Abstract
A large haplotype on chromosome 19p13.11 tagged by rs10401969 in intron 8 of SURP and G patch domain containing 1 (SUGP1) is associated with coronary artery disease (CAD), plasma LDL cholesterol levels, and other energy metabolism phenotypes. Recent studies have suggested that TM6SF2 is the causal gene within the locus, but we postulated that this locus could harbor additional CAD risk genes, including the putative splicing factor SUGP1. Indeed, we found that rs10401969 regulates SUGP1 exon 8 skipping, causing non-sense-mediated mRNA decay. Hepatic Sugp1 overexpression in CD1 male mice increased plasma cholesterol levels 20–50%. In human hepatoma cell lines, SUGP1 knockdown stimulated 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) alternative splicing and decreased HMGCR transcript stability, thus reducing cholesterol synthesis and increasing LDL uptake. Our results strongly support a role for SUGP1 as a novel regulator of cholesterol metabolism and suggest that it contributes to the relationship between rs10401969 and plasma cholesterol.
Introduction
In the largest genome-wide association study meta-analysis of lipids published to date, only ten loci were associated with both LDL cholesterol (LDLC) and triglycerides (TG) levels significantly (1). Of these, a locus on Chr19p13.11 has also been associated with incidence of coronary artery disease (CAD), as shown in both case–control and prospective cohort studies of Caucasians and Asians (1–3). The haplotype in this locus tagged by the C minor allele of rs10401969 has been associated with reduced plasma LDLC and/or TG in African American, Hispanic and Native American populations (3–10), as well as in several populations of European ancestry (1,4,11–14). This locus has also been associated with Type 2 diabetes (11,15,16), hepatic steatosis, non-alcoholic fatty liver disease (17–20) and body mass index (21), suggesting that genetic variation within this region has a broad impact on human metabolism.
Recently, three studies have implicated TM6SF2 as the causative gene within the Chr19p13.11 locus. Exome sequencing studies identified an association between rs58542926, a non-synonymous (Glu167Lys) variant in TM6SF2, and risk of myocardial infarction and non-alcoholic fatty liver disease (22,23). Expression of the recombinant TM6SF2 variant encoding p.Glu167Lys in HepG2 cells resulted in 50% less protein compared to wildtype. Tm6sf2 knockdown in an AAV murine model (23) increased hepatic TG and decreased very low-density lipoprotein (VLDL) secretion, consistent with the effects reported in vitro (24).
Notably, rs58542926 is in very strong linkage disequilibrium (LD) with rs10401969 in Europeans (r2 = 0.95 and D′ = 1 in 503 EUR, 1000 Genomes Project, Phase 3). In fact, in individuals of European descent, rs10401969 (EUR MAF = 7.1%) resides in a large LD block that extends ∼360 kb across at least 14 genes and contains 32 other genetic variants that are all in strong LD (r2 > 0.8) with rs10410969. Although it may be simplest to attribute all of the phenotypic associations with the 19p13.11 haplotype described above to one causal gene, such as TM6SF2, the complex genetic architecture of this region suggests other possibilities. For instance, any number of the 33 variants in strong LD (and perhaps rarer SNPs, via synthetic association) (25) could have functional effects that contribute to the associated phenotypes, perhaps by acting on different genes. In addition, even a single variant could impact multiple genes that contribute to the same phenotype, especially because functionally related genes are often clustered in the genome. Consistent with these possibilities, Blattmann et al. (26) reported that knockdown of at least six genes in the Chr19p13.11 locus in Hela-Kyoto cells impacted DiI-LDL uptake, intracellular cholesterol levels or both. rs10401969 is located within an intron of SURP and G patch domain containing 1 (SUGP1), previously known as splicing factor 4 (SF4). Because splicing is a mechanism that regulates cholesterol homeostasis (27) and genetic variants associated with variation in plasma LDLC levels can influence splicing of the key cholesterol metabolism genes 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) and low-density lipoprotein receptor (LDLR) (28,29), we sought to investigate whether genetic variation within a splicing factor itself (30) can affect cellular cholesterol homeostasis. Thus, here we investigate whether SUGP1 may contribute to the relationship between rs10401969 and plasma lipids through the regulation of alternative splicing.
Results
SUGP1 impacts cholesterol metabolism in vitro
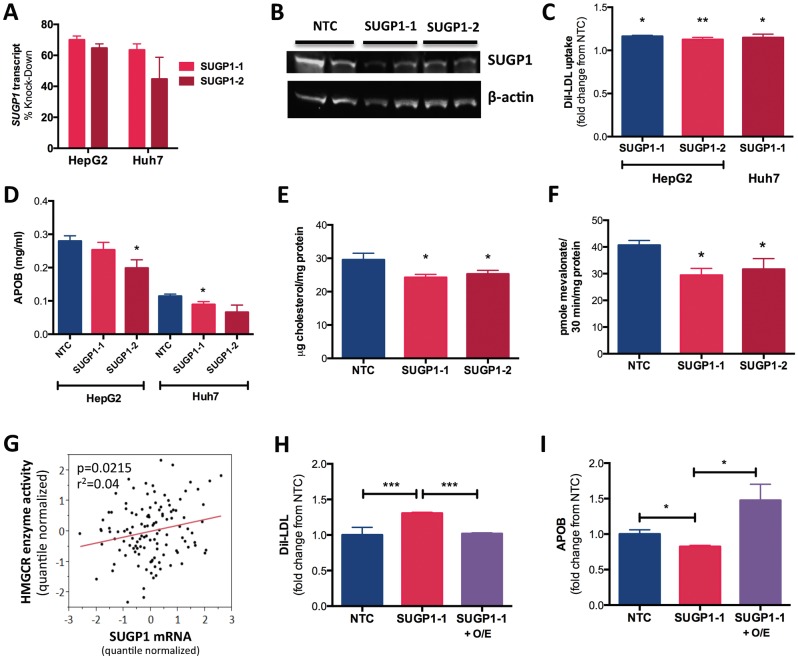
To determine whether SUGP1 impacts cellular cholesterol metabolism, we transfected human hepatoma HepG2 and Huh7 cell lines with siRNAs targeting SUGP1, which reduced SUGP1 transcript levels by 45–70% (n = 12) and protein levels by 72–91% (n = 2) (Fig. 1A and B). SUGP1 knockdown increased rates of DiI-LDL uptake 10–15% (Fig. 1C) and reduced APOB in the cell culture media (Fig. 1D), but did not alter APOE concentrations (Supplementary Material, Fig. S1A). SUGP1 knockdown also reduced rates of cholesterol synthesis (Fig. 1E) and HMGCR enzyme activity ∼20% (Fig. 1F). Consistent with this observation, we also found that SUGP1 transcript levels were positively correlated with variation in HMGCR enzyme activity in 118 human lymphoblastoid cell lines (P = 0.0215, r2 = 0.04; Fig. 1G). To demonstrate the specificity of these effects, we performed a rescue experiment in which SUGP1 was overexpressed in HepG2 cells after SUGP1 knockdown (Supplementary Material, Fig. S1B and S1C). As expected, SUGP1 overexpression was able to reverse the effects of SUGP1 knockdown on rates of DiI-LDL uptake (Fig. 1H) and APOB in the media (Fig. 1I). Similarly, SUGP1 overexpression alone also reduced rates of DiI-LDL uptake (Supplementary Material, Fig. S1D). In contrast, knockdown of ATPase type 13A1 (ATP13A1) and MAU2 sister chromatid cohesion factor (MAU2), two additional genes within the Chr19p13.11 locus (Supplementary Material, Fig. S1E) whose expression levels are associated with rs10401969 (31), did not produce consistent and statistically significant changes in measures of cellular cholesterol metabolism (Supplementary Material, Fig. S1F and S1G).
Figure 1.
SUGP1 expression level impacts cellular cholesterol metabolism. HepG2 and Huh7 cells were reverse transfected with an NTC siRNA or Silence Select siRNAs against SUGP1, SUGP1-1 and SUGP1-2. After 48 hours, cells were collected, and various cellular phenotypes were measured. (A) SUGP1 transcript levels were quantified by quantitative PCR (n = 6). (B) SUGP1 protein was detected by immunoblot and band density was quantified using Image J (n = 4), one representative blot shown. (C) DiI-LDL uptake was quantified by Fluorescence-activated cell sorting (n = 6). (D) APOB in the conditioned media was quantified by ELISA and normalized to total cellular protein quantified by a bicinchoninic acid assay (n = 9). (E) Rates of cholesterol biosynthesis were quantified using a radiolabeled precursor in Huh7 cells (n = 3), with values normalized to total cellular protein. (F) HMGCR activity in HepG2 cells was quantified by incubating lysed cells with 14C-HMG-CoA (precursor) to directly assess incorporation rates into 14C-mevalonate (product). Pmol of mevalonate formed was normalized to total protein (n = 6). (G) SUGP1 transcript levels were quantified by expression array in 118 LCLs, normalized by array batch and corresponding covariates. HMGCR enzyme activity was measured in these same LCLs and tested for association with SUGP1 transcript levels using linear regression in JMP 9.0. (H and I) HepG2 cells were reverse transfected with SUGP1-1 siRNA or NTC, and after 24 hours, cells were transfected with an SUGP1 overexpression construct (SUGP1-1 + O/E) or empty vector (SUGP1-1) and incubated for an additional 48 hours. LDL-uptake and APOB in the culture media were quantified (n = 6). All statistical analyses were performed in JMP 9.0. With the exception of I, ANOVA with post hoc two-tailed t-tests were used to identify differences between treatments. *P < 0.05, **P < 0.01, ***P < 0.001. All Ns reported are per cell type/per condition.
SUGP1 alters cholesterol metabolism in vivo
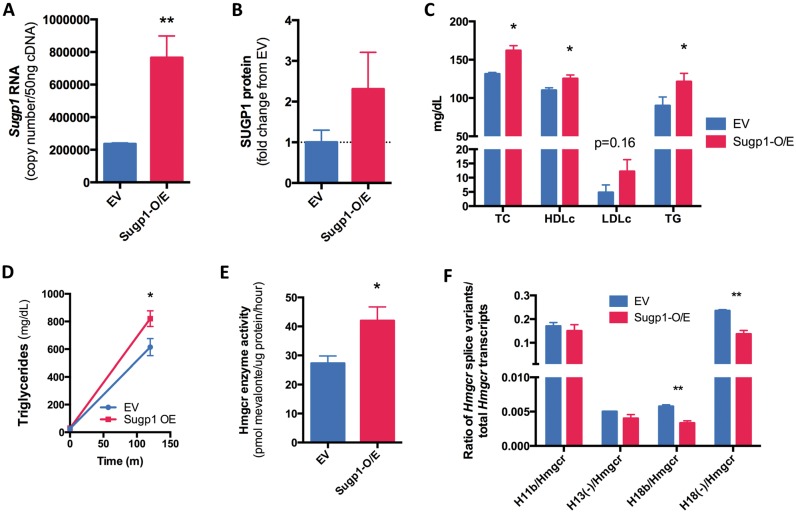
To test the physiological relevance of SUGP1, we delivered murine Sugp1 to the liver by hydrodynamic tail vein injection in CD-1 mice. After 7 days, hepatic overexpression of Sugp1 was assessed. Sugp1 transcript and protein levels were increased (Fig. 2A and B and Supplementary Material, Fig. S2A). Fasting plasma lipids were measured and total cholesterol (TC), triglyceride and high-density lipoprotein (HDL) cholesterol were significantly increased, with a trend of elevated LDLC observed (Fig. 2C). Similar effects were also observed after 28 days of Sugp1 overexpression (Supplementary Material, Fig. S2B–S2D), with greater hepatic Sugp1 expression correlated with greater increases in TC (P = 0.01, r2 = 0.53 and Supplementary Material, Fig. S2E). In addition, we found elevated triglyceride secretion in the overexpressing animals after Polyoxamer 407 treatment (Fig. 2D). Consistent with in vitro results, Sugp1 overexpression resulted in greater hepatic Hmgcr enzyme activity (Fig. 2E), whereas we observed no change in hepatic Hmgcr transcript levels (Supplementary Material, Fig. S2F).
Figure 2.
Hepatic Supg1 overexpression increases plasma cholesterol in mice. An Sugp1 overexpression (O/E) vector or an empty vector control was injected into the tail veins of CD-1 male mice (n = 4 or 5/condition). (A) Sugp1 mRNA was quantified by quantitative PCR (qPCR) 7 days post injection. (B) SUGP1 protein levels in mouse livers were determined by western blot and normalized against glyceraldehyde-3-phosphate dehydrogenase as a loading control. (C) Plasma lipids were quantified by colorimetric assay, adjusted for percent change in body weight over time, and unpaired two-tailed t-tests were used to identify statistically significant differences between the residuals. (D) Plasma triglycerides were quantified by colorimetric assay before and 3 hours after injection with Polyoxamer 407. (E) Hepatic Hmgcr enzyme activity was quantified as described in Figure 1. (F) Hepatic Hmgcr splice variants were quantified by qPCR. Unpaired two-tailed t-tests were used to identify statistically significant differences *P < 0.05, **P < 0.01.
SUGP1 knockdown increases expression of alternatively spliced HMGCR transcripts
Given the effects of SUGP1 knockdown on LDL uptake and HMGCR enzyme activity, and its putative role as a splicing factor, we postulated that SUGP1 might directly affect LDLR or HMGCR transcript structure. Because alternative splicing of LDLR is extensive and highly varied, we tested the four major LDLR splice variants, specifically, those lacking exons 3, 4, 12 or 14 (28). SUGP1 knockdown did not produce consistent changes in expression levels of any of these LDLR transcripts in vitro (Supplementary Material, Fig. S3A).
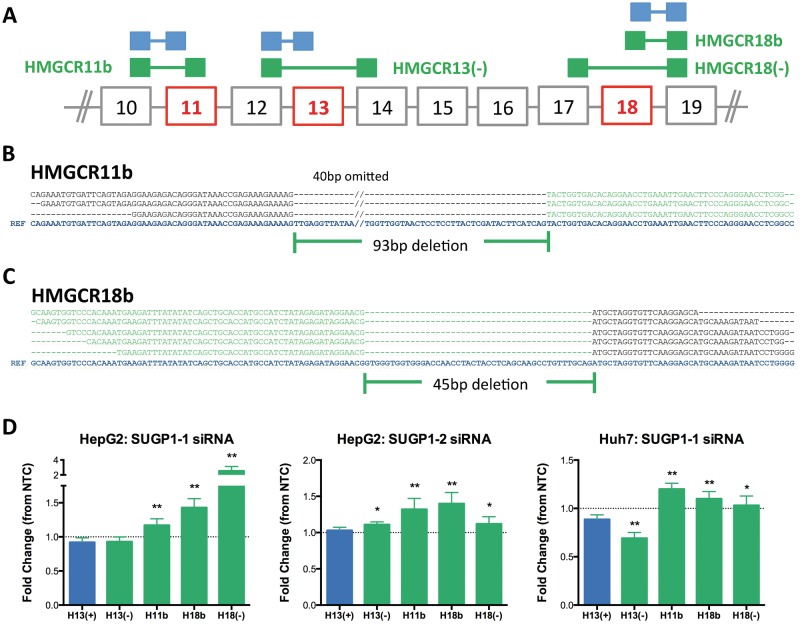
To assess effects of SUGP1 on HMGCR structure, we first identified HMGCR splice variants expressed in primary liver tissue derived from human donors (n = 3), human hepatoma cell lines (HepG2, Hep3B and Huh7) and lymphoblastoid cell lines (LCLs) (n = 8) by RNAseq. In addition to the known HMGCR splice variants, which lack exon 13, HMGCR13(−), or exon 18, HMGCR18(−), we also detected transcripts with shortened versions of exons 11 (ex11b) and 18 (ex18b). Ex11b is missing 93 bp at its 5′ end, whereas ex18b is missing 45 bp at its 3′ end (Fig. 3A–C). Although HMGCR11b and HMGCR18b have a similar level of abundance, RT-PCR with primers specific for ex11b to ex18b failed to amplify a product (Supplementary Material, Fig. S3B), suggesting that these two splice events do not occur on the same transcript. SUPG1 knockdown resulted in statistically significant increases (14–49%) in HMGCR11b, HMGCR18b and HMGCR18(−) in human hepatoma cell lines transfected with SUGP1 siRNAs (Fig. 3D). In contrast, there were no consistent changes in HMGCR13(−), the most abundant HMGCR splice variant, and a trend of increased HMGCR13(+). We found detectable expression levels of these splice variants in the mouse, and consistent with these effects, increased Hmgcr alternative splicing was observed after Sugp1 overexpression in vivo (Fig. 2F).
Figure 3.
SUGP1 knockdown increases expression levels of the known and novel HMGCR splice variants. Size selected polyA-purified RNA-seq libraries were prepared from human hepatoma cell lines (HepG2, Hep3B and Huh7), primary human liver tissue (n = 3 donors), and lymphoblastoid cell lines (n = 8 African American donors), and sequenced using paired-end sequencing. Sequences were aligned to the genome and the known transcriptome using TopHat, and instances of alternative splicing were identified at HMGCR exons 11, 13 and 18. Neither the HMGCR11b nor the HMGCR18b splice variant has been previously identified. Exon 11b is missing 93 bp at the 5′ end of exon 11, and exon 18b is missing 45 bp at the 3′ end of exon 18. (A) Diagram of canonical versus alternatively spliced junctions. Canonical splice junctions are shown in blue, and alternatively spliced junctions in green. Exons affected by alternative splicing are in red. Sequence alignment of junction spanning reads identifying HMGCR11b (B) and HMGCR18b (C). The reference sequence (blue) is shown at the bottom in bolded blue text. The canonical exon sequences are shown in black, and the novel exon 11b and 18b are in green. Due to space constraints, 40 bp of skipped sequence was omitted from the reference sequence for exon 11. (D) HMGCR splice variants quantified by real-time qPCR (n = 12) after 48 hours of SUGP1 knockdown by siRNA (SUGP1-1, SUGP1-2 or NTC control) in HepG2 and Huh7. Data are shown as relative fold change compared to NTC control. Student’s t-test comparing the effect of SUGP1 knock-down on HMGCR splice variants compared to HMGCR13(+). *P < 0.05; **P < 0.01.
To determine whether these novel HMGCR splice variants are subject to non-sense-mediated decay (NMD), we treated HepG2 cells with cycloheximide, an inhibitor of NMD, and quantified HMGCR splice variants over 3 hours. HMGCR18(−) levels dramatically increased, whereas HMGCR11b and HMGCR18b transcripts levels remained unchanged, indicating that HMGCR18(−) abundance is likely regulated by NMD (Supplementary Material, Fig. S3C). These findings suggest that the reduction in HMGCR enzyme activity observed after SUGP1 knockdown may be attributed to greater expression levels of the novel HMGCR splice variants.
SUGP1 regulates HMGCR alternative splicing and transcript stability
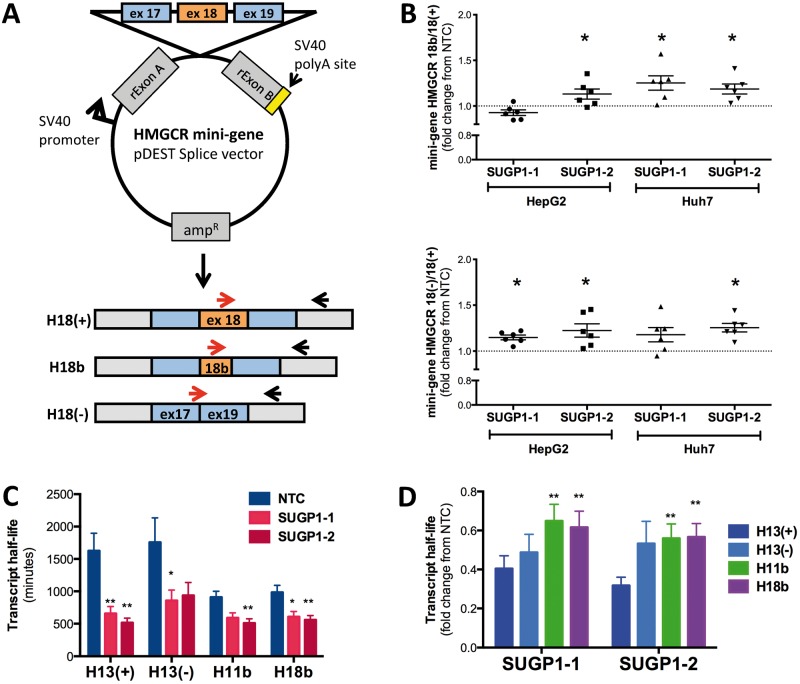
SUGP1 is predicted to function as a splicing factor (30). However, to date, the function of SUGP1 has not yet been validated. To determine whether SUGP1 regulates HMGCR alternative splicing, HepG2 and Huh7 cell lines were transfected with a mini-gene containing the HMGCR genomic sequence from intron 17 to 19 (mHMGCR17-19) following SUGP1 knockdown (Fig. 4A). HMGCR transcripts derived from the mini-gene were quantified by quantitative PCR. SUGP1 knockdown increased the relative expression levels of the mini-gene-derived HMGCR18b and HMGCR18(−) transcripts, compared to HMGCR18(+) transcripts in vitro (Fig. 4B).
Figure 4.
SUGP1 knockdown modulates HMGCR alternative splicing and transcript half-life. (A) Diagram of the mini-gene construct and quantitative PCR (qPCR) primers used to directly test SUGP1 effects on HMGCR exon 18 alternative splicing. Red arrows indicate different forward primers within the cDNA sequence used to assay for alternative splice variants. (B) Mini-gene-derived HMGCR transcript ratios show an increase in alternative splice variants upon SUGP1 knockdown. HepG2 and Huh7 cells were first transfected with SUGP1-1, SUGP1-3 or NTC siRNAs. After 24 hours, cells were transfected with the HMGCR exon 18 containing mini-gene construct and incubated for an additional 24 hours. Mini-gene-derived HMGCR transcripts were quantified by qPCR (n = 6 per condition). (C) SUGP1 knockdown decreases transcript half-life. HepG2 cells were transfected with SUGP1 or NTC siRNAs, and actinomycin D was added after 48 hours to halt cellular transcription. HMGCR transcript levels were quantified over 24 hours, and half-life calculated as previously described. Data are represented as mean ± SEM. (D) Relative changes in HMGCR transcript half-life in cells transfected with one of the two SUGP1 siRNAs compared to NTC. *P < 0.05, **P < 0.01 indicate effects of SUGP1 knockdown decrease HMGCR11b and HMGCR18b transcript half-life compared to HMGCR13(+). *P < 0.05; **P < 0.01.
We previously reported that HNRNPA1, another RNA binding protein, increases the relative abundance of HMGCR13(−) transcript compared to HMGCR13(+) transcript by promoting alternative splicing as well as preferentially stabilizing the HMGCR13(−) transcript (32). To test whether SUGP1 also impacts HMGCR transcript stability, we treated HepG2 and Huh7 cell lines with actinomycin D upon SUGP1 knockdown and measured endogenous HMGCR transcripts. SUGP1 knockdown dramatically reduced HMGCR transcript half-lives (Fig. 4C and Supplementary Material, Fig. S4A); however, this effect was less pronounced for HMGCR11b and HMGCR18b than for HMGCR13(+) or HMGCR13(−) in HepG2 cells (Fig. 4D). No decay of HMGCR18(−) was detected after actinomycin D treatment, and thus half-life was not determined (Supplementary Material, Fig. S4B). These results suggest that despite the overall reduction in HMGCR transcript stability, the increase in the relative ratio of the HMGCR11b and HMGCR18b transcripts over the canonical HMGCR13(+) after SUGP1 knockdown is likely due to both promotion of HMGCR alternative splicing and smaller reductions in the stability of the alternatively spliced transcripts.
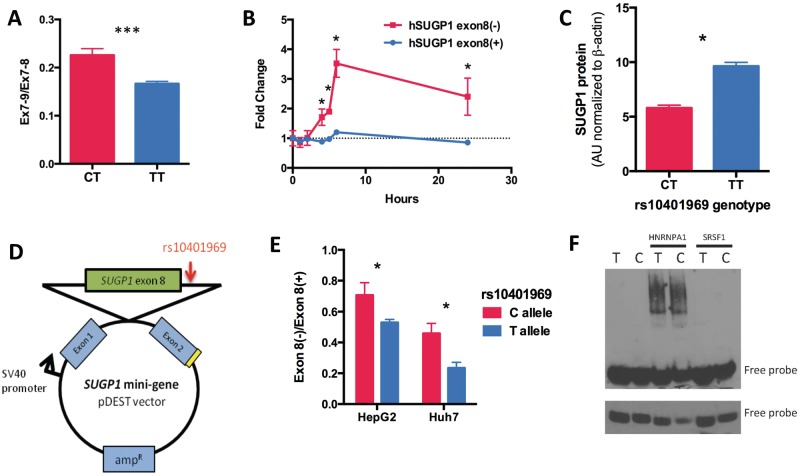
rs10401969 modulates SUGP1 alternative splicing and protein levels
rs10401969, despite being located in SUGP1 intron 8, has not been associated with SUGP1 transcript levels in any publically available eQTL dataset, including liver (33). Thus, we postulated that rs10401969 may influence SUGP1 transcript structure. Using RT-PCR and Sanger sequencing of cDNA from human hepatoma RNA, we detected a novel SUGP1 splice variant lacking exon 8, SUGP1 exon 8(−) (Supplementary Material, Fig. S5A). The rs10401969 C allele was associated with 20% greater SUGP1 exon 8 skipping in LCLs derived from participants of the Cholesterol and Pharmacogenetics (CAP) clinical trial (Fig. 5A), whereas no difference in overall SUGP1 expression levels was detected. Similar trends were detected in human livers derived from the St. Jude’s Liver Repository (Supplementary Material, Fig. S5B). Importantly, exon 8 skipping disrupts the open reading frame, introducing a stop codon after 27 base pairs, which we confirmed triggers non-sense-mediated decay because SUGP1 exon 8(–) levels increase in cycloheximide-treated HepG2 cells (Fig. 5B). In addition, we found that the rs10401969 C allele carriers also had reduced levels of SUGP1 protein compared to TT homozygotes (P = 0.01, n = 60, Fig. 5C and Supplementary Material, Fig. S5C). However, there was no evidence that reduced SUGP1 protein in C allele carriers was due to greater protein decay rates (Supplementary Material, Fig. S5D and S5E).
Figure 5.
rs10401969 regulates SUGP1 exon 8 skipping by modulating HNRNPA1 binding. (A) The rs10401969 C allele modulates increases in SUGP1 exon 8 skipping compared to the reference allele by ∼20%. SUGP1 transcript levels were quantified by qPCR in LCLs genotyped for rs10401969 (CT n = 18; TT n = 30). Total SUGP1 was quantified by qPCR assays for exon 7–9 (Ex7-9) and exon 7–8 (Ex7-8) junctions. The relative ratio of Ex7-9 and Ex7-8 is shown. (B) To measure non-sense-medicated mRNA decay, HepG2 cells were treated with cycloheximide (1 μg/ml), and SUGP1 8(−) and 8(+) transcripts were quantified over 24 hours by qPCR. *P < 0.05, two-tailed paired t-test, n = 3. (C) SUGP1 protein levels from CAP LCLs genotyped for the rs10401696 SNP (CT n = 28, TT n = 32) were measured by western blot. Band densities were calculated by Image J and normalized to a β-actin protein levels. (D) Scheme of the SUGP1 mini-gene (mSUGP1) construct containing the genomic fragment of SUGP1 introns 7–8 with the rs10401969 minor allele (C) introduced through site directed mutagenesis. (E) HepG2 and Huh7 (n = 3) cell lines were transfected with either the rs10401969 ‘T’ or ‘C’ allele mSUGP1 construct and mini-gene-derived transcripts quantified by qPCR after 48 hours. Values shown are the relative ratios of mSUGP1 exon 8(−) to mSUGP1 exon 8(+) transcripts are shown. (F) RNA gel shift assay was performed in triplicate with rs10401969 3’ biotin-labeled RNA oligos and either His-tagged HNRNPA1 or SRSF1 proteins. The upper panel shows 30-minute exposure image of a representative gel while the lower panel is a 2-minute exposure of the same gel.
GWAS-identified SNPs are often assumed to be in LD with functionally relevant SNPs and not the causal variant themselves. Although rs10401969 is 80 base pairs downstream from the exon 8 splice donor, there are no other SNPs in high LD (r2 > 0.9) with rs10401969 that are predicted to modulate exon 8 skipping. We tested whether rs10401969 directly alters exon 8 alternative splicing using an SUGP1 mini-gene (mSUGP1) construct (Fig. 5D). Transcripts derived from the mini-gene with the rs10401969 C allele had a higher frequency of exon 8 skipping than those expressed from the T allele (Fig. 5E), consistent with our observations in the CAP LCLs. Using Human Splicing Finder (34), we noted that rs10401969 lies immediately adjacent to a predicted binding site for HNRNPA1, a splicing factor that we have previously implicated in cholesterol metabolism (32). Using an RNA electrophoretic mobility shift assay with probes containing SUGP1 intron 8 with either the rs10401969 C or T allele, we found that the C allele had 27% stronger binding affinity to HNRNPA1 protein compared to the T allele as indicated by reduced intensity of the free probe (Fig. 5F and Supplementary Material, Fig. S5F and S5G). As a control, we also performed RNA electrophoretic mobility gel shift assay (RNA EMSA) with serine/arginine-rich splicing factor 1 (SRSF1), a splicing factor whose binding to SUGP1 is not predicted to be disrupted by rs10401969; and as expected, observed no differences in SRSF1 binding between alleles (Fig. 5F and Supplementary Material, Fig. S5G).
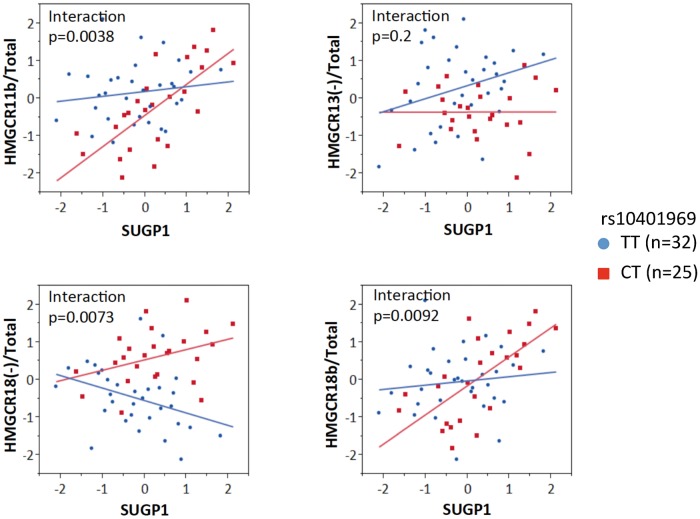
Inter-individual variation in SUGP1 transcript levels is correlated with variation in HMGCR alternative splicing in an rs10401969 allele-dependent fashion
To determine whether endogenous variation in SUGP1 expression was associated with variation in HMGCR alternative splicing, we quantified SUGP1 and HMGCR splice variants in 57 LCLs. We found evidence of a positive correlation between SUGP1 transcript levels with variation in HMGCR11b/Total HMGCR (P = 0.0045, r2 = 0.14) and HMGCR18b/Total HMGCR ratios (P = 0.0018, r2 = 0.16), whereas there was no association with either HMGCR13(−)/Total HMGCR or HMGCR18(−)/Total HMGCR ratios. Interestingly, we found that this relationship was modified by rs10401969. As shown in Figure 6, correlations between SUGP1 and HMGCR11b/Total (P = 6.3e − 5, r2 = 0.51) or HMGCR18b/Total (P = 0.0002, r2 = 0.46) were observed in rs10401969 minor allele carriers only (n = 25), with statistically significant interaction terms.
Figure 6.
SUGP1 and HMGCR alternative splicing are correlated in an rs10401969-dependent manner. SUGP1 and HMGCR transcripts, H11b, H18b, H18(+), H13(−) and total HMGCR, were quantified in LCLs by quantitative PCR, and linear regression was used to identify the relationship between quantile normalized SUGP1 transcript levels and HMGCR alternative splicing. All statistical analyses were performed in JMP 9.0 using a general linear model.
Discussion
Here, we sought to determine whether SUGP1 contributes to the relationship between rs10401969 and plasma lipids through regulation of alternative splicing. Through cellular and animal studies, we found that SUGP1 is a novel regulator of cholesterol metabolism that post-transcriptionally modifies HMGCR, impacting cholesterol synthesis. In addition, we show that rs10401969 is a functional variant, regulating SUGP1 alternative splicing. Together, these results implicate SUGP1 as a contributor to inter-individual variation in plasma cholesterol.
SUGP1 was first identified as a putative splicing factor based on domain composition analysis (30). The SUGP1 protein is predicted to contain two SURP motifs, domains of alternative splicing regulators thought to be involved in RNA binding (35), as well as a G-patch domain, a region of six highly conserved glycine residues commonly found in eukaryotic RNA-processing proteins (36). Although SUGP1 has been shown to co-purify with components of the spliceosome in multiple studies (37–39), prior to this report, no functional studies of SUGP1 have been published. Here, we confirm that manipulation of SUGP1 does lead to changes in the structure and stability of some transcripts, consistent with its annotation as an RNA regulatory protein.
Importantly, we found that one of SUGP1’s targets is HMGCR, a key cholesterol regulatory gene. SUGP1 knockdown increased expression levels of several alternatively spliced HMGCR transcripts, HMGCR11b, HMGCR18b and HMGCR18(−) by both promoting exon skipping and reducing transcript stability in a splice variant-dependent fashion. Although the open reading frame is retained in these splice variants, they omit a portion of the catalytic domain, and if translated we predict these variants to encode HMGCR isoforms with reduced or abolished enzyme activity. We found that in vitro SUGP1 knockdown reduced HMGCR enzyme activity, rate of cholesterol synthesis, and APOB in the culture media, as well as increased LDL uptake. Consistent with these effects, we observed elevated plasma cholesterol and hepatic Hmgcr expression, which could be attributed to the increased hepatic triglyceride secretion, elevated Hmgcr enzyme activity and reduced expression of Hmgcr splice variants after Sugp1 overexpression in vivo. Together, these data strongly support the likelihood that SUGP1 is a novel cholesterol regulatory gene.
We also demonstrate that rs10401969, a GWAS identified SNP associated with plasma lipids, CAD and hepatic steatosis (4,22,23,40,41), regulates SUGP1 alternative splicing. Located 80 bp upstream of the exon 8 splice donor, we found that the rs10401969 C allele promotes binding of HNRNPA1, a well known factor that stimulates alternative splicing, and induces skipping of SUGP1 exon 8. Loss of exon 8 disrupts the open reading frame, stimulating non-sense-mediated decay of the alternatively spliced transcript. SUGP1 knockdown both increases HMGCR alternative splicing and reduces overall HMGCR transcript stability, consistent with our observation that the correlation between inter-individual variation in SUGP1 expression quantified in the LCLs with variation in HMGCR alternative splicing differs by rs10401969 genotype. Together, these observations support the likelihood that rs10401969 itself is a functional variant that affects SUGP1 regulation of HMGCR. Because HMGCR encodes the rate-limiting step of the cholesterol biosynthesis pathway, knockdown also reduced cholesterol synthesis in vitro, whereas overexpression increased plasma cholesterol in vivo. These relationships are consistent with many GWAS studies showing an association between the rs10401969 C allele and reduced plasma cholesterol (Supplementary Material, Fig. S6).
These results suggest that SUGP1-induced changes in cholesterol metabolism are mediated by the regulation of HMGCR; however, RNA-binding proteins are not usually specific for a single target gene. For example, although it is tempting to conclude that the reduced cholesterol synthesis stimulates LDL uptake by promoting SREBF2 activation, there is no evidence to suggest that these are cause and effect. We failed to observe increased expression levels of SREBF2 target genes including MVK, LDLR and PCSK9 (data not shown) after SUGP1 knockdown in vitro, indicating a lack of SREBF2 activation. Although greater hepatic Hmgcr transcript levels (another SREBF2 target gene) were detected after Sugp1 overexpression in vivo, this up-regulation could be attributed to direct effects of Sugp1 on Hmgcr transcript stability. Further study is required to determine whether SUGP1 targets other key cholesterol regulators beyond HMGCR and contributes to the overall cholesterol phenotype observed.
As previously described, rs10401969 is in a large LD block containing many genetic variants in individuals of European ancestry, and one or more of these variants could have a functional impact. Additional fine mapping and conditional association analyses in ethnic populations with smaller haplotypic blocks in this region could help reveal the number of functional variants that contribute to the genetic association signals in individuals of European ancestry. In particular, rs10401969 is in strong LD with a functional variant in TM6SF2, p.Glu167Lys. A trio of recent studies utilizing human genetics and functional studies in cellular and animal models all implicate TM6SF2 as a causative functional gene within the Chr19.13p11 locus. Using an AAV model, Holzmen et al.(2014) found that overexpression of human TM6SF2 in C57BL/6J mice increased plasma TC, LDLC and triglycerides and reduced HDL cholesterol after 5 days. Consistent with these findings, Kozlitina et al.(23) reported that Tm6sf2 knockdown reduced plasma cholesterol and triglycerides after 8 weeks, and the effect was attributed to a defect in VLDL secretion. TM6SF2 knockdown was also shown to reduce secretion of triglyceride rich lipoproteins (24), whereas TM6SF2 overexpression was reported to increase cholesterol synthesis (42) in human hepatoma cell lines. Notably, these studies suggest that TM6SF2 is the only causative gene within the locus. Conditional analyses found that the TM6SF2 p.Glu167Lys variant remained significantly associated with plasma LDLC and triglycerides when controlled for other SNPs within the locus, whereas the signal of these other SNPs (including rs10401969) was abolished after controlling for the TM6SF2 variant (22,23). In addition, using a panel of >200 human livers, variation in the expression of TM6SF2, but not any of the other 18 genes tested within the locus, was correlated with both rs10401969 genotype and variation in hepatic triglyceride content (24).
Given these findings, it is challenging to interpret our results and the relevance of SUGP1 and rs10401969. Because the function of TM6SF2 has not been fully established, it is possible that the effect of TM6SF2 on lipid metabolism may be mediated (at least in part) by SUGP1 or vice versa. However, in preliminary studies, we have failed to identify any direct effect of TM6SF2 knockdown on SUGP1 expression or vice versa (data not shown). In addition, LCLs do not express TM6SF2. Thus, the relationships we observed between SUGP1 and HMGCR enzyme activity and alternative splicing using these cells cannot be attributed to a potential effect of either the TM6SF2 SNP or gene. In addition, although our results do not dispute the importance of the TM6SF2 gene or variant as a causative gene within the 19p13.11 locus, the consistency in our findings across cellular, animal and genetic analyses supports the conclusion that SUGP1 is a novel cholesterol regulator. Finally, although the conditional genetic analyses suggest that the effect of rs10401969 alone is not sufficient to impact plasma lipids in vivo, they do not discount the possibility that rs10401969 may act in an additive or synergistic manner with the TM6SF2 SNP, especially given the high LD of the two SNPs. The minor allele of each SNP reduces SUGP1 or TM6SF2 protein levels, and these two proteins have concordant effects on plasma cholesterol as overexpression of each gene alone increases plasma cholesterol. Thus, the relationships observed between genetic variation within this locus and plasma cholesterol may be due to the combined effect of SUGP1 and TM6SF2.
In summary, our results have shown SUGP1 to be a multi-functional RNA regulatory protein that modulates expression levels of several HMGCR splice variants, altering HMGCR enzyme activity and impacting both cellular and in vivo cholesterol metabolism. Furthermore, the finding that rs10401969, a GWAS-identified SNP associated with plasma LDLC and CAD, regulates SUGP1 implicates SUGP1 as a potential contributor to this genetic association. Overall, the present results reinforce the role of genetic regulation of transcript structure in pathways affecting cholesterol metabolism and cardiovascular disease risk.
Materials and Methods
Cell culture
HepG2, Hep3B and Huh7 cells were obtained from the American Type Culture Collection and grown under standard culture conditions. LDL was isolated as previously described (43). Cells were exposed in replicate to conditions of sterol depletion (10% lipoprotein deficient serum and 2 µm activated simvastatin), and after 24 hours, 50 µg/ml LDLC was added for an additional 24 hours. Immortalized LCLs (n = 54) were derived from donors of the CAP clinical trial and grown as previously described (44).
siRNA transfections and cellular measurements
SUGP1, ATP13A1 and MAU2 knockdown were achieved using the Ambion Silence Select siRNAs (SUGP1-1: s33721, SUGP1-3: s33722, ATP13A1: s32750, MAU2: s225955) or non-targeting control (NTC: AM4611) according to the manufacturer’s protocol. Forty-eight hours after knockdown, RNA was isolated. cDNA was synthesized and transcripts were quantified by quantitative PCR with assays shown in Supplementary Table S1. Values were normalized to CLPTM as described (44). For rescue experiments, cells were transfected in replicate with SUGP1-1 siRNA or NTC as described above. Forty-eight hours post transfection, cells SUGP1-1 siRNA treated cells were transfected with either the SUGP1 overexpression plasmid (pCMV-SUGP1) or an empty vector control (pCMV) and incubated for an additional 48 hours. APOB and APOE were quantified in triplicate by sandwich-style ELISA in culture media. HMGCR enzyme activity and DiI-LDL uptake were quantified as previously described (44,45). Cholesterol synthesis was measured by the conversion of [1-13C] sodium acetate to 13C-labeled cholesterol as described (46). Analysis of variance with post hoc two-tailed paired t-tests were used to determine statistically significant differences between treatments.
In vivo study
An Sugp1 overexpression construct was created by subcloning the murine Sugp1 gene (purchased from Origene) into the pLIVE-MCS vector (Mirus Bio). Sugp1-pLIVE was injected (25 μg) into the tail vein of 6-week-old CD-1 male mice (n = 10; Charles River Laboratories) using the TransIT-EE hydrodynamic gene delivery system (Mirus Bio), as previously described (47). In a second set of CD-1 male mice (n = 10), the empty vector was injected to serve as the negative control. Five mice for each construct were euthanized at 7 and 28 days after injection by cardiac puncture. Animal studies were approved by the Children’s Hospital Oakland Research Institute Animal Care and Use Committee and conform to all regulatory standards. Whole blood, liver and plasma were collected. Plasma cholesterol was quantified using a LIASYS 330 chemistry analyzer (AMS Diagnostics). Hepatic gene expression levels were quantified as described above, with values normalized to the geomean of Clptm1 and Rplp0. All RNAs were DNaseI treated during isolation. Hepatic Sugp1 protein levels were quantified by Western blot analysis.
RNA electrophoretic mobility gel shift assay (RNA EMSA)
3′ end-biotinylated RNA oligonucleotides were synthesized from Sigma-Aldrich. Purified His-tagged human recombinant HNRNPA1 and SRSF1 proteins were purchased from Prospec and ProteinOne. RNA gel shift assay was performed using LightShift Chemiluminescent RNA EMSA kit (Thermo Scientific). RNA oligos with either the SUGP1 rs10401969 ‘T’ or ‘C’ allele were incubated for 30 minutes in the presence or in the absence of either His-HNRNPA1 or His-SRSF1 protein in 1x binding buffer, 5% glycerol, 2 µg tRNA, and 6.25 nm biotin-labeled RNA oligos. 5x loading dye was added to each reaction, loaded onto a 4–20% Novex® TBE Gels (Life Technologies), and run at 100 V for 2 hours in 0.5x Novex® TBE Running Buffer (Invitrogen) at 4 °C. The RNA–protein complexes were transferred from the gel to a nylon membrane (Thermo Scientific) at 35V for 40 minutes in a 4 °C cold room, and crosslinked using the Stratalinker UV crosslinker (Stratagene). Biotin-labeled RNA oligos were detected by the Chemiluminescent Nucleic Acid detection Module (Thermo Scientific), and quantified using the GelQuant.NET software.
SUGP1 and HMGCR mini-gene construct
To create the SUGP1 and HMGCR mini-genes, SUGP1 introns 7–8 and HMGCR introns 16–19 were cloned into pDEST exon-trap vector provided by Dr. Stephan Stamm. The plasmids were DNA sequence verified. The SUGP1 mini-gene was co-transfected into HepG2 and Huh7 cells with the HNRNPA1 overexpression plasmid. The HMGCR mini-gene was co-transfected one of the two SUGP1 targeting siRNA or NTC.
Identification of novel HMGCR splice variants
Whole transcriptome sequencing (RNA-seq) was performed on LCLs from eight African American individuals (seven female and one male) as previously described (48). Libraries were sequenced on an Illumina GAII machine and aligned to hg19 using TopHat v1.2.0 and Bowtie v0.12.7 (49,50). Splice junctions detected by TopHat were compared to Ensembl v61 annotations to identify potentially novel junctions in HMGCR. From this analysis, two novel splice variants were identified with truncated versions of exons 11 and 18 using both direct evidence of junction spanning reads, and indirect evidence based the distance of paired end reads. Novel junctions were validated using RT-PCR with Sanger sequencing. The effect of SUGP1 on the stability of these HMGCR transcripts was assessed in HepG2 and Huh7 cells incubated with actinomycin D, with transcript half-life calculated as previously described (48).
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
The CAP LCLs used in these studies were provided by Sheila Pressman and Dr. Jerome I. Rotter. Human liver RNA-seq data was generated through the PGRN Network-wide RNA-Seq Project. We thank Dr. Ronald M. Krauss for his scientific discussion and critical review of this manuscript.
Conflict of Interest statement. None declared.
Funding
This work was supported by the National Institutes of Health [R01 HL104133], [U19 HL069757], [P50 GM115318], and the American Heart Association [12POST10430005]. Funding to pay the Open Access publication charges for this article was provided by NIH R01 HL104133.
References
- 1.Global Lipids Genetics Consortium, Willer C.J., Schmidt E.M., Sengupta S., Peloso G.M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M.L., More S., et al. (2013) Discovery and refinement of loci associated with lipid levels. Nat. Genet., 45, 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhuang K., Zhang W., Zhang X., Wu F., Cheng L. (2011) Effects of SNPs at newly identified lipids loci on blood lipid levels and risk of coronary heart disease in Chinese Han population: a case control study. J. Huazhong Univ. Sci. Technol., 31, 452–456. [DOI] [PubMed] [Google Scholar]
- 3.Zhou L., Ding H., Zhang X., He M., Huang S., Xu Y., Shi Y., Cui G., Cheng L., Wang Q.K., et al. (2011) Genetic variants at newly identified lipid loci are associated with coronary heart disease in a Chinese Han population. PLoS One, 6, e27481.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N.P., Rieder M.J., Cooper G.M., Roos C., Voight B.F., Havulinna A.S., et al. (2008) Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet., 40, 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakayama K., Bayasgalan T., Yamanaka K., Kumada M., Gotoh T., Utsumi N., Yanagisawa Y., Okayama M., Kajii E., Ishibashi S., et al. (2009) Large scale replication analysis of loci associated with lipid concentrations in a Japanese population. J. Med. Genet., 46, 370–374. [DOI] [PubMed] [Google Scholar]
- 6.Yan T.T., Yin R.X., Li Q., Huang P., Zeng X.N., Huang K.K., Aung L.H., Wu D.F., Liu C.W., Pan S.L. (2011) Sex-specific association of rs16996148 SNP in the NCAN/CILP2/PBX4 and serum lipid levels in the Mulao and Han populations. Lipids Health Dis., 10, 248.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adeyemo A., Bentley A.R., Meilleur K.G., Doumatey A.P., Chen G., Zhou J., Shriner D., Huang H., Herbert A., Gerry N.P., et al. (2012) Transferability and Fine Mapping of genome-wide associated loci for lipids in African Americans. BMC Med. Genet., 13, 88.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comuzzie A.G., Cole S.A., Laston S.L., Voruganti V.S., Haack K., Gibbs R.A., Butte N.F. (2012) Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One, 7, e51954.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weissglas-Volkov D., Aguilar-Salinas C.A., Nikkola E., Deere K.A., Cruz-Bautista I., Arellano-Campos O., Munoz-Hernandez L.L., Gomez-Munguia L., Ordonez-Sanchez M.L., Reddy P.M., et al. (2013) Genomic study in Mexicans identifies a new locus for triglycerides and refines European lipid loci. J. Med. Genet., 50, 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor K.C., Carty C.L., Dumitrescu L., Buzkova P., Cole S.A., Hindorff L., Schumacher F.R., Wilkens L.R., Shohet R.V., Quibrera P.M., et al. (2013) Investigation of gene-by-sex interactions for lipid traits in diverse populations from the population architecture using genomics and epidemiology study. BMC Genet., 14, 33.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segre A.V., Steinthorsdottir V., Strawbridge R.J., Khan H., Grallert H., Mahajan A., et al. (2012) Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet., 44, 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Talmud P.J., Drenos F., Shah S., Shah T., Palmen J., Verzilli C., Gaunt T.R., Pallas J., Lovering R., Li K., et al. (2009) Gene-centric association signals for lipids and apolipoproteins identified via the HumanCVD BeadChip. Am. J. Hum. Genet., 85, 628–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kathiresan S., Willer C.J., Peloso G.M., Demissie S., Musunuru K., Schadt E.E., Kaplan L., Bennett D., Li Y., Tanaka T., et al. (2009) Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet., 41, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aulchenko Y.S., Ripatti S., Lindqvist I., Boomsma D., Heid I.M., Pramstaller P.P., Penninx B.W., Janssens A.C., Wilson J.F., Spector T., et al. (2009) Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet., 41, 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harder M.N., Ribel-Madsen R., Justesen J.M., Sparso T., Andersson E.A., Grarup N., Jorgensen T., Linneberg A., Hansen T., Pedersen O. (2013) Type 2 diabetes risk alleles near BCAR1 and in ANK1 associate with decreased beta-cell function whereas risk alleles near ANKRD55 and GRB14 associate with decreased insulin sensitivity in the Danish Inter99 cohort. J. Clin. Endocrinol. Metab., 98, E801–E806. [DOI] [PubMed] [Google Scholar]
- 16.Saxena R., Elbers C.C., Guo Y., Peter I., Gaunt T.R., Mega J.L., Lanktree M.B., Tare A., Castillo B.A., Li Y.R., et al. (2012) Large-scale gene-centric meta-analysis across 39 studies identifies type 2 diabetes loci. Am. J. Hum. Genet., 90, 410–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hernaez R., McLean J., Lazo M., Brancati F.L., Hirschhorn J.N., Borecki I.B., Harris T.B., Genetics of Obesity-Related Liver Disease C., Nguyen T., Kamel I.R., et al. (2013) Association between variants in or near PNPLA3, GCKR, and PPP1R3B with ultrasound-defined steatosis based on data from the Third National Health and Nutrition Examination Survey. Clin. Gastroenterol. Hepatol., 11, 1183–1190. e1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorden A., Yang R., Yerges-Armstrong L.M., Ryan K.A., Speliotes E., Borecki I.B., Harris T.B., Chu X., Wood G.C., Still C.D., et al. (2013) Genetic variation at NCAN locus is associated with inflammation and fibrosis in non-alcoholic fatty liver disease in morbid obesity. Hum. Heredity, 75, 34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmer N.D., Musani S.K., Yerges-Armstrong L.M., Feitosa M.F., Bielak L.F., Hernaez R., Kahali B., Carr J.J., Harris T.B., Jhun M.A., et al. (2013) Characterization of european ancestry nonalcoholic fatty liver disease-associated variants in individuals of african and hispanic descent. Hepatology, 58, 966–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Speliotes E.K., Yerges-Armstrong L.M., Wu J., Hernaez R., Kim L.J., Palmer C.D., Gudnason V., Eiriksdottir G., Garcia M.E., Launer L.J., et al. (2011) Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet., 7, e1001324.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westra H.J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E., et al. (2013) Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet., 45, 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmen O.L., Zhang H., Fan Y., Hovelson D.H., Schmidt E.M., Zhou W., Guo Y., Zhang J., Langhammer A., Lochen M.L., et al. (2014) Systematic evaluation of coding variation identifies a candidate causal variant in TM6SF2 influencing total cholesterol and myocardial infarction risk. Nat. Genet., 46, 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozlitina J., Smagris E., Stender S., Nordestgaard B.G., Zhou H.H., Tybjaerg-Hansen A., Vogt T.F., Hobbs H.H., Cohen J.C. (2014) Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet., 46, 352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahdessian H., Taxiarchis A., Popov S., Silveira A., Franco-Cereceda A., Hamsten A., Eriksson P., van't Hooft F. (2014) TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc. Natl Acad. Sci. U. S. A., 111, 8913–8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K., Dickson S.P., Stolle C.A., Krantz I.D., Goldstein D.B., Hakonarson H. (2010) Interpretation of association signals and identification of causal variants from genome-wide association studies. Am. J. Hum. Genet., 86, 730–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blattmann P., Schuberth C., Pepperkok R., Runz H. (2013) RNAi-based functional profiling of loci from blood lipid genome-wide association studies identifies genes with cholesterol-regulatory function. PLoS Genet., 9, e1003338.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Medina M.W., Gao F., Naidoo D., Rudel L.L., Temel R.E., McDaniel A.L., Marshall S.M., Krauss R.M. (2011) Coordinately regulated alternative splicing of genes involved in cholesterol biosynthesis and uptake. PLoS One, 6, e19420.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tveten K., Ranheim T., Berge K.E., Leren T.P., Kulseth M.A. (2006) Analysis of alternatively spliced isoforms of human LDL receptor mRNA. Clin. Chim. Acta, 373, 151–157. [DOI] [PubMed] [Google Scholar]
- 29.Burkhardt R., Kenny E.E., Lowe J.K., Birkeland A., Josowitz R., Noel M., Salit J., Maller J.B., Pe'er I., Daly M.J., et al. (2008) Common SNPs in HMGCR in micronesians and whites associated with LDL-cholesterol levels affect alternative splicing of exon13. Arterioscler. Thromb. Vasc. Biol., 28, 2078–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sampson N.D., Hewitt J.E. (2003) SF4 and SFRS14, two related putative splicing factors on human chromosome 19p13.11. Gene, 305, 91–100. [DOI] [PubMed] [Google Scholar]
- 31.Zeller T., Wild P., Szymczak S., Rotival M., Schillert A., Castagne R., Maouche S., Germain M., Lackner K., Rossmann H., et al. (2010) Genetics and beyond–the transcriptome of human monocytes and disease susceptibility. PLoS One, 5, e10693.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu C.Y., Theusch E., Lo K., Mangravite L.M., Naidoo D., Kutilova M., Medina M.W. (2014) HNRNPA1 regulates HMGCR alternative splicing and modulates cellular cholesterol metabolism. Hum. Mol. Genet., 23, 319–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Innocenti F., Cooper G.M., Stanaway I.B., Gamazon E.R., Smith J.D., Mirkov S., Ramirez J., Liu W., Lin Y.S., Moloney C., et al. (2011) Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet., 7, e1002078.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desmet F.O., Hamroun D., Lalande M., Collod-Beroud G., Claustres M., Beroud C. (2009) Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res., 37, e67.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spikes D.A., Kramer J., Bingham P.M., Van Doren K. (1994) SWAP pre-mRNA splicing regulators are a novel, ancient protein family sharing a highly conserved sequence motif with the prp21 family of constitutive splicing proteins. Nucleic Acids Res., 22, 4510–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aravind L., Koonin E.V. (1999) G-patch: a new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci., 24, 342–344. [DOI] [PubMed] [Google Scholar]
- 37.Hegele A., Kamburov A., Grossmann A., Sourlis C., Wowro S., Weimann M., Will C.L., Pena V., Luhrmann R., Stelzl U. (2012) Dynamic protein-protein interaction wiring of the human spliceosome. Mol. Cell, 45, 567–580. [DOI] [PubMed] [Google Scholar]
- 38.Behzadnia N., Golas M.M., Hartmuth K., Sander B., Kastner B., Deckert J., Dube P., Will C.L., Urlaub H., Stark H., et al. (2007) Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J., 26, 1737–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agafonov D.E., Deckert J., Wolf E., Odenwalder P., Bessonov S., Will C.L., Urlaub H., Luhrmann R. (2011) Semiquantitative proteomic analysis of the human spliceosome via a novel two-dimensional gel electrophoresis method. Mol .Cell. Biol., 31, 2667–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waterworth D.M., Ricketts S.L., Song K., Chen L., Zhao J.H., Ripatti S., Aulchenko Y.S., Zhang W., Yuan X., Lim N., et al. (2010) Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler. Thromb. Vasc. Biol., 30, 2264–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J., et al. (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature, 466, 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fan Y., Lu H., Guo Y., Zhu T., Garcia-Barrio M.T., Jiang Z., Willer C.J., Zhang J., Chen Y.E. (2016) Hepatic transmembrane 6 superfamily member 2 regulates cholesterol metabolism in mice. Gastroenterology, 150, 1208–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen M.M., Krauss R.M., Lindgren F.T., Forte T.M. (1981) Heterogeneity of serum low density lipoproteins in normal human subjects. J. Lipid Res., 22, 236–244. [PubMed] [Google Scholar]
- 44.Medina M.W., Gao F., Ruan W., Rotter J.I., Krauss R.M. (2008) Alternative splicing of 3-hydroxy-3-methylglutaryl coenzyme A reductase is associated with plasma low-density lipoprotein cholesterol response to simvastatin. Circulation, 118, 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Medina M.W., Bauzon F., Naidoo D., Theusch E., Stevens K., Schilde J., Schubert C., Mangravite L.M., Rudel L.L., Temel R.E., et al. (2014) Transmembrane protein 55B is a novel regulator of cellular cholesterol metabolism. Arterioscler. Thromb. Vasc. Biol., 34, 1917–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Serra M., Matabosch X., Ying L., Watson G., Shackleton C. (2010) Hair and skin sterols in normal mice and those with deficient dehydrosterol reductase (DHCR7), the enzyme associated with Smith-Lemli-Opitz syndrome. J. Steroid Biochem. Mol. Biol., 122, 318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim M.J., Skewes-Cox P., Fukushima H., Hesselson S., Yee S.W., Ramsey L.B., Nguyen L., Eshragh J.L., Castro R.A., Wen C.C., et al. (2011) Functional characterization of liver enhancers that regulate drug-associated transporters. Clin. Pharmacol. Therap., 89, 571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Medina M.W., Theusch E., Naidoo D., Bauzon F., Stevens K., Mangravite L.M., Kuang Y.L., Krauss R.M. (2012) RHOA is a modulator of the cholesterol-lowering effects of statin. PLoS Genet., 8, e1003058.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trapnell C., Pachter L., Salzberg S.L. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics, 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol., 10, R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.