Abstract
Motivation: The identification of functional modules from protein–protein interaction (PPI) networks is an important step toward understanding the biological features of PPI networks. The detection of functional modules in PPI networks is often performed by identifying internally densely connected subnetworks, and often produces modules with “core” and “peripheral” proteins. The core proteins are the ones having dense connections to each other in a module. The difference between core and peripheral proteins is important to understand the functional roles of proteins in modules, but there are few methods to explicitly elucidate the internal structure of functional modules at gene level.
Results: We propose NCMine, which is a novel network clustering method and visualization tool for the core-peripheral structure of functional modules. It extracts near-complete subgraphs from networks based on a node-weighting scheme using degree centrality, and reports subgroups as functional modules. We implemented this method as a plugin of Cytoscape, which is widely used to visualize and analyze biological networks. The plugin allows users to extract functional modules from PPI networks and interactively filter modules of interest. We applied the method to human PPI networks, and found several examples with the core-peripheral structure of modules that may be related to cancer development.
Availability and Implementation: The Cytoscape plugin and tutorial are available at Cytoscape AppStore. (http://apps.cytoscape.org/apps/ncmine).
Contact: kengo@ecei.tohoku.ac.jp
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Biological relationships among genes or proteins, such as gene coexpression and protein–protein interactions (PPI), are modeled as interaction networks or graphs, where a node corresponds to a gene or a protein and an edge represents an interaction between the connected node pair. Interaction networks sometimes contain several densely connected subnetworks, which are recognized as functional modules (Gao et al., 2009; Hartwell et al., 1999; Spirin and Mirny, 2003). Especially, Hartwell et al. emphasized the importance of understanding of interactions between modules when phenomenological analysis is performed. Proteins in the same subnetwork are considered to perform a specific biological task and tend to have related functions. Therefore, the detection of functional modules within a large interaction network is one of the fundamental steps to understand the biological functions of genes and proteins.
The task to find functional modules in interaction networks is called the network clustering problem and many algorithms have been proposed for this purpose. For example, Newman (2006) proposed a measure of modularity called ‘Q’ to quantify how well a network is divided into a number of subnetworks globally, and suggested a method to map the network clustering problem to the problem of maximizing Q. They tried to identify community structures from various social and biological networks. In the biological fields, Bader and Hogue (2003) developed the MCODE method to find protein complexes in PPI networks. The method first defines a score for each node, which reflects the connectivity of the node, and searches for groups of nodes with higher scores. With a different approach, Adamcsek et al. (2006) developed CFinder, which uses the clique-percolation method (CPM) (Rivera et al., 2010). This method searches for k-cliques (complete graphs with k nodes) and then combines the k-cliques. It can detect more dense subnetworks compared with other methods, like MCODE. Rivera et al. (2010) proposed NeMo, which employs a hierarchy-based clustering strategy and uses a log odds score reflecting the degree of sharing neighboring nodes. Yanjun et al. (Qi et al., 2008) and Lei et al. (Shi et al., 2011) used machine learning techniques to detect protein complexes or functional modules by a combination of topological of networks given as input data and features of known protein complexes. Graph clustering problem is one of the most hot topics in the field of computer science, and several algorithms are being developed recently (Andersen et al., 2016; Seok-Joo et al., 2016; Uno et al., 2015; Zhang and Zou, 2015). All of these methods have distinct characteristics and could produce interesting biological modules, but they are unable to handle an important feature of biological networks, that is, overlap among modules. Careful investigations of biological networks have suggested that some functional modules in an interaction network share the same nodes (Palla et al., 2005; Rives and Galitski, 2003). In other words, some functional modules have overlapping regions. In the field of graph theory, this problem is called the ‘soft-clustering’ problem.
The importance of the soft clustering problem in biological networks may be rephrased as the ‘core-peripheral structure’ of functional modules (Gavin et al., 2006; Kuhner et al., 2009). When two tightly related protein groups share many proteins, which is typical in a signal network cascade, the two groups will be identified as a single functional module using the current hard clustering methods. From the viewpoint of a biological network, the core proteins play an important role in the module and peripheral proteins (with fewer edges) interact with a subset of core proteins and may be involved in a specific function, such as the regulation of the network. Previously proposed methods cannot effectively elucidate the relationships between clusters that share proteins and produce many independent clusters when overlaps are permitted. There exists few graph clustering methods that also enables visualization of relationships between clusters.
To overcome these limitations of previous methods, we developed a method called NCMine, which focuses on the identification of modules in biological networks and the extraction of biologically meaningful clusters. NCMine detects functional modules from biological interaction networks by constructing ‘local near-cliques’ using a node-weighting scheme based on degree centrality, and by iteratively merging local near-cliques according to cluster overlap. The combination of the local-cluster construction phase and the merge phase enables the detection of overlapping proteins between modules and reveals relationships among modules at the same time. The ‘near-clique’ is a graph structure lacking a few edges compared with the complete graph. Clique or complete graphs are mathematically well studied and easy to enumerate, but near-clique searches are quite difficult to implement. Several previous studies have used clique detection algorithms to find functional modules (Li et al., 2005; Liu et al., 2009), but the clique is a stringent requirement for biological phenomena because some elements of functional modules may interact with only the part of the module (i.e. a subset of its members) and because experimental nose will hide some existing edges in the modules. Wu et al. first tried to find core-peripheral structure for biological networks (Wu et al., 2009). Wu et al. first searched for clusters and extended them to find peripheral nodes, while our methods first searched for clusters, merged the found clusters and identified core part of the overlapping clusters as described later.
2 Methods
2.1 NCMine algorithm
The proposed method has two phases. In the first phase, several ‘local’ near-clique clusters are constructed according to connectivity between nodes. In the second phase, the local clusters are iteratively merged based on the degree of overlap of the clusters.
2.1.1 Phase 1: local cluster construction
We first assigned weights to each node based on the connectivity of each node. In this clustering method, node degree centrality was used to assign node weight. Then, we selected the node with the highest node weight value, which we called the seed node, and considered it a local cluster with a single node. Next, we added nodes adjacent to the initial cluster to the local cluster. All adjacent nodes were examined for addition according to weight order, and nodes were added if the cliqueness of the new cluster exceeded a pre-defined threshold and if the cliqueness-change between the new and old cluster was lower than a threshold, where cliqueness was a measure of the similarity between a graph and the complete graph with the same number of nodes, defined as follows:
Cliqueness is an important factor to evaluate the goodness of a cluster, but heavy reliance on cliqueness may be inadequate in biological networks. Accordingly, we also introduced cliqueness-change as another indicator of local cluster expansion. Cliqueness-change avoids the inclusion of a ‘tail’ node, which is a node that has only a few edges with other nodes in a cluster. In our expansion algorithm, some tail nodes were included when clusters became large, unless cliqueness-change was introduced.
Local clusters were constructed from every node in the network in decreasing order of node weight, and they were merged in the second phase as described in the next section. The cost of calculation for the first phase of the NCMine algorithm is approximately , where n represents the number of nodes in a graph.
2.1.2 Phase 2: merge of local clusters
The local clusters constructed in the previous phase were iteratively merged according to the overlap of clusters. The degree of overlap was measured by the Jaccard coefficient when a cluster and a node were considered a set and an element. If the overlap between two clusters was greater than a pre-defined overlap threshold and the cliqueness of the merged cluster was greater than a pre-defined threshold, the two clusters were merged. However, if the overlap between a pair of clusters exceeded the pre-defined threshold and if the cliqueness of the merged cluster did not satisfy the threshold, they were not merged and treated as a cluster hierarchy in the final sets of clusters, where the overlapping region of the two clusters was considered a parent cluster and each element cluster was depicted as a child clusters The local clusters were merged according to the overlapping region of two clusters, and the method could assign a node to multiple clusters; thus, it was a type of soft-clustering method. It should be noted that the merging of clusters was performed according to the order of cluster generation because it was expected that nodes in a near-clique cluster tend to have similar node degrees and similar node degree centrality, and similar clusters tend to be close in the order of cluster construction. Based on the nature of node degree centrality, we could reduce the number of comparisons necessary to check cluster overlap.
After the merge phase, we obtained the final set of clusters. The calculation cost for the second phase is approximately , where n represents the number of nodes in a graph, because the maximum number of clusters generated in the first phase is n. Therefore, the overall cost is also . We implemented our algorithm as a Cytoscape (Smoot et al., 2011) plugin, as described later. In this implementation, the default threshold values for cliqueness-change, overlap and cliqueness are set to 0.2, 0.6 and 0.6, respectively, according to the results of our experiments described in the next section. In addition, pseudo code and flowchart of the NCMine algorithm are available in supplementary material (Supplementary Fig. S1).
It is noteworthy that the final solution returned by our method is not necessarily a globally optimal solution. There are two types of clustering methods with respect to clustering results: methods that return globally optimal solutions and those that generate local optimum solutions. The global optimum algorithms, like Newman’s Q described in the previous section, use a measure that indicates how isolated one cluster is from other clusters, and the clustering is performed by the optimization of the measure. The clustering results contain clusters that are well isolated; therefore, the method is a global optimum method. In contrast, our method only considers limited parts of the whole network when constructing each local cluster and thus it should be referred to as a locally optimal method.
2.2 Evaluation of clustering methods by artificially generated networks
To evaluate our algorithm, we generated artificial networks in two steps. First, we generated a number of near-cliques with random cliqueness values ranging from 0.6 to 1.0; second, we established edges between randomly chosen pairs of nodes among all near-clique clusters generated in the first step.
We applied our method and previously proposed methods [MCODE (Bader and Hogue, 2003), CFinder (Adamcsek et al., 2006) and NeMo (Rivera et al., 2010)] for a comparative analysis. We compared extracted clusters with embedded clusters and then calculated precision and recall. A cluster identified by a method was judged to be included in one of the embedded clusters if the pair of two clusters was the best pair in a bidirectional manner. The similarity of clusters was calculated by the Jaccard coefficient, and if the similarity for a pair of two clusters was higher than 0.6, the clusters were considered the same cluster.
The recall and precision were calculated by the following formulae:
Precision = True_Positive/(True_Positive + False_Positive)
Recall = True_Positive/(True_Positive + False_Negative),
where True_Positive represents clusters that are included in the set of extracted clusters and embedded clusters, False_Positive represents clusters that are included in extracted clusters, but not in embedded clusters and False_Negative represents clusters that are included in embedded clusters, but not in extracted clusters.
We compared our method with three established methods, MCODE (Bader and Hogue, 2003), CFinder (Adamcsek et al., 2006) and NeMo (Rivera et al., 2010). The details of the methods are described in the original articles, but for convenience, we briefly describe some features of the algorithms.
The MCODE algorithm contains three phases: (1) vertex weighting, (2) cluster construction based on vertex weights and (3) post-processing. In the first phase, the weight of each node is calculated, where vertex weight is defined as node-density * degree. In the second phase, a cluster is constructed from the node with the highest weight by adding neighboring nodes with higher weights. In the MCODE algorithm, one protein may be assigned to multiple clusters; however, the relationship among clusters that share some number of proteins cannot be determined. The local cluster construction phase of NCMine employs a similar strategy to that of MCODE; however, evaluation functions that are used to construct local clusters are different, and MCODE tends to include hub proteins in a cluster and makes relatively larger clusters, as shown in subsequent sections.
CFinder employs the CPM (Reid et al., 2012) to detect functional modules. The basis for the CPM algorithm is the detection of maximal cliques from networks, which are combined according to their overlap; it detects k-cliques that share k − 1 nodes. When we apply this method to biological interaction networks, the clique is a hard requirement because some members of functional modules may interact with only a subset of module members. Another problem is that nodes that do not participate any k-clique cannot be contained within any clusters, and a k-clique community cannot be constructed depending on nodes and edges in a graph.
In the NeMo algorithm, a log odds score called rab is used to observe an edge between two nodes, i.e. node a and node b. The rab scores are assumed to follow a Poisson distribution, and rab approximately equals the log odds ratio between the probability of rab under the alternative hypotheses and null hypotheses (Rivera et al., 2010). In the NeMo clustering process, log odds scores for all node pairs in a network are calculated, followed by hierarchical agglomerative clustering using the scores. Several previous studies (Palla et al., 2005; Rives and Galitski, 2003) have shown that the form or composition of a protein complex may differ depending on conditions; however, NeMo cannot assign a protein to multiple clusters and accordingly cannot account for this observed property of biological networks.
2.3 Evaluation of clustering methods by human protein–protein interaction networks
We also evaluated our method using a real biological network. We used human PPI data obtained from the Human Protein Reference database (HPRD, Release 9) (Keshava Prasad et al., 2009). The data obtained from HPRD contained (i) binary PPI data and (ii) protein complex data. We applied clustering methods to binary PPI data and evaluated the results by comparisons with the protein complex data. In addition, we checked the biological relevance of the extracted clusters using Gene Ontology (GO) enrichment tests. GO annotations were obtained from the Gene Ontology Consortium (http://www.geneontology.org) in September 2012. Enrichment was evaluated using Fisher’s exact test and a P-value of 0.05 was used as the threshold for significance. To correct for multiple comparisons, the Bonferroni correction method was used. We considered a cluster ‘biologically meaningful’ if one or more GO terms were shared among more than 60% of cluster members. These evaluations were also performed using MCODE (Bader and Hogue, 2003), CFinder (Adamcsek et al., 2006) and NeMo (Rivera et al., 2010).
2.4 Materials
The Kyoto Encyclopedia and Genes and Genomes (KEGG) (Kanehisa et al., 2014) pathway annotation information for genes or proteins was retrieved from the KEGG pathway website (http://www.genome.jp/kegg/pathway.html).
The following clustering methods were used: MCODE 1.4.0.beta2 (released December 2012) implemented in Cytoscape (Smoot et al., 2011) 3.0.0, CFinder 2.0.5 (released April 2011) and NeMo 1.4 (released February 2010) implemented in Cytoscape 2.7. All experiments related to the cluster extraction process were performed using the same desktop computer (Apple iMac, 3.4-GHz Intel Core i7 CPU, 32-GB RAM).
Results
3.1 Evaluation of clustering methods by artificially generated networks
We generated artificial networks with 10 000 nodes and with 500 near-cliques embedded, where the 500 near-cliques in each network had a size of 7.2 ± 2.83 and a cliqueness of 0.80 ± 0.11. Table 1 shows the comparative performance results. All methods ran in a comparable period of time (≤10 s), except NeMo, possibly owing to implementation problems. Based on the algorithms, large differences were not expected.
Table 1.
Summary of extracted clusters from an artificially generated network
| Running time | # of found clusters | Cluster size (avg, SD) | Cluster cliqueness (avg, SD) | |
|---|---|---|---|---|
| NCMine | 10 s | 706 | 4–14 (6.77, 1.95) | 0.60–0.97(0.67, 0.06) |
| MCODE | 8 s | 370 | 3–53 (7.72, 5.84) | 0.09–1.00(0.77, 0.24) |
| CFinder | 10 s | 1340 | 3–13 (8.41, 2.52) | 0.57–1.00(0.87, 0.10) |
| NeMo | 1 m 30 s | 953 | 4–13 (6.54, 2.31) | 0.00–1.00(0.57, 0.37) |
CFinder extracted the most clusters among the methods, and MCODE identified the fewest clusters. Since CFinder invoked the CPM algorithm with various ‘k-core’ size parameters and returns all clusters, CFinder was expected to produce the highest number of clusters. In the artificial network, this was true, but as shown in Table 1, the actual number of clusters depended on network structure. MCODE identified the fewest clusters. MCODE finds clusters based on local node-density (node-degree * core-density), and the evaluation function tended to combine multiple blocks into a single cluster. The evaluation function of MCODE also tended to produce clusters with radically connected nodes, and thus MCODE produced relatively bigger clusters and a smaller number of clusters. This tendency was consistent using the human PPI network (Table 3).
Table 3.
Summary of extracted clusters from the HPRD human protein–protein interaction (PPI) network using each method
| Running time | # of extracted clusters (A) | Cluster size (avg, SD) | Cluster cliqueness (avg, SD) | # of biologically meaningful clusters (B) | Ratio (B/A) | |
|---|---|---|---|---|---|---|
| NCMine | 15 s | 2309 (102*) | 3–21 | 0.600–1.000 | 1259 | 0.54 |
| (4.880, 2.887) | (0.764,0.162) | |||||
| MCODE | 7 s | 250 | 2–160 | 0.016–1.000 | 93 | 0.45 |
| (9.359,19.945) | (0.654, 0.337) | |||||
| CFinder | 28 s (10 s**) | 764 | 3–1010 | 0.017–1.000 | 315 | 0.41 |
| (7.055,38.954) | (0.900,0.164) | |||||
| NeMo | 180 s | 1510 | 4–68 | 0.000–1.000 | 784 | 0.52 |
| (8.940, 7.456) | (0.128, 0.244) |
*The number in the parenthesis is the number of clusters that are involved in core-peripheral structures.
**The number in the parenthesis is calculation time with approx. option = 0.1 s.
Regarding the size of clusters, MCODE produced clusters of various sizes ranging from 3 to 53 (average ± SD = 7.72 ± 5.85), while NCMine, CFinder and NeMo extracted similarly sized clusters. The higher variance observed using MCODE reflects the evaluation function described above. MCODE produced clusters with a wide range of cliqueness (0.09–1.0) compared with NCMine and CFinder. We could not explain the observation, but NeMo also produced clusters with a wide range of cliqueness (0.0–1.00). It should be noted that the SD of cliqueness for clusters extracted by NCMine was extremely low because NCMine specialized in grabbing near-cliques in networks, and attempted to find the most ‘loose’ local clusters that satisfied a pre-defined cliqueness threshold. As a result, we can expect that the average cluster size for NCMine will be much smaller than those of the other methods.
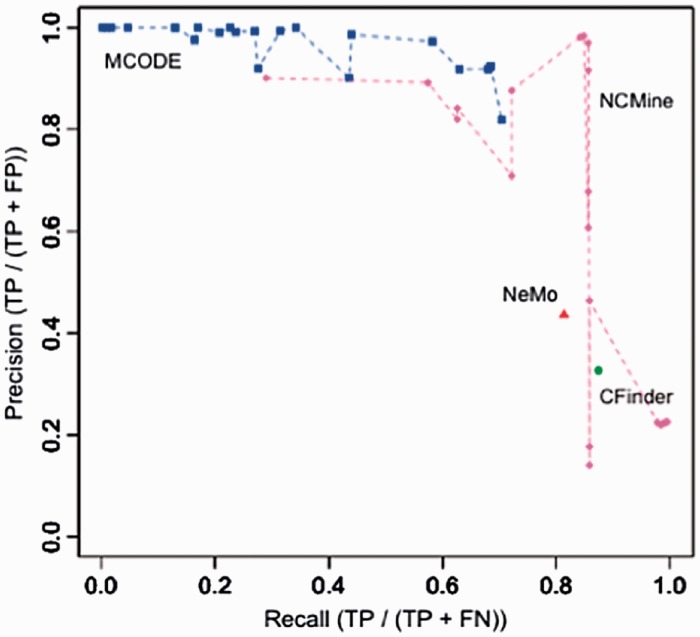
We also compared extracted clusters and 500 embedded near-clique clusters by changing the clustering parameters for each method and calculating the recall and precision for each set of parameters (Fig. 1). A dot in Figure 1 corresponds to a pair of recall and precision values calculated for a set of parameters. MCODE and NCMine had 4 and 3 parameters, respectively. To calculate the recall and precision values, we fixed one parameter and changed the other parameters in the ranges and widths shown in Table 2. In this evaluation, we generated 1000 artificial networks and calculated the recall and precision values for all networks. We found that the precision and recall curves were very similar for all cases, and thus one typical example is shown in Figure 1. NeMo and CFinder have no adjustable parameters, and they were executed only once with the pre-defined configuration.
Fig. 1.
Recall and precision calculated from cluster extraction results by comparing extracted clusters and actual embedded clusters in artificial networks. Higher recall and precision indicate better performance
Table 2.
Parameter settings for MCODE and NCMine used for performance tests with artificially generated networks
| Default value | Range of value used in tests (step) | |
|---|---|---|
| MCODE | ||
| Degree cutoff | 2 | 2 – 10 (2) |
| Node score cutoff | 0.2 | 0.0–1.0 (0.2) |
| K-core cutoff | 2 | 2–10 (2) |
| Degree cutoff | 2 | 2–10 (2) |
| NCMine | ||
| Cliqueness threshold | 0.6 | 0.0–1.0 (0.2) |
| Merge threshold | 0.6 | 0.0–1.0 (0.2) |
| Cliqueness-change threshold | 0.2 | 0.0–1.0 (0.2) |
As shown in Figure 1, NCMine achieved a strong tradeoff between recall and precision with respect to near-clique extraction when we selected appropriate parameters. We obtained the highest precision using the following settings: cliqueness threshold = 0.6, merge threshold = 0.6 and cliqueness change = 0.6. MCODE achieved high precision, but its recall values were low. MCODE tended to extract larger clusters, and thus the high precision indicated that almost all embedded clusters were included in the extracted clusters, and the low recall implied that only parts of embedded clusters were found by MCODE. In contrast, NeMo and CFinder had high recall values and low precision values, indicating that too many false positive clusters were generated using these methods, consistent with the large number of extracted clusters shown in Table 1.
3.2 Application to the human protein–protein interaction network
We applied our method to the human PPI network, and evaluated the results by calculating the proportion of clusters that were ‘biologically meaningful’, i.e. those that had one or more significantly enriched GO terms. The PPI network used in this study were taken from HPRD release 9, where 9670 proteins (nodes) and 3922 interactions (edges) were available. The results are summarized in Table 3. All methods, except NeMo, were completed in comparative lengths of time. This observation was consistent with the results obtained for the artificial network. Time complexity of the core clustering parts of MCMine, CFinder and NeMo are O(n2), and that of MCODE is O(nmh3). Therefore, the difference of running time of each method would be due to the pre- and post-process of clustering or the difference of implementations. MCODE extracted the fewest clusters, consistent with the artificially generated networks, but NCMine extracted far more clusters that the other methods using the PPI network. NCMine tended to extract near-cliques that satisfy given thresholds as much as possible, indicating that the real PPI network contained a high number of clique-like sub-graphs and that it is important to detect clique-like sub-graphs. The importance of clique-like sub-graphs was also confirmed by the higher percentage of biologically meaningful clusters, as described by the ‘Ratio’ estimate in Table 3 for our method. NCMine detected a much larger number of clique-like clusters and more than half (54%) of the clusters were biologically meaningful. Note that CFinder extracted clusters with higher cliqueness values (0.900, on average) than those of NCMine (0.764, on average), but the proportion of clusters that were biologically meaningful for CFinder was relatively low, which may indicate that near cliques with modest cliqueness were more important than those with higher cliqueness. MCODE extracted the smallest number of biologically meaningful clusters and the ratio of meaningful clusters to the total number of extracted clusters was relatively low. This might be explained by the MCODE evaluation function, which tended to allow multiple clusters to be combined into a single large cluster; clusters with no functional relationships might be merged in PPI networks.
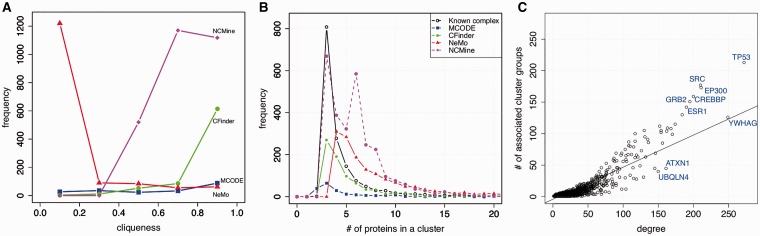
We also checked the concordance of extracted clusters with known protein complexes. For this purpose, the distributions of cluster sizes and the cliqueness of known protein complexes and extracted clusters were compared (Fig. 2A andB) using protein complexes obtained from the ‘protein complex dataset’ in HPRD. We observed that the sizes of known protein complexes were widely distributed, and the distribution of clusters extracted by NCMine had two peaks. When we neglected the second peak, the distribution obtained using NCMine was similar to that of known complexes (Fig. 2A). With respect to cliqueness, we observed extensive differences in the distribution between the clustering methods and known complexes (Fig. 2B), where the cliqueness of a known protein complex was calculated using binary PPI data in HPRD. Experimental methods to detect interactions between proteins and methods to identify protein complex members are typically different.
Fig. 2.
(A) Comparison of cliqueness of clusters extracted by each method. (B) Comparison of the sizes of extracted clusters and known protein complexes (C) Relationship between node degree and cluster membership calculated from the NCMine cluster extraction results. Genes included in the plot are listed in Supplementary Table S1(A) and (B)
One important feature of NCMine is soft clustering. Accordingly, we observed the number of cluster groups associated with proteins and its relationship to the number of interactions (Fig. 2C). On average, using the NCMine clustering algorithm, the number of proteins belonging to clusters of nodes tended to increase as node interactions increased, but there were some exceptional cases, as shown in Figure 2C. Interestingly, most of the exceptional cases were related to cancer development (these points were above the regression line in Fig. 2C). Proteins below the regression line, such as ATXN1 and UBQLN4, had relatively few cluster memberships and little relationship with cancer development. This observation suggests that a higher degree of cluster membership is a good indicator of the importance of proteins involved in cancer.
3.3 Examples of clusters detected by NCMine from the human PPI network
To illustrate the biological meaning of the clusters obtained by NCMine, we selected two examples.
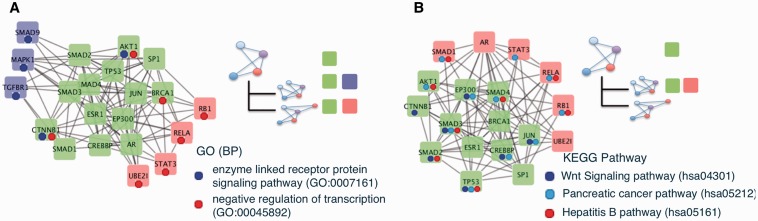
Example A is summarized in Figure 3A. The cluster was extracted by NCMine from the HRPD PPI network, which revealed relationships between two clusters related to cancer development. The nodes shown in green in Figure 3A represent the proteins in a parent cluster (i.e. a core cluster) and red nodes were in child clusters (i.e. peripheral clusters). A visual representation of the cluster hierarchy is shown on the upper right-hand portion of the figure. Based on the KEGG pathway database (Kanehisa et al., 2014), proteins shown with blue, light blue, and red dots had functions in the Wnt signaling pathway, pancreatic cancer, and the hepatitis B pathway, respectively. The Wnt signaling pathway is related to embryonic development and cancer development, and is considered a key pathway in the development of various cancer types (Nusse and Nusse, 2005). The pancreatic cancer and hepatitis B pathways are specific. The relationships among these core proteins and peripheral proteins can be clearly seen in the cluster relationships shown in Figure 3A, as indicated by the limited distribution of blue dots, indicating the Wnt signaling pathway, in the core cluster (genes in green boxes).
Fig. 3.
(A) Two sets of peripheral proteins (blue and red) with the same core proteins (green) are shown. In this example, most of the proteins were related to cancer development. However, the proteins that participate in blue peripherals and red peripherals had different functions. Genes in the clusters: AKT1: v-akt murine thymoma viral oncogene homolog 1; AR: Androgen receptor; BRCA1: Breast cancer 1, early onset; CREBBP: CREB-binding protein; CTNNB1: Catenin (cadherin-associated protein), beta 1, 88 kDa; EP300: E1A-binding protein p300; ESR1: Estrogen receptor 1; JUN: Jun proto-oncogene; RB1: Retinoblastoma 1; RELA: v-rel avian reticuloendotheliosis viral oncogene homolog A; SMAD1: SMAD family member 1; SMAD2: SMAD family member 2; SMAD3: SMAD family member 3; SMAD4: SMAD family member 4; SP1: Sp1 transcription factor; STAT3: Signal transducer and activator of transcription 3 (acute-phase response factor); TP53: Tumor protein p53; UBE2I: Ubiquitin-conjugating enzyme E2I (B) Most of the core-proteins (green) were involved in the Wnt signaling pathway, which is related to cancer, generally; however, peripheral-proteins involved specific cancer development pathways. Genes in the clusters: AKT1: v-akt murine thymoma viral oncogene homolog 1; AR: Androgen receptor; BRCA1: Breast cancer 1, early onset; CREBBP: CREB-binding protein; CTNNB1: Catenin (cadherin-associated protein), beta 1, 88 kDa; EP300: E1A-binding protein p300; ESR1: Estrogen receptor 1; JUN: Jun proto-oncogene; MAPK1: Mitogen-activated protein kinase 1; RB1: Retinoblastoma 1; SMAD1: SMAD family member 1; SMAD2: SMAD family member 2; SMAD3: SMAD family member 3; SMAD4: SMAD family member 4; SMAD9: SMAD family member 9; SP1: Sp1 transcription factor; STAT3: Signal transducer and activator of transcription 3 (acute-phase response factor); TGFBR1: Transforming growth factor, beta receptor 1; TP53: Tumor protein p53; UBE2I: Ubiquitin-conjugating enzyme E2I
Example B is summarized in Figure 3B, which shows another example of extracted clusters. The set of blue-colored proteins and green-colored proteins in Figure 3B represent one of the clusters found by NCMine, and the set of green-colored proteins along with red-colored proteins is another single cluster. The set of green-colored proteins were the intersection of two clusters and NCMine automatically detected these three clusters and their relationship, as shown in the hierarchy of clusters at the upper right of Figure 3B. Again, by inspecting the KEGG (Kanehisa et al., 2014) pathway database, we found that all proteins in Figure 3B were related to the pancreatic cancer pathway (hsa05212); however, GO terms associated with the ‘enzyme linked receptor protein signaling pathway’ (GO:0007167) and ‘negative regulation of transcription’ (GO:0017148) were differently distributed and only found in the peripheral clusters, as indicated by blue and red dots. This indicated that proteins in the green box may play a central role in pancreatic cancer development and participated in other pathways along with the peripheral proteins shown in blue boxes and red boxes.
3.4 Implementation and availability
Cyotoscape (http://cytoscape.org/) is an open source platform for analysis and visualization of networks. Cytoscape core provides basic functions for handling network (interaction) data, for example, parsing and loading network files, layout nodes in networks, querying networks, and so on. Cytoscape also provides several APIs (Application Programming Interface) to manipulate network data loaded into Cytoscape core, which can be used to extend the Cytoscape functions.
Using the APIs, we implemented our network analysis method and original network visualization. NCMine Cytoscape plugin is built on top of Cytoscape core and exposed APIs. NCMine plugin interacts with Cytoscape core and obtain network data, and then it carries out NCMine algorithm and returns cluster extraction result to Cytoscape core for visualization. The NCMine plugin was implemented in Java, and the plugin is distributed through Cytoscape App Store (http://apps.cytoscape.org/), which is an official repository of Cytoscape plugins (apps). Plugins in the repository can be installed via Cytoscape’s AppManager.
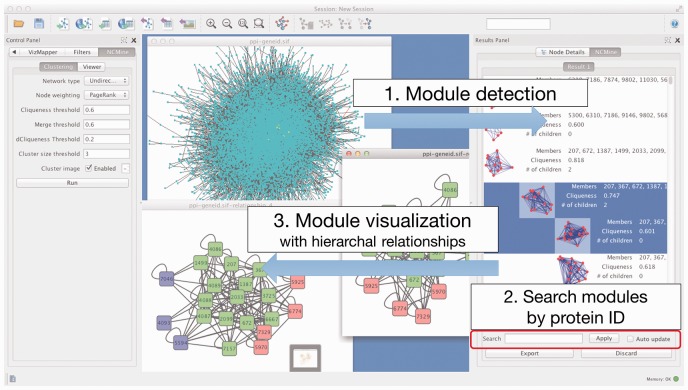
Figure 4 shows a basic workflow with NCMine. The NCMine plugin can detect near-clique clusters from input data, and can be used to visualize their hieratical relationships. Quick tutorial of the plugin is also available from Cytoscape App Store page.
Fig. 4.
Cytoscape plugin for NCMine, and basic workflow
4 Conclusion
We developed a new method, NCMine, to identify and visualize functional modules with consideration of the core-peripheral structure of modules. The core idea of the NCMine algorithm is (i) the extraction of complete subgraph-like structures from networks based on a node-weighting scheme and (ii) merging local clusters based on module overlap. In a comparative analysis with other methods, NCMine achieved better performance. We also showed a good example of hierarchical structures in functional modules by an analysis of a human PPI network. However, based on the comparative analysis, each method had unique characteristics, and it might suggest that it is important to select a clustering method suitable for the purpose.
Supplementary Material
Acknowledgements
The super-computing resources were provided by the Human Genome Center (the University of Tokyo) and the National Institute of Genetics, Research Organization of Information and Systems, Japan.
Funding
This research was partially supported by the Platform Project for Supporting in Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science) from the Japan Agency for Medical Research and Development (AMED). K.K. was supported by JSPS KAKENHI Grant Number 15H02773.
Conflicts of Interest: none declared.
References
- Adamcsek B. et al. (2006) CFinder: locating cliques and overlapping modules in biological networks. Bioinformatics, 22, 1021–1023. [DOI] [PubMed] [Google Scholar]
- Andersen R. et al. (2016) Almost optimal local graph clustering using evolving sets. J. ACM, 63, 1–31. [Google Scholar]
- Bader G., Hogue C. (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics, 4, 2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L. et al. (2009) Clustering algorithms for detecting functional modules in protein interaction networks. J. Bioinform. Comput. Biol., 7, 217–242. [DOI] [PubMed] [Google Scholar]
- Gavin A.C. et al. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature, 440, 631–636. [DOI] [PubMed] [Google Scholar]
- Hartwell L.H. et al. (1999) From molecular to modular cell biology. Nature, 402, C47–C52. [DOI] [PubMed] [Google Scholar]
- Kanehisa M. et al. (2014) Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res., 42, D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshava Prasad T.S. et al. (2009) Human Protein Reference Database–2009 update. Nucleic Acids Res., 37, D767–D772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhner S. et al. (2009) Proteome organization in a genome-reduced bacterium. Science, 326, 1235–1240. [DOI] [PubMed] [Google Scholar]
- Li X.L. et al. (2005) Interaction graph mining for protein complexes using local clique merging. Genome Inform., 16, 260–269. [PubMed] [Google Scholar]
- Liu G. et al. (2009) Complex discovery from weighted PPI networks. Bioinformatics, 25, 1891–1897. [DOI] [PubMed] [Google Scholar]
- Newman M.E.J. (2006) Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA, 103, 8577–8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R., Nusse R. (2005) Wnt signaling in disease and in development. Cell Res., 15, 28–32. [DOI] [PubMed] [Google Scholar]
- Palla G. et al. (2005) Uncovering the overlapping community structure of complex networks in nature and society. Nature, 435, 814–818. [DOI] [PubMed] [Google Scholar]
- Qi Y. et al. (2008) Protein complex identification by supervised graph local clustering. Bioinformatics, 24, i250–i258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid F. et al. (2012) Percolation computation in complex networks. In: Proceedings of the 2012 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining, ASONAM 2012. Kadir Has University, Istanbul, Turkey, pp. 274–281.
- Rivera C.G. et al. (2010) NeMo: network module identification in cytoscape. BMC Bioinformatics, 11 Suppl 1, S61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rives A.W., Galitski T. (2003) Modular organization of cellular networks. Proc. Natl. Acad. Sci. USA, 100, 1128–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok-Joo L. et al. (2016) An efficient parallel graph clustering technique using Pregel. In, International Conference on Big Data and Smart Computing (BigComp). IEEE, Hong Kong, pp. 370–373.
- Shi L. et al. (2011) Protein complex detection with semi-supervised learning in protein interaction networks. Proteome Sci, 9, S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot M.E. et al. (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics, 27, 431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirin V., Mirny L.A. (2003) Protein complexes and functional modules in molecular networks. Proc. Natl. Acad. Sci. USA, 100, 12123–12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno T. et al. (2015) Micro-clustering: finding small clusters in large diversity. http://arxiv.org/abs/1507.03067.
- Wu M. et al. (2009) A core-attachment based method to detect protein complexes in PPI networks. BMC Bioinformatics, 10, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Zou X. (2015) A new method for detecting protein complexes based on the three node cliques. IEEE/ACM Trans. Comput. Biol. Bioinform., 12, 879–386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.