Abstract
The analysis of time-lapse images showing cells dividing to produce clones of related cells is an important application in biological microscopy. Imaging at the temporal resolution required to establish accurate tracking for vertebrate stem or cancer cells often requires the use of transmitted light or phase-contrast microscopy. Processing these images requires automated segmentation, tracking and lineaging algorithms. There is also a need for any errors in the automated processing to be easily identified and quickly corrected. We have developed LEVER, an open source software tool that combines the automated image analysis for phase-contrast microscopy movies with an easy-to-use interface for validating the results and correcting any errors.
Availability and Implementation: LEVER is available free and open source, licensed under the GNU GPLv3. Details on obtaining and using LEVER are available at http://n2t.net/ark:/87918/d9rp4t.
Contact: acohen@coe.drexel.edu
New and rapid development of microscope imaging technologies have enabled biological researchers to focus on the analysis of living cell behaviors. Manual analysis of the data is difficult and time consuming. Behavioral changes in cells may also be subtle and not easily identified by a human observer. These challenges necessitate the development of computational tools (Cohen, 2014).
LEVER combines lineage analysis with the segmentation and tracking of individual cells. This allows the quantification of individual cellular properties and their changes across generations or between cells of different fate. In a twist on the usual processing flow, LEVER uses population information from the lineage tree to refine the segmentation results, informing the number of cells in each connected component of foreground pixels and greatly reducing error rates (Winter et al., 2015). LEVER is designed so that errors in the automated results can be easily corrected.
Several tools provide features for lineage identification and analysis, but there is a lack of tools that support phase contrast microscopy. CellProfiler contains modules for tracking proliferating cells automatically (Bray and Carpenter, 2015). The Icy bioinformatics platform supports several active contour algorithms for cell segmentation and tracking (De Chaumont et al., 2012). Both Icy and CellProfiler focus on fluorescent image data and do not support correction of lineaging or tracking results. Amat et al.(2014) provide a tool for semi-automated lineage analysis of 3-D fluorescent microscopy. This approach is similar to LEVER, but is designed specifically for 3-D fluorescence images. Extensions to LEVER supporting 2-D and 3-D multichannel fluorescence time-lapse images (Mankowski et al., 2014; Wait et al., 2014) are under active development.
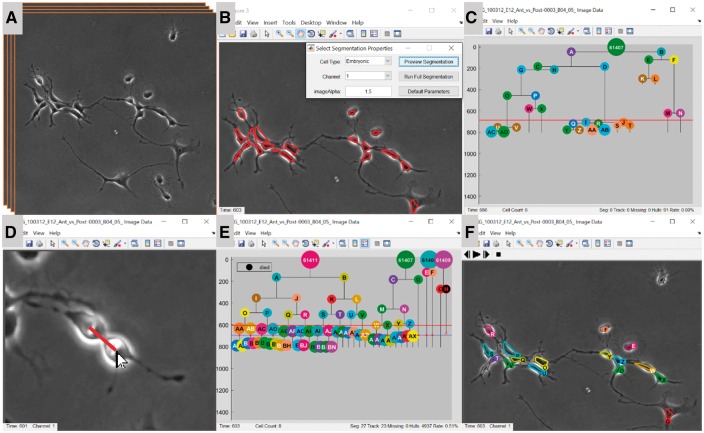
LEVER images can be imported using the integrated OME bioformats library (Linkert et al., 2010), or read directly (Fig. 1A). The segmentation preview window allows the user to compare results from several cell detection algorithms (Fig. 1B). Next, segmentation results are associated between frames using multitemporal association tracking (MAT). MAT has been effectively applied in a variety of applications (Chenouard et al., 2014; Mankowski et al., 2014; Winter et al., 2011, 2012, 2015), all using the same implementation. Tracking results are next used to form the lineage tree (Fig. 1C), or the tree can be specified by manually identifying mitosis events (Fig. 1D).
Fig. 1.
LEVER processing pipeline. Cells in the input image sequence (A) are automatically identified. Cell detection algorithms and parameters are tested on single image frames using the segmentation previewer (B). LEVER automatically creates an initial lineage tree based on cell tracking (C). The user quickly identifies all mitotic events to specify a corrected lineage tree in minutes (D). Using four interacting lineages (E) LEVER segmentation refinement can automatically correct most errors. Remaining errors are corrected manually during the refinement process (F) (Color version of this figure is available at Bioinformatics online.)
Once the lineage tree is complete, the initial segmentation and tracking can be refined. In practice, we recommend this task be done in conjunction with validation, so any errors can be corrected online. Segmentation refinement processes each frame in sequence making sure that every cell on the active lineage trees has a corresponding segmentation. As LEVER automatically corrects segmentation, tracking results are also updated automatically. Figure 1E and F shows a set of four interacting trees corrected using the segmentation refinement interface. Error rates are estimated from the number of edits required to fully correct the analysis. Once a tree has been fully validated, descriptive statistics such as phenotype tags, cycle times, average and standard deviations of size and speed for each cell can be exported to any spreadsheet.
The LEVER executables require 64-bit Windows; other platforms would require compiling the source code. Automated analysis generally takes 1–2 h for large image datasets. Manual lineage identification generally takes 15–30 min even for highly proliferative cells. Detailed documentation for using and extending LEVER is available at http://n2t.net/ark:/87918/d9rp4t.
Funding
Portions of this research were supported by the NIH NINDS (R01NS076709), and by the NIH NIA (R01AG041861).
Conflict of Interest: none declared.
References
- Amat F. et al. , (2014) Fast, accurate reconstruction of cell lineages from large-scale fluorescence microscopy data. Nat. Methods, 11, 951–958. [DOI] [PubMed] [Google Scholar]
- Bray M.A., Carpenter A.E. (2015) CellProfiler Tracer: exploring and validating high-throughput, time-lapse microscopy image data. BMC Bioinformatics, 16, 368.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenouard N. et al. (2014) Objective comparison of particle tracking methods. Nat. Methods, 11, 281–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A.R. (2014) Extracting meaning from biological imaging data. Mol. Biol. Cell, 25, 3470–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Chaumont F. et al. (2012) Icy: an open bioimage informatics platform for extended reproducible research. Nat. Methods, 9, 690–696. [DOI] [PubMed] [Google Scholar]
- Linkert M. et al. (2010) Metadata matters: access to image data in the real world. J. Cell Biol., 189, 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankowski W.C. et al. (2014). Segmentation of occluded hematopoietic stem cells from tracking. Paper presented at the Engineering in Medicine and Biology Society (EMBC), 2014 36th Annual International Conference of the IEEE. Chicago, IL: IEEE Press [DOI] [PMC free article] [PubMed]
- Wait E. et al. (2014) Visualization and correction of automated segmentation, tracking and lineaging from 5-D stem cell image sequences. BMC Bioinformatics, 15, 328.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter M. et al. (2011) Vertebrate neural stem cell segmentation, tracking and lineaging with validation and editing. Nat. Protoc., 6, 1942–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter M.R. et al. (2012) Axonal transport analysis using Multitemporal Association Tracking. Int. J. Comput. Biol. Drug Des., 5, 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter M.R. et al. (2015) Computational image analysis reveals intrinsic multigenerational differences between anterior and posterior cerebral cortex neural progenitor cells. Stem Cell Rep., 5, 609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]