbiol. Rev. 2011;35:912–935. doi: 10.1111/j.1574-6976.2011.00294.x. [DOI] [PubMed] [Google Scholar]
- 143.Toussaint A, Merlin C. Mobile elements as a combination of functional modules. Plasmid. 2002;47:26–35. doi: 10.1006/plas.2001.1552. [DOI] [PubMed] [Google Scholar]
- 144.Trokter M, Felisberto-Rodrigues C, Christie PJ, Waksman G. Recent advances in the structural and molecular biology of type IV secretion systems. Curr. Opin. Struct. Biol. 2014;27:16–23. doi: 10.1016/j.sbi.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Val M-E, Bouvier M, Campos J, Sherratt D, Cornet F, et al. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae . Mol. Cell. 2005;19:559–566. doi: 10.1016/j.molcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 146.Valentine PJ, Shoemaker NB, Salyers AA. Mobilization of Bacteroides plasmids by Bacteroides conjugal elements. J. Bacteriol. 1988;170:1319–1324. doi: 10.1128/jb.170.3.1319-1324.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Waksman G, Orlova EV. Structural organisation of the type IV secretion systems. Curr. Opin. Microbiol. 2014;17:24–31. doi: 10.1016/j.mib.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang H, Mullany P. The large resolvase TndX is required and sufficient for integration and excision of derivatives of the novel conjugative transposon Tn 5397 . J. Bacteriol. 2000;182:6577–6583. doi: 10.1128/jb.182.23.6577-6583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang Y, Rotman ER, Shoemaker NB, Salyers AA. Translational control of tetracycline resistance and conjugation in the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 2005;187:2673–2680. doi: 10.1128/JB.187.8.2673-2680.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang Y, Shoemaker NB, Salyers AA. Regulation of a Bacteroides operon that controls excision and transfer of the conjugative transposon CTnDOT. J. Bacteriol. 2004;186:2548–2557. doi: 10.1128/JB.186.9.2548-2557.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Waters JL, Salyers AA. Regulation of CTnDOT conjugative transfer is a complex and highly coordinated series of events. mBio. 2013;4:e00569-13. doi: 10.1128/mBio.00569-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Weiser JN, Rubens CE. Transposon mutagenesis of group B Streptococcus beta-hemolysin biosynthesis. Infect. Immun. 1987;55:2314–2316. doi: 10.1128/iai.55.9.2314-2316.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Whitaker N, Chen Y, Jakubowski SJ, Sarkar MK, Li F, Christie PJ. The all-alpha domains of coupling proteins from the Agrobacterium tumefaciens VirB/VirD4 and Enterococcus faecalis pCF10-encoded type IV secretion systems confer specificity to binding of cognate DNA substrates. J. Bacteriol. 2015;197:2335–2349. doi: 10.1128/JB.00189-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wickner S, Hurwitz J. Association of phiX174 DNA-dependent ATPase activity with an Escherichia coli protein, replication factor Y, required for in vitro synthesis of phiX174 DNA. PNAS. 1975;72:3342–3346. doi: 10.1073/pnas.72.9.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wilkins BM, Boulnois GJ, Lanka E. A plasmid DNA primase active in discontinuous bacterial DNA replication. Nature. 1981;290:217–221. doi: 10.1038/290217a0. [DOI] [PubMed] [Google Scholar]
- 156.Wong JJ, Lu J, Glover JN. Relaxosome function and conjugation regulation in F-like plasmids - a structural biology perspective. Mol. Microbiol. 2012;85:602–617. doi: 10.1111/j.1365-2958.2012.08131.x. [DOI] [PubMed] [Google Scholar]
- 157.Wozniak RA, Fouts DE, Spagnoletti M, Colombo MM, Ceccarelli D, et al. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLOS Genet. 2009;5:e1000786. doi: 10.1371/journal.pgen.1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wozniak RA, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 2010;8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 159.Wozniak RAF, Waldor MK. A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLOS Genet. 2009;5:e1000439. doi: 10.1371/journal.pgen.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wright LD, Johnson CM, Grossman AD. Identification of a single strand origin of replication in the integrative and conjugative element ICE Bs1 of Bacillus subtilis . PLOS Genet. 2015;11:e10055561. doi: 10.1371/journal.pgen.1005556. [DOI] [PMC free article] [PubMed] [Google Scholar]