Highlight
We reveal that ABA affects flowering through two independent regulatory mechanisms: the activation of GI and CO functions upstream of the florigen genes and the down-regulation of SOC1 signalling.
Key words: Abscisic acid (ABA), adaptation, drought stress, florigen expression, flowering, photoperiod.
Abstract
One strategy deployed by plants to endure water scarcity is to accelerate the transition to flowering adaptively via the drought escape (DE) response. In Arabidopsis thaliana, activation of the DE response requires the photoperiodic response gene GIGANTEA (GI) and the florigen genes FLOWERING LOCUS T (FT) and TWIN SISTER OF FT (TSF). The phytohormone abscisic acid (ABA) is also required for the DE response, by promoting the transcriptional up-regulation of the florigen genes. The mode of interaction between ABA and the photoperiodic genes remains obscure. In this work we use a genetic approach to demonstrate that ABA modulates GI signalling and consequently its ability to activate the florigen genes. We also reveal that the ABA-dependent activation of FT, but not TSF, requires CONSTANS (CO) and that impairing ABA signalling dramatically reduces the expression of florigen genes with little effect on the CO transcript profile. ABA signalling thus has an impact on the core genes of photoperiodic signalling GI and CO by modulating their downstream function and/or activities rather than their transcript accumulation. In addition, we show that as well as promoting flowering, ABA simultaneously represses flowering, independent of the florigen genes. Genetic analysis indicates that the target of the repressive function of ABA is the flowering-promoting gene SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), a transcription factor integrating floral cues in the shoot meristem. Our study suggests that variations in ABA signalling provide different developmental information that allows plants to co-ordinate the onset of the reproductive phase according to the available water resources.
Introduction
Water deprivation triggers several physiological adjustments at the cellular and organ levels (Shinozaki and Yamaguchi-Shinozaki, 2007). Depending on the intensity and duration of drought episodes, some plants can also respond adaptively, by activating the drought escape (DE) response (Franks, 2011; Riboni et al., 2013, 2014; Kazan and Lyons, 2016). DE allows plants to accelerate the floral transition and set seeds before drought conditions become too severe. While escaping the potentially lethal effects of drought, plants undergoing DE usually produce fewer fruits and seeds, indicating a trade-off between plant survival and successful seed set (Su et al., 2013; Kenney et al., 2014). Therefore, a more precise understanding of the mechanisms leading to DE is of fundamental importance to assess the diverse modes of adaptations of natural plant populations as well as to produce crops with increased productivity under water deprivation (Lovell et al., 2013; Kooyers, 2015).
Arabidopsis thaliana is a facultative long-day (LD) plant, flowering much earlier under LDs, typical of spring/summer compared with short days (SDs). The DE response occurs under LDs, but not SDs, indicating an interdependence between DE and photoperiod signalling in Arabidopsis (Han et al., 2013; Riboni et al., 2013). The photoperiodic pathway comprises three key genes, whose regulation and activity are required for the correct interpretation of day length: GIGANTEA (GI), CONSTANS (CO), and FLOWERING LOCUS T (FT) (Putterill et al., 1995; Fowler et al., 1999; Kardailsky et al., 1999; Kobayashi et al., 1999; Park et al., 1999). CO encodes a nuclear protein (Putterill et al., 1995; Samach et al., 2000) able to induce the transcriptional activation of the florigen genes FT and TWIN SISTER OF FT (TSF) (An et al., 2004; Yamaguchi et al., 2005; Jang et al., 2009). Accumulation of the CO transcript during the day depends on LIGHT OXYGEN VOLTAGE (LOV) domain-containing, blue light receptor FLAVIN-BINDING, KELCH REPEAT F-BOX 1 (FKF1), and GI (Imaizumi et al., 2003, 2005; Sawa et al., 2007; Fornara et al., 2009; Song et al., 2012). Formation of a GI–FKF1 complex is stimulated by blue light and leads to degradation of the CO transcriptional repressors CYCLING DOF FACTORs (CDFs) (Imaizumi et al., 2005; Fornara et al., 2009), allowing CO transcription. While CO transcript accumulation broadly occurs under both LDs and SDs, CO protein is activated to promote flowering only under LDs when CO mRNA peaks in the light phase at the end of the day (Suarez-Lopez et al., 2001). Such a daily pattern of CO protein accumulation is controlled by several types of photoreceptors, which generate a peak of CO abundance in coincidence with dusk under LDs (Valverde et al., 2004; Jang et al., 2008; Liu et al., 2008; Zuo et al., 2011; Lazaro et al., 2012; Song et al., 2012).
CO promotes FT transcription in the phloem companion cells (Adrian et al., 2010). However, FT protein acts as a florigenic signal, moving long distance to the shoot apical meristem (SAM), where it interacts with the bZIP transcription factors FLOWERING LOCUS D (FD) and FD PARALOGUE (FDP) to orchestrate the floral transition (Abe et al., 2005; Wigge et al., 2005; Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007; Jaeger et al., 2013). Amongst the early targets of the FT–FD complex is SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), a MADS box transcription factor, which integrates several floral pathways in the SAM (Borner et al., 2000; Lee et al., 2000; Samach et al., 2000; Moon et al., 2003; Searle et al., 2006; Jang et al., 2009; Wang et al., 2009; Lee and Lee, 2010).
Besides photoperiod, FT activation is modulated by several environmental cues (Pin and Nilsson, 2012), including drought stress (Riboni et al., 2013). The activation of FT by drought requires abscisic acid (ABA), a key hormone mediating water stress stimuli (Riboni et al., 2013). ABA derives from the carotenoid zeaxanthin synthetized in chloroplasts. Here, different enzymes, including ABA1, transform zeaxanthin into xanthoxin prior to its translocation to the cytoplasm where another set of enzymes, namely ABA2, complete the last biosynthetic steps leading to bioactive ABA (Nambara and Marion-Poll, 2005). Three signalling proteins form the core ABA signalling, including the PYRABACTIN RESISTANCE (PYR)/REGULATORY COMPONENT OF ABA RECEPTOR (RCAR), the PROTEIN PHOSPHATASE 2Cs (PP2Cs), and SNF1-RELATED PROTEIN KINASE 2s (SnRK2s) (Cutler et al., 2010). The PYR/RCARs are the ABA receptors, the PP2Cs [e.g. the ABA INSENSITIVE 1 (ABI1) gene] act as negative regulators of the pathway, and the SnRK2s act as positive regulators of downstream signalling (Ma et al., 2009; Park et al., 2009).
ABA-deficient mutants aba1 and aba2 display a general delay in flowering in LDs, which is more evident under drought conditions as well as reduced florigen transcript accumulation. Similar to aba1, mutants of GI are impaired in DE, and display no florigen up-regulation under drought conditions (Riboni et al., 2013). The nature of GI signalling upstream of the florigen activation during DE is however unclear. Because no DE occurs in wild-type plants under SDs, one can conclude that GI activates DE by mediating photoperiodic signals. However, such a mechanism does not appear to require CO activity, since co mutants display a normal DE response (Han et al., 2013; Riboni et al., 2013). Modes of GI-dependent but CO-independent pathways include the activation of a class of miRNA, the miR172, which targets the APETALA 2-like factors that repress FT and other flowering genes (Jung et al., 2007; Mathieu et al., 2009). The role of GI in DE may also be indirect and/or biochemically distinct from its role in photoperiodic flowering. For example, GI affects phytochrome signalling (Huq et al., 2000; Martin-Tryon et al., 2007; Oliverio et al., 2007), clock function (Park et al., 1999; Fowler et al., 1999; Mizoguchi et al., 2005), and several plant–environment responses, namely salinity and freezing tolerance (Han et al., 2013; Kim et al., 2013b ; Fornara et al., 2015; Xie et al., 2015), through mechanisms which cannot be fully ascribed to the canonical photoperiodic signalling cascade.
In this study, tests were carried out to elucidate the role of GI signalling in the DE response. We analysed the DE response and patterns of florigen accumulation in Arabidopsis mutant backgrounds with varying levels of CO and in the presence or absence of GI. To assess the role of ABA in the GI-mediated pathway, we combined mutants impaired in ABA signalling with transgenic plants overexpressing GI. We show that impaired ABA signalling affects GI downstream functions and/or activity, thus causing reduced accumulation of florigen genes, but no effects on CO accumulation. Our results also clarify the relationship between GI and CO in the context of DE response by showing that the drought/ABA-dependent activation of FT requires CO. In contrast, up-regulation of TSF under drought stress can occur without CO, thus expanding the repertoire of regulatory mechanisms of florigen gene activation in plants. Alongside these results, we also demonstrate a florigen-independent floral repressive role for ABA in flowering, which requires SOC1. The transition to flowering under drought conditions thus depends on activation of separate ABA-dependent developmental programmes.
Materials and methods
Plant materials and growing conditions
In this study, we used wild-type Arabidopsis thaliana plants, ecotype Columbia (Col-0) or Landsberg erecta (Ler). Mutant or transgenic lines were obtained from the Nottingham Arabidopsis Stock Centre or other laboratories as detailed in Supplementary Table S3 at JXB online. Seeds were stratified in the dark at 4 °C for 2 d before sowing, and plants grown in a controlled-environment cabinet at a temperature of 18–23 °C, 65% relative humidity, under either LD (16 h light/8 h dark) or SD (8 h light/16 h dark) photoperiods. Light was provided by cool white fluorescent tubes (Philips Lighting, F36W/33-640 36W) at a fluence of 120–150 μmol m−2 s−1 (photosynthetically active radiation). The procedures to impose drought stress, and perform photoperiod shift experiments were previously detailed (Riboni et al., 2013).
Experiments in Fig. 1B were performed in a greenhouse, with a semi-controlled climate. Temperature was 19–23 °C and relative humidity was set at 65%. Natural light was supplemented by incandescent (metal halide) lamps when external light was <150 μmol m−2 s−1 (photosynthetically active radiation) in an LD photo cycle. Two independent greenhouse experiments were performed (autumn 2015 in Milan). ABA application experiments were performed by daily supplying 2 ml of ABA (25 μM) or mock solutions (0.025% v/v ethanol) 7 h after dawn. ABA applications started 3 d after germination and continued for 21 d. Each Arabasket pot was fitted with a pipette tip to facilitate the application of the solutions directly in the soil and thus in contact with roots (Supplementary Fig. S1).
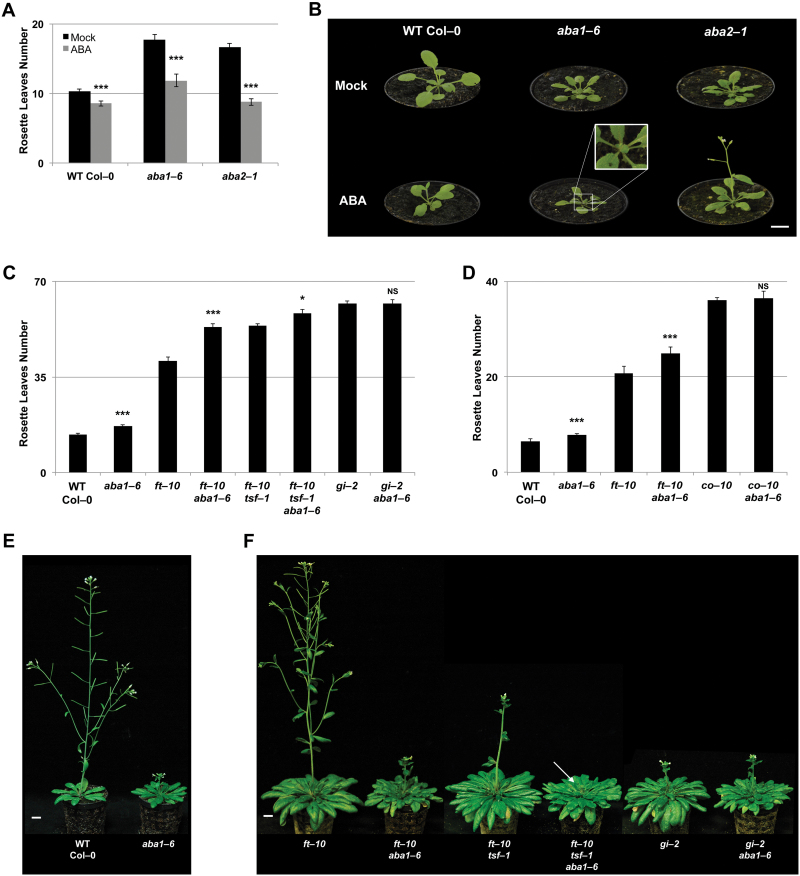
Fig. 1.
ABA activates flowering through GI, CO, and the florigen genes. (A) Mean number of rosette leaves of the wild type (Col-0) and ABA-deficient mutant plants grown under LDs and treated with ABA or mock treated. Error bars represent ±SE, n=15. Student’s t-test P-values ≤0.001 (***) compared with mock treatment. (B) Images of representative 24-day-old plants of the indicated genotypes grown under LDs and treated with ABA or mock treated. Inset of aba1-6 shows a visible inflorescence. (C and D) Mean number of rosette leaves of the wild type (Col-0) and flowering time mutants grown under LDs. Error bars represent ±SE, n=15. Student’s t-test P-values ≤0.05 (*), ≤0.001 (***), >0.05 not significant (NS) are shown to indicate differences between mutants and the corresponding mutant containing the aba1-6 allele. The experiment in (D) was performed under semi-controlled greenhouse conditions. (E) and (F), Images of representative plants of the indicated genotypes grown under LDs. (E) Wild-type Col-0 and aba1-6 mutant plants are 4 weeks old, (F) ft-10, ft-10 aba1-6, ft-10 tsf-1, ft-10 tsf-1 aba1-6, gi-2, and gi-2 aba1-6 mutant plants are 14 weeks old. The arrow indicates the visible bolt in ft-10 tsf-1 aba1-6. Scale bars=1 cm. (This figure is available in colour at JXB online.)
Isolation of double mutants and genotyping
Mutant combinations were generated by crossing. The aba1-6 mutation was genotyped as described in Riboni et al. (2013). ft-10 mutants were selected on Murashige and Skoog plates containing Sulafadiazide as described (Rosso et al., 2003). abi1-1 mutants were selected by genomic PCR amplification with primers flanking the abi1-1 polymorphism followed by digestion with NcoI. Genotyping primers for tsf1-1, co-10, and abi1-1 are listed in Supplementary Table S4. Plants carrying the gi-2 and soc1-1 alleles were selected based on their late flowering phenotype, while elf3-1 mutants were selected on the basis of their early flowering and long hypocotyl.
RNA extraction and real-time qPCR
Total RNA was extracted with TRIzol reagent (Invitrogen). A 1.5 µg aliquot of total RNA was used for cDNA synthesis with the SuperScript VILO cDNA Synthesis Kit (Invitrogen). Quantitative real-time PCR was performed as previously detailed (Riboni et al., 2013) and PCR primers are provided in Supplementary Table S4.
Molecular cloning and plant transformation
A 2.2 kbp promoter region upstream of the ABI1 coding sequence was cloned using the Gateway cloning technology (Invitrogen) with specifics primers (Supplementary Table S4). The promoter was cloned into the pDONR207 entry vector (Invitrogen) and moved into the pBGWFS7 destination vector (Karimi et al., 2002). The resulting plasmid was introduced into Agrobacterium strain GV3101 (pMP90RK) (Koncz and Schell, 1986) and transformed in wild-type Col-0 plants by floral dip (Clough and Bent, 1998). Six independent transgenic plants were selected based on the segregation of Basta resistance in a Mendellian 3:1 ratio in the T2 generation and analysed for β-glucuronidase (GUS) staining.
GUS assay
Plants were grown under LDs and sampled at the indicated Zeitgeber time (ZT) time. Tissue was fixed for 30 min at 0 °C with 90% (v/v) acetone. After being washed in 50 mM sodium phosphate buffer, pH 7.0 they were incubated for 14 h at 37 °C in staining solution [0.5 mg ml–1 X-Gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronide), 50 mM sodium phosphate buffer, pH 7.0, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, and 0.1% (v/v) Triton X-100]. Samples were cleared with a chloral hydrate:glycerol:water solution (8:1:2, v/v/v) for 3 h and then stored in 70% (v/v) ethanol before GUS histochemical reactions were visualized under a stereomicroscope.
Results
ABA promotes FT expression through CO
Mutants of aba1-6 were later flowering compared with the wild type under LDs (Fig. 1A–C). We confirmed a similar late flowering phenotype in aba2-1 mutants, defective in the final steps of ABA biosynthesis (Finkelstein, 2013). Soil applications of ABA could accelerate flowering in wild-type plants, reminiscent of DE response (Fig. 1A; Supplementary Table S1) (Koops et al., 2011). Using this experimental set-up, we could also largely rescue the late flowering of aba1-6 and aba2-1 mutants, indicating a role for ABA as an activator of flowering (Fig. 1A, B).
We have previously demonstrated that ABA activates flowering under LDs but not SDs and that ABA affects photoperiodic signalling upstream of FT expression (Riboni et al., 2013). To understand how ABA interacts with photoperiod signalling to affect flowering, we generated combinations of ABA-deficient (aba1-6) and photoperiodic pathway mutants (Fig. 1C, D; Supplementary Table S1). Consistent with lack of flowering defects of aba1-6 under SDs (Riboni et al., 2013), double mutants of gi-2 aba1-6 displayed a similar flowering time compared with gi-2 single mutants under LDs (Fig. 1C, F). Since double mutants of ft-10 aba1-6 were later flowering than ft-10 single mutants, ABA could affect flowering time via other florigen genes, namely TSF (Fig. 1C, F). The tsf-1 ft-10 aba1-6 triple mutants were slightly later flowering than tsf-1 ft-10 double mutants (Fig. 1C, F). TSF thus contributes to the late flowering phenotype of ft-10 aba1-6 plants although ABA also appears to have an effect on other floral pathways, independent of FT and TSF. Interestingly, double mutants of co-10 aba1-6 were similar to co-10 single mutants, indicating that CO is also required for the late flowering phenotype of aba1-6 mutants (Fig. 1D).
Unlike gi, co mutants generate a DE response, indicating that high levels of ABA accumulation, as a result of drought stress, may eventually overcome CO function to activate flowering (Riboni et al., 2013). To test whether drought could activate the florigen genes in the absence of a functional CO protein we grew wild-type and co-10 mutant plants under control or water stress conditions in SDs before shifting to LDs to induce a photoperiodic response. As expected, in wild-type plants FT expression was strongly up-regulated during the photo-extension period and even further increased under low watering conditions (Fig. 2A). In the co-10 mutants, the levels of FT transcripts were barely detectable at any time point, independent of the watering regime, indicating that drought stress cannot cause FT up-regulation in the absence of a functional CO (Fig. 2B). The pattern of accumulation of TSF showed diurnal oscillations similar to those of FT in wild-type plants, peaking at dusk during the photo-extension period (Fig. 2A, B). Similar to FT, TSF expression was increased in coincidence with the photo-extension period under drought conditions. Furthermore in co-10 mutants, TSF levels were much lower compared with the wild type under normal watering conditions, confirming a role for CO in TSF transcriptional activation (Yamaguchi et al., 2005; Jang et al., 2009). Surprisingly, drought stress caused TSF up-regulation in the co-10 background, partially resuming its diurnal cycle with peaks at ZT8 under the SD part of the experiment and at ZT15 following a photo-extension. Slightly increased TSF levels were observed during SDs under drought conditions (on average 3.8 ± 1.6-fold) but this was not correlated with a DE phenotype under SDs in co-10 mutants (Fig. 2B, D). Thus, unlike FT, TSF can be up-regulated under drought conditions in a CO-independent manner.
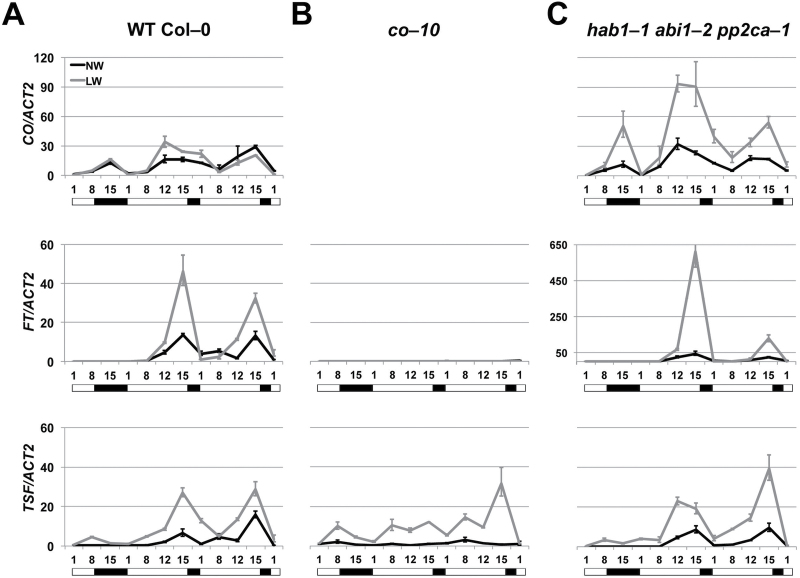
Fig. 2.
CO is required for the activation of FT under drought stress. (A–C) Real-time qPCR of CO, FT, and TSF transcripts in 3-week-old wild-type (Col-0) (A), co-10, (B) and hab1-1 abi1-2 pp2ca-1 (C) seedlings. Plants were subject to normal watering (NW; black lines) or low watering (LW; grey lines) regimes and harvested at the indicated time points in coincidence with the light phase (open bar) or in the dark (black bar) during an SD to LD shift. At each time point, values represent fold change variations of CO, FT, and TSF transcript levels relative to Col-0 under NW. ACT2 expression was used for normalization; error bars represent the SD of two technical replicates. A representative experiment of two biological replicates is shown.
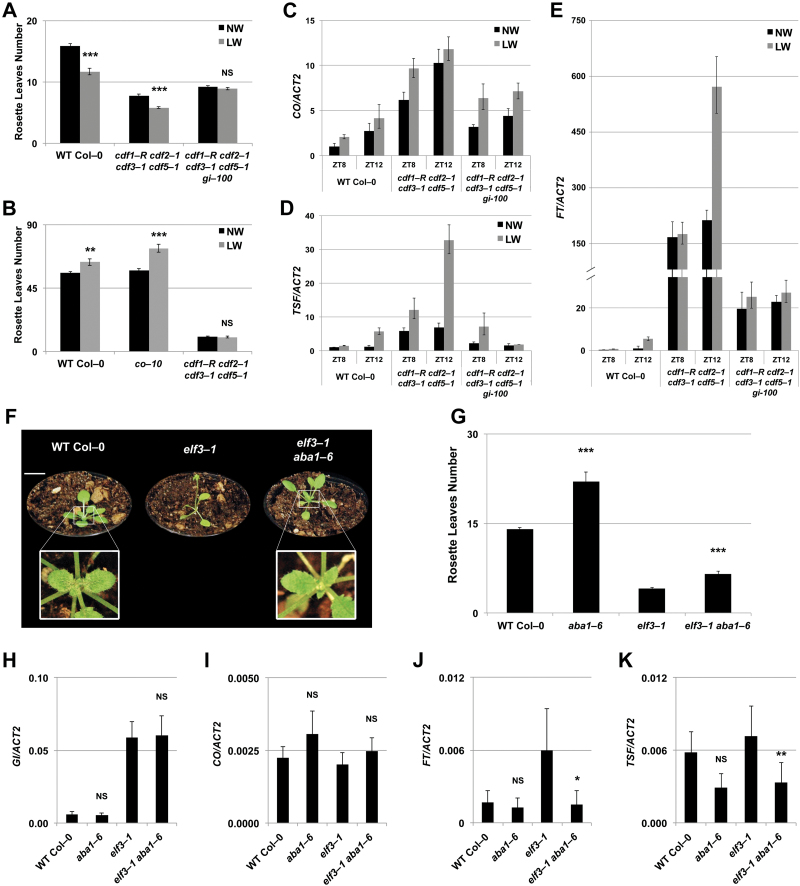
GI is required for DE downstream of CO transcriptional activation
Our experiments indicate that ABA promotes FT transcript accumulation through CO. However, CO transcript levels are not greatly affected by drought stress or when ABA level are reduced (Han et al., 2013; Riboni et al., 2014). Here we wanted to test whether drought could affect flowering downstream of CO transcriptional activation events, by analysing mutants of cdf1-R cdf2-1 cdf3-1 cdf5-1, hereafter referred to as cdf1235, characterized by constitutively elevated CO levels (Fornara et al., 2009). The cdf1235 mutants flowered early and produced a DE response quantitatively similar to that of the wild type under LDs (Fig. 3A). Despite their early flowering phenotype under SDs, cdf1235 plants did not produce any DE response (Fig. 3B), suggesting a requirement for LDs in DE response, even when CO levels are elevated (Fornara et al., 2009) (Fig. 3C). We therefore compared the flowering time and DE response of the quadruple cdf1235 mutant with that of gi cdf1235 quintuple mutants under LDs (Fig. 3A). As previously shown, mutants of cdf1235 are slightly earlier flowering than gi cdf1235 under normal watering conditions (Fornara et al., 2009). However, while the cdf1235 mutants produced a DE response, the gi cdf1235 did not (Fig. 3A). We next sought to ascertain if the lack of DE response in the gi cdf1235 mutants was correlated with impaired transcriptional up-regulation of the florigen genes under drought stress. Control and water-stressed wild-type, cdf1235 and gi cdf1235 plants were grown under SD conditions for 2 weeks before shifting to LDs, and transcript levels were analysed at ZT8 (corresponding to dusk in the SDs) and ZT12 (4 h after the photo-extension) (Fig. 3C–E). As expected, the levels of CO transcript were generally higher in cdf1235 and gi cdf1235 mutants as compared with the wild type. Under drought conditions, we observed a small increase in CO transcript abundance in all the genotypes analysed at any time point, suggesting a contribution of drought stress in CO transcript accumulation (Fig. 3C). We finally determined how different patterns of CO transcript were correlated with accumulation of florigen genes (Fig. 3D, E). Under well-watered conditions, mutants of cdf1235 showed the largest FT and TSF transcript accumulations before and after the photo-extension period. Mutants of gi cdf1235 displayed levels of FT and TSF intermediate between the wild type and the cdf1235 mutants. This is probably as a result of residual CDF-mediated repression in cdf1235 on both CO and FT promoters (Fornara et al., 2009; Song et al., 2012). However, while both the wild type and the cdf1235 mutants showed a significant and similar up-regulation of FT and TSF under drought stress conditions at ZT12 (2- to 4-fold, respectively), no such up-regulation occurred in the gi cdf1235 mutants (Fig. 3D, E).
Fig. 3.
ABA promotes GI and CO functions to activate the florigen genes. (A and B) Mean number of rosette leaves of the wild type (Col-0) and flowering time mutants subject to normal watering (NW; black bars) or low watering (LW; grey bars) regimes grown under LDs (A) and SDs (B). Error bars represent ±SE n=15. Student’s t-test P values ≤0.001 (***), >0.05 not significant (NS) compared with NW. (C–E) Real-time qPCR of CO (C), TSF (D), and FT (E) transcripts in 2-week-old wild-type (Col-0), cdf1-R cdf2-1 cdf3-1 cdf5-1, and cdf1-R cdf2-1 cdf3-1 cdf5-1 gi-100 seedlings. Plants were subject to NW (black columns) or LW (grey columns) regimes and harvested at the indicated Zeitgeber time during a shift from SDs to LDs. ZT8 represents dusk in SDs and ZT12 represents 4 h of photo-extension. At each time point, values represent fold change variations of CO, FT, and TSF transcript levels relative to the wild type at ZT8 under NW. ACT2 expression was used for normalization; error bars represent the SD of two technical replicates. A representative experiment of two biological replicates is shown. (F) Images of representative plants grown under LDs for 27 d. Insets shows a visible inflorescence in elf3-1 aba1-6 double mutants, which is not visible in the wild type. (G) Mean numbers of rosette leaves of the wild type (Col-0) and mutants under LDs. Error bars represent ±SE, n=5–12. Student’s t-test P-values ≤0.001 (***) are shown to indicate differences between mutants and the corresponding mutant containing the aba1-6 allele. (H–K) Real-time qPCR of GI (H), CO (I), FT (J), and TSF (K) transcripts in 12-day-old seedlings grown under LDs and sampled at ZT16. Data shown are from 5–6 biological replicates. Error bars represent ±SD. Differences between the wild type versus aba1-6 and elf3-1 versus elf3-1 aba1-6 double mutants are here highlighted with P-values ≤0.01 (**), ≤0.05 (*), >0.05 not significant (NS), one-way ANOVA with Tukey’s HSD (honestly significant difference) test. (This figure is available in colour at JXB online.)
In a complementary approach, we asked whether ABA production might be required for FT transcriptional activation when GI levels are increased. Mutants of early flowering 3 (elf3) are extremely early flowering, accumulate high levels of FT, and present increased accumulation of GI transcript and GI protein (Fowler et al., 1999; Kim et al., 2005; Yu et al., 2008). This early flowering phenotype requires ABA since elf3-1 aba1-6 double mutants were significantly later flowering compared with elf3-1 single mutants (Fig. 3F, G). FT and TSF transcript levels were slightly but not significantly reduced in aba1-6 mutants compared with the wild type at this early developmental stage (Fig. 3J, K; Supplementary Table S2). However, double mutants of elf3-1 aba1-6 had a significant reduction in both FT and TSF levels compared with the elf3-1 single mutants (Fig. 3J, K; Supplementary Table S2). The reduced levels of FT and TSF in elf3-1 aba1-6 compared with elf3-1 mutants were not caused by diminished GI or CO transcript accumulations (Fig. 3H, I; Supplementary Table S2), indicating that ABA might be required for the activation of GI and CO signalling.
ABA signalling genes control FT transcript accumulation with little effect on CO
We analysed ABA-hypersensitive mutants plants hab1-1 abi1-2 pp2ca-1, impaired in three ABA-related PP2C phosphate genes, under different watering and photoperiodic conditions (Rubio et al., 2009). Consistent with previous observations, mutants of hab1-1 abi1-2 pp2ca-1 had much increased (up to 6-fold) levels of FT compared with the wild type under LDs (Riboni et al., 2013) (Fig. 2C). The experiment in Fig. 2C also shows that FT expression was even further activated under drought conditions compared with the wild type (up to 13.3-fold). In contrast, TSF expression was not clearly increased in hab1-1 abi1-2 pp2ca-1 plants compared with the wild type under any watering condition. No FT or TSF up-regulation occurred under SDs in the hab1-1 abi1-2 pp2ca-1 mutants under any watering condition.
Under control conditions the strong up-regulation of FT in hab1-1 abi1-2 pp2ca-1 plants was not caused by increased CO levels, which were comparable with those observed in the wild type (Fig. 2C). Increased levels of CO were, however, observed in the hab1-1 abi1-2 pp2ca-1 mutants under drought stress, indicating that high levels of ABA signalling can ultimately induce the transcriptional activation of CO (Koops et al., 2011; Yoshida et al., 2014).
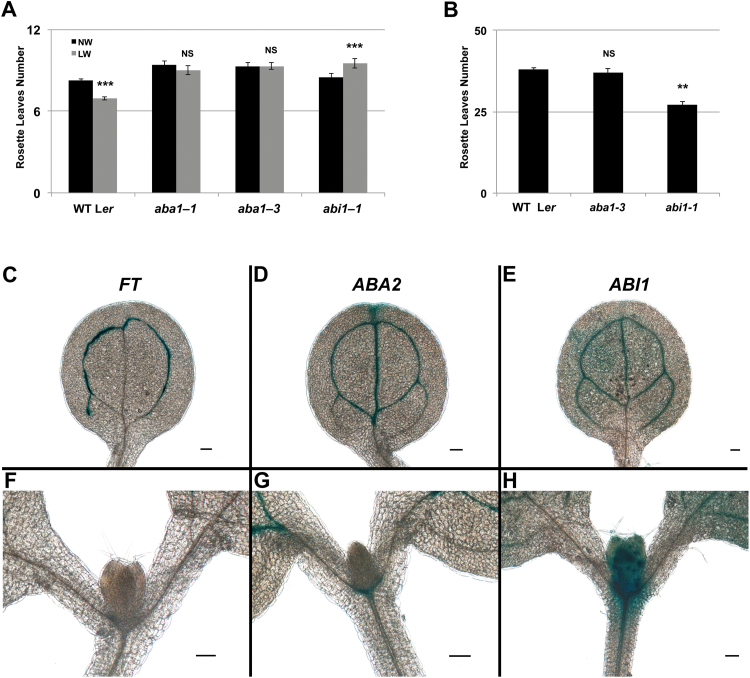
To explore further the role of ABA signalling in the transcriptional control of FT, we analysed abi1-1 mutant plants (Ler background), carrying a dominant mutation in the PP2C phosphatase ABI1 (Koornneef et al., 1984) which results in severely reduced ABA responses. abi1-1 mutant plants did not show flowering defects under LDs, but exhibited an early flowering phenotype under SDs, consistent with previous observations (Martínez-Zapater et al., 1994; Chandler et al., 2000) (Fig. 4A, B). Ruling out an ecotype-specific effect for ABA action in flowering, the ABA biosynthetic mutants aba1-1 and aba1-3 (Ler background) showed a marginal late flowering phenotype compared with the wild type under LDs (ANOVA P<0.01 and P<0.05, respectively), but no defects under SDs (Fig. 4A, B). The late flowering phenotype of these aba1 mutants was more pronounced under drought conditions and LDs, indicative of a reduced DE response (Fig. 4A). Mutants of abi1-1 were even more impaired in the DE response compared with the aba1 alleles, producing on average 14 ± 2% more leaves (n = 8 independent experiments, 15 plants each), relative to the untreated control.
Fig. 4.
A negative role for ABA in flowering. (A and B) Mean number of rosette leaves of the wild type (Ler) and ABA-deficient or signalling mutants grown under LDs and subject to normal watering (NW; black bars) or low watering (LW; grey bars) regimes (A), or under SDs in NW regime (B). Error bars represent ±SE n=15. Student’s t-test P-values ≤0.001 (***), >0.05 not significant (NS), compared with NW (A). One-way ANOVA with Tukey’s HSD (honestly significant difference) test P-values ≤0.01 (**), >0.05 not significant (NS), compared with the wild type (B). (C–H) Histochemical GUS detection in transgenic seedlings expressing pFT::GUS (C) and (F), pABA2::GUS (D) and (G), and pABI1::GUS (line # 1) (E) and (H) in the Col-0 background, scale bars=100 µm. (This figure is available in colour at JXB online.)
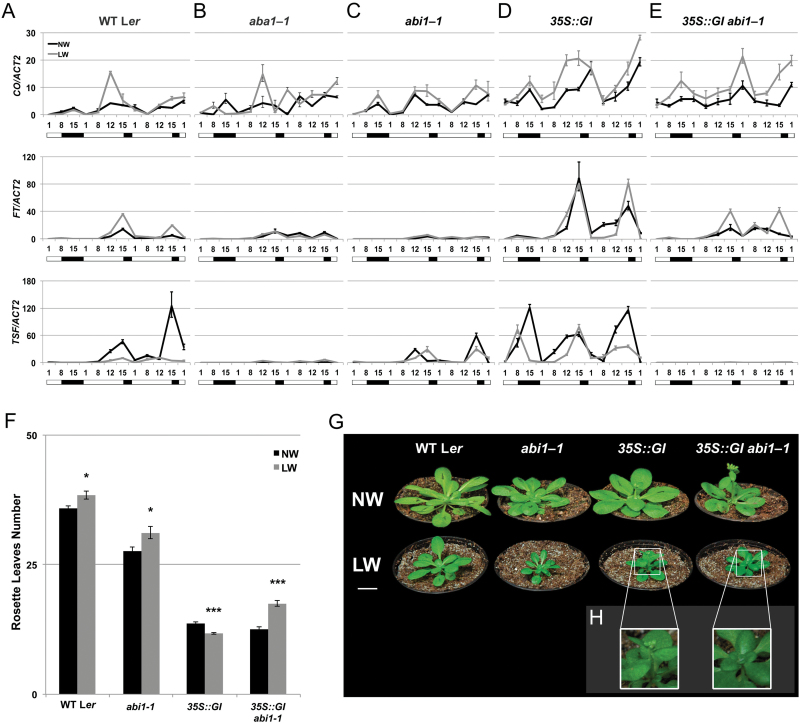
We next analysed the pattern of accumulation of the florigen genes in abi1-1 plants. As expected, in wild-type plants, the accumulation of FT was strongly induced under drought conditions in a photoperiod-dependent manner (Fig. 5A). TSF expression was instead down-regulated under drought conditions in the Ler background, revealing an ecotype-specific effect for TSF regulation under drought (Fig. 5A). Lower levels of FT and TSF were observed in the aba1-1 mutants compared with the wild type under both normal watering (TSF) and drought conditions (FT and TSF), confirming the contribution of ABA in both FT and TSF regulation (Fig. 5B) (Riboni et al., 2013). Strikingly, in abi1-1 plants the levels of FT were dramatically reduced compared not only with the wild type but also with aba1-1 plants, under any watering condition analysed (Fig. 5C). Such low expression of the florigen genes did not depend on reduced CO transcript accumulation in abi1-1 which was, if anything, up-regulated (Fig. 5C). Taken together, our data point to a model where ABA affects accumulation of florigen genes without an effect on CO expression.
Fig. 5.
ABA activates GI signalling and florigen expression with little effect on CO transcript accumulation. (A–E) Real-time qPCR of CO, FT, and TSF transcripts in 2-week-old wild-type (Ler) (A), aba1-1 (B), abi1-1 (C), 35S::GI (D), and 35S::GI abi1-1 (E) seedlings. Plants were subject to normal watering (NW; black lines) or low watering (LW; grey lines) regimes and harvested at the indicated time points in coincidence with the light phase (open bar) or in the dark (black bar) during an SD to LD shift. At each time point, values represent fold change variations of CO, FT, and TSF transcript levels relative to Ler under NW. ACT2 expression was used for normalization; error bars represent the SD of two technical replicates. A representative experiment of two biological replicates is shown. (F) Mean number of rosette leaves of the wild type (Ler) and mutants grown under SDs and subject to NW (black bars) or LW (grey bars) regimes, Error bars represent ±SE n=15. Student’s t-test P-values ≤0.05 (*), ≤0.001 (***) compared with NW. (G) Images of representative 5-week-old plants of the indicated genotypes grown under SDs and subject to NW or LW regimes. Scale bar=1 cm. (H) Higher magnification of LW 35S::GI and 35S::GI abi1-1 plants shown in (G). Note the appearance of a bolt in 35S::GI but not in 35S::GI abi1-1.
Loss of PP2C function (as in hab1-1 abi1-2 pp2ca-1) results in increased FT transcript accumulation, while expression of a gain-of-function form of ABI1 (as in abi1-1) leads to reduced FT activation. To determine whether the negative regulation of ABI1 on FT expression could be exerted in the cells expressing FT, we fused a 2.2 kb promoter region of ABI1 to the GUS reporter. We detected GUS staining in several independent transgenic T2 plants (n = 6) with comparable results, at ZT8, where ABI1 transcript accumulation is highly abundant according to a publicly available data set (http://diurnal.mocklerlab.org; Mockler et al., 2007). For comparison, we also studied the pattern of GUS activity in Arabidopsis transgenic lines marking the FT expression domain; the ABA2 (Lin et al., 2006; Kuromori et al., 2014) and the FT promoter itself (Notaguchi et al., 2008). Histochemical detection in young seedlings revealed that ABI1 expression (Fig. 4E) occurred in the vasculature of cotyledons in a pattern similar to ABA2 and FT (Fig. 4C, D), demonstrating an overlap between ABA biosynthesis and signalling genes in the tissue known to be the source of FT protein production. Broadly distributed GUS staining was also observed in the apical region of ABI1::GUS transgenic plants (Fig. 4H). This pattern of expression may also indicate a role for ABA signalling in the shoot apex.
Impaired ABA signalling negatively affects GI action
Whether impairing ABA signalling affects GI action was tested by generating abi1-1 35S::GI plants. As previously observed, 35S::GI plants had increased levels of FT under both SDs and LDs compared with the wild type (Mizoguchi et al., 2005). Under drought conditions, FT expression was generally less responsive to drought in the 35S::GI background compared with the wild type (Fig. 5D). The levels of TSF were much more increased in 35S::GI plants compared with the wild type during the SD part of the experiment. However, no further up-regulation of TSF occurred as a result of drought stress compared with normal watering (Fig. 5D). The overaccumulation of FT observed in 35S::GI plants was strongly rescued in the abi1-1 35S::GI mutants under any watering conditions (Fig. 5E). The levels of TSF transcript fell even more severely in abi1-1 35S::GI plants compared with 35S::GI. Such reductions in florigen accumulation in abi1-1 35S::GI plants were not related to decreased CO levels as these were much higher than in the wild type (Fig. 5A, E). Interestingly the levels of CO in abi1-1 35S::GI plants were only mildly reduced compared with 35S::GI, which could suggest that the negative role exerted by abi1-1 protein on GI signalling is more related to FT and TSF regulation rather than to CO (Fig. 5D, E).
Our data describe a regulatory role of ABA in GI signalling. Such ABA-mediated post-transcriptional activation of GI is consistent with previous observations on 35S::GI plants showing a DE-responsive phenotype under SDs (Riboni et al., 2013). In contrast, no DE response occurred in abi1-1 35S::GI mutants, which flowered much later compared with well-watered plants of the same genotype, although still earlier than abi1-1 plants (Fig. 5F). Under normal watering conditions, double mutants of abi1-1 35S::GI had a similar flowering phenotype to 35S::GI plants, despite showing reduced accumulation of the florigen genes (Fig. 5E, F). A similar observation could be made for abi1-1 plants, which did not show flowering defects under LDs compared with the wild type, but had reduced florigen expression (Fig. 5A, C). We conclude that late flowering of abi1-1 or abi1-1 35S::GI plants under drought stress cannot be solely ascribed to reduced florigen up-regulation.
A negative role for ABA signalling in flowering
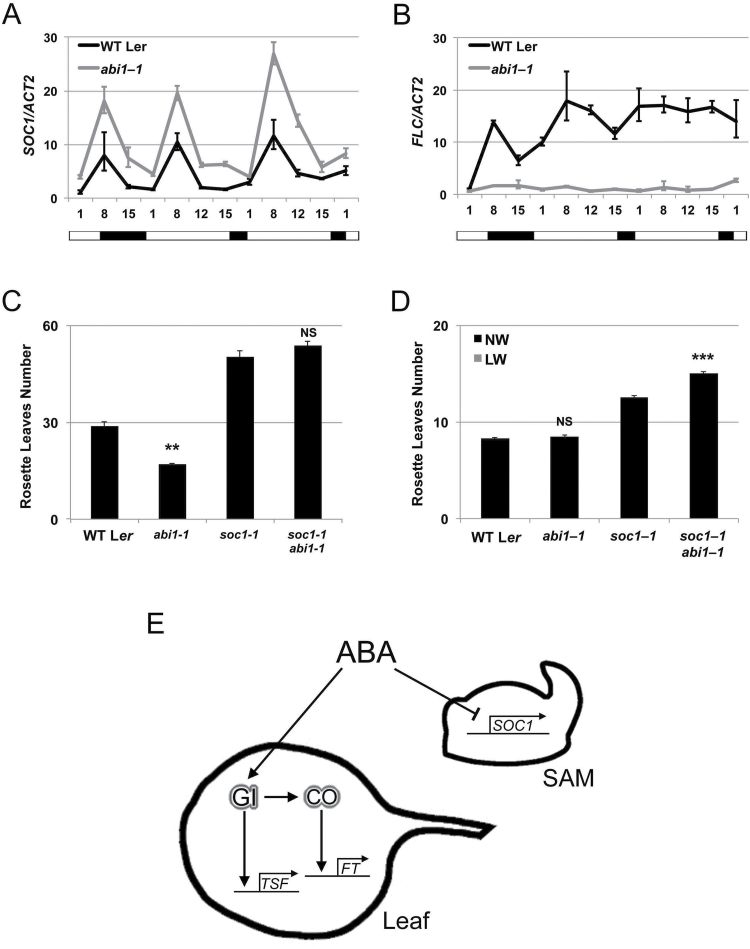
The early flowering of abi1-1 plants under SDs (Fig. 4B) implies that ABA signalling also exerts a negative role in flowering, which is usually undetectable under LDs or in ABA biosynthetic mutants (Fig. 4A). Supporting this model, we have previously reported a delay of flowering time under SDs in mutants of hab1-1 abi1-2 pp2ca-1 and observed a similar phenotype also in hab1-1 abi1-2 abi2-2 plants (Riboni et al., 2013) (Supplementary Fig. S2). abi1-1 mutants showed no increase in FT and TSF levels under SDs (Fig. 5B). In contrast, the accumulation of another floral integrator, SOC1, was increased in abi1-1 plants as compared with the wild type under any photoperiodic condition (Fig. 6A). Mutants of abi1-1 also had strongly reduced levels of FLOWERING LOCUS C (FLC) (Fig. 6B), a transcriptional repressor of SOC1 which contributes to delaying flowering under drought condition (Riboni et al., 2013; Y. Wang et al., 2013; Shu et al., 2016). Since SOC1 integrates different floral pathways in the SAM (Moon et al., 2003; Wang et al., 2009; Song et al., 2012, 2014) which promote flowering under SDs we created the abi1-1 soc1-1 double mutants. Under SDs, these plants displayed a flowering time similar to the soc1-1 single mutants. With respect to flowering time, soc1-1 is thus completely epistatic to abi1-1, indicating that SOC1 activity is required for the early flowering of abi1-1 mutants under SDs (Fig. 6C).
Fig. 6.
The inhibitory role of ABA in flowering requires SOC1. (A and B) Real-time qPCR of SOC1 (A) and FLC (B) transcripts in 2-week-old wild-type (Ler) and abi1-1 seedlings. The experimental conditions were described in Fig. 5. ACT2 expression was used for normalization; error bars represent the SD of two technical replicates. A representative experiment of two biological replicates is shown. (C and D) Mean number of rosette leaves of the wild type (Ler) and mutants grown under SDs (C) or LDs (D). Error bars represent ±SE n=15. Differences between the wild type versus abi1-1 and soc1-1 versus soc1-1 abi1-1 double mutants are here highlighted with P-values ≤0.001 (***), ≤0.01 (**), >0.05 not significant (NS), one-way ANOVA with Tukey’s HSD (honestly significant difference) test. (E) Model summarizing the proposed modes of ABA action in flowering. In the leaves, under LDs, drought promotes ABA accumulation leading to enhanced GI signalling and activation of florigen genes. CO protein is required for FT up-regulation, but not TSF. At the same time, at the shoot apex ABA represses flowering, downplaying SOC1 signalling, independent of photoperiodic conditions.
Under LDs, abi1-1 soc1-1 double mutants were later flowering than soc1-1 single mutants (Fig. 6D). Thus, the knocking out of SOC1 produces a novel flowering phenotype in the abi1-1 background, consistent with ABA being able to affect flowering differentially in different domains of the plant; by promoting FT expression in the leaves and negatively regulating floral stimuli in the SAM (Fig. 6E).
Discussion
A fundamental question related to the DE mechanism is how ABA signals are integrated in the photoperiodic flowering network. Here we provide evidence for how ABA controls FT gene expression under normal and drought stress conditions by affecting photoperiodic signalling. We also highlight a negative effect of ABA during the floral transition of Arabidopsis, which is independent of the photoperiodic pathway.
ABA requires both GI and CO to regulate FT
Our genetic data point to a model where ABA requires both GI and CO to affect flowering under LDs through the transcriptional activation of the florigen genes. Since mutants of ft-10 tsf-1 aba1-6 were still slightly later flowering than ft-10 tsf-1, it is possible that ABA may act on other pathways or through activation of MFT, a third florigen gene with a marginal role in flowering (Kim et al., 2013a ).
Expression and phenotypic analyses of cdf1235, gi cdf1235, as well as aba1 elf3 mutants collectively suggest that ABA promotes GI and CO signalling upstream of the florigen genes. CO function is essential for the drought-dependent activation of FT (but not TSF) as demonstrated by the lack of FT accumulation in co mutants under drought conditions. Therefore, although we could not resolve the underlying molecular mechanism, our data underscore a regulatory role for ABA in stimulating photoperiodic signalling. In further support of this model, 35S::GI plants under SDs generate a DE response, suggesting drought/ABA acting independently of GI transcript accumulation. Secondly, we observe a strong reduction in accumulation of florigen transcripts in abi1-1 35S::GI compared with 35S::GI plants. Thirdly, the pattern of CO accumulation in abi1-1 or abi1-1 35S::GI plants is unaltered compared with their respective controls, as opposed to the florigen levels, which are very low. In the light of our results, abi1-1 protein appears to affect a specific aspect of GI function (the activation of FT) without producing significant effects on the transcriptional profile of CO accumulation. Previous studies have demonstrated genetically separable roles for GI in regulating the circadian clock and flowering (Mizoguchi et al., 2005; Martin-Tryon et al., 2007) which could reflect distinct biochemical activities for GI in these two pathways. ABA might thus control a novel biochemical function of GI.
GI is found at different promoter locations of FT in association with transcriptional repressors including SHORT VEGETATIVE PHASE and TEMPRANILLO (Sawa and Kay, 2011). A substrate of the GI–FKF1 complex, CDF1, also binds to the FT promoter and acts as a transcriptional repressor (Sawa et al., 2007). Furthermore, by activating miR172 expression, GI directs post-transcriptional gene silencing of the AP2-type transcriptional repressors of FT (Jung et al., 2007). Overexpression of a miR172-related miRNA of soybean facilitates the DE response, promotes FT accumulation under drought conditions, and increases ABA sensitivity of Arabidopsis (Li et al., 2016). Thus, one role of GI could be to favour the recruitment of CO at the FT promoter by promoting the proteasome-dependent degradation or the post-transcriptional gene silencing of transcriptional repressors (such as AP2-like) in an ABA-dependent manner. Another, not mutually exclusive, model is that the combined presence of GI and ABA alters the pattern of CO protein accumulation during the day through an unknown mechanism. In addition to these post-transcriptional effects, there is evidence for other layers of transcriptional regulation of CO exerted by drought/ABA (Fig. 2C) (Koops et al., 2011; Ito et al., 2012; P. Wang et al., 2013; Yoshida et al., 2014). The contribution of these regulatory nodes to DE will require further studies. Regardless of the mechanisms involved and considering the role of the circadian clock in the control of ABA accumulation and response (Fukushima et al., 2009), our results suggest that daily variations in ABA signalling may represent a further layer of regulation of CO protein function.
Different modes of regulation of FT and TSF by drought
While FT and TSF share a common mechanism of transcriptional regulation through the photoperiodic pathway (Yamaguchi et al., 2005; Jang et al., 2009), they also display clear differences in their pattern of expression (Yamaguchi et al., 2005), response to ambient temperature (Blázquez et al., 2003), and other kinds of regulation (Michaels et al., 2005; D’Aloia et al., 2011; Liu et al., 2014). In this work, we report variations in the transcriptional activations of TSF and FT in response to drought. Our expression studies on co-10 mutants revealed that the expression of TSF, but not FT, is strongly induced by drought, even in the absence of functional CO. Previously we proposed a model whereby photoperiod-stimulated GI protein triggers a DE response via activation of the florigen genes, independent of CO (Riboni et al., 2013). Based on our new results, this model only applies to TSF regulation, not FT. The DE response observed in the co mutants could therefore derive from residual TSF expression, which still depends on GI (Riboni et al., 2013). Examples of GI acting independently of CO in activating the florigen genes have been described in the literature, but how these mechanisms are related to ABA signalling is unknown (Kim et al., 2005; Mizoguchi et al., 2005; Sawa and Kay, 2011). Other hormones modulate the expression of the florigen genes without an apparent contribution of CO. Cytokinin can induce the transcriptional activation of TSF, but not FT, irrespective of photoperiod conditions (D’Aloia et al., 2011). Foliar applications of gibberellins under SDs promote flowering, at least in part through FT ad TSF and without a clear effect on CO transcript accumulation (Porri et al., 2012). Similarly, there are examples of environmental cues activating FT, which do not fully require the activity of CO or GI, namely under elevated ambient temperature (Balasubramanian et al., 2006). Here, we demonstrate that the activation of TSF can occur in the absence of CO under drought conditions but, unlike the previous examples, such activation requires GI (Riboni et al., 2013).
Multiple and contrasting roles of ABA in flowering
The role of ABA during the floral transitions is controversial, as both positive and negative effects of ABA have been reported (Domagalska et al., 2010; Conti et al., 2014). Depending on the site of application, ABA exerts opposite roles in flowering. Unlike leaf applications, we show that root applications of ABA promote flowering, consistent with previous data (Koops et al., 2011). Also, this treatment largely rescues the late flowering of ABA biosynthetic mutants. In the light of these results, root applications fully mimic the positive role of endogenous ABA in flowering.
Impairing the function of ABA-activated kinases SnRK2.2/2.3/2.6 results in early flowering, especially under SDs, supporting a negative role for ABA in flowering (P. Wang et al., 2013). Arguing against a direct negative role of the SnRK2s in the flowering network, overexpression of SnRK2.6/OST1 causes a small flowering acceleration under LDs, not a delay (Zheng et al., 2010). The negative role of ABA in flowering has been linked to the direct activation of FLC by ABA-stimulated bZIP transcriptional factor ABSCISIC ACID-INSENSITIVE 5 (ABI5) and AP2/ERF domain-containing transcription factor ABSCISIC ACID-INSENSITIVE 4 (ABI4) (Y. Wang et al., 2013; Shu et al., 2016). Such activation of FLC may account for the general reduction in FT transcript accumulation following exogenous ABA applications on leaves (Hoth et al., 2002). The study of abi1-1 plants under SDs supports this negative effect of ABA in flowering. ABA-deficient mutants do not produce similar flowering alterations under SDs, which could depend on ABA biosynthetic mutants still producing a sufficient amount of biologically active ABA (Léon-Kloosterziel et al., 1996). The early flowering of abi1-1 plants in SDs can be completely suppressed by mutations in SOC1, a floral integrator activating flowering in the SAM (Searle et al., 2006). Elevated levels of SOC1 transcript in abi1-1 mutants also suggest a negative role for ABA in SOC1 expression, perhaps mediated by FLC (Fig. 6A, B). The proposed positive role of ABA-activated ABI5 on FLC transcriptional activation is consistent with this model (Y. Wang et al., 2013).
abi1-1 plants do not present obvious flowering phenotypes under LDs despite impaired photoperiod-dependent accumulation of FT. We thus propose that the abi1-1 mutants compensate for their defects in FT up-regulation with increased SOC1 signalling. The late flowering phenotype of abi1-1 soc1-1 compared with soc1-1 under LDs is consistent with ABA playing antagonistic and spatially distinct roles in flowering, through the transcriptional activation of the florigen genes in the leaves and the repression of SOC1 action in the shoot.
In addition to the ABA-dependent negative regulation of flowering, an ABA-independent floral repression mechanism emerged from the study of abi1-1 plants. Under LDs, mutants of abi1-1 exhibit a late flowering phenotype under drought stress, which is even more severe than aba1 plants. We observed an even more pronounced delay in flowering under SDs in abi1-1 35S::GI plants upon drought stress compared with 35S::GI. We interpret these results to indicate that the defects in florigen up-regulation of abi1-1 contribute to the late flowering of abi1-1 under drought stress. However, the levels of florigen expression in abi1-1 were generally also low under normal watering conditions. Therefore, we hypothesize a further layer of negative regulation of flowering, which is triggered by drought stress and is probably independent of ABA (as it occurs in abi1-1 plants). Both flowering-repressive mechanisms, the ABA-dependent and the ABA-independent mechanism, can be largely overcome under LDs, upon migration of the florigen protein in the SAM.
In conclusion, Arabidopsis plants have independent and contrasting mechanisms to modulate flowering according to water inputs; ABA stimulates GI and CO signalling to boost FT activation. Under drought conditions TSF activation is independent of CO and requires photoactivated GI. Simultaneously, ABA negatively regulates flowering through a pathway that requires SOC1 (Fig. 6E), perhaps in conjunction with an ABA-independent type of regulation. Integration of these pathways in the SAM may provide plants with a flexible control of reproductive development under water stress and maximization of reproductive success.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Method used for root applications of ABA.
Figure S2. Activated ABA signalling inhibits flowering under SDs.
Table S1. Flowering time of mutant and transgenic plants used in this study.
Table S2. Expression analysis of aba elf3 mutant plants.
Table S3. Genotypes used in this study and references.
Table S4. Primers used in this study.
Acknowledgements
We thank Drs George Coupland (Max Planck Institute for Breeding Research), Pedro Rodriguez (Consejo Superior de Investigaciones Científicas), Takashi Araki (Kyoto University), and the Nottingham Arabidopsis Stock Centre for providing seed lines. We also thank Dr Fabio Fornara and Dr Sara Castelletti (University of Milan) for insightful comments on the manuscript. This work was supported by Fondazione Umberto Veronesi per il Progresso delle Scienze (AGRISOST), Milano, Italy, Università degli Studi di Milano for a PhD studentship to ART, and in part by the MIUR PRIN project (2010–2011 prot. 2010HEBBB8_006).
Glossary
Abbreviations:
- DE
drought escape
- LD
long day
- SD
short day.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. 2005. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. [DOI] [PubMed] [Google Scholar]
- Adrian J, Farrona S, Reimer JJ, Albani MC, Coupland G, Turck F. 2010. cis-regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. The Plant Cell 22, 1425–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, Roussot C, Suarez-Lopez P, et al. 2004. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131, 3615–3626. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Sureshkumar S, Lempe J, Weigel D. 2006. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genetics 2, e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez MA, Ahn JH, Weigel D. 2003. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nature Genetics 33, 168–171. [DOI] [PubMed] [Google Scholar]
- Borner R, Kampmann G, Chandler J, Gleissner R, Wisman E, Apel K, Melzer S. 2000. A MADS domain gene involved in the transition to flowering in Arabidopsis. The Plant Journal 24, 591–599. [DOI] [PubMed] [Google Scholar]
- Chandler J, Martínez-Zapater JM, Dean C. 2000. Mutations causing defects in the biosynthesis and response to gibberellins, abscisic acid and phytochrome B do not inhibit vernalization in Arabidopsis fca-1. Planta 210, 677–682. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Conti L, Galbiati M, Tonelli C. 2014. ABA and the floral transition. In: Zhang D-P. ed. Abscisic acid: metabolism, transport and signaling. Dordrecht, The Netherlands: Springer, 365–384. [Google Scholar]
- Corbesier L, Vincent C, Jang S, et al. 2007. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. 2010. Abscisic acid: emergence of a core signaling network. Annual Review of Plant Biology 61, 651–679. [DOI] [PubMed] [Google Scholar]
- D’Aloia M, Bonhomme D, Bouché F, Tamseddak K, Ormenese S, Torti S, Coupland G, Périlleux C. 2011. Cytokinin promotes flowering of Arabidopsis via transcriptional activation of the FT paralogue TSF. The Plant Journal 65, 972–979. [DOI] [PubMed] [Google Scholar]
- Domagalska MA, Sarnowska E, Nagy F, Davis SJ. 2010. Genetic analyses of interactions among gibberellin, abscisic acid, and brassinosteroids in the control of flowering time in Arabidopsis thaliana. PLoS One 5, e14012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. 2013. Abscisic acid synthesis and response. The Arabidopsis Book 11, e0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, de Montaigu A, Sánchez-Villarreal A, Takahashi Y, Ver Loren van Themaat E, Huettel B, Davis SJ, Coupland G. 2015. The GI–CDF module of Arabidopsis affects freezing tolerance and growth as well as flowering. The Plant Journal 81, 695–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, Panigrahi KCS, Gissot L, Sauerbrunn N, RUhl M, Jarillo JA, Coupland G. 2009. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Developmental Cell 17, 75–86. [DOI] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J. 1999. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO Journal 18, 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks SJ. 2011. Plasticity and evolution in drought avoidance and escape in the annual plant Brassica rapa. New Phytologist 190, 249–257. [DOI] [PubMed] [Google Scholar]
- Fukushima A, Kusano M, Nakamichi N, Kobayashi M, Hayashi N, Sakakibara H, Mizuno T, Saito K. 2009. Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proceedings of the National Academy of Sciences, USA 106, 7251–7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Zhang X, Wang Y, Ming F. 2013. The suppression of WRKY44 by GIGANTEA–miR172 pathway is involved in drought response of Arabidopsis thaliana. PLoS One 8, e73541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoth S, Morgante M, Sanchez J-P, Hanafey MK, Tingey SV, Chua N-H. 2002. Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. Journal of Cell Science 115, 4891–4900. [DOI] [PubMed] [Google Scholar]
- Huq E, Tepperman JM, Quail PH. 2000. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proceedings of the National Academy of Sciences, USA 97, 9789–9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. 2005. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297. [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA. 2003. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426, 302–306. [DOI] [PubMed] [Google Scholar]
- Ito S, Song YH, Josephson-Day AR, Miller RJ, Breton G, Olmstead RG, Imaizumi T. 2012. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proceedings of the National Academy of Sciences, USA 109, 3582–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Pullen N, Lamzin S, Morris RJ, Wigge PA. 2013. Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. The Plant Cell 25, 820–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA. 2007. FT protein acts as a long-range signal in Arabidopsis. Current Biology 17, 1050–1054. [DOI] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KCS, Wenkel S, Soppe W, Deng X-W, Valverde F, Coupland G. 2008. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO Journal 27, 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Torti S, Coupland G. 2009. Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. The Plant Journal 60, 614–625. [DOI] [PubMed] [Google Scholar]
- Jung J-H, Seo Y-H, Seo PJ, Reyes JL, Yun J, Chua N-H, Park C-M. 2007. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. The Plant Cell 19, 2736–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. 1999. Activation tagging of the floral inducer FT. Science 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kazan K, Lyons R. 2016. The link between flowering time and stress tolerance. Journal of Experimental Botany 67, 47–60. [DOI] [PubMed] [Google Scholar]
- Kenney AM, McKay JK, Richards JH, Juenger TE. 2014. Direct and indirect selection on flowering time, water-use efficiency (WUE, δ (13)C), and WUE plasticity to drought in Arabidopsis thaliana. Ecology and Evolution 4, 4505–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Park TI, Yoo SJ, Jun AR, Ahn JH. 2013. a Generation and analysis of a complete mutant set for the Arabidopsis FT/TFL1 family shows specific effects on thermo-sensitive flowering regulation. Journal of Experimental Botany 64, 1715–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W-Y, Ali Z, Park H-J, et al. 2013. b Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nature Communications 4, 1352. [DOI] [PubMed] [Google Scholar]
- Kim W-Y, Hicks KA, Somers DE. 2005. Independent roles for EARLY FLOWERING 3 and ZEITLUPE in the control of circadian timing, hypocotyl length, and flowering time. Plant Physiology 139, 1557–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. 1999. A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. 1986. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Molecular and General Genetics 204, 383–396. [Google Scholar]
- Koops P, Pelser S, Ignatz M, Klose C, Marrocco-Selden K, Kretsch T. 2011. EDL3 is an F-box protein involved in the regulation of abscisic acid signalling in Arabidopsis thaliana. Journal of Experimental Botany 62, 5547–5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM. 1984. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiologia Plantarum 61, 377–383. [Google Scholar]
- Kooyers NJ. 2015. The evolution of drought escape and avoidance in natural herbaceous populations. Plant Science 234, 155–162. [DOI] [PubMed] [Google Scholar]
- Kuromori T, Sugimoto E, Shinozaki K. 2014. Inter-tissue signal transfer of abscisic acid from vascular cells to guard cells. Plant Physiology 164, 1587–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A, Valverde F, Pineiro M, Jarillo JA. 2012. The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. The Plant Cell 24, 982–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Suh S, Park E, Cho E, Ahn J, Kim S, Lee J, Kwon Y, Lee I. 2000. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes and Development 14, 2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lee I. 2010. Regulation and function of SOC1, a flowering pathway integrator. Journal of Experimental Botany 61, 2247–2254. [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JAD, Koornneef M. 1996. Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. The Plant Journal 10, 655–661. [DOI] [PubMed] [Google Scholar]
- Li W, Wang T, Zhang Y, Li Y. 2016. Overexpression of soybean miR172c confers tolerance to water deficit and salt stress, but increases ABA sensitivity in transgenic Arabidopsis thaliana. Journal of Experimental Botany 67, 175–194. [DOI] [PubMed] [Google Scholar]
- Lin P-C, Hwang S-G, Endo A, Okamoto M, Koshiba T, Cheng W-H. 2006. Ectopic expression of ABSCISIC ACID 2/GLUCOSE INSENSITIVE 1 in Arabidopsis promotes seed dormancy and stress tolerance. Plant Physiology 143, 745–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang J, Adrian J, Gissot L, Coupland G, Yu D, Turck F. 2014. Elevated levels of MYB30 in the phloem accelerate flowering in Arabidopsis through the regulation of FLOWERING LOCUS T. PLoS One 9, e89799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L-J, Zhang Y-C, Li Q-H, Sang Y, Mao J, Lian H-L, Wang L, Yang H-Q. 2008. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. The Plant Cell 20, 292–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell JT, Juenger TE, Michaels SD, Lasky JR, Platt A, Richards JH, Yu X, Easlon HM, Sen S, McKay JK. 2013. Pleiotropy of FRIGIDA enhances the potential for multivariate adaptation. Proceedings of the Royal Society B: Biological Sciences 280, 20131043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. 2009. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. [DOI] [PubMed] [Google Scholar]
- Martin-Tryon EL, Kreps JA, Harmer SL. 2007. GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiology 143, 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Zapater JM, Coupland G, Dean C, Koornneef M. 1994. The transition to flowering in Arabidopsis. Cold Spring Harbor Monograph Archive 27, 403–433. [Google Scholar]
- Mathieu J, Warthmann N, Küttner F, Schmid M. 2007. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Current Biology 17, 1055–1060. [DOI] [PubMed] [Google Scholar]
- Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M. 2009. Repression of flowering by the miR172 target SMZ. PLoS Biology 7, e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Himelblau E, Kim SY, Schomburg FM, Amasino RM. 2005. Integration of flowering signals in winter-annual Arabidopsis. Plant Physiology 137, 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wright L, Fujiwara S, et al. 2005. Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. The Plant Cell 17, 2255–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM, Givan SA, McEntee C, Kay SA, Chory J. 2007. The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harbor Symposia on Quantitative Biology 72, 353–363. [DOI] [PubMed] [Google Scholar]
- Moon J, Suh S-S, Lee H, Choi K-R, Hong CB, Paek N-C, Kim S-G, Lee I. 2003. The SOC1MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. The Plant Journal 35, 613–623. [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. 2005. Abscisic acid biosynthesis and catabolism. Annual Review of Plant Biology 56, 165–185. [DOI] [PubMed] [Google Scholar]
- Notaguchi M, Abe M, Kimura T, Daimon Y, Kobayashi T, Yamaguchi A, Tomita Y, Dohi K, Mori M, Araki T. 2008. Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant and Cell Physiology 49, 1645–1658. [DOI] [PubMed] [Google Scholar]
- Oliverio KA, Crepy M, Martin-Tryon EL, Milich R, Harmer SL, Putterill J, Yanovsky MJ, Casal JJ. 2007. GIGANTEA regulates phytochrome A-mediated photomorphogenesis independently of its role in the circadian clock. Plant Physiology 144, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh M-S, Kim HJ, Kay SA, Nam HG. 1999. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285, 1579–1582. [DOI] [PubMed] [Google Scholar]
- Park S-Y, Fung P, Nishimura N, et al. 2009. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin PA, Nilsson O. 2012. The multifaceted roles of FLOWERING LOCUS T in plant development. Plant, Cell and Environment 35, 1742–1755. [DOI] [PubMed] [Google Scholar]
- Porri A, Torti S, Romera-Branchat M, Coupland G. 2012. Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139, 2198–2209. [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. 1995. The Constans gene of arabidopsis promotes flowering and encodes a protein showing similarities to zinc-finger transcription factors. Cell 80, 847–857. [DOI] [PubMed] [Google Scholar]
- Riboni M, Galbiati M, Tonelli C, Conti L. 2013. GIGANTEA enables drought escape response via abscisic acid-dependent activation of the florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1. Plant Physiology 162, 1706–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riboni M, Robustelli Test A, Galbiati M, Tonelli C, Conti L. 2014. Environmental stress and flowering time. The photoperiodic connection. Plant Signaling and Behavior 9, e29036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso M, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. 2003. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Molecular Biology 53, 247–259. [DOI] [PubMed] [Google Scholar]
- Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim T-H, Santiago J, Flexas J, Schroeder JI, Rodriguez PL. 2009. Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiology 150, 1345–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. 2000. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616. [DOI] [PubMed] [Google Scholar]
- Sawa M, Kay SA. 2011. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 108, 11698–11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T. 2007. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G. 2006. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes and Development 20, 898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. 2007. Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany 58, 221–227. [DOI] [PubMed] [Google Scholar]
- Shu K, Chen Q, Wu Y, Liu R, Zhang H, Wang S, Tang S, Yang W, Xie Q. 2016. ABSCISIC ACID-INSENSITIVE 4 negatively regulates flowering through directly promoting Arabidopsis FLOWERING LOCUS C transcription. Journal of Experimental Botany 67, 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Estrada DA, Johnson RS, Kim SK, Lee SY, MacCoss MJ, Imaizumi T. 2014. Distinct roles of FKF1, GIGANTEA, and ZEITLUPE proteins in the regulation of CONSTANS stability in Arabidopsis photoperiodic flowering. Proceedings of the National Academy of Sciences, USA 111, 17672–17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T. 2012. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336, 1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Ma X, Guo H, Sukiran NL, Guo B, Assmann SM, Ma H. 2013. Flower development under drought stress: morphological and transcriptomic analyses reveal acute responses and long-term acclimation in Arabidopsis. The Plant Cell 25, 3785–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. 2001. CONSTANS: mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120. [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. 2004. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006. [DOI] [PubMed] [Google Scholar]
- Wang J-W, Czech B, Weigel D. 2009. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749. [DOI] [PubMed] [Google Scholar]
- Wang P, Xue L, Batelli G, Lee S, Hou Y-J, Van Oosten MJ, Zhang H, Tao WA, Zhu J-K. 2013. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proceedings of the National Academy of Sciences, USA 110, 11205–11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li L, Ye T, Lu Y, Chen X, Wu Y. 2013. The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. Journal of Experimental Botany 64, 675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. 2005. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. [DOI] [PubMed] [Google Scholar]
- Xie Q, Lou P, Hermand V, et al. 2015. Allelic polymorphism of GIGANTEA is responsible for naturally occurring variation in circadian period in Brassica rapa. Proceedings of the National Academy of Sciences, USA 112, 3829–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T. 2005. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant and Cell Physiology 46, 1175–1189. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Maruyama K, Mogami J, Todaka D, Shinozaki K, Yamaguchi-Shinozaki K. 2014. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant, Cell and Environment 38, 35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J-W, Rubio V, Lee N-Y, et al. 2008. COP1 and ELF3 GI stability. Molecular Cell 32, 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Xu X, Crosley RA, et al. 2010. The protein kinase SnRK2.6 mediates the regulation of sucrose metabolism and plant growth in Arabidopsis. Plant Physiology 153, 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X, Lin C. 2011. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Current Biology 21, 841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.