Abstract
Summary: The Poisson Random Field (PRF) model has become an important tool in population genetics to study weakly deleterious genetic variation under complicated demographic scenarios. Currently, there are no freely available software applications that allow simulation of genetic variation data under this model. Here we present PReFerSim, an ANSI C program that performs forward simulations under the PRF model. PReFerSim models changes in population size, arbitrary amounts of inbreeding, dominance and distributions of selective effects. Users can track summaries of genetic variation over time and output trajectories of selected alleles.
Availability and Implementation: PReFerSim is freely available at: https://github.com/LohmuellerLab/PReFerSim
Contact: klohmueller@ucla.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Understanding the behavior of selected mutations in finite populations is important in population, medical and conservation genetics. Two main computational approaches have been used to model weakly deleterious mutations. First, under the Poisson Random Field (PRF) framework (Hartl et al., 1994; Sawyer and Hartl, 1992), a Poisson number of independent mutations enter the population each generation, with subsequent changes in allele frequency due to selection and drift. The PRF framework is a mainstay in population genetic inference of demography and selection (Akashi and Schaeffer, 1997; Boyko et al., 2008; Bustamante et al., 2001; Desai and Plotkin, 2008; Eyre-Walker et al., 2006; Gutenkunst et al., 2009; Huerta-Sanchez et al., 2008; Williamson et al., 2005). However, existing implementations of the PRF model do not provide the ages or selection coefficients of individual mutations or permit the user to follow allele frequency trajectories over time. Moreover, including biological complications, like inbreeding at certain time points, or dominance effects, are difficult for the average user within existing software (Boyko et al., 2008; Gutenkunst et al., 2009).
The second approach involves simulation of entire populations forward in time. Here, individuals reproduce each generation in proportion to their fitness. Several software programs exist to perform such simulations, including SFS_CODE (Hernandez, 2008), SLiM (Messer, 2013) and fwdpp (Thornton, 2014). These programs are designed to simulate large regions of genetic variation with linkage and arbitrary recombination and they track chromosomal regions through time, rather than the behavior of individual mutations. Thus, unless the recombination rate is exceptionally large, they are not directly comparable to the PRF model which assumes free recombination. Further, these simulations may not be ideal for generating trajectories of selected alleles because the trajectories of individual alleles may be altered through linkage to other selected sites (Felsenstein, 1965). Finally, some features, such as arbitrary amounts of inbreeding, are not included in SFS_CODE and SLiM. The recently released GO Fish (Lawrie, 2016) also models individual mutations, but currently does not model distributions of fitness effects nor does it have a complete stand-alone software package.
Here, we present a new program, PReFerSim, which combines these approaches and performs forward simulations under the PRF model for a single population. Two features distinguish PReFerSim from existing implementations of the PRF model and forward simulation programs. First, PReFerSim can track allele frequencies over time, and the ages and trajectories of individual mutations. Second, PReFerSim can simulate large amounts of genetic variation data with free recombination quickly.
2 Implementation
PReFerSim will simulate genetic variation data under the Poisson Random Field model. Each generation a Poisson number (with mean θ/2, where θ is the population-scaled mutation rate) of mutations enter the population. Information about each mutation is stored in a linked list. Mutations change frequency every generation due to natural selection and genetic drift. Genetic drift is implemented through a binomial sampling step. The program in written in ANSI C and utilizes the GNU Scientific Library for random number generation.
PReFerSim allows users to consider a variety of models of selection and demography. The program accepts demographic models with an arbitrary number of population size changes. Inbreeding can be modeled by specifying an inbreeding coefficient, F, which can be constant or change over time. Variable inbreeding is useful to simulate domestication events, or inbreeding following population decline, and is a unique feature of this software. Six distributions of fitness effects are included and the program allows selected mutations to have arbitrary dominance. PReFerSim will output summaries of genetic variation in the present day as well as at various times throughout the simulation. Trajectories of selected alleles can be generated and used in downstream coalescent simulations to model neutral genetic variation linked to the selected site (Ewing and Hermisson, 2010; Przeworski et al., 2005). Importantly, trajectories from PReFerSim can be conditioned to have any present-day frequency desired by the user. Such simulations can be used to estimate the ages of selected alleles and/or strength of selection (Ormond et al., 2016; Przeworski, 2003; Tishkoff et al., 2007), to estimate the rate of selective sweeps (Jensen et al., 2008), or to distinguish between hard and soft selective sweeps (Garud et al., 2015; Peter et al., 2012).
3 Results
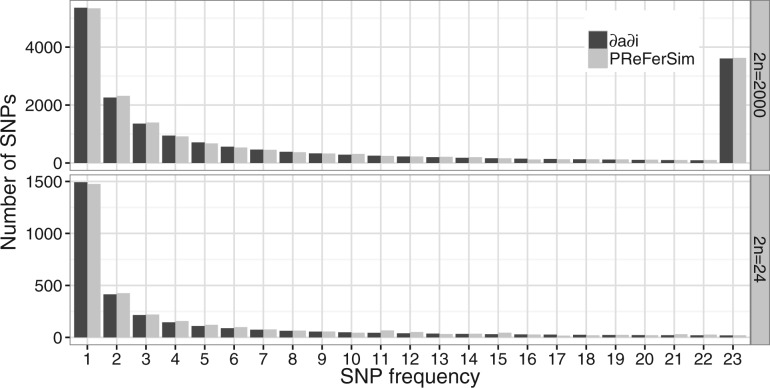
To demonstrate the performance of PReFerSim, we generated the site frequency spectrum (SFS) under a population expansion demographic model with a gamma distribution of fitness effects (Boyko et al., 2008). The SFS produced by PReFerSim is comparable to that generated using δaδi (Fig. 1). Additionally, ages of deleterious alleles obtained from PReFerSim are comparable to classical theoretical predictions made by Maruyama (1974; Supplementary Table S1). In sum, these results demonstrate that PReFerSim can generate predictions about the frequency spectrum and allele ages under complex models of demography and selection. Example applications of PReFerSim can be found in Marsden et al. (2016) and Robinson et al. (2016).
Fig. 1.
Comparison of the SFS between PReFerSim (gray) and the diffusion approximation (black; δaδi) for complex demographic models with purifying selection. Top: the SFS are similar in a sample of 2000 chromosomes. SNPs with counts greater than 22 are combined in the last bin. Bottom: SFS for a sample of 24 chromosomes. See Supplementary Text for a description of the model parameters
4 Conclusion
PReFerSim is a forward-in-time population genetic simulation program that can generate data under the PRF model considering complex demography and arbitrary models of selection and dominance. Given the growing interest in studying how demography affects deleterious variation across a wide range of taxa (Brandvain and Wright, 2016; Henn et al., 2015; Marsden et al., 2016; Schubert et al., 2014), there is a need for population genetic software that can easily simulate large amounts of data under complex evolutionary models. As such, we anticipate that PReFerSim will play an important role in future studies.
Supplementary Material
Acknowledgements
We thank Bernard Kim for the preparation of Figure 1.
Funding
This work was supported by the National Institutes of Health [R01 HG007089]; the Alfred P. Sloan Foundation [BR2014-009]; UC MEXUS-CONACYT doctoral fellowship [213627]; and a UCLA Dissertation Year Fellowship.
Conflict of Interest: none declared.
References
- Akashi H., Schaeffer S.W. (1997) Natural selection and the frequency distributions of “silent” DNA polymorphism in Drosophila. Genetics, 146, 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko A.R. et al. (2008) Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet., 4, e1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandvain Y., Wright S.I. (2016) The limits of natural selection in a nonequilibrium world. Trends Genet., 32, 201–210. [DOI] [PubMed] [Google Scholar]
- Bustamante C.D. et al. (2001) Directional selection and the site-frequency spectrum. Genetics, 159, 1779–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M.M., Plotkin J.B. (2008) The polymorphism frequency spectrum of finitely many sites under selection. Genetics, 180, 2175–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing G., Hermisson J. (2010) MSMS: a coalescent simulation program including recombination, demographic structure and selection at a single locus. Bioinf. Oxf. Engl., 26, 2064–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A. et al. (2006) The distribution of fitness effects of new deleterious amino acid mutations in humans. Genetics, 173, 891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1965) The effect of linkage on directional selection. Genetics, 52, 349–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garud N.R. et al. (2015) Recent selective sweeps in North American Drosophila melanogaster show signatures of soft sweeps. PLoS Genet., 11, e1005004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutenkunst R.N. et al. (2009) Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet., 5, e1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D.L. et al. (1994) Selection intensity for codon bias. Genetics, 138, 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn B.M. et al. (2015) Estimating the mutation load in human genomes. Nat. Rev. Genet., 16, 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez R.D. (2008) A flexible forward simulator for populations subject to selection and demography. Bioinf. Oxf. Engl., 24, 2786–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Sanchez E. et al. (2008) Population genetics of polymorphism and divergence under fluctuating selection. Genetics, 178, 325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J.D. et al. (2008) An approximate Bayesian estimator suggests strong, recurrent selective sweeps in Drosophila. PLoS Genet., 4, e1000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie D.S. (2016) Accelerating Wright-Fisher forward simulations on the graphics processing unit. bioRxiv, doi:10.1101/042622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden C.D. et al. (2016) Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proc. Natl. Acad. Sci., 113, 152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T. (1974) The age of an allele in a finite population. Genet. Res., 23, 137–143. [DOI] [PubMed] [Google Scholar]
- Messer P.W. (2013) SLiM: simulating evolution with selection and linkage. Genetics, 194, 1037–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormond L. et al. (2016) Inferring the age of a fixed beneficial allele. Mol. Ecol., 25, 157–169. [DOI] [PubMed] [Google Scholar]
- Peter B.M. et al. (2012) Distinguishing between selective sweeps from standing variation and from a de novo mutation. PLoS Genet., 8, e1003011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M. (2003) Estimating the time since the fixation of a beneficial allele. Genetics, 164, 1667–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M. et al. (2005) The signature of positive selection on standing genetic variation. Evol. Int. J. Org. Evol., 59, 2312–2323. [PubMed] [Google Scholar]
- Robinson J.A. et al. (2016) Genomic flatlining in the endangered Island Fox. Curr. Biol., 26, 1183–1189. [DOI] [PubMed] [Google Scholar]
- Sawyer S.A., Hartl D.L. (1992) Population genetics of polymorphism and divergence. Genetics, 132, 1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert M. et al. (2014) Prehistoric genomes reveal the genetic foundation and cost of horse domestication. Proc. Natl. Acad. Sci., 111, E5661–E5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K.R. (2014) A C ++ template library for efficient forward-time population genetic simulation of large populations. Genetics, 198, 157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff S.A. et al. (2007) Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet., 39, 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson S.H. et al. (2005) Simultaneous inference of selection and population growth from patterns of variation in the human genome. Proc. Natl. Acad. Sci. U. S. A., 102, 7882–7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.