Abstract
Motivation: Recent studies reveal an important role of non-coding circular RNA (circRNA) in the control of cellular processes. Because of differences in the organization of plant and mammal genomes, the sensitivity and accuracy of circRNA prediction programs using algorithms developed for animals and humans perform poorly for plants.
Results: A circRNA prediction software for plants (termed PcircRNA_finder) was developed that is more sensitive in detecting circRNAs than other frequently used programs (such as find_circ and CIRCexplorer), Based on analysis of simulated and real rRNA-/RNAase R RNA-Seq data from Arabidopsis thaliana and rice PcircRNA_finder provides a more comprehensive sensitive, precise prediction method for plants circRNAs.
Availability and Implementation: http://ibi.zju.edu.cn/bioinplant/tools/manual.htm.
Contact: fanlj@zju.edu.cn
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Non-coding circular RNA (circRNA) is a covalently continuous closed loop that usually originates from exonic regions (named exonic circRNA), but can also arise from intronic and intergenic regions. CircRNAs can function as a miRNA sponge (Hansen et al., 2013; Memczak et al., 2013) and have the potential to enhance transcription of their host genes (Li et al., 2015; Zhang et al., 2013). The emergence of rRNA-depleted high-throughput RNA-Seq technology provides a revolutionary approach for the systematic discovery of circRNAs in various species, including human, mouse, Arabidopsis and rice (Lu et al., 2015; Ye et al., 2015).
A robust method for circRNA identification is an important tool for investigating the role of these molecules. The available circRNA prediction methods (e.g. find_circ and CIRCexplorer) were primarily developed for use with human or animal datasets (Memczak et al., 2013; Pan and Xiong, 2015; Salzman et al., 2013; Szabo et al., 2015; Zhang et al., 2014). There are large differences between mammal and plant genomes and therefore the prediction accuracy and sensitivity of detecting circRNAs in plants using the currently available methods are relatively low (Ye et al., 2015). In this study, we developed a software (termed PcircRNA_finder) that shows a more comprehensive ability and greater sensitivity and precision in predicting circRNAs in plants.
2 Materials and Methods
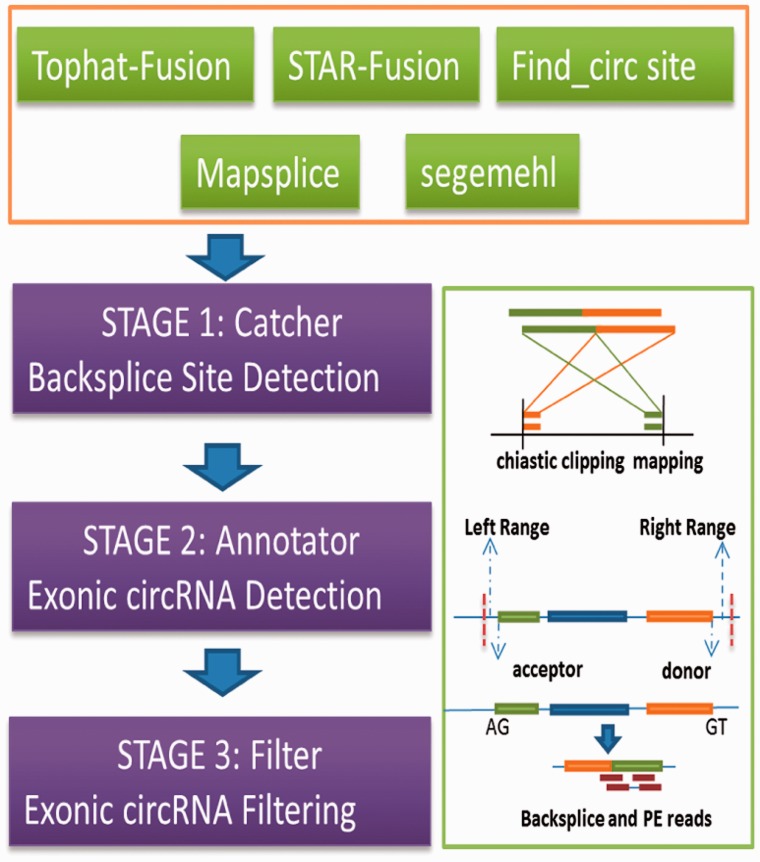
PcircRNA_finder is mainly designed for exonic circRNA prediction and consists of three modules as shown in Figure 1. These modules are: (i) Catcher, which is used to collect all backsplice sites by chiastic clipping mapping of PE reads based on available main fusion detection methods, including Tophat-Fusion (Kim and Salzberg, 2011), STAR-Fusion (Dobin, et al., 2013), find_circ (Memczak et al., 2013), Mapsplice (Wang et al., 2010) and segemehl (Hoffmann et al., 2014). Among these candidate backsplice sites, false positive sites will be filtered out in the Filter module. The increased read mapping accuracy in our program excludes some false predictions dues to the high copy number of genes in plants (Supplementary Data). (ii) Annotator, that can be used to annotate the candidate exonic backsplice sites based on available gene annotation. Recent studies have demonstrated that circRNA's backsplicing site is flexible and alternative splicing of circRNAs is prevalent (Starke et al., 2015; Szabo et al., 2015). Much of the alternative splicing of circRNAs occurred near by canonical splicing sites (Szaboet al., 2015; Starke et al., 2015) and therefore, 5-bp flanking the two canonical backsplice sites (acceptor and donor) were allowed for our candidate backsplice sites and (iii) Filter, which is a quality control module for the above candidate circRNAs. It creates a pseudoRef file with the flanking sequences of chiastic backsplice sites and then maps raw reads to it and confirms the backsplice sites. It also requires that the candidate circRNAs contain at least one of two kinds of splicing signals, either a U2 based spliceosome (usually with a consensus sequence of GT-AG and GC-AG) and a U12-based minor spliceosome (usually with a consensus sequence of AT-AC) (Reddy et al., 2013; Staiger and Brown, 2013).
Fig. 1.
The flowchart of PcircRNA_finder for circRNA prediction. It consists of three modules (stages)
3 Benchmark
To test the performance of PcircRNA_finder, we first compare it with two popular circRNA finding algorithms (find_circ and CIRCexplorer) using a simulation dataset for the analysis. Simulated RNA-Seq data (paired end reads, 100 ;bp and 6000 backsplicing reads for each sample) were generated by randomly choosing 200 chiastic transcripts based upon the Arabidopsis thaliana and rice genome annotations, respectively (Supplementary Data). The sensitivity, precision and sensitivity ;+ ;precision (a comprehensive value) (Chuang et al., 2016) was used to evaluate the performance of the three methods. The results indicate that PcircRNA_finder has a higher sensitivity (74–88%) than either find_circ or CIRCexplorer (each about 20%) and better precision (63–67%) compared to find_circ and CIRCexplorer, (72 and 100%, respectively) in the two test genomes (Supplementary Data). Finally, PcircRNA_finder obtained a significantly higher comprehensive value in the two test plant species (68–76%), compared to the other two methods (each ;<35%).
Transcriptomic data were generated from three RNA-Seq libraries (‘RNAase R’, ‘rRNA-’ and ‘polyA’) of rice seedlings (Supplementary Data). ‘RNAase R’ refers to linear mRNAs isolated from the rice seedlings that were degraded by RNAase R treatment (Circle-Seq, Jeck and Sharpless, 2014). CircRNAs in the various samples were predicted using all three circRNA prediction methods. Using PcircRNA_finder, we found 1,113 circRNAs in the RNAase R sample compared to 915 and 933 predicted by find_circ and CIRCexplorer, respectively. Of the circRNAs detected by PcircRNA_finder, 567 were not found using the other prediction programs. We define high-confidence circRNAs as those predicted circRNAs found in common between the ‘RNAase R’ and ‘rRNA-’ libraries, but not present in the ‘polyA’ library. Based on this definition, PcircRNA_finder predicted more high-confidence circRNAs from the rice RNA-Seq data sample (117) than either of the other two methods (104 and 74) (Supplementary Data).
Supplementary Material
Funding
This work was supported by the National Basic Research Program of China (2015CB150200), the National Science Foundation of China (91435111) and Jiangsu Collaborative Innovation Center for Modern Crop Production (JCIC-MCP).
Conflict of Interest: none declared.
References
- Chuang T.J. et al. (2016) NCLscan: accurate identification of non-co-linear transcripts (fusion, trans-splicing and circular RNA) with a good balance between sensitivity and precision. Nucleic Acids Res, 44, e29., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A. et al. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T.B. et al. (2013) Natural RNA circles function as efficient microRNA sponges. Nature, 495, 384–388. [DOI] [PubMed] [Google Scholar]
- Hoffmann S. et al. (2014) A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol., 15, R34., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W.R., Sharpless N.E. (2014) Detecting and characterizing circular RNAs. Nat. Biotechnol., 32, 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Salzberg S.L. (2011) TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol., 12, R72.,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. et al. (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol., 22, 256–264., [DOI] [PubMed] [Google Scholar]
- Lu T. et al. (2015) Transcriptome-wide investigation of circular RNAs in rice. RNA, 21, 2076–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S. et al. (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature, 495, 333–338. [DOI] [PubMed] [Google Scholar]
- Pan X., Xiong K. (2015) PredcircRNA: computational classification of circular RNA from other long non-coding RNA using hybrid features. Mol. Biosyst., 11, 2219–2226., [DOI] [PubMed] [Google Scholar]
- Reddy A.S. et al. (2013) Complexity of the alternative splicing landscape in plants. Plant Cell, 25, 3657–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J. et al. (2013) Cell-type specific features of circular RNA expression. PLoS Genet., 9, e1003777., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D., Brown J.W. (2013) Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell, 25, 3640–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starke S. et al. (2015) Exon circularization requires canonical splice signals. Cell Rep., 10, 103–111., [DOI] [PubMed] [Google Scholar]
- Szabo L. et al. (2015) Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol., 16, 126., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. et al. (2010) MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res., 38, e178., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C.Y. et al. (2015) Widespread noncoding circular RNAs in plants. New Phytol., 208, 88–95., [DOI] [PubMed] [Google Scholar]
- Zhang X.O. et al. (2014) Complementary sequence-mediated exon circularization. Cell, 159, 134–147. [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. (2013) Circular intronic long noncoding RNAs. Mol. Cell, 51, 792–806. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.