Highlight
RBE functions with microRNA319 to control the growth of petals by regulating the transcription of TCP4 in Arabidopsis.
Key words: Arabidopsis, organ growth, petal development, RABBIT EARS, TCP4, transcription.
Abstract
Plant organ growth requires the proper transition from cell proliferation to cell expansion and differentiation. The CIN-TCP transcription factor gene TCP4 and its post-transcriptional regulator microRNA319 play a pivotal role in this process. In this study, we identified a pathway in which the product of the C2H2 zinc finger gene RABBIT EARS (RBE) regulates the transcription of TCP4 during Arabidopsis (Arabidopsis thaliana) petal development. RBE directly represses TCP4 during the early stages of petal development; this contributes to the role of RBE in controlling the growth of petal primordia. We also found that the rbe-1 mutant strongly enhanced the petal phenotypes of tcp4soj6 and mir319a, two mutants with compromised miR319 regulation of TCP4. Our results show that transcriptional and post-transcriptional regulation function together to pattern the spatial and temporal expression of TCP4. This in turn controls petal size and shape in Arabidopsis.
Introduction
Plant lateral organs, such as leaves and flowers, initiate on the flanks of the shoot apical meristem as a peg-like primordium (Ha et al., 2010). At early developmental stages, the organ primordium consists of cells that undergo active cell division, which marks the period of ‘growth by cell proliferation’. As the organ grows, cells gradually cease division, generally in a basipetal fashion and enter a phase of post-mitotic expansion, which is described as ‘growth by cell expansion’. In addition, during growth by cell expansion, cells differentiate to form specific structures to allow the organ to carry out its biological functions (Breuninger and Lenhard, 2012; Powell and Lenhard, 2012).
The transition from the period of cell proliferation to expansion and differentiation is precisely controlled by spatial and temporal specific molecular mechanisms, which largely determine the final form of an organ. A number of genes and pathways have been characterized in the regulation of this critical step during organ growth (Powell and Lenhard, 2012). Among these regulators, a suite of genes from the TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) gene family play a key role (Efroni et al., 2008; Martin-Trillo and Cubas, 2010; Powell and Lenhard, 2012). TCP family members encode products that contain a basic helix–loop–helix (bHLH) TCP domain that functions in DNA-binding and protein dimerization. The TCP family is categorized into two classes according to differences within this domain (Kosugi and Ohashi, 2002; Martin-Trillo and Cubas, 2010). Class I TCP genes have been proposed to mainly function to promote cell proliferation and concomitant organ growth, while in contrast, Class II TCP genes often act as repressors of plant organ growth (Nath et al., 2003; Li et al., 2005; Efroni et al., 2013; Daviere et al., 2014; Schommer et al., 2014).
The Arabidopsis thaliana genome contains twenty-four TCP genes (Martin-Trillo and Cubas, 2010). Five of them, TCP2, TCP3, TCP4, TCP10, and TCP24 are post-transcriptionally regulated by microRNA319 (Palatnik et al., 2003; Nag et al., 2009). These genes are closely related members in the CIN clade of the Class II TCP family and function together to control a variety of processes in plant development (Koyama et al., 2007; Koyama et al., 2010; Koyama et al., 2011). Among these miR319-regulated genes, TCP4 has been extensively studied: ectopic expression of a miR319-insensitive TCP4 (mTCP4) gene during early stages of leaf development resulted in the formation of miniature leaves, presumably due to the early onset of a maturation program that precociously terminates cell division in developing leaf primordia (Efroni et al., 2008). Similar phenotypes were also observed in floral organs when mTCP4 was expressed using a flower-specific promoter (Nag et al., 2009). These phenotypes suggest that TCP4 represses cell proliferation and promotes post-mitotic differentiation during organ development. Furthermore, it has been suggested that TCP4 might carry out this role in part by directly activating repressors of cell proliferation, such as the cell cycle inhibitor ICK1/KRP1 and the miRNA gene MIR396b (Rodriguez et al., 2010; Schommer et al., 2014). It has also been shown that TCP4 regulates the action of the plant hormones auxin, cytokinin and jasmonate, which are implicated in plant growth (Schommer et al., 2008; Efroni et al., 2013).
Apart from miR319 regulation, comparatively little is known of how TCP4 expression itself is regulated. It has been proposed that the relative levels and domains of expression of TCP4 and its regulator miR319 are critical for defining TCP4 activity (Palatnik et al., 2003; Palatnik et al., 2007), indicating that transcriptional and post-transcriptional regulation are both important in controlling the function of TCP4. It has also been suggested that the product of the C2H2 zinc finger gene JAGGED (JAG) might modulate TCP4 expression but a detailed analysis of this interaction is lacking (Schiessl et al., 2014). In this study, we show that RABBIT EARS (RBE), which encodes a C2H2 zinc finger transcription factor closely related to JAG, directly regulates the transcription of TCP4 during early petal development in Arabidopsis. RBE is specifically expressed in petal primordia at early floral stages. It directly represses the expression of the miRNA gene MIR164c that controls the organ boundary regulators CUP SHAPED COTYLEDON 1 (CUC1) and CUP SHAPED COTYLEDON 2 (CUC2) to effect the establishment of petal primordia (Huang et al., 2012). RBE also promotes petal primordium growth by directly and negatively regulating the TCP5 growth repressor gene, which also belongs to the CIN clade of the Class II TCP family (Huang and Irish, 2015). TCP5 has a similar function to TCP4 in repressing cell proliferation, but is not a target of miR319. During early petal development, RBE inhibits the expression of TCP5 to promote cell proliferation and petal growth (Huang and Irish, 2015). In this study, we show that RBE directly associates with the promoter of TCP4 and acts in concert with miR319 to control TCP4 expression during early petal development.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana plants were grown under long day conditions (16-hour day/8-hour night) at 22 °C. The rbe-1 (Takeda et al., 2004) and mir319a 129 (Nag et al., 2009; abbreviated as mir319a hereafter) mutants are in the Landsberg erecta (L er) background. tcp4soj6 (Palatnik et al., 2007; Nag et al., 2009) and tcp4-1 mutants (Koyama et al., 2010) were originally in the Columbia (Col) background and were backcrossed with L er four times. Homozygous tcp4soj6 and tcp4 mutants were identified by genotyping the progeny of the fourth backcross and crossing with rbe-1 to generate rbe-1 tcp4soj6/+, rbe-1 tcp4soj6 and rbe-1 tcp4. The rbe-1 mir319a double mutant was made by conventional breeding of both parental lines and confirmed with PCR. Both mir319a and tcp4soj6 seeds were gifts from Dr. Thomas Jack (Dartmouth College, Hanover, NH, USA). tcp4-1 (GK_363H08) was obtained from the Arabidopsis Biology Resource Center (ABRC). Primers used in genotyping all the mutants are listed in Supplementary Table S1 at JXB online.
35S:GR-RBE transgenic plants were described previously (Huang et al., 2012). TCP4p:uidA is an enhancer trap line (ET5977) in the L er background (Sarvepalli and Nath, 2011). This transgenic line was kindly provided by the Cold Spring Harbor Laboratory (http://genetrap.cshl.edu/). TCP4p:uidA was introduced into rbe-1 by crossing to generate rbe-1 TCP4p:uidA.
Dexamethasone (DEX) induction, total RNA extraction and qRT-PCR
In order to induce the function of GR-RBE, DEX (10 μM dexamethasone, 0.1% ethanol, 0.015% silwet) or mock (0.1% ethanol, 0.015% silwet) solution was applied to 35S:GR-RBE young floral buds. After a 4 hour treatment, floral tissues were harvested and snap-frozen with liquid nitrogen. RNA was extracted with Trizol (Life Technologies), purified using TURBO DNA-free Kit (Life Technologies), and reverse transcribed with Multiscribe reverse transcriptase (Life Technologies) following the manufacturer’s protocols. qRT-PCR was carried out using the Taqman gene expression assay (Life Technologies). Gene expression levels were calculated from three biological replicates using the 2–ΔΔC T method (Livak and Schmittgen, 2001). The relative RNA levels were normalized to the value of ACTIN 2 (ACT2).
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) was performed according to the previously described protocol (Huang et al., 2012). The 35S:GR-RBE floral tissues treated with 4 hour DEX or mock were harvested and crosslinked with 1% formaldehyde. Extraction and sonication of nuclei were conducted as in (Huang et al., 2012). In the next step, 10 µL of anti-GR P-20 antibody (Santa Cruz Biotechnology, Dallas, TX, USA) was added to each sample to immunoprecipitate GR-RBE protein. Following reverse-crosslinking and DNA purification, semi-quantitative PCR and qPCR were performed to quantitate the ChIP result. Three regions of the promoter of TCP4 were examined and the APETALA3 (AP3) exon was used as a negative control. Primers used for PCR are listed in Supplementary Table S1. Semi-quantitative PCR conditions were: 33 cycles, 94 °C for 30 seconds, 55 °C for 30 seconds and 72 °C for 30 seconds. qPCR was carried out using SYBR Green PCR Master Mix (Life Technologies). The fold enrichment of a specific promoter region was determined by first calculating the abundance ratio in DEX- versus mock-treated ChIP samples normalized by the negative control (AP3 exon) and then divided by the normalized ratio of DEX- to mock-treated input values. Three biological replicates were used for each ChIP experiment.
Histology and in situ hybridization
Detection of β-Glucuronidase activity was conducted as described previously (Nakayama et al., 2005). Whole inflorescences were fixed in 80% acetone at –20 °C for 20 min, then treated with 2 mM X-Gluc in uidA-staining buffer (50 mM sodium phosphate buffer, 10 mM EDTA, 0.1% Triton X-100, 6 mM potassium ferrocyanide and potassium ferricyanide) overnight at 37 °C. After removing the chlorophyll with an ethanol series, inflorescences or individual flowers were mounted in 30% glycerol and examined under a dissecting microscope for whole mount images.
For in situ hybridization, the uidA coding region was amplified and cloned into pGEM-T Easy vector (Promega, Madison, MI, USA) using primers listed in Supplementary Table S1. The DIG RNA Labeling kit (Roche, Mannheim, Germany) was utilized to synthesize the uidA probe. Floral tissues were fixed in freshly made FAA and embedded in Paraplast (Fisher Scientific). 6µm sections were fixed to Probe-on-Plus slides (Fisher Scientific) at 42 °C. In situ hybridization and detection all follow the protocols in (Mara and Irish, 2008; Mara et al., 2010).
Phenotypic analyses
Floral organ number and size were analyzed using the fifth to the twentieth flowers on the inflorescence. Individual floral organs were dissected and photographed for measuring organ width and length in ImageJ. Thirty flowers were counted for floral organ number. Twenty sepals or petals from 10 plants were measured for organ size. Average petal cell sizes were calculated from the number of cells per unit area in ImageJ. Ten petals from 5 plants were used for the cell size measurement. Statistical analyses are described in detail in figure legends.
Results
RBE regulates the transcription of TCP4
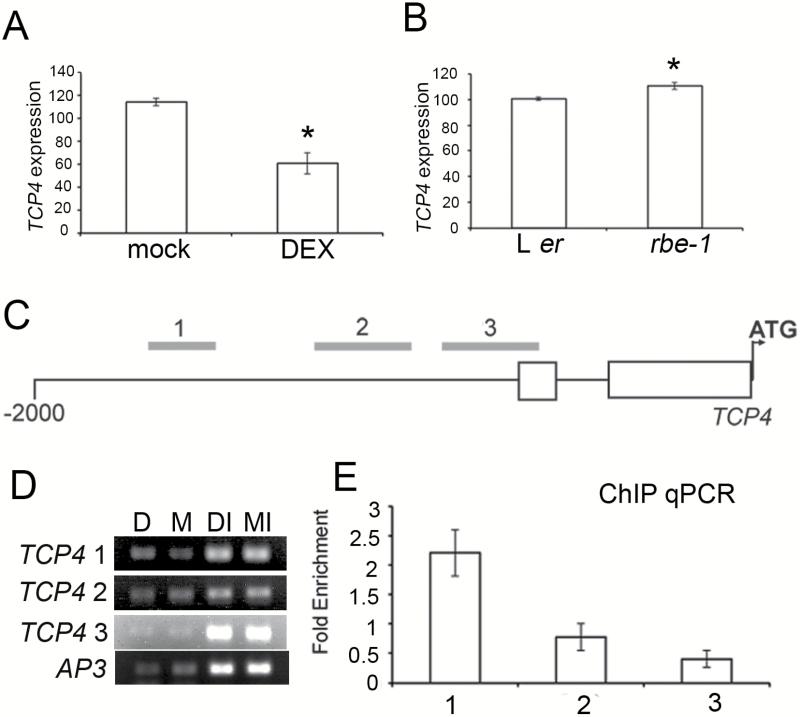
RBE is a key transcriptional regulator of Arabidopsis petal initiation and growth (Takeda et al., 2004; Krizek et al., 2006). We previously performed RNA-sequencing on steroid-inducible 35S:GR-RBE transgenic plants and characterized components of the downstream gene network responsive to RBE activity (Huang et al., 2012). Among the genes whose expression was significantly reduced upon the induction of GR-RBE, we identified TCP4 as a likely candidate for direct targeting by RBE because of its important function in plant organ growth. We first confirmed the RNA-seq result using qRT-PCR to show that the expression of TCP4 is decreased in 35S:GR-RBE floral tissues in the dexamethasone (DEX) treatment as compared to the mock control (Fig. 1A). We also found that the transcript level of TCP4 is modestly but significantly elevated in the flowers of the rbe-1 mutant as compared to wild type, which is consistent with a repressive role of RBE on TCP4 expression (Fig. 1B).
Fig. 1.
RBE negatively regulates TCP4 by directly interacting with its promoter. (A) Expression of TCP4 examined by qRT-PCR in 4 hour DEX- and mock-treated 35S:GR-RBE flowers. Note that TCP4 expression is reduced in DEX-treated flowers. (B) Expression of TCP4 examined by qRT-PCR in L er and rbe-1 flowers. Note that TCP4 expression is increased in rbe-1 compared with L er. In (A) and (B) the y-axis shows relative expression levels normalized to the value of ACT2. Error bars represent stand error of the mean (SEM). Asterisks show a significant difference from the control (P < 0.05, Student t test). (C) Relative positions of fragment 1, 2, and 3 used for ChIP in the -2000 region upstream of the start codon of TCP4. Open boxes indicate 5 prime non-coding exons. (D) Results of semi-quantitative PCR to amplify fragment 1, 2, and 3 in (C) and the control gene (AP3). D: DEX-treated samples; M: mock-treated samples. DI and MI indicate input controls. (E) Fold enrichment of fragment 1, 2 and 3 in (C) in DEX- versus mock-treated flowers assessed by ChIP-qPCR after normalizing with input controls. Fragment 1 is approximately 2.5-fold enriched in DEX- versus mock-treatment, consistent with the semi-qPCR result. Error bars represent SEM for three biological replicates.
TCP4 is one of the five TCP genes that are post-transcriptionally regulated by microRNA319 (Palatnik et al., 2003; Nag et al., 2009). In order to test whether RBE also influences other miR319 target genes, we examined the expression of TCP2, TCP3, TCP10, and TCP24 in the floral tissues of DEX- and mock-treated 35S:GR-RBE plants, as well as in rbe-1 and wild type flowers (see Supplementary Fig. S1). qRT-PCR results showed that DEX-treatment slightly reduced the expression of TCP3, TCP10, and TCP24 in 35S:GR-RBE but had no significant impact on the level of TCP2 (Supplementary Fig. S1). Furthermore, none of the four TCP genes showed a significant change of expression in rbe-1 as compared to wild type. This indicates that RBE does not regulate, or at most plays a minor role, in the regulation of these TCP genes (Supplementary Fig. S1). These results all suggested that TCP4 is a major target of RBE among the miR319-regulated TCP genes and that RBE likely regulates TCP4 independent of miR319. This is also consistent with our RNA-seq results in that none of the three primary miR319 genes (MIR319a/At4g23713, MIR319b/At5g41663, and MIR319c/At2g40805) were expressed at statistically different levels in 35S:GR-RBE upon DEX- versus mock-treatment (Huang et al., 2012).
In order to further examine the possible direct association of RBE with the regulatory regions of TCP4, we carried out chromatin immunoprecipitation (ChIP) and compared the binding of GR-RBE to the 5′- promoter region of TCP4 in DEX- and mock-treated conditions. Both semi-quantitative PCR and qPCR revealed a statistically significant enrichment of TCP4 regulatory sequences in the DEX-treated 35S:GR-RBE plant extract that was immunoprecipitated with GR antibodies, suggesting that RBE is directly associated with the 5′- regulatory region of TCP4 and represses the transcription of TCP4 (Fig. 1D, E).
RBE controls the temporal expression pattern of TCP4
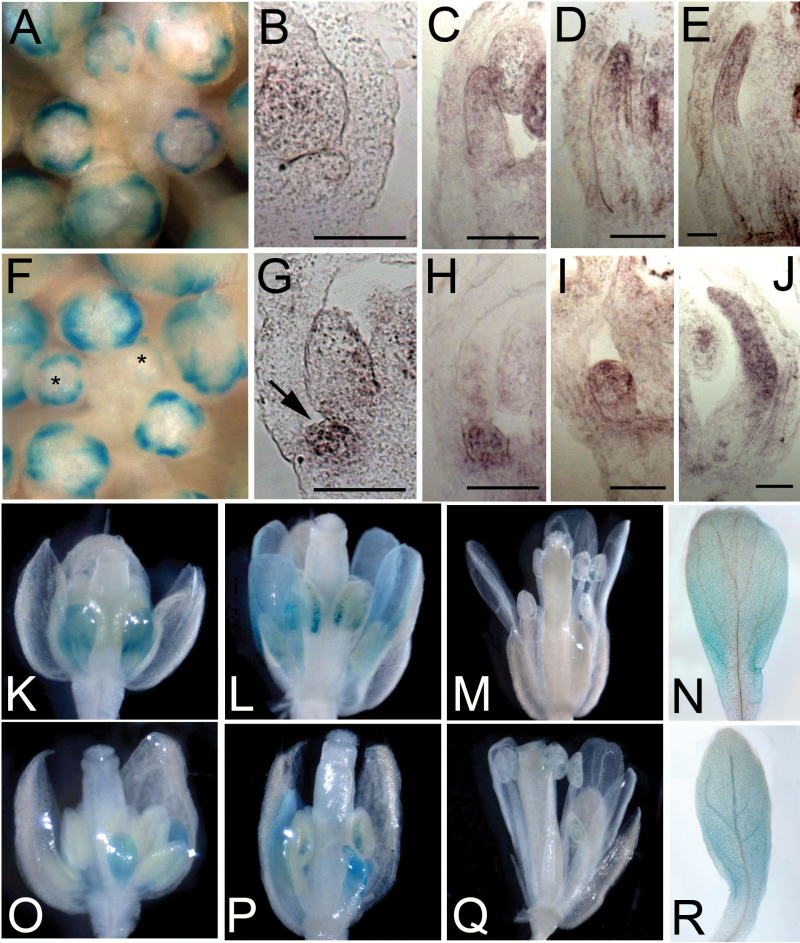
The temporal expression pattern of TCP4 has been described in detail in leaves and reproductive organs (Palatnik et al., 2003; Sarvepalli and Nath, 2011) but its expression pattern in perianth organs has not been well studied. In order to learn more about the temporal pattern of TCP4 expression during petal development, we examined the activity of the TCP4p:uidA reporter in petal primordia at different floral stages. In wild type L er flowers, expression of TCP4p:uidA was not detected in the early petal primordia until late stage 8 (Fig. 2A to 2C). From stage 9 to stage 12, TCP4p:uidA expression was predominantly localized in the distal region of petals and expression was undetectable in open flowers (Fig. 2A, 2D, 2E, 2K to 2N). In contrast, precocious TCP4p:uidA expression in the petal primordia was observed in the rbe-1 mutant from stage 6 and was relatively stronger when compared to the expression in L er prior to stage 9 (Fig. 2F to 2H). However, at later stages of petal development, the expression of TCP4p:uidA in the rbe-1 mutant and L er was similar, even in the small and deformed petals (Fig. 2F, 2I, 2J, 2O to 2R). These analyses suggest that RBE specifically represses TCP4 at early stages of petal development.
Fig. 2.
RBE controls TCP4 promoter activities during petal development. (A) and (F) Whole mount β-Glucoronidase staining shows patterns of TCP4p:uidA expressed in the early stage flowers in wild type L er (A) and rbe-1 (F). Note that β-Glucoronidase staining was detected in the presumptive petal primordia at earlier stages (asterisks) in rbe-1 as compared with wild type. (B) to (E) in situ hybridization using the probe that recognizes the uidA gene shows the expression of TCP4p:uidA in stage 6 (B), 8 (C), 9 (D), 10 (E) petals in L er. Note that TCP4p:uidA activity starts to be visible in stage 8 petals and is predominantly localized at the distal end of the petal. Bar=50μm. (G) to (J) in situ hybridization shows TCP4p:uidA expression in stage 6 (G), 8 (H), 9 (I), 10 (J) rbe-1 petals. Note that TCP4p:uidA is precociously expressed in the stage 6 petal (arrow in G) and at a higher level in a stage 8 petal (H) whose growth was strongly reduced. However, TCP4p:uidA expression appears to be similar in rbe-1 and L er at later stages of petal development. Bar=50μm. (K) to (M) TCP4p:uidA expression shown by whole mount β-Glucoronidase staining in late L er flowers at stage 10 (K), 12 (L) and in the open flower (M). (N) Individual petal dissected from (L) shows TCP4p:uidA expression at the distal end. (O) to (Q) TCP4p:uidA expression shown by whole mount β-Glucoronidase staining in rbe-1 flowers at stage 10 (O), 12 (P) and in the open flower (Q). Note that TCP4p:uidA expression is similar in rbe-1 and L er at these stages. (R) A narrow petal dissected from (P) shows expression of TCP4p:uidA at the distal region.
(This figure is available in colour at JXB online.)
tcp4 partially rescues the mutant phenotypes of rbe-1 in petal growth
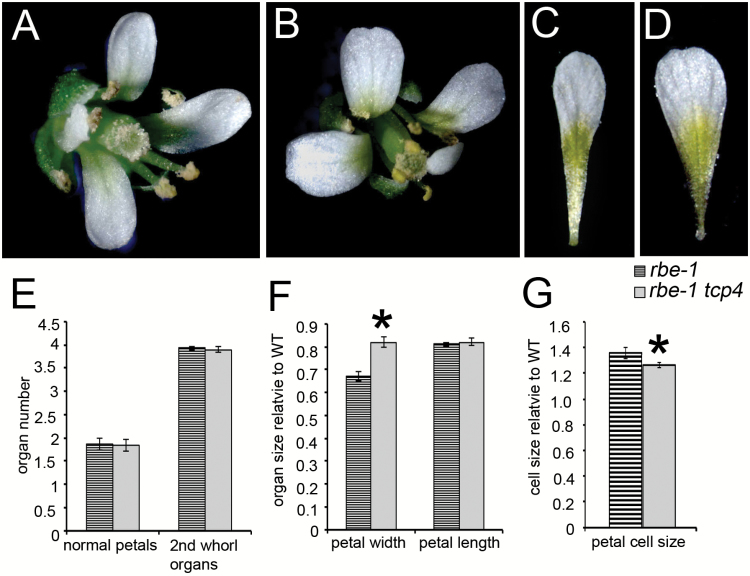
To test whether the repression of transcription of TCP4 is involved in the function of RBE in promoting petal growth, we characterized the petal phenotypes of the rbe-1 tcp4 double mutant. Although the number of ‘normal petals’ - that is petals that have the typical spoon-shape, are white, and are of approximately typical wild type size - and the number of total second whorl organs were similar in rbe-1 and rbe-1 tcp4, the narrow petal phenotype of rbe-1 was obviously ameliorated in the double mutant (Fig. 3, Supplementary Table S2). Petal width in rbe-1 tcp4 was significantly increased compared with that of rbe-1, which is also consistent with the expression of TCP4 at the distal region of the petal (Fig. 3C, 3D, 3F, Supplementary Table S2). We also examined cell size in the distal region of petals. rbe-1 has larger petal cells than wild type (Fig. 3G), which is probably due to compensation for the reduction of cell proliferation in the mutant (Huang and Irish, 2015). In rbe-1 tcp4, cell size is slightly but significantly decreased compared with that of rbe-1, suggesting that increased cell proliferation is the main reason for the restoration of the petal growth defect in the double mutant (Fig. 3G). These results are consistent with the function of RBE and TCP4 as positive and negative regulators of cell proliferation, respectively (Rodriguez et al., 2010; Schommer et al., 2014; Huang and Irish, 2015) and suggest that the transcriptional repression of TCP4 plays a role in mediating the function of RBE in petal development, especially the lateral growth of the petal blade.
Fig. 3.
tcp4 partially rescues the petal phenotypes of rbe-1. (A) a rbe-1 flower, (B) a rbe-1 tcp4 flower, (C) a rbe-1 petal, (D) a rbe-1 tcp4 petal. (E) Numbers of normal petals and second whorl organs in flowers 5–20 for rbe-1 and rbe-1 tcp4 (n=30; mean±SEM). (F) Measurements of petal width and length in flowers 5–20 for rbe-1 and rbe-1 tcp4. Petal sizes were normalized to the values of the L er control (n=20; mean±SEM). Asterisks indicate a significant difference between the single and double mutants (P < 0.05; one-way ANOVA with Tukey test). See Supplementary Table S2 for the details of statistical analyses. (G) Measurements of petal cell size of rbe-1 and rbe-1tcp4. Cell size was normalized to the wild type value (n=10; mean±SEM). Asterisks indicate a significant difference between the single and double mutants (P < 0.05, Student t test).
(This figure is available in colour at JXB online.)
rbe-1 enhances the effect of TCP4 overexpression and miR319 loss-of-function in floral organ development
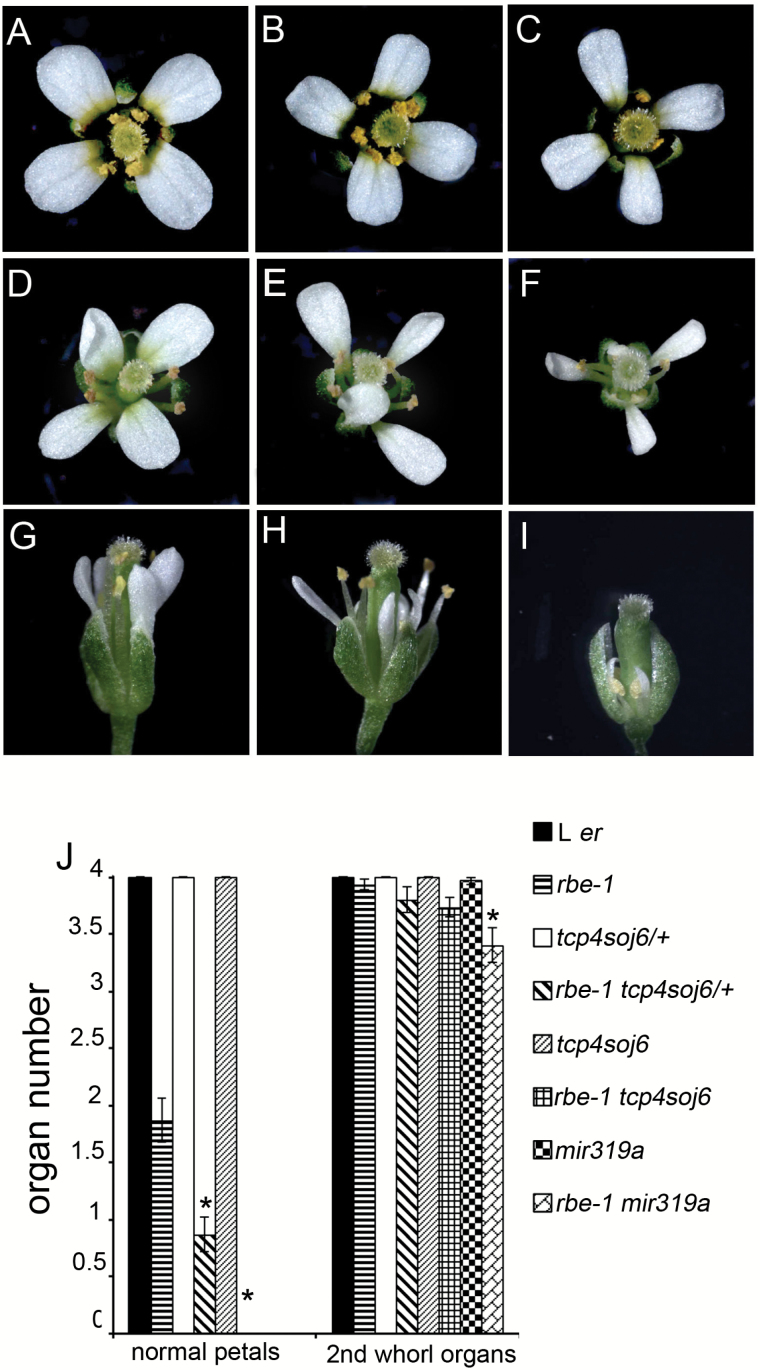
The function of TCP4 is controlled by the combined effect of transcriptional and post-transcriptional regulation. This was shown by a partially miR319-resistant form of TCP4 driven by promoters with different strengths leading to different severities of morphological defects (Palatnik et al., 2007). To test how the ectopic expression of TCP4 in rbe-1 influences the effect of TCP4 in controlling petal growth, we introduced a mutated form of TCP4 into rbe-1. The mutant TCP4, tcp4soj6, contained a single base pair change that partially disrupts cleavage by miR319 (Palatnik et al., 2007; Nag et al., 2009). Plants heterozygous or homozygous for tcp4soj6 produced flowers that were morphologically similar to wild type [Fig. 4A to 4C, Supplementary Table S3, (Palatnik et al., 2007; Nag et al., 2009)]. We only observed a slight reduction in petal size in these mutants (Fig. 4A to 4C, Fig. 5A, 5C, 5E, 5I, Supplementary Table S4 at JXB online). However, when combined with rbe-1, the double mutants displayed much stronger defects in petal development: both rbe-1 tcp4soj6/+ and rbe-1 tcp4soj6 had a significantly decreased number of normal petals compared with their parental lines (Fig. 4B to 4F, 4J, Supplementary Table S3). The measurements of petal size also showed that petal width and length were both reduced in these double mutants (Fig. 5B to 5F, 5I, Supplementary Table S4). In addition, sepal width and length were decreased as well, albeit to a lesser extent, in rbe-1 tcp4soj6 (Supplementary Fig. S2B to S2F, S2I), supporting a previously suggested non-cell-autonomous function of TCP4 in growth repression (Nag et al., 2009). Furthermore, petal number and size, and sepal size were all more dramatically affected in rbe-1 tcp4soj6 compared with rbe-1 tcp4soj6/+ (Fig. 4E, 4F, 4J, Fig 5D, 5F, 5I, Supplementary Fig. S2D, S2F, S2I, Supplementary Table S3, Supplementary Table S4). This showed that the expression level of TCP4 is critical for its function in regulating organ growth.
Fig. 4.
rbe-1 enhances the petal phenotypes of tcp4soj6 and mir319a. (A) to (C) Flowers of L er (A), heterozygous tcp4soj6 (tcp4soj6/+) (B) and homozygous tcp4soj6 (C). (D) to (F) Flowers of rbe-1 (D), rbe-1 tcp4soj6/+ (E) and rbe-1 tcp4soj6. (F). (G) to (I) Flowers of mir319a (G), rbe-1 mir319a with a weaker phenotype (H) and rbe-1 mir319a with a strong phenotype (I). All the flowers in (A) to (I) are the fifth flower formed on the inflorescence. (J) Numbers of normal petals and second whorl organs in flowers 5–20 for L er, rbe-1, tcp4soj6/+, rbe-1 tcp4soj6/+, tcp4soj6, rbe-1 tcp4soj6, mir319a, and rbe-1 mir319a (n=30; mean±SEM). Asterisks indicate a significant difference of the double mutant from the corresponding tcp4soj6/+, tcp4soj6 or mir319a single mutant (P < 0.05; one-way ANOVA with Tukey test). See Supplementary Table S3 for details of the statistical analyses.
(This figure is available in colour at JXB online.)
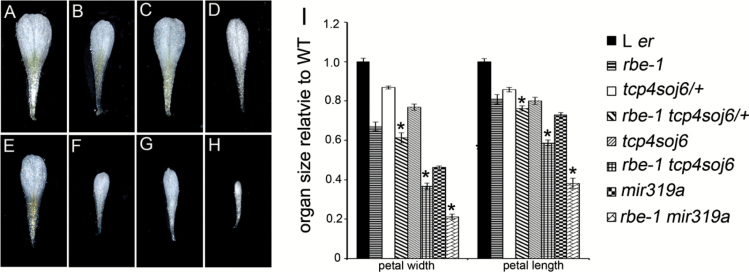
Fig. 5.
Reduction of petal size in tcp4soj6 and mir319a mutants is largely amplified by the loss of function of RBE. Individual petals of L er (A), rbe-1 (B), tcp4soj6/+(C), rbe-1 tcp4soj6/+ (D), tcp4soj6 (E), rbe-1 tcp4soj6 (F), mir319a (G), and rbe-1 mir319a (H). (I) Measurements of petal width and length in flowers 5–20 for L er, rbe-1,tcp4soj6/+, rbe-1 tcp4soj6/+, tcp4soj6, rbe-1 tcp4soj6, mir319a, and rbe-1 mir319a. Petal sizes were normalized to the values of the L er control (n=20; mean±SEM). Asterisks indicate a significant difference of the double mutant from the corresponding tcp4soj6/+, tcp4soj6 or mir319a single mutant (P < 0.05; one-way ANOVA with Tukey test). See Supplementary Table S4 for details of the statistical analyses.
(This figure is available in colour at JXB online.)
Compared with the weak floral phenotypes of tcp4soj6 mutants, plants mutant for MIR319a showed more severe morphological changes in the flower, particularly in the petals [Fig. 4G, 4J, Fig. 5G, 5I, Supplementary Fig. S2G, S2I, Supplementary Table S3, Supplementary Table S4, (Nag et al., 2009)]. In the double mutant rbe-1 mir319a, the floral defects were even more dramatic. Not only did all the petals become small and skinny, the petal loss phenotype was also obvious (Fig. 4G to 4J, Fig. 5G to 5I, Supplementary Table S3, Supplementary Table S4). Petal size was drastically decreased compared with miR319a; petals and sepals also became smaller than those from both parental genotypes (Fig. 4G to 4J, Fig. 5G to 5I, Supplementary Table S3, Supplementary Table S4). In some flowers with an extreme phenotype, all the floral organs became small and stunted (Fig. 4I). These results all suggested a strong enhancement of mir319a mutant phenotypes by rbe-1, which is likely attributable to the alleviation of both transcriptional and post-transcriptional repression of TCP4 in flowers.
Discussion
TCP4 is a major target gene of miR319 in the CIN-TCP family. In petal development, overexpression of miR319-resistant mTCP4 and a mutation in the miR319a 129 allele that reduces the targeting of TCP4 both resulted in a dramatic reduction of petal growth (Nag et al., 2009). Moreover, the tcp4soj6 mutant with a mutation in the TCP4 coding sequence that complements the miR319a 129 allele could largely rescue the miR319a 129 phenotypes, suggesting that TCP4 is a major downstream effector of miR319 in petal development (Nag et al., 2009). Among the three genes that encode mature miR319, MIR319a was thought to play a more prominent role in the petal. The expression of MIR319a partly overlaps with that of TCP4 in developing petals (Nag et al., 2009), suggesting that miR319 functions to dampen rather than eliminate the transcription of TCP4, thus fine-tuning the function of TCP4 during petal growth.
In this study, we identified another upstream regulator of TCP4, RBE, which acts in concert with miR319 to control the expression of TCP4 in petal development. RBE encodes a C2H2 zinc finger transcriptional repressor, so it presumably functions via down-regulating the transcription of TCP4; this was confirmed by our results that TCP4 was more highly expressed in the flowers of rbe-1 compared with wild type (Fig 1). The increase of TCP4 expression in rbe-1 is modest, probably because it only occurs at early developmental stages (Fig. 2). However, this specific ectopic expression of TCP4 might in part be responsible for the petal growth defects in rbe-1, as tcp4 partially rescues the mutant phenotype of rbe-1 (Fig. 3). This is also consistent with the observation in leaves that excess expression of TCP4 at early developmental stages results in the reduction of organ growth in the plant (Efroni et al., 2008). Furthermore, we also found that rbe-1 dramatically enhanced the phenotypes of tcp4soj6 and miR319a 129, mutants in which the post-transcriptional regulation of TCP4 was compromised. The strong defects in rbe-1 tcp4soj6 and rbe-1 miR319a further confirmed previous studies showing that transcriptional and post-transcriptional regulation function together to control the level of TCP4 (Palatnik et al., 2007). A similar means of regulation was also proposed as a key mechanism determining the expression of other miRNA-targeting genes, including HD-ZIP III and CUC transcription factors (Bao et al., 2004; Sieber et al., 2007).
Despite the prominent effects of miRNA on its targets, identification of the upstream transcriptional regulators is also critical in unraveling the spatial and temporal regulation that patterns the expression of these miRNA-targeting genes. To our knowledge, RBE is one of the few transcriptional regulators of TCP4 that has been functionally characterized in detail. We hope our findings will provide novel insights in uncovering the genetic network that controls this key growth regulating gene and its related developmental processes.
Supplementary Data
Supplementary data are available at JXB online.
Fig. S1: RBE has minor and indirect effects on other miR319-regulating TCP genes.
Fig. S2: Sepal growth is affected in tcp4soj6 and mir319a mutants and further impaired when combined with rbe-1.
Table S1: Primers used in this study.
Table S2: Statistical analyses of the petal size of rbe-1 and rbe-1 tcp4.
Table S3: Statistical analyses of the floral organ number of the single and double mutants of rbe-1, tcp4soj6/+, tcp4soj6 and mir319a.
Table S4: Statistical analyses of the floral organ size of the single and double mutants of rbe-1, tcp4soj6/+, tcp4soj6 and mir319a.
Acknowledgements
We thank Dr. Thomas Jack for providing the seeds of mir319a and tcp4soj6 and the Cold Spring Harbor Laboratory for the ET5977 line. This work is supported by the National Natural Science Foundation of China (31571252), Guangdong Innovation Research Team Fund (2014ZT05S078) and Natural Science Startup Foundation of Shenzhen University (2016101).
References
- Bao N, Lye KW, Barton MK. 2004. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Developmental Cell 7, 653–662. [DOI] [PubMed] [Google Scholar]
- Breuninger H, Lenhard M. 2012. Expression of the central growth regulator BIG BROTHER is regulated by multiple cis-elements. BMC Plant Biology 12, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daviere JM, Wild M, Regnault T, Baumberger N, Eisler H, Genschik P, Achard P. 2014. Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Current Biology 24, 1923–1928. [DOI] [PubMed] [Google Scholar]
- Efroni I, Blum E, Goldshmidt A, Eshed Y. 2008. A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. The Plant Cell 20, 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Han SK, Kim HJ, Wu MF, Steiner E, Birnbaum KD, Hong JC, Eshed Y, Wagner D. 2013. Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Developmental Cell 24, 438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha CM, Jun JH, Fletcher JC. 2010. Shoot apical meristem form and function. Current Topics in Developmental Biology 91, 103–140. [DOI] [PubMed] [Google Scholar]
- Huang T, Irish VF. 2015. Temporal Control of Plant Organ Growth by TCP Transcription Factors. Current Biology 25, 1765–1770. [DOI] [PubMed] [Google Scholar]
- Huang T, Lopez-Giraldez F, Townsend JP, Irish VF. 2012. RBE controls microRNA164 expression to effect floral organogenesis. Development 139, 2161–2169. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. 2002. DNA binding and dimerization specificity and potential targets for the TCP protein family. The Plant Journal 30, 337–348. [DOI] [PubMed] [Google Scholar]
- Koyama T, Furutani M, Tasaka M, Ohme-Takagi M. 2007. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. The Plant Cell 19, 473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. 2010. TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. The Plant Cell 22, 3574–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Ohme-Takagi M, Sato F. 2011. Generation of serrated and wavy petals by inhibition of the activity of TCP transcription factors in Arabidopsis thaliana. Plant Signaling & Behavior 6, 697–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Lewis MW, Fletcher JC. 2006. RABBIT EARS is a second-whorl repressor of AGAMOUS that maintains spatial boundaries in Arabidopsis flowers. The Plant Journal 45, 369–383. [DOI] [PubMed] [Google Scholar]
- Li C, Potuschak T, Colon-Carmona A, Gutierrez RA, Doerner P. 2005. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proceedings of the National Academy of Sciences, USA 102, 12978–12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Mara CD, Huang T, Irish VF. 2010. The Arabidopsis floral homeotic proteins APETALA3 and PISTILLATA negatively regulate the BANQUO genes implicated in light signaling. The Plant Cell 22, 690–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mara CD, Irish VF. 2008. Two GATA transcription factors are downstream effectors of floral homeotic gene action in Arabidopsis. Plant Physiology 147, 707–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Trillo M, Cubas P. 2010. TCP genes: a family snapshot ten years later. Trends in Plant Science 15, 31–39. [DOI] [PubMed] [Google Scholar]
- Nag A, King S, Jack T. 2009. miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proceedings of the National Academy of Sciences, USA 106, 22534–22539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama N, Arroyo JM, Simorowski J, May B, Martienssen R, Irish VF. 2005. Gene trap lines define domains of gene regulation in Arabidopsis petals and stamens. The Plant Cell 17, 2486–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath U, Crawford BC, Carpenter R, Coen E. 2003. Genetic control of surface curvature. Science 299, 1404–1407. [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. 2003. Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Wollmann H, Schommer C, et al. 2007. Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Developmental Cell 13, 115–125. [DOI] [PubMed] [Google Scholar]
- Powell AE, Lenhard M. 2012. Control of organ size in plants. Current Biology 22, R360–R367. [DOI] [PubMed] [Google Scholar]
- Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF. 2010. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvepalli K, Nath U. 2011. Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. The Plant Journal 67, 595–607. [DOI] [PubMed] [Google Scholar]
- Schiessl K, Muino JM, Sablowski R. 2014. Arabidopsis JAGGED links floral organ patterning to tissue growth by repressing Kip-related cell cycle inhibitors. Proceedings of the National Academy of Sciences, USA 111, 2830–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C, Debernardi JM, Bresso EG, Rodriguez RE, Palatnik JF. 2014. Repression of cell proliferation by miR319-regulated TCP4. Molecular Plant 7, 1533–1544. [DOI] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, Chetelat A, Cubas P, Farmer EE, Nath U, Weigel D. 2008. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biology 6, e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM. 2007. Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development 134, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Takeda S, Matsumoto N, Okada K. 2004. RABBIT EARS, encoding a SUPERMAN-like zinc finger protein, regulates petal development in Arabidopsis thaliana. Development 131, 425–434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.