Abstract
Long intergenic noncoding RNAs (lincRNAs) play important roles in disease, but the vast majority of these transcripts remain uncharacterized. We defined a set of 54 944 human lincRNAs by drawing on four publicly available lincRNA datasets, and annotated ∼2.5 million single nucleotide polymorphisms (SNPs) from each of 15 cardiometabolic genome-wide association study datasets into these lincRNAs. We identified hundreds of lincRNAs with at least one trait-associated SNP: 898 SNPs in 343 unique lincRNAs at 5% false discovery rate, and 469 SNPs in 146 unique lincRNAs meeting Bonferroni-corrected P < 0.05. An additional 64 trait-associated lincRNAs were identified using a class-level testing strategy at Bonferroni-corrected P < 0.05. To better understand the genomic context and prioritize trait-associated lincRNAs, we examined the pattern of linkage disequilibrium between SNPs in the lincRNAs and SNPs that met genome-wide-significance in the region (±500 kb of lincRNAs). A subset of the lincRNA-trait association findings was replicated in independent Genome-wide association studies data from the Pakistan Risk of Myocardial Infarction Study study. For trait-associated lincRNAs, we also investigated synteny and conservation relative to mouse, expression patterns in five cardiometabolic-relevant tissues, and allele-specific expression in RNA sequencing data for adipose tissue and leukocytes. Finally, we revealed a functional role in human adipocytes for linc-NFE2L3-1, which is expressed in adipose and is associated with waist-hip ratio adjusted for BMI. This comprehensive profile of trait-associated lincRNAs provides novel insights into disease mechanism and serves as a launching point for interrogation of the biology of specific lincRNAs in cardiometabolic disease.
Introduction
Genome-wide association studies (GWAS) have identified many candidate loci for complex cardiometabolic traits, yet a large portion of the heritability of these traits has yet to be discovered (1,2). A substantial proportion of these GWAS findings fall in regulatory or intergenic regions, and genome-wide regulatory mapping (e.g. ENCODE) is annotating a remarkable extent of tissue and cell-specific regulatory domains in regions that were previously considered silent (3–5). Many novel non-coding transcripts are also being annotated and described. It is thus likely that causal variations in promoters, enhancers and non-coding RNAs account for a substantial proportion of the regulatory and intergenic GWAS signals for complex traits.
Recently, there has been an explosion of interest in long non-coding RNAs (lncRNAs) and their roles in homeostasis and disease. LncRNAs are transcripts at least 200 base pairs in length that do not code for proteins. LncRNAs are typically expressed at lower levels than protein-coding genes, and are more tissue-specific (6). Although some annotated lncRNAs may be transcriptional noise (6,7), there are already many examples of lncRNAs involved in important biological processes through a variety of mechanisms. XIST, which is required for X-chromosome inactivation, uses the three-dimensional chromatin structure to identify initial binding sites, from which it spreads to coat and inactivate the X chromosome (8). Jiang et al. recently demonstrated that insertion of XIST into an extra copy of chromosome 21 in Down’s syndrome pluripotent stem cells leads to silencing of the extra chromosome (9). HOTAIR (10) and HOTTIP (11) act as scaffolds, recruiting chromatin modifying complexes to alter expression of target genes. NEAT1 (12) binds proteins to form and maintain nuclear paraspeckles (13), and represses transcription of several genes (12). FIRRE, located on the X chromosome, interacts with hnRNPU to act in cis and trans in modulating nuclear architecture across chromosomes (14). Sauvageau et al. (15) recently demonstrated that knocking out specific lncRNAs in mice can lead to severe developmental defects and death.
In spite of the clear importance of specific lncRNAs, there are surprisingly few examples of GWAS follow-up focused on individual lncRNAs. This is partially due to a deliberate focus on protein-coding genes in the initial translation of GWAS results. To date, there have been few efforts to interrogate lncRNAs as a group. LincPoly is a database of 712 394 single nucleotide polymorphisms (SNPs) in 4662 human lncRNAs, including 1408 trait-associated SNPs (16). LincSNP is a database of 128 407 SNP-phenotype associations annotated to 5804 lncRNAs (17). Atlhough useful, these databases are limited, because they do not include complete GWAS SNP data, and focus on only a few thousand lncRNAs. To date, no analysis has been performed on a comprehensive set of lncRNAs in complete cardiometabolic GWAS datasets.
Here we address the potential role of lncRNAs in the genetics of several cardiometabolic phenotypes, including coronary artery disease (CAD), plasma lipoproteins, type 2 diabetes, glucometabolic traits, and obesity. Since intergenic lncRNAs (lincRNAs) make up the majority of lncRNAs and it is difficult to study SNPs residing in lncRNAs that overlap protein-coding genes, we focused our analyses exclusively on lincRNAs. Specifically, we (i) define a comprehensive genome-wide set of 54 944 lincRNAs using publically available resources, (ii) investigate lincRNA enrichment and association with fifteen GWAS studies for cardiometabolic traits using SNP and class-based approaches, (iii) use regional trait associations and linkage disequilibrium (LD) patterns to prioritize trait-associated lincRNAs, (iv) assess replication patterns for a subset of traits in independent GWAS datasets, (v) examine synteny and conservation for trait-associated lincRNAs, (vi) probe lincRNA tissue expression patterns and allelic specific expression (ASE) and (vii) perform translational and functional studies for an illustrative adipocyte-expressed lincRNA.
Results
Characteristics of the combined lincRNA dataset
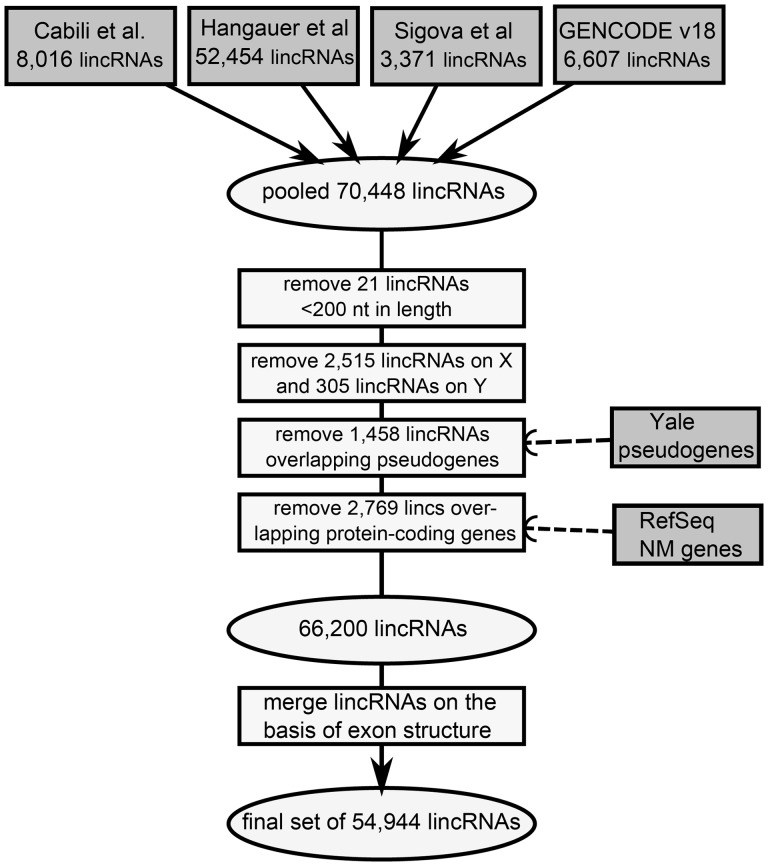
Human lincRNA annotations were drawn from four published catalogs based on human RNA-seq data, version 18 of Gencode, Hangauer et al. (18), Cabili et al. (19) and Sigova et al. (20) (details in Materials and Methods section). The pipeline to combine lincRNAs from all four datasets is presented in Figure 1.
Figure 1.
Filtering and merging pipeline to create a comprehensive human lincRNA dataset. After pooling the four original datasets, lincRNAs were filtered based on length and overlap with pseudogenes and protein-coding genes. After filtering, they were merged on the basis of exon structure, to produce a final comprehensive set of 54 944 lincRNAs that was used in all subsequent analyses.
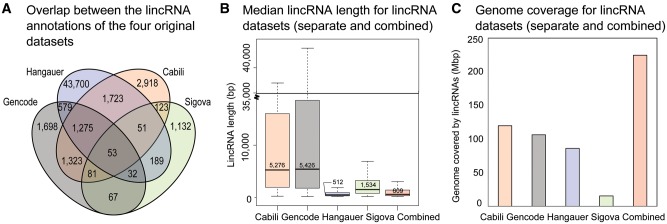
Only 10% of lincRNAs in our final ‘combined dataset’ originated from more than one RNA-seq catalog, indicating the distinctiveness of each original dataset (Fig. 2A). As might be expected based on their selection criteria (e.g. multi-exon lincRNAs required in Cabili et al.) and sequencing strategies, the Cabili and GENCODE datasets contained most of the very long lincRNAs (Fig. 2B). The value of drawing on multiple human lincRNA annotation databases is apparent also in the genome coverage of the separate versus combined datasets (Fig. 2C). For example, our final combined dataset contains fewer individual lincRNAs than the original Hangauer dataset, yet it covers 2.6-times more bases of the genome. This is because of the merging process; often, many small lincRNAs (e.g. from the Hangauer dataset) were combined with one larger lincRNA (e.g. from the Cabili dataset) perhaps consistent with some smaller annotated lincRNAs belonging to a larger lincRNAs defined by deeper RNA-seq. Although a portion of single-exon lincRNAs that are annotated in the source RNA-seq datasets may be spurious non-functional sequences (7), we did not exclude single-exon lincRNAs because there are several examples of such lincRNAs having important biological actions e.g. NEAT1, MALAT1 and PAUPAR (13,22–26). Rather, we highlight single-exon versus multi-exon lincRNA in results so that investigators can use this information in their prioritizations.
Figure 2.
Characteristics of the original and combined lincRNA datasets. Various features of the original lincRNA datasets and the final combined dataset are highlighted. (A) Overlap between lincRNA annotations: Each lincRNA in the final combined dataset is counted once in this diagram, depending on which combination of datasets it originated from. The majority of lincRNAs in the combined dataset were lifted directly from one of the original datasets unchanged, but 10% of the lincRNAs in the combined dataset did result from merging of overlapping annotations between datasets. The most overlap occurred between Cabili, Gencode, and Hangauer datasets, typically with smaller Hangauer lincRNAs being engulfed by larger Cabili or Gencode lincRNAs. Fifty-three lincRNAs were found in some form in all four datasets. (B) Median lincRNA length: Each of the individual datasets was processed by the filtering pipeline before collecting these summary statistics, in order to make a consistent comparison with the combined dataset. LincRNA length includes introns. The Gencode and Cabili datasets contained the greatest number of very long lincRNAs. Many of these very long lincRNAs are still present in the Combined dataset, but the interquartile range of the combined dataset appears much smaller due to the high number of small lincRNAs from the Hangauer dataset that are also present. Outliers are not shown. (C) The coverage calculation includes introns and exons of lincRNAs. The entire human genome is about 3137 Mb, meaning that the combined dataset covers 7.16% of the genome
Hundreds of lincRNAs are associated with cardiometabolic traits
A summary of GWAS datasets examined in discovery and replication analyses is presented in Table 1 (details in Materials and Methods section and in Supplementary Materials).
Table 1.
Description of the 20 GWAS datasets
| GWAS or consortium name | Trait | GWAS size (no. of subjects) | Total no. of SNPs |
|---|---|---|---|
| Discovery Studies | |||
| CARDIoGRAM Consortium (Schunkert et al.66) | CAD | 22 233 cases 64 762 controls | 2 407 380 |
| MI | 2 361 556 | ||
| MI in setting of Angiographic CAD (Reilly et al.67) | MI in subjects with CAD | 5783 cases 3644 controls | 2 503 078 |
| GLGC: Global Lipids Genetics Consortium (Teslovich et al.29) | Total cholesterol | 100 184 | 2 606 897 |
| LDL-C | 95 454 | 2 607 046 | |
| Triglycerides (TGs) | 96 598 | 2 607 042 | |
| HDL-C | 99 900 | 2 606 916 | |
| DIAGRAM: Diabetes Genetics Replication and Meta-analysis (Morris et al.65) | Type 2 diabetes (T2DM) | 12 171 cases 56 862 controls | 2 451 781 |
| MAGIC: Meta-Analysis of Glucose and Insulin-related Traits Consortium (Dupuis et al.64) | Fasting glucose | 46 186 | 2 457 436 |
| Glycated hemoglobin | 46 368 | 2 543 467 | |
| Fasting insulin | 38 238 | 2 448 111 | |
| HOMA-IR, a measure of insulin resistance | 37 037 | 2 445 117 | |
| HOMA-B, a measure of beta-cell function | 36 466 | 2 444 006 | |
| GIANT: Genetic Investigation of Anthropometric Traits Consortium (Speliotes et al.24, Heid et al.27) | Body mass index (BMI) | 123 865 | 2 458 315 |
| Waist-hip ratio adjusted for BMI | 77 167 | 2,469,665 | |
| Replication Studies | |||
| PROMIS (Saleheen et al.68) | HDL-C | 16 674 | 2 530 948 |
| LDL-C | 16 995 | 2 530,816 | |
| Total cholesterol | 17 212 | 2 530 769 | |
| Triglycerides (TG) | 17 134 | 2 531 043 | |
| MI | 9029 cases | 2 531 273 | |
| 8379 controls | |||
Details about the fifteen discovery GWAS datasets and five replication datasets used to determine lincRNA-trait associations. The study name and relevant trait(s) are listed. For case-control studies, the numbers of cases and controls are indicated separately. The total number of SNPs refers to the total number of SNPs remaining in each dataset after our filtering steps.
In overview, detailed findings of all SNP and lincRNA class association analyses are presented in Supplementary Material, Table S1 (for every trait-associated lincRNA at 5% false discovery rate (FDR) SNP level and also for GenCAT analyses). Top prioritized findings are discussed below. Supplementary Material, Table S2 summarizes lincRNAs that did not contain the strongest signal in their region but did contain one or more Bonferroni-significant SNPs, and Supplementary Material, Table S3 presents lincRNAs that only had SNP trait associations at the 5% FDR SNP level. Enrichment analysis is shown in Supplementary Material, Table S4.
SNP-based lincRNA trait associations
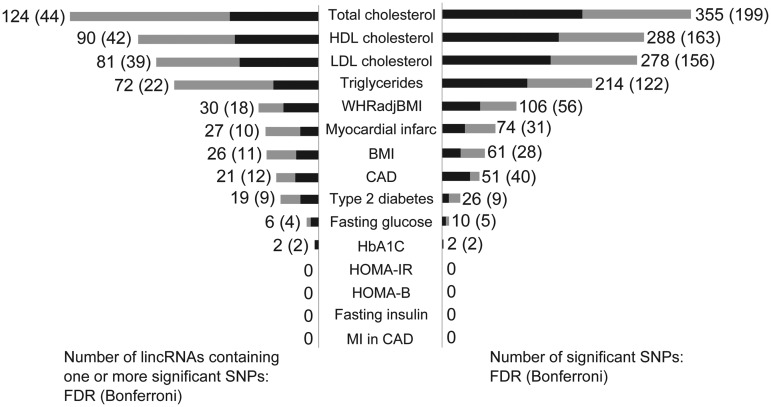
Of the ∼226 000 SNPs annotated to lincRNAs for each trait, 898 SNPs in 343 unique lincRNAs were significant at 5% FDR level (total of 498 individual lincRNA-trait associations), and 469 SNPs in 146 unique lincRNAs reached the more stringent Bonferroni-corrected P < 0.05 threshold (uncorrected P ∼2.2 × 10−7) (total of 232 individual lincRNA-trait associations). Lipid trait datasets had the greatest number of significant SNPs at both FDR and Bonferroni thresholds (Fig. 3). The number of lincRNA SNP associations with individual traits is likely to reflect larger sample sizes for some GWAS traits (e.g. total cholesterol and BMI) but also a stronger genetic underpinning for specific traits (e.g. waist-hip ratio adjusted for BMI vs. BMI despite larger sample size for BMI, n = 123 865 versus 77 167) (27–29). Of the 343 unique lincRNAs with at least one significant SNP at the 5% FDR SNP level, 122 were associated with more than one trait and the majority of multiple associations occurred with lipid traits (Supplementary Material, Figure S1). In resampling-based enrichment analyses, lincRNAs were enriched for SNPs with low P-values relative to other intergenic regions for multiple traits including CAD, myocardial infarction (MI), waist-hip ratio adjusted for BMI, total cholesterol, low-density lipoprotein cholesterol (LDL-C) and high-density lipoprotein cholesterol (HDL-C) (Supplementary Material, Table S4).
Figure 3.
Number of significant SNPs and the lincRNAs containing at least one significant SNP for 14 GWAS datasets. The bars on the right indicate the number of lincRNA-annotated significant SNPs for the trait, while the bars on the left indicate the number of lincRNAs containing at least one of the significant SNPs. The grey bars and the first numbers indicate results of the FDR significance threshold, while the smaller black bars and parenthetical numbers indicate results of the more stringent Bonferroni significance threshold. Abbreviations: WHRadjBMI, waist-hip ratio adjusted for BMI; BMI, body mass index; CAD, coronary artery disease; HbA1C, glycated hemoglobin; HOMA-IR, insulin resistance; HOMA-B, pancreatic β cell function; MI in CAD, myocardial infarction in coronary artery disease.
Class-level lincRNA associations
GenCAT revealed an additional 79 lincRNA-trait associations (64 unique lincRNAs) for which the lincRNA did not contain any individual SNP that reach Bonferroni level of association. In total, GenCAT identified 275 individual significant lincRNA associations for 180 unique lincRNAs, ranging from two lincRNAs for insulin resistance to 61 lincRNAs for total cholesterol (Supplementary Material, Table S1).
Prioritization of trait-associated lincRNAs
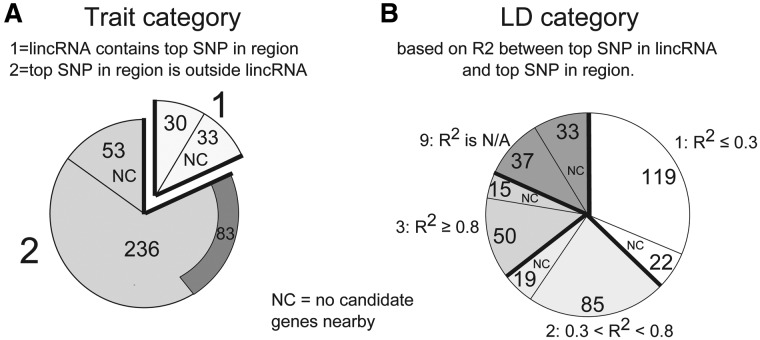
Of the 343 significant lincRNAs based on FDR-corrected SNP analysis, 63 contained the strongest trait-associated SNP in their region (Trait category 1) (Fig. 4A) and 14 of these contained one or more SNPs meeting Bonferroni significance (Table 2). Of these 14 lincRNAs, three are at chr. 9p21 and, as published (30–34), overlap intergenic regions that show associations with CAD and type 2 diabetes. One lincRNA, FPKM1_GROUP_7090_TRANSCRIPT_1, had no candidate genes nearby and was associated with fasting glucose levels. The 10 remaining lincRNAs did have candidate genes nearby, although some of these candidates had no proven functional relationship to the trait (e.g. see linc-NFE2L3-1, below). For the 280 lincRNAs with SNPs meeting the 5% FDR threshold but not containing the strongest regional SNP signal for the trait (Fig. 4B), a large proportion (141 lincRNAs) had low LD (R2 < 0.3) with the strongest trait-associated SNP in the region (±500 kb) raising the possibility of independent regulatory effects for these lincRNAs at these regions.
Figure 4.
Distribution of lincRNAs across trait categories and LD categories. Counts of lincRNAs in different Trait and LD Categories are shown. The proportion of lincRNAs within each category that had no candidates in the region is indicated by ‘NC’. (A) Trait category. Sixty-three lincRNAs were assigned trait category 1, indicating that they contained the strongest SNP in their 1 MB region. Of the 236 lincRNAs that did not contain the strongest SNP in the region and did have candidate genes nearby, 83 still had a stronger SNP than their neighboring candidate genes, indicated by the dark arc. (B) LD category is based on the LD between the top SNP in the lincRNA and the top SNP in the region. The 63 lincRNAs that already contained the top SNP in the region were placed in LD category 9. The remaining lincRNAs were placed in either category 1, 2 or 3 depending on the R2 value, with about half showing low LD (R2 ≤ 0.3) with the top SNP in the region, suggesting that the lincRNA may contain an independent signal.
Table 2.
LincRNAs that contain the strongest signal in the region and contain one or more Bonferroni-significant SNPs
| LincRNA | LincRNA location | no. Exons | GWAS trait | Top lincRNA SNP, P-value | no. Bonf. significant SNPs | Candidate gene information | ||
|---|---|---|---|---|---|---|---|---|
| Name | Top SNP, P-value | LD | ||||||
| GS1-122H1.2 | chr1:182159255-182283196 | 1 | HDL-C | rs1689800, 3.18 × 10−10 | 12 | ZNF648 | rs41326547, 4.190 × 10−2 | NA |
| linc-GRB14 | chr2:165509841-165522523 | 2 | Waist-hip ratio adjusted for BMI | rs10195252, 3.23 × 10−10 | 2 | GRB14 | rs2892923, 7.800 × 10−3 | 0.004 |
| Triglycerides | rs10195252, 1.63 × 10−10 | 2 | – | – | – | |||
| linc-DTNBP1-4 | chr6:16126537-16129130 | 2 | LDL-C | rs3757354, 1.16 × 10−11 | 2 | MYLIP | rs9370867, 5.876 × 10−6 | 0.191 |
| Total cholesterol | rs3757354, 2.78 × 10−9 | 2 | MYLIP | rs9370867, 1.555 × 10−4 | 0.191 | |||
| RP1-80N2.2 | chr6:6692977-6745648 | 4 | Waist-hip ratio adjusted for BMI | rs1294421, 6.31 × 10−9 | 4 | LY86 | rs3804462, 4.400 × 10−3 | 0 |
| linc-NPVF-3 | chr7:25870838-25872690 | 3 | Waist-hip ratio adjusted for BMI | rs1055144, 1.49 × 10−8 | 1 | NFE2L3 | rs2040786, 1.700 × 10−1 | 0.0018 |
| RP11-136O12.2 | chr8:126479045-126553615 | 1 | HDL-C | rs10808546b, 6.34 × 10−19 | 30 | TRIB1 | rs2385114, 1.1610 × 10−5 | 0.4845 |
| LDL-C | rs2954022b, 2.59 × 10−29 | 46 | TRIB1 | rs2385114, 1.956 × 10−12 | 0.412 | |||
| Total cholesterol | rs2954022a, 5.02 × 10−36 | 46 | TRIB1 | rs2385114, 2.6770 × 10−15 | 0.412 | |||
| Triglycerides | rs2954029a, 3.29 × 10−55 | 50 | TRIB1 | rs2385114, 1.954 × 10−19 | 0.421 | |||
| RP11-115J16.1 | chr8:9106927-9271224 | 1 | HDL-C | rs9987289a, 6.40 × 10−25 | 31 | PPP1R3B | rs330927, 3.00 × 10−2 | 0.028 |
| LDL-C | rs2126259a, 7.43 × 10−15 | 23 | PPP1R3B | rs330927, 7.368 × 10−3 | 0.020 | |||
| Total cholesterol | rs2126259a, 8.98 × 10−24 | 43 | PPP1R3B | rs330927, 1.710 × 10−2 | 0.020 | |||
| FPKM1_group_33469_ transcript_1 | chr9:22111740-22128078 | 4 | CAD | rs1333045b, 4.63 × 10−32 | 18 | – | – | – |
| MI | rs1333049a, 3.01 × 10−41 | 18 | – | – | – | |||
| FPKM1_group_33469_ transcript_2 | chr9:22126860-22135488 | 2 | Type 2 diabetes | rs2383208, 6.00 × 10−14 | 4 | CDKN2A | rs3731246, 1.2 × 10−2 | 0.001 |
| CDKN2B | rs1063192, 1.2 × 10−5 | 0.002 | ||||||
| FPKM1_group_33469_ transcript_6 | chr9:22131766-22132847 | 1 | Type 2 diabetes | rs2383208, 6.00 × 10−14 | 1 | CDKN2A | rs3731246, 1.2 × 10−2 | 0.001 |
| CDKN2B | rs1063192, 1.2 × 10−5 | 0.002 | ||||||
| FPKM1_group_3827_ transcript_1 | chr10:94839485-94839724 | 1 | Triglycerides | rs2068888, 2.38 × 10−8 | 2 | CYP26A1 | rs35630863, 2.384 × 10−1 | 0.032 |
| lncRNAlocus2194chr12p64501347p64502165 | chr12:66215082-66215898 | 1 | Type 2 diabetes | rs2612069, 7.70 × 10−8 | 1 | HMGA2 | rs2854603, 1.999 × 10−4 | 0.695 |
| FPKM1_group_7090_ transcript_1 | chr12:97868487-97869951 | 1 | Fasting glucose | rs17390909, 3.40 × 10−8 | 1 | - | – | – |
| linc-LACTB | chr15:63383481-63402781 | 2 | HDL-C | rs2652834, 8.75 × 10−9 | 11 | LACTB | rs2729823, 1.547 × 10−6 | 0.655 |
Fourteen lincRNAs contained both the strongest signal in their region (trait category 1) and ≥1 SNP meeting the Bonferroni significance threshold. Information about the genomic location, most significant SNP P-value, and nearby candidate protein-coding genes is provided below for these lincRNAs. ‘LD’ indicates the linkage disequilibrium measured by R2 between the SNP with the lowest P-value in the lincRNA and the SNP with the lowest P-value in the nearby candidate gene. Abbreviations: HDL-C, HDL cholesterol; BMI, body mass index; LDL-C, LDL cholesterol; NA, not available.
aSNPs exhibiting GWAS association with the same cardiometabolic traits in independent PROMIS GWAS dataset.
bSNPs exhibiting GWAS association with related cardiometabolic traits in independent PROMIS GWAS dataset.
Finally, for GenCAT analyses we filtered findings to include trait-associated lincRNAs that had at least three SNPs in GWAS datasets because GenCAT is designed to identify lincRNAs specifically with multiple SNP associations of moderate strength. Of the 64 unique lincRNAs significant in GenCAT that did not contain any individual SNP that reached the Bonferroni threshold (Supplementary Material, Table S1), 26 lincRNAs contained three or more SNPs (Supplementary Material, Table S5); of these lincRNAs, several had the strongest GenCAT signal of all protein-coding genes and lincRNAs in the region (±500 kb) (Table 3).
Table 3.
LincRNAs with the most significant GenCAT P-values without trait-associated SNPs at the Bonferroni-corrected threshold and with no protein coding genes with stronger GenCAT signal in the region (±500 kb of the lincRNA)
| LincRNA Name | LincRNA Location | No. Exons | GWAS Trait | No. SNPs (total) | No. SNPs (Independent) | GenCAT Csum | GenCAT Raw P value | Top LincRNA SNP, P value | Candidate Gene Information | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | GenCAT Raw P value | LD Max | |||||||||
| linc-ZMYM6a | chr1:35606940-35611998 | 3 | Triglycerides | 3 | 2 | 37.36 | 7.70 × 10−9 | rs2101970, 0.04 | – | – | – |
| RP11-316I3.2a | chr1:188838608-188856565 | 1 | Fasting Insulin | 3 | 2 | 29.24 | 4.48 × 10−7 | rs16829499, 0.06 | – | – | – |
| AC010745.3a | chr2:16408295-16409462 | 1 | Triglycerides | 3 | 3 | 29.52 | 1.74 × 10−6 | rs1429409, 0.03 | – | – | – |
| linc-ST3GAL5-1a | chr2:86209699-86225242 | 3 | LDL-C | 7 | 2 | 29.90 | 3.21 × 10−7 | rs4346389, 0.02 | GGCX | 0.06 | 0.04 |
| VAMP5 | 0.66 | 0.04 | |||||||||
| VAMP8 | 0.63 | 0.03 | |||||||||
| FPKM1_group_22516_ transcript_3a | chr3:142657679-142658088 | 1 | LDL-C | 3 | 3 | 35.53 | 9.40 × 10−8 | rs6770560, 5.00 × 10−3 | – | – | – |
| Total cholesterol | 3 | 3 | 54.92 | 7.14 × 10−12 | rs9852373, 3.00 × 10−3 | ||||||
| FPKM1_group_3023_ transcript_2 | chr10:17259481-17261980 | 1 | Total cholesterol | 4 | 2 | 27.48 | 1.08 × 10−6 | rs7903259, 7.23 × 10−7 | CUBN | 0.75 | 0.1609 |
| VIM | 2.80 × 10−5 | 0.8706 | |||||||||
| FPKM1_group_26174_ transcript_1 | chr5:76425326-76427732 | 1 | Type 2 diabetes | 7 | 3 | 31.53 | 6.59 × 10−7 | rs7708285, 1.10 × 10−6 | ZBED3 | 0.01 | 0.08 |
| FPKM1_group_28834_ transcript_2 | chr6:116385049-116387244 | 1 | LDL-C | 3 | 3 | 37.71 | 3.25 × 10−8 | rs1999930, 6.36 × 10−6 | FRK | 4.72 × 10−6 | 1 |
| Total cholesterol | 3 | 3 | 40.20 | 9.68 × 10−9 | rs1999930, 1.95 × 10−6 | ||||||
| FPKM1_group_13482_ transcript_1 | chr17:68723150-68724281 | 1 | HDL-C | 3 | 3 | 50.29 | 6.94 × 10−11 | rs7213806, 2.64 × 10−4 | – | – | – |
| linc-OPRM1-1a | chr6:153860946-153862090 | 2 | Hemoglobin A1C | 3 | 2 | 28.57 | 6.26 × 10−7 | rs1575106, 0.02 | – | – | – |
| FPKM1_group_29919_ transcript_2 | chr7:21557500-21558289 | 1 | Total cholesterol | 3 | 2 | 29.41 | 4.11 × 10−7 | rs10224283, 7.73 × 10−7 | DNAH11 | 4.00 × 10−4 | 0.67 |
| linc-GPR26-2a | chr10:125319367-125321719 | 4 | MI | 8 | 4 | 32.78 | 1.33 × 10−6 | rs1878165, 4.59 × 10−5 | – | – | – |
| FPKM1_group_3023_ transcript_2 | chr10:17259481-17261980 | 1 | Total cholesterol | 4 | 2 | 27.48 | 1.08 × 10−6 | rs7903259, 7.23 × 10−7 | CUBN | 7.46 × 10−1 | 0.16 |
| VIM | 2.80 × 10−5 | 0.87 | |||||||||
| RP11-534L6.2a | chr10:49872244-49880469 | 1 | MI | 4 | 4 | 34.83 | 5.04 × 10−7 | rs7911516, 7.36 × 10−5 | – | – | – |
| FPKM1_group_13482_ transcript_1a | chr17:68723150-68724281 | 1 | HDL-C | 3 | 3 | 50.29 | 6.94 × 10−11 | rs7213806, 3.00 × 10−4 | – | – | – |
Because GenCAT is designed to identify lincRNAs with multiple SNP associations of moderate strength, we present findings here for lincRNAs that had at least three significant SNPs in GWAS datasets.
alincRNAs (nine in total) with a significant GenCAT P-value without trait-associated SNPs even at the 5% FDR threshold.
LincRNA-trait associations are replicated in independent PROMIS GWAS data
For SNP-based replication in Pakistan Risk of Myocardial Infarction Study (PROMIS) GWAS datasets, between 14% (for LDL-C) and 81% (for MI) of SNPs that were Bonferroni-significant in the original GWAS were also significant in PROMIS according to replication thresholds (from 2.54 × 10−4 for total cholesterol to 1.61 × 10−3 for MI) (Table 4). Detailed SNP-level results of replication analyses are provided in Supplementary Material, Table S6. Also consistent with initial GWAS findings, lincRNAs were enriched for SNPs with low P-values relative to other intergenic regions for PROMIS traits (Supplementary Material, Table S4). However, these replication analyses should be considered preliminary because our main analyses accessed the largest consortia datasets available and similarly large datasets were not available for replication, thus limiting power (see Table 1 for sample sizes; e.g. for LDL-C, sample sizes of 95 454 and 16 995 for discovery versus replication, respectively).
Table 4.
Replication of SNP-level lincRNA associations in PROMIS GWAS datasets
| Trait | Original GWAS | Replication in PROMIS | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Bonferroni significance threshold | Number of SNPs meeting Bonferroni significance threshold | Number of lincRNAs with ≥1 SNP meeting the Bonferroni significance threshold | Number of original significant SNPs also present in PROMIS data | Replication significance threshold | Number of PROMIS SNPs meeting replication significance threshold | Number of PROMIS lincRNAs with ≥1 SNP meeting the replication- significant SNP | Percent of original Bonferroni- significant SNPs also significant in PROMIS | Percent of original significant lincRNAs also significant in PROMIS | |
| MI | 2.34 × 10−7 | 31 | 10 | 31 | 1.61 × 10−3 | 25 | 8 | 81% | 80% |
| HDL-C | 2.12 × 10−7 | 163 | 42 | 162 | 3.09 × 10−4 | 37 | 8 | 23% | 19% |
| LDL-C | 2.12 × 10−7 | 156 | 39 | 155 | 3.23 × 10−4 | 21 | 7 | 14% | 18% |
| Total cholesterol | 2.12 × 10−7 | 199 | 44 | 197 | 2.54 × 10−4 | 52 | 9 | 26% | 21% |
| Triglycerides | 2.12 × 10−7 | 122 | 22 | 121 | 4.13 × 10−4 | 69 | 12 | 57% | 55% |
The replication results are shown here for five cardiometabolic traits. The number of SNPs and lincRNAs meeting the specified significance threshold are shown for the original GWAS and the corresponding PROMIS replication dataset. The percentage of originally significant SNPs and lincRNAs significant in PROMIS are also reported.
Trait-associated lincRNAs have greater synteny than non-associated lincRNAs
Although lincRNAs in primate and human are undergoing expansion and rapid evolution (35,36), conservation with lower species are often used to infer functional importance and to prioritize for follow-up study. Notably, conserved lincRNAs have very limited sequence similarity with their species-orthologs but exhibit strong conservation in their genomic locations during evolution (35,36). Of lincRNA loci with at least one FDR-significant SNP or GenCAT trait-associations, 72.8% were syntenic in mouse. Of the non-trait associated lincRNA loci, 55.6% were syntenic in mouse. Thus, trait-associated lincRNAs are more likely to be syntenic than non-trait associated lincRNAs (P = 1.09 × 10−11). Overall, however, only 21.6% of syntenic trait-associated lincRNAs had transcripts documented at the syntenic position in Gencode M4 mouse lincRNA annotation and only 23.7% of trait-associated lincRNAs had sequence conservation at E-value threshold of < 1 × 10−10 using reciprocal BLASTN. Thus, although trait-associated human lincRNAs are more likely to be syntenic, a substantial subset appears to be either non-syntenic or syntenic but not transcribed in mouse suggesting that mouse models may have limited utility in functional genomic studies of many human-trait associated lincRNAs.
Tissue-specific expression pattern and ASE of trait-associated lincRNAs
Interrogation of five cardiometabolic-relevant tissues in the Human Body Map lincRNAs revealed a tissue-specific pattern for the majority of trait-associated lincRNA (Supplementary Material, Figure S2 and Supplementary Material, Table S7) consistent with prior reports for human lincRNAs (6). We further examined expression of lincRNAs in our own deeply sequenced human PBMCs and adipose, blood cells and metabolic tissues of specific relevance to cardiometabolic traits. Of trait-associated lincRNAs, 12% were expressed in all 15 PBMC samples and 21.2% were expressed in all 25 adipose samples. Of the non-trait-associated lincRNAs, 6.8% were expressed in all PBMCs and 12.7% were expressed in all adipose samples. Thus, trait-associated lincRNAs are almost 2-fold more likely to be expressed in PBMC (P = 1.99 × 10−4) and adipose (P = 4.28 × 10−6).
Next, we probed whether trait associated SNPs in lincRNAs had evidence for ASE in our PBMC and adipose RNA-seq data. For all trait-associated lincRNAs (i.e. SNPs significant at 5% FDR level or GenCAT significant), there were 188 heterozygous SNPs in 34 unique lincRNAs that had at least 10 reads in PBMC RNA-seq data and 12 of these SNPs in nine unique lincRNAs met the PBMC ASE Bonferroni threshold (P < 0.00027, i.e. 0.05/188 SNPs). In adipose RNA-seq data, 686 heterozygous SNPs in 76 unique trait-associated lincRNAs were covered by at least 10 reads and 58 of these SNPs in 28 unique lincRNAs passed the adipose ASE Bonferroni threshold (P < 7.3 × 10−5, i.e. 0.05/686 SNPs). Table 5 presents the subset of these PBMC and adipose ASE SNPs in trait-associated lincRNAs that also had LD R2 > 0.5 with trait-associated SNP in the lincRNA. Haplotype analysis revealed that multiple trait-associated lincRNAs contained ASE SNPs in which the over-expressed allele (i.e. allelic ratio >0.5) was on the same haplotype with the trait-increasing allele. These results support the concept that the GWAS signal at these loci may act via modulating expression of the corresponding lincRNAs.
Table 5.
ASE of trait-associated lincRNAs in PBMCs and adipose
| GWAS Trait | Top Trait SNP rs number (trait-increasing allele) | Top Trait SNP Position | Top Trait SNP P-value | LincRNA Name | LincRNA ASE SNP rs (allele on the same haplotype as the trait- increasing allele) | LincRNA ASE SNP Position | LD between top trait SNP and ASE SNP | No. Subjects with ASE | Top ASE P-value | Reads (allele 1/allele 2) | Allelic ratio of allele 1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PBMCs | |||||||||||
| LDL | rs3177647 (T) | chr12:112277576 | 2.596 × 10−06 | linc-BRAP-2 | rs2879603 (G) | chr12:112278025 | 1 | 3 | 2.837 × 10−18 | 0/76 | 0 |
| TC | rs3177647 (T) | chr12:112277576 | 1.507 × 10−05 | linc-BRAP-2 | rs2879603 (G) | chr12:112278025 | 1 | 3 | 2.837 × 10−18 | 0/76 | 0 |
| HDL | rs9432453 (A) | chr1:93771629 | 1.251 × 10−06 | linc-TMED5 | rs4847408 (C) | chr1:93791437 | 1 | 3 | 9.635 × 10−5 | 25/4 | 0.86 |
| TC | rs7903259 (C) | chr10:17259642 | 7.221 × 10−07 | FPKM1_group_3023_ transcript_2 | rs7903402 (C) | chr10:17259772 | 0.995 | 3 | 1.828 × 10−4 | 14/0 | 1 |
| Adipose | |||||||||||
| HDL | rs9432453 (A) | chr1:93771629 | 1.251 × 10−06 | linc-TMED5 | rs1535699 (A) | chr1:93804952 | 1 | 2 | 1.799 × 10−20 | 0/86 | 0 |
| HDL | rs942196 (A) | chr1:93802264 | 9.946 × 10−07 | linc-DR1 | rs1535699 (A) | chr1:93804952 | 1 | 2 | 1.799 × 10−20 | 0/86 | 0 |
| LDL | rs3177647 (T) | chr12:112277576 | 2.596 × 10−06 | linc-BRAP-2 | rs2879603 (G) | chr12:112278025 | 1 | 3 | 6.211 × 10−19 | 0/79 | 0 |
| TC | rs3177647 (T) | chr12:112277576 | 1.507 × 10−05 | linc-BRAP-2 | rs2879603 (G) | chr12:112278025 | 1 | 3 | 6.211 × 10−19 | 0/79 | 0 |
| WHRadjBMI | rs12747505 (G) | chr1:219618035 | 3.84 × 10−06 | linc-IARS2-2 | rs61838776 (A) | chr1:219618564 | 1 | 1 | 3.297 × 10−09 | 0/35 | 0 |
| HbA1C | rs2257084 (C) | chr17:80687475 | 4.948 × 10−21 | RP11-388C12.5 | rs3803771 (G) | chr17:80687758 | 0.914 | 1 | 7.238 × 10−08 | 29/0 | 1 |
| WHRadjBMI | rs6694509 (T) | chr1:219631981 | 3.46 × 10−09 | RP11-392O17.1 | rs6679091 (T) | chr1:219622509 | 0.548 | 5 | 5.733 × 10−07 | 25/0 | 1 |
| HDL | rs9432453 (A) | chr1:93771629 | 1.251 × 10−06 | linc-TMED5 | rs7524964 (T) | chr1:93801092 | 0.994 | 5 | 1.504 × 10−6 | 10/46 | 0.18 |
| HDL | rs942196 (A) | chr1:93802264 | 9.946 × 10−07 | linc-DR1 | rs7524964 (T) | chr1:93801092 | 0.994 | 5 | 1.504 × 10−6 | 10/46 | 0.18 |
| HDL | rs7838829 (T) | chr8:19780630 | 1.076 × 10−20 | linc-CSGALNACT1-2 | rs28371439 (A) | chr8:19782676 | 1 | 4 | 1.62 × 10−6 | 23/0 | 1 |
| TG | rs7838829 (T) | chr8:19780630 | 5.242 × 10−15 | linc-CSGALNACT1-2 | rs28371439 (A) | chr8:19782676 | 1 | 4 | 1.62 × 10−6 | 23/0 | 1 |
| HDL | rs9432453 (A) | chr1:93771629 | 1.251 × 10−06 | linc-TMED5 | rs942196 (A) | chr1:93802264 | 1 | 1 | 2.727 × 10−06 | 0/22 | 0 |
| WHRadjBMI | rs9987000 (G) | chr7:25883207 | 3.03 × 10−08 | linc-NFE2L3-1 | rs979165 (G) | chr7:25886478 | 0.797 | 1 | 4.291 × 10−06 | 42/96 | 0.30 |
| LDL | rs7255 (T) | chr2:20878820 | 1.929 × 10−06 | AC012065.7 | rs3072 (C) | chr2:20878406 | 0.576 | 2 | 2.209 × 10−5 | 0/18 | 0 |
| TC | rs7255 (T) | chr2:20878820 | 5.771 × 10−06 | AC012065.7 | rs3072 (C) | chr2:20878406 | 0.576 | 2 | 2.209 × 10−5 | 0/18 | 0 |
| HDL | rs9895554 (A) | chr17:46123642 | 5.538 × 10−05 | linc-COPZ2 | rs9900074 (G) | chr17:46124326 | 1 | 4 | 2.669 × 10−5 | 23/2 | 0.92 |
All heterozygous SNPs within trait-associated lincRNAs were tested for ASE in adipose from 21 subjects and in PBMCs from 15 subjects. The trait-increasing allele of the top trait SNP and the ASE allele that was on the same haplotype are shown. Top ASE P-values show the most significant case of ASE among subjects tested. Allele 1 is the allele on the same haplotype as the trait-increasing allele, and allele 2 is the other allele. Allelic ratio was calculated as the number of allele 1 over the total number of alleles for each ASE SNP.
Linc-NFE2L3-1, an obesity associated lincRNA, modulates adipocyte phenotypes
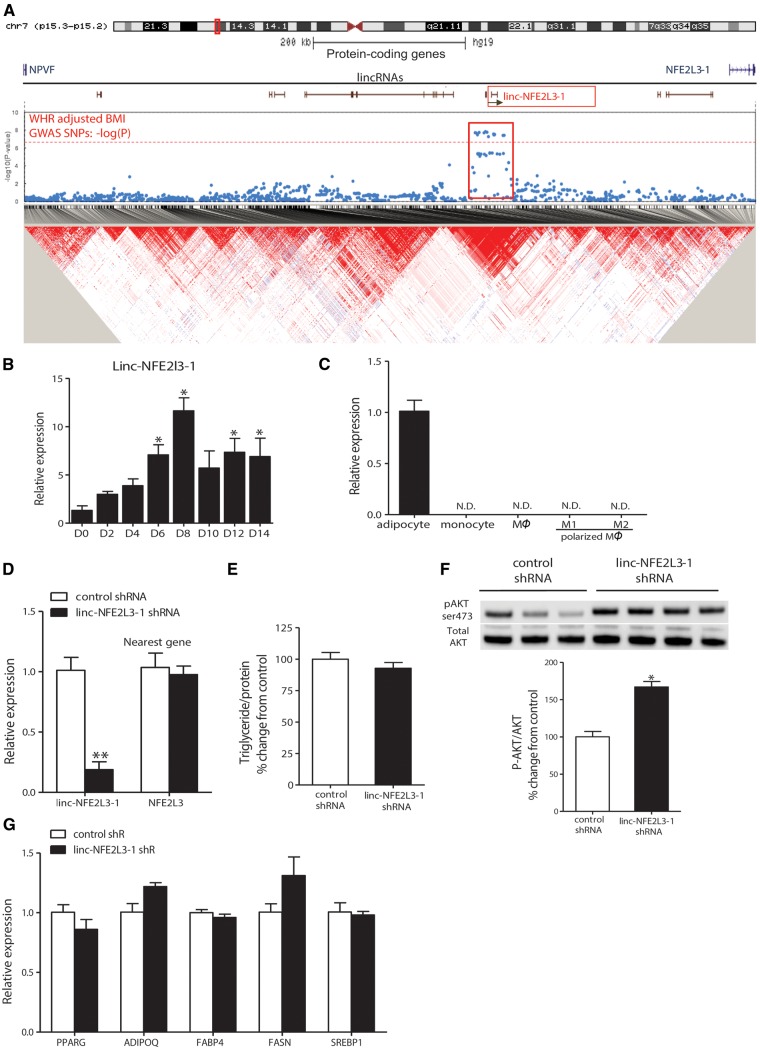
We selected linc-NFE2L3-1 on chromosome 7 as an example for preliminary translational and functional studies in human adipocytes. Interestingly, linc-NPVF-3 not linc-NFE2L3-1 harbors the strongest SNP signal for central obesity (WHRadjBMI) in this region but linc-NPVF-3 is expressed only in testes in Human Body Map data and is not expressed at all in our deeply sequenced adipose RNA-seq data (Supplementary Material, Figure S3A). In fact, of the seven annotated lincRNAs in this region, only linc-NFE2L3-1 is expressed in our adipose RNA-seq data. Supplementary Material, Figure S3B summarizes additional rationale for selection of linc-NFE2L3-1. Briefly, it contains two exons and multiple intronic SNPs that met genome-wide significance (P < 5 × 10−8) for central obesity (WHRadjBMI). An additional 8 obesity-associated SNPs are located 5′ of linc-NFE2L3-1, including rs1055144, the leading GWAS SNP (P = 1.49 × 10−8) at the locus (Fig. 5A and Supplementary Material, Figure S3A). Furthermore, linc-NFE2L3-1 also was significant in our GenCAT gene-based analysis (unadjusted P = 4.4 × 10−7, Bonferroni corrected P =0.01; Supplementary Material, Table S1). Importantly, two neighboring protein-coding genes, NFE2L3-1 (nuclear factor, erythroid 2-like 3) and NPVF (neuropeptide VF precursor), contain no significant GWAS SNPs (Fig. 5A). NPVF is not expressed in human adipose and NFE2L3-1 has no known biological association with adipose biology or obesity. Furthermore, all GWAS SNPs in the region that linc-NFE2L3-1 resides are in high LD with each other but not with SNPs in these two nearest protein-coding genes (R2 < 0.001) (Fig. 5A, LD heatmap). The enrichment of GWAS SNPs in 5′ upstream and intronic regions suggests these genetic variants may regulate linc-NFE2L3-1 expression. Indeed, our adipose RNA-seq data provide direct evidence that obesity-associated SNPs at this locus are associated with ASE of linc-NFE2L3-1 (Table 5) while interrogation of adipose eQTL data from the MuTHER resource (37) revealed no evidence of association between GWAS SNPs for WHRadjBMI and expression levels of NPVF or NFE2L3-1 (Table S8).
Figure 5.
Linc-NFE2L3-1, a lincRNA harboring obesity-associated GWAS SNPs, modulates human adipocyte metabolism. (A) GWAS SNPs associated with WHRadjBMI, and LD heat map at linc-NFE2L3-1 locus. (B) RNA abundances of linc-NFE2L3-1 were significantly increased during differentiation of primary human ASCs to adipocytes. (C) Linc-NFE2L3-1 was detected in human primary adipocytes, but not in human primary monocytes, macrophage or polarized macrophages. *P < 0.05 compared with differentiation Day 0; n = 6/group; N.D., not detected. (D) Lentivirus based shRNA achieved efficient knockdown of linc-NFE2L3-1 in differentiated ASC-derived adipocytes, but not the nearest neighbor protein-coding genes. Linc-NFE2L3-1 knockdown (E) had no effects on triglyceride accumulation in differentiated adipocytes; (F) in mature adipocytes resulted in a significant increase in phosphorylated AKT signal induced by 15-minute insulin treatment (100 nM). **P < 0.01; *P < 0.01 compared with control shRNAs; (G) had no impact on lipogenic gene expression in differentiated adipocytes. Data are combined from three to four independent experiments (n = 3–4 replicates per group).
Based on Human Body Map lincRNA data (18) and our own adipose RNA-seq data (Supplementary Material, Figure S3A), linc-NFE2L3-1 expression is almost entirely restricted to two major metabolic and insulin-responsive tissues, adipose and skeletal muscle. Linc-NFE2L3-1 is barely detected in adipose stromal cells (ASCs) pre-adipocytes, but is strongly induced during in vitro ASC differentiation to mature adipocytes (Fig. 5B) and it is not found in primary human monocytes or macrophages (Fig. 5C), suggesting that linc-NFE2L3-1 is restricted to the mature adipocyte fraction of human adipose. To investigate biological actions of linc-NFE2L3-1 in human adipocytes, we utilized lentivirus-based shRNAs and achieved over 80% knocked-down in ASC-derived adipocytes (Fig. 5D). Knock-down had no effect on adipocyte triglyceride accumulation (Fig. 5E), but significantly enhanced insulin-induced phosphorylation of AKT in mature adipocytes (Fig. 5F). In support of possible trans rather than cis actions, shRNAs targeting linc-NFE2L3-1 did not alter expressions of NFE2L3, the nearest protein-coding gene (Fig. 5D). Arguing against a non-specific effect on adipocyte differentiation, mRNA levels of adiponectin (ADIPOQ), peroxisome proliferator-activated receptor gamma (PPARG), fatty acid binding protein 4 (FABP4), fatty acid synthase (FASN) and sterol regulatory element binding transcription factor 1 (SREBF1) were not impacted by linc-NFE2L3-1 knock-down in mature adipocytes (Fig. 5G). Collectively, our results suggest that linc-NFE2L3-1 might be a causal element at this central obesity locus, may play regulatory roles in insulin-mediated adipocyte metabolism and that genetic variation affecting its expression might affect adipocyte functions, thus contributing to cardiometabolic phenotypes.
Discussion
This is the first systematic interrogation of human lincRNAs for their association with cardiometabolic traits on a genome-wide scale. A complete annotation of all human lincRNAs in all cells and tissues has yet to be determined, but the dataset of human lincRNAs compiled here is one of the most comprehensive currently available. In this paper, we identified hundreds of suggestive lincRNA associations with multiple cardiometabolic traits, performed replications, outlined an approach for prioritizing lincRNAs likely to have independent and causal effects in a region, and provided an illustrative example of a trait-relevant, adipocyte-specific functional follow-up study for a central obesity-associated lincRNA.
Most interpretation and follow-up of GWAS results have focused on protein-coding genes, but it is apparent given the intergenic distribution of most GWAS signals for complex traits that it is essential to consider the regulatory, non-coding elements of the genome. This includes the rapidly evolving catalog of non-coding RNA species such as lincRNAs. Because lincRNAs are only partly characterized in terms of evolution, secondary structure and functions, there is limited understanding of their role in human physiology and disease. Despite debate regarding the proportion of annotated lincRNAs that are truly functional (7), most do have messenger RNA-like features (e.g. polyA tail and exons/introns) and a rapidly-expanding list of examples illustrates their critical functions in genome regulation, cell biology, development, cancer, and disease (e.g. XIST, MALAT1, FIRRE, LINCRNA-COX2 and LNC-DC.)
To date, there are surprisingly few data implicating lincRNAs in complex diseases based on GWAS. One recent study, examining the association of SNPs with lincRNAs expression in human blood, identified 112 cis-regulated lincRNAs, many of which were regulated by disease-associated SNPs (38). A SNP associated with papillary thyroid carcinoma lies in the promoter of a thyroid-expressed lincRNA PTCSC3, and differentially affects the expression of this lincRNA (39,40). Similarly, colorectal cancer risk SNPs at 8q24.21 led to the discovery of CCAT2, a novel lincRNA contributing to cancer pathogenesis (41).
Even less is known of lncRNAs associated with cardiometabolic traits. A multi-isoform lncRNA, ANRIL, that overlaps the protein-coding gene CDKN2B, is proposed as the causal ‘gene’ (32,33) at chr9p21 the strongest GWAS locus for CAD and MI (42). Despite excluding ANRIL in our analysis (because of its overlap with CDKN2B), we identified multiple lincRNAs in this chr9p21 region (e.g. FPKM1_GROUP_33469 _TRANSCRIPT_1, chr9:22111740-22128078) that overlap some ANRIL isoforms, but not CDKN2B, and these show strong associations with CAD and MI (Supplementary Material, Figure S4A and B). We have not performed deep genomic and transcriptomic mapping at this locus and cannot define the exact lincRNAs and isoforms with strongest trait-signals. We do not propose that our annotated chr9p21 lincRNAs provide an alternative explanation to ANRIL for CAD and MI association—rather, our findings are consistent with studies that have interrogated the locus in greater depth and implicated the ANRIL family of lncRNAs (32,33). Of note, distinct non-coding variation at chr9p21 that does not overlap the CAD and MI locus has been associated with type 2 diabetes (34,43–45) (Supplementary Material, Figure S4B). Furthermore, these CAD-associated and T2D-associated SNPs fall into distinct LD blocks (Supplementary Material, Figure S4C). We found that several chr9p21 lincRNAs (e.g. FPKM1_GROUP_33469 _TRANSCRIPT_2, chr9:22126860-22135488 and FPKM1_GROUP_33469 _TRANSCRIPT_6, chr9:22131766-22132847) have associations with type 2 diabetes (Supplementary Material, Figure S4A and B) and these do not overlap ANRIL or the CAD-associated SNPs. Our results of strong association of multiple lincRNAs at chr9p21 with MI and CAD as well as with type 2 diabetes highlight the need for additional genomic, translational and functional studies to define more precisely the lincRNA structures and the tissue-specific role of lncRNAs in both MI/CAD and type 2 diabetes associations at chr9p21.
LincRNAs are increasingly implicated in key cellular functions important for cardiometabolic traits, such as cardiac lineage commitment during heart development (46), liver lipoprotein ApoC2 production (47), cell differentiation and proliferation (e.g. T cell (48), dendritic cell (49) and endothelial cell (50)), vascular smooth muscle contraction (51), monocyte and macrophage immune responses (52,53). In addition, it has been shown that hundreds of lncRNAs are induced during adipogenesis in mouse (54). Indeed, several lincRNAs, such as lincRNA FIRRE (14), ADNIR (55), BLNC1 (56) and Lnc-BATE1 (57) have demonstrated specific regulatory roles in white or brown adipocyte differentiation.
Our study reveals several noteworthy findings in prioritization of trait-associated lincRNAs. Approximately one-quarter of all trait-associated lincRNAs we identified has no previously proposed candidate genes in their region and thus hold a specific interest for follow-up, particularly when they contain the strongest trait-associated SNP or GenCAT signal in the region. Four lincRNAs without a previously proposed candidate gene nearby contain one or more SNPs significant at the SNP Bonferroni threshold (P < 2.2 × 10−7): U47924.27 (chr12) for HbA1c, FPKM1_GROUP_840_TRANSCRIPT_1 (chr1) for BMI, AC068138.1 (chr2) for HDL-C and triglyceride levels and FPKM1_GROUP_7090_TRANSCRIPT_1 (chr12) for fasting glucose. Although the majority of trait-associated lincRNAs do have a candidate gene within 500 kb, few of these lincRNAs have strong LD (R2 > 0.8) with the proposed candidate gene at the locus. Even a well-established biological connection between a protein-coding gene and a trait does not exclude a potential functional role for a nearby lincRNA. For instance, the lincRNA may regulate its neighboring protein-coding gene(s) in cis, as is the case for HOTTIP, DBE-T and MISTRAL (58). Examples from this study of trait-associated lincRNAs near well-known candidate genes include linc-CCBE1 (chr18) and linc-CDH20-2 (chr18) near MC4R associated with BMI, triglycerides and HDL-C and lincRNA AP000770.1 (chr11) near APOA1 and APOA5 associated with triglycerides, HDL-C, LDL-C and total cholesterol. Furthermore, patterns of ASE suggest that some trait-associated SNPs at lincRNA loci may indeed act via modulating expression of the lincRNA at that GWAS locus. Finally, ∼20% of trait-associated lincRNAs harbor the strongest signal in the surrounding 1MB region; thus it is imperative to investigate these lincRNAs and not just their protein-coding neighbors when experimentally evaluating the GWAS signals.
Our work identified many lincRNAs with association signals near putative candidate protein-coding genes that lack clear functional relevance to the phenotype. Such genes often were chosen as candidate genes at the locus simply because they were the nearest protein-coding gene. For example, linc-NFE2L3-1, an adipose-expressed lincRNA associated with central obesity, contains multiple genome-wide significant SNPs spanning the lincRNA and its 5′ region. The two closest neighboring protein-coding genes at the locus, NFE2L3 and NPVF, contain no significant GWAS SNPs yet NFE2L3 was credited as the possible causal gene at this GWAS locus although there is no functional data supporting NFE2L3 in adipocyte or adipose biology. Furthermore, NFE2L3 and NPVF SNPs have no LD with the obesity-associated SNPs at linc-NFE2L3-1 and adipose eQTL datasets reveal that the obesity GWAS SNPs at this locus are not associated with adipose mRNA levels of either NFE2L3 or NPVF. In contrast, our adipose RNA-seq data suggests ASE of linc-NFE2L3-1 for obesity-related SNPs at this locus.
Because of our interest in adipose biology, we selected linc-NFE2L3-1 for proof of principle functional studies. Linc-NFE2L3-1, expressed in adipose and skeletal muscle, was absent in pre-adipocytes but markedly induced during adipocyte differentiation. It was not detected in monocytes or macrophages, other cell constituents of adipose tissue. Knockdown of linc-NFE2L3-1 enhanced insulin-stimulated activation of AKT signaling, suggesting it may be a negative regulator of insulin signaling in adipocytes and thus contribute to metabolic traits including central obesity. By way of precedence, haploinsufficiency mutations in PTEN, a well-known negative modulator for insulin pathways, are associated with increased obesity in humans (59). Thus, our functional studies further support linc-NFE2L3-1 as a possible causal element at this central obesity locus.
There is substantial interest in gene- and class-level testing in complex traits, e.g. focusing on classes of regulatory elements or particular gene families. These approaches serve as a complement, rather than alternative, to a SNP-focused strategy. Several tools are available. We applied GenCAT, a novel approach we recently developed (60) that is similar to the quadratic test (QT) and versatile gene-based association study approaches (61,62), because it takes into account local SNP correlation patterns so that all SNP-level P-values within an element (e.g. a lincRNA) are accounted for optimally. Using GenCAT, we found trait-association with multiple lincRNAs that also showed SNP-level trait association, but we also identified many additional lincRNAs showing a significant GenCAT association but not individually significant SNP association. Thus, GenCAT analyses provide substantial novel and complementary insights into the landscape of lincRNAs in cardiometabolic pathophysiology.
Our study has several unique strengths but also limitations that highlight the need for additional genomic interrogation and experimental follow-up. We examined the SNPs between the start and end positions of lincRNAs, but excluded the promoter and 3′ regulatory regions which may harbor genetic variation that regulates lincRNA expression and therefore function. We chose this narrow focus to be conservative and limit overlap with protein-coding genes and to enhance statistical power by reducing the number of SNPs being considered. Furthermore, we did not consider each lincRNA transcript separately, but rather we only considered the region specified by the start and end positions of each lincRNA in our merged dataset. These lincRNA sequences are not complete and a more comprehensive analysis of all isoforms of lincRNAs was not possible. Because comprehensive tissue expression data are not yet available for all the lincRNAs included in this study, further work is needed to leverage genomic regulatory data resources (including the ENCODE project, the NIH Roadmap Epigenetic project, the Genotype-Tissue Expression Project Portal and comprehensive RNA-seq datasets) in order to assign appropriate promoter and enhancer domains that coincide with the cells and tissues where the lincRNAs are expressed.
Many recent analyses, including ours, have focused on the subset of lncRNAs that are intergenic (lincRNAs) and do not overlap exons of protein-coding genes. This excludes the analysis of antisense RNAs, an important class of lncRNAs that are especially involved in regulation of their ‘sense’ protein-coding partners. Indeed, as noted above this filter excluded the MI-associated ANRIL, perhaps the best-known lncRNA linked to a complex cardiometabolic trait. Unfortunately, current datasets and bioinformatics strategies limit an independent evaluation of association for SNPs that fall in overlapping protein-coding genes and lncRNAs. Our analysis lacked SNP data (∼50% of lincRNAs did not harbor HAPMAP-imputed SNPs) for a substantial proportion of our fully annotated set of lincRNAs, most likely for the following reasons. First, lincRNAs are shorter in size than protein coding genes (median size 0.6 versus 2.4 kb respectively). Second, as noted above, we restricted our analysis to SNPs that fell within the start and end position of lincRNAs. Third, our SNP interrogations were restricted to publicly available HapMap-imputed datasets. As they become publicly available, 1000-Genomes-imputed disease association datasets as well as whole genome datasets will permit analysis of lincRNAs that could not be included here, as well as a more comprehensive analysis of all lincRNAs and their transcripts.
In conclusion, using SNP- and class-level testing in existing GWAS datasets, we identified hundreds of suggestive lincRNA associations with multiple cardiometabolic traits, performed preliminary replications and functional studies, and outlined an approach for prioritizing lincRNAs more likely to have independent effects at a locus. Additional experimental follow-up of promising trait-associated lincRNAs is warranted to identify causal pathways and therapeutic opportunities in cardiometabolic diseases.
Materials and Methods
Defining the lincRNA dataset and cardiometabolic trait GWAS summary datasets
Human lincRNA annotations were drawn from four published catalogs based on human RNA-seq data derived from a total of 25 human tissues and cell lines (Supplementary Material, Table S9): 6763 lincRNAs from version 18 of Gencode, 54 776 from Hangauer et al., 8255 from Cabili et al. and 3474 from Sigova et al. for a total of 73 268 lincRNAs (18–21). The UCSC LiftOver tool was used to map the hg18 annotations in the Hangauer data (S3 dataset) (20) and Sigova data (21) into human genome build 19.
The processing methods applied to the original datasets varied considerably and are summarized in detail in the Supplementary Materials. Because some lincRNA annotations overlapped between the datasets, and because different processing steps were applied originally to generate each dataset, a filtering and merging pipeline was developed to clean and combine lincRNAs from all four datasets in a standardized manner (Fig. 1). LincRNA exclusion filters were applied first to remove all 2515 lincRNAs on the X chromosome, 305 lincRNAs on the Y chromosome, 1458 lincRNAs overlapping a pseudogene (63) by one or more bases, and 21 lincRNAs less than 200 base pairs in length. 2769 lincRNAs were removed due to overlap or proximity to a RefSeq protein-coding gene (within 1000 bases). Briefly, lincRNAs with strand information (∼30%) were removed if they were within 1000 bases of a protein-coding gene on the same strand, or overlapped a protein-coding gene on the opposite strand. LincRNAs without strand information were removed if they were within 1000 bases of a protein-coding gene. After all filtering steps, 66 200 lincRNAs remained.
Next, overlapping lincRNAs from the four original datasets were merged together on the basis of location and exon structure. Two lincRNAs were merged when one lincRNA had an exon that was overlapped 50% or more by an exon of another lincRNA, the same merging criterion used by Hangauer et al. (20). The newly created lincRNA was assigned an exon structure in which any base belonging to an exon of either of its parent lincRNAs was classified as exonic. LincRNAs with strand information were never merged with a lincRNA known to be on the opposite strand. When two lincRNAs with different names were merged, for ease of reference the newly annotated lincRNA received whichever name was shorter. After merging, there remained 54 944 distinct autosomal lincRNAs, covering 224 456 248 bases (7.16%) of the genome.
In order to interrogate lincRNA associations with cardiometabolic traits, fifteen HapMap-SNP-imputed GWAS summary datasets with hg18 mapped positions (Table 1) were obtained and processed such that all SNPs were assigned updated rs numbers and updated chromosomal positions in hg19 (27–29, 64–68) (see Supplementary Materials).
SNP-based association of lincRNAs with traits
To pinpoint specific lincRNAs of interest, GWAS SNPs were annotated to lincRNAs on the basis of position. Of the 54 944 lincRNAs considered, ∼50% were represented by SNPs in the HapMap-based GWAS datasets (range: 214 039 SNPs in 25 726 lincRNAs in the CARDIoGRAM myocardial infarction (MI) to 236 016 SNPs in 27,449 lincRNAs in the GLGC triglycerides). The SNP P-values for each trait were then corrected for multiple testing based on the total number of SNPs annotated to the full set of lincRNAs, i.e. the total number of SNP tests under consideration for each trait. A Bonferroni correction (P < 0.05, equivalent to uncorrected P < 2.2 × 10−7) as well as a Benjamini and Hochberg 5% FDR correction were both applied separately to unadjusted P-values. LincRNAs containing one or more SNPs with a corrected P-value < 0.05, by either correction method, were noted. Although Bonferroni correction for SNP testing is conservative in this setting because many SNPs within lincRNAs are in LD and therefore not independent, we used this in order to prioritize lincRNAs with strong evidence for trait association. We also present lincRNAs with SNPs reaching the 5% FDR level, to provide a comprehensive insight into lincRNA association patterns with cardiometabolic traits. Because we focused on a ‘candidate’ set of lincRNAs for related cardiometabolic traits, we did not use a genome-wide correction threshold of 5 × 10−8 or correct for the multiple correlated traits examined. Our approach allows investigators to focus on specific lincRNAs of interest depending on the strength of association as well as other evidence supporting the lincRNA in the trait biology.
Finally, to determine whether lincRNAs as a group were enriched for trait associated SNPs relative to non lincRNA intergenic regions, the distributions of SNP P-values in lincRNAs were compared with those in non lincRNA intergenic regions using a resampling-based approach (see Supplementary Materials).
Class-level selection of additional lincRNAs with trait associations
The GenCAT framework (60) uses a quadratic form to test for associations between lincRNAs and traits, based on SNP-level association test statistics within lincRNAs, and after transformation that takes into account the local SNP correlation structure and data redundancies. The resulting chi-square (χ2) statistic for each lincRNA has degrees of freedom equal to the number of transformed test statistics, and thus accounts for the number of SNPs per lincRNA. As a complement to single SNP-based approaches, the method is particularly useful for selecting genomic classes (e.g. protein-coding genes or lincRNAs) that may contain multiple SNP associations of moderate strength that would be missed by relying solely on single-SNP tests. The GenCAT algorithm was applied to each GWAS dataset to discover additional lincRNAs with trait associations. Because GenCAT generates a single test statistic for each lincRNA, a Bonferroni adjustment based on the total number of lincRNAs is applied. This approach assumes independent signal across lincRNAs. Although stringent, this assumption is less conservative than for SNP testing (where many SNPs within lincRNAs are in LD). For GenCAT, we do not report less conservative FDR-corrected P-values.
Prioritization of trait-associated lincRNAs
Some lincRNAs with trait associations may be near candidate protein-coding genes for the trait, or near intergenic SNPs with stronger association signals than SNPs in the lincRNAs. The true trait effect(s) at such loci could be via (i) the lincRNA itself, (ii) the protein-coding gene, (iii) both the lincRNA and the protein-coding gene, e.g. cis-regulatory actions of the lincRNA on local protein coding gene or (iv) other regulatory features at the locus independent of the lincRNA. To explore which trait-associated lincRNAs are most likely to have independent effects, first we characterized the patterns of SNP association strengths for each trait at each lincRNA region using ±500 kb of lincRNA as the regions of interest. Since lincRNAs are generally shorter than protein coding genes, we chose ±500 kb as an appropriate threshold that balances our primary focus on identifying lincRNAs for further prioritizations vs. identifying the influence of more distant regional signals and protein coding genes. If the lincRNA contained the most significant SNP in this 500 kb region, it was assigned to ‘trait category’ 1, otherwise it was assigned to trait category 2. If the lincRNA did not contain the strongest SNP signal in the region for a given trait, then we calculated the degree of LD using R2 between the most significant SNP in the lincRNA and the most significant SNP in the region. We used the LD between the top SNP in the lincRNA and the top SNP in the region to designate an ‘LD category’ for these lincRNAs. For R2 ≤ 0.3, the lincRNA was assigned LD category 1; for 0.3 < R2 < 0.8, LD category 2; for R2 ≥ 0.8, LD category 3. For each trait, we also calculated R2 between the most significant SNP in the lincRNA and the most significant SNP in any putative candidate protein-coding genes in the region. The software PLINK (69) and the 1000 Genomes Phase 2 European dataset (70) were used for all R2 calculations. Each trait was examined separately and the complete lists of genome-wide significant SNPs as well as proposed candidate protein-coding genes for each trait were extracted from the original GWAS datasets and publications for the 15 cardiometabolic traits (Supplementary Material, Table S10).
Replication in independent GWAS data
Our primary analyses accessed the largest consortia datasets available thus limiting availability of additional large datasets for replication. Therefore, our replication analyses, performed in a South Asian sample, should be considered preliminary and illustrative rather than definitive. GWAS summary data from the PROMIS study were used for preliminary replication for several traits, including MI and plasma lipids. A focused SNP-based replication was conducted, in which SNPs that were Bonferroni-significant in the ‘original’ GWAS and also present in the corresponding PROMIS dataset were used to calculate a replication significance threshold (e.g. P < 0.05/197 = 2.54 × 10−4 for 197 SNPs for total cholesterol, to P < 0.05/31 = 1.61 × 10−3 for 31 SNPs for MI). The PROMIS SNPs meeting this replication threshold were extracted and the lincRNAs containing these SNPs were noted. In addition, to examine the generalizability of enrichment findings, lincRNA enrichment in trait associated SNPs was also analyzed via resampling in PROMIS (see Supplementary Materials).
Synteny and conservation of trait-associated lincRNAs
Synteny and sequence conservation are hallmarks of functionally conserved genomic elements. Synteny of lincRNA loci across species was described by Ulitsky et al. (71), in which many zebrafish lincRNA loci are found to have conserved genomic location in mammalian genomes while only limited zebrafish lincRNAs are conserved at the level of primary sequence. Additionally, two recent articles (35,36) examining lincRNA conservation in mammalian species demonstrate that most conserved lincRNAs are syntenic and have poor primary sequence conservation across species. In the absence of detailed knowledge of expressed lincRNAs in all tissues across species, we analyzed genomic synteny, an important feature of conserved lincRNAs, as well as sequence conservation for trait-associated human lincRNAs.
For synteny analysis, we obtained all coding genes within 500 kb of a human lincRNA and searched for homologous genes in mouse, using HomoloGene from NCBI to determine homology. A lincRNA was considered syntenic if its two neighboring genes had homologs in mouse. Next, to determine conservation, the human sequences of the syntenic lincRNAs were extracted from human genome build 19 and compared with the mouse genome build mm9 using BLASTN. A threshold of Expect Value (E-value) ≤ 1e-10 was applied. Conservation hits in mouse were required to be within the syntenic mouse lincRNAs. These mouse sequence hits were then compared with the human genome using BLASTN with the same threshold. Only the hits that could be mapped again to the original sequences in human were considered conserved. The proportion of syntenic trait-associated lincRNAs was compared with the proportion of syntenic non-trait-associated lincRNAs using the test.
Tissue expression patterns and ASE for trait associated lincRNAs
LincRNAs are known to be tissue specific (18). First, we utilized the Human Body Map dataset (18) to examine the expression of trait-associated lincRNAs in five cardiometabolic-relevant tissues: adipose, blood, brain, liver and muscle. Since the expression levels of lincRNAs are generally low, to aid visualization of the expression of lincRNAs across tissues, we set the FPKM intervals with upper limit to 0.001, 0.01, 0.1, 1 and >1 and generated a color heat map of each expressed lincRNA for each tissue.
Next, we examined lincRNA expression in our own deeply sequenced RNA-seq data (72) for peripheral blood mononuclear cells (PBMCs; n = 15 samples; ∼369 million reads per sample) and adipose tissue (n = 25 samples; ∼223 million reads per sample). In each sample, we identified lincRNAs that were expressed above 1% FPKM among all lincRNAs and RefSeq coding genes, and considered a lincRNA as fully ‘expressed’ only when it was present in all PBMC or all adipose tissue samples. The proportion of trait-associated lincRNAs expressed in PBMC and adipose was compared with the proportion of non-trait-associated lincRNAs expressed in PBMC and adipose using the test.
Finally, to test ASE, we first identified all heterozygous SNPs within each lincRNA. For each SNP covered by at least 10 reads in our human PBMC and adipose tissue RNA-seq data, we counted the number of reads mapped for each allele. The allele counts were then tested against a null of 1:1 ratio for each allele by test. P-values were adjusted for the total number of heterozygous SNPs meeting the analysis criteria. For SNPs with ASE, we performed haplotype analysis to examine whether the over-expressed allele (i.e. allelic ratio > 0.5) at the ASE SNP resides on the same haplotype with the risk allele or trait increasing allele at the top trait-associated SNP. To obtain haplotype information, we downloaded 1000 Genomes phase 3 data (ftp://ftp.1000genomes.ebi.ac.uk/Vol02514/ftp/release/20130502/) for all subjects of European ancestry, and calculated haplotype frequency for each top trait-associated SNP and ASE SNP pair.
Adipocyte functional studies of linc-NFE2L3-1 an obesity-associated lincRNAs
Because we have interest in novel genomic pathways that regulate adipocyte functions and cardiometabolic traits in humans, we selected linc-NFE2L3-1, an adipose-expressed lincRNA associated with waist-hip ratio adjusted for body mass index (‘WHRadjBMI’), as an example for preliminary functional study in adipocytes. Detailed methods for adipocyte culture and insulin stimulation, shRNA knockdown of lincRNAs, and adipocyte lipid and metabolic assays are provided in Supplementary Materials. Statistical analyses of adipocyte data were performed using Graphpad Prism. Conditions were compared using unpaired Student’s t-test (for two groups) or analysis of variance followed by Tukey's test for pairwise comparisons (for more than two conditions).
Statistical power calculations
Statistical power calculations for SNP-based analyses of lincRNAs of binary traits (CAD, MI, type 2 diabetes) and quantitative traits (plasma lipids, plasma metabolic parameters, and anthropometric characteristics) are presented in Supplementary Material, Table S11. Power was calculated using the Genetic Power Calculator (http://pngu.mgh.harvard.edu/∼purcell/gpc) assuming additive genetic effect, correcting for 226 000 SNPs tested in all lincRNAs. For each trait, we calculated power at various sample sizes and effect sizes (genotype relative risk for a binary trait; percent variation explained by a SNP for a quantitative trait).
Web resources
All computer code for defining the lincRNA dataset, annotating SNPs to lincRNAs, applying the Bonferroni and FDR corrections to lincRNA SNP P-values, performing resampling for enrichment, and analyzing LD around each lincRNA are available on GitHub at: https://github.com/rachellea/lincRNAs/tree/master#lincrnas, [last accessed date July 2015].
Additional software resources:
PLINK: http://pngu.mgh.harvard.edu/∼purcell/plink/, [last accessed date August 2014]
Human Body Map: http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-513/, [last accessed date June 2015]
GenCAT: https://cran.r-project.org/web/packages/GenCAT/index.html, [last accessed date November 2014]
BLASTN: https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch, [last accessed date June 2015]
GTEx: http://www.gtexportal.org/home/, [last accessed date June 2015]
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
Data on coronary artery disease (CAD)/myocardial infarction (MI) have been contributed by CARDIoGRAMplusC4D investigators and have been downloaded from www.CARDIOGRAMPLUSC4D.ORG. Data on LDL-C, HDL-C, triglycerides and total cholesterol have been contributed by GLGC, the Global Lipids Genetics Consortium, and have been downloaded from http://csg.sph.umich.edu//abecasis/public/lipids2013/. Data on type II diabetes have been contributed by DIAGRAM, the Diabetes Genetics Replication and Meta-analysis consortium, and have been downloaded from http://diagram-consortium.org/downloads.html. Data on glycaemic traits have been contributed by MAGIC investigators and have been downloaded from www.magicinvestigators.org. Data on body mass index and waist-hip ratio adjusted for BMI have been contributed by GIANT, the Genetic Investigation of Anthropometric Traits Consortium, and have been downloaded from https://www.broadinstitute.org/collaboration/giant/index.php/Main_Page.
Conflict of Interest statement. None declared.
Funding
This work was supported by National Institutes of Health (NIH) R01-HL113147 (to M.P.R. and M.L.), by K24-HL107643 (to M.P.R.), by R01-GM108600 (to M.L.) and by R01-HL107196 (to A.S.F.). MPR is also funded in genomic studies of cardiometabolic traits by R01-HL111694 and R01-DK090505.
References
- 1.Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A. et al. (2009) Finding the missing heritability of complex diseases. Nature, 461, 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eichler E.E., Flint J., Gibson G., Kong A., Leal S.M., Moore J.H., Nadeau J.H. (2010) Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet., 11, 446–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neph S., Vierstra J., Stergachis A.B., Reynolds A.P., Haugen E., Vernot B., Thurman R.E., John S., Sandstrom R., Johnson A.K. et al. (2012) An expansive human regulatory lexicon encoded in transcription factor footprints. Nature, 489, 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F. et al. (2012) Landscape of transcription in human cells. Nature, 489, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Consortium, E.P. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ulitsky I., Bartel D.P. (2013) lincRNAs: genomics, evolution, and mechanisms. Cell, 154, 26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haerty W., Ponting C.P. (2015) Unexpected selection to retain high GC content and splicing enhancers within exons of multiexonic lncRNA loci. RNA, 21, 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engreitz J.M., Pandya-Jones A., McDonel P., Shishkin A., Sirokman K., Surka C., Kadri S., Xing J., Goren A., Lander E.S. et al. (2013) The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science, 341, 1237973.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang J., Jing Y., Cost G.J., Chiang J.C., Kolpa H.J., Cotton A.M., Carone D.M., Carone B.R., Shivak D.A., Guschin D.Y. et al. (2013) Translating dosage compensation to trisomy 21. Nature, 500, 296–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. (2010) Long noncoding RNA as modular scaffold of histone modification complexes. Science, 329, 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A. et al. (2011) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature, 472, 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirose T., Virnicchi G., Tanigawa A., Naganuma T., Li R., Kimura H., Yokoi T., Nakagawa S., Benard M., Fox A.H. et al. (2014) NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell, 25, 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clemson C.M., Hutchinson J.N., Sara S.A., Ensminger A.W., Fox A.H., Chess A., Lawrence J.B. (2009) An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell, 33, 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hacisuleyman E., Goff L.A., Trapnell C., Williams A., Henao-Mejia J., Sun L., McClanahan P., Hendrickson D.G., Sauvageau M., Kelley D.R. et al. (2014) Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol., 21, 198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sauvageau M., Goff L.A., Lodato S., Bonev B., Groff A.F., Gerhardinger C., Sanchez-Gomez D.B., Hacisuleyman E., Li E., Spence M. et al. (2013) Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife, 2, e01749.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ning S., Wang P., Ye J., Li X., Li R., Zhao Z., Huo X., Wang L., Li F., Li X. (2013) A global map for dissecting phenotypic variants in human lincRNAs. Eur. J. Hum. Genet., 21, 1128–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ning S., Zhao Z., Ye J., Wang P., Zhi H., Li R., Wang T., Li X. (2014) LincSNP: a database of linking disease-associated SNPs to human large intergenic non-coding RNAs. BMC Bioinformatics, 15, 152.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L. (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev, 25, 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. et al. (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res., 22, 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hangauer M.J., Vaughn I.W., McManus M.T. (2013) Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet., 9, e1003569.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sigova A.A., Mullen A.C., Molinie B., Gupta S., Orlando D.A., Guenther M.G., Almada A.E., Lin C., Sharp P.A., Giallourakis C.C. et al. (2013) Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc. Natl. Acad. Sci. USA, 110, 2876–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji P., Diederichs S., Wang W., Boing S., Metzger R., Schneider P.M., Tidow N., Brandt B., Buerger H., Bulk E. et al. (2003) MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene, 22, 8031–8041. [DOI] [PubMed] [Google Scholar]
- 23.Hutchinson J.N., Ensminger A.W., Clemson C.M., Lynch C.R., Lawrence J.B., Chess A. (2007) A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics, 8, 39.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernard D., Prasanth K.V., Tripathi V., Colasse S., Nakamura T., Xuan Z., Zhang M.Q., Sedel F., Jourdren L., Coulpier F. et al. (2010) A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. Embo J., 29, 3082–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A. et al. (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell, 39, 925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vance K.W., Sansom S.N., Lee S., Chalei V., Kong L., Cooper S.E., Oliver P.L., Ponting C.P. (2014) The long non-coding RNA Paupar regulates the expression of both local and distal genes. Embo J., 33, 296–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heid I.M., Jackson A.U., Randall J.C., Winkler T.W., Qi L., Steinthorsdottir V., Thorleifsson G., Zillikens M.C., Speliotes E.K., Magi R. et al. (2010) Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet., 42, 949–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U., Lango Allen H., Lindgren C.M., Luan J., Magi R. et al. (2010) Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet., 42, 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J. et al. (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature, 466, 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Broadbent H.M., Peden J.F., Lorkowski S., Goel A., Ongen H., Green F., Clarke R., Collins R., Franzosi M.G., Tognoni G. et al. (2008) Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum. Mol. Genet., 17, 806–814. [DOI] [PubMed] [Google Scholar]
- 31.Cunnington M.S., Santibanez Koref M., Mayosi B.M., Burn J., Keavney B. (2010) Chromosome 9p21 SNPs Associated with Multiple Disease Phenotypes Correlate with ANRIL Expression. PLoS Genet., 6, e1000899.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holdt L.M., Beutner F., Scholz M., Gielen S., Gabel G., Bergert H., Schuler G., Thiery J., Teupser D. (2010) ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler. Thromb. Vasc. Biol., 30, 620–627. [DOI] [PubMed] [Google Scholar]
- 33.Holdt L.M., Hoffmann S., Sass K., Langenberger D., Scholz M., Krohn K., Finstermeier K., Stahringer A., Wilfert W., Beutner F. et al. (2013) Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet., 9, e1003588.,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cugino D., Gianfagna F., Santimone I., de Gaetano G., Donati M.B., Iacoviello L., Di Castelnuovo A. (2012) Type 2 diabetes and polymorphisms on chromosome 9p21: a meta-analysis. Nutr. Metab. Cardiovasc. Dis., 22, 619–625. [DOI] [PubMed] [Google Scholar]
- 35.Hezroni H., Koppstein D., Schwartz M.G., Avrutin A., Bartel D.P., Ulitsky I. (2015) Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep., 11, 1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Necsulea A., Soumillon M., Warnefors M., Liechti A., Daish T., Zeller U., Baker J.C., Grutzner F., Kaessmann H. (2014) The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature, 505, 635–640. [DOI] [PubMed] [Google Scholar]
- 37.Nica A.C., Parts L., Glass D., Nisbet J., Barrett A., Sekowska M., Travers M., Potter S., Grundberg E., Small K. et al. (2011) The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet., 7, e1002003.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar V., Westra H.J., Karjalainen J., Zhernakova D.V., Esko T., Hrdlickova B., Almeida R., Zhernakova A., Reinmaa E., Vosa U. et al. (2013) Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS Genet., 9, e1003201.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He H., Li W., Liyanarachchi S., Jendrzejewski J., Srinivas M., Davuluri R.V., Nagy R., de la Chapelle A. (2015) Genetic predisposition to papillary thyroid carcinoma: involvement of FOXE1, TSHR, and a novel lincRNA gene, PTCSC2. J. Clin. Endocrinol. Metab., 100, E164–E172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jendrzejewski J., He H., Radomska H.S., Li W., Tomsic J., Liyanarachchi S., Davuluri R.V., Nagy R., de la Chapelle A. (2012) The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc. Natl. Acad. Sci. USA, 109, 8646–8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ling H., Spizzo R., Atlasi Y., Nicoloso M., Shimizu M., Redis R.S., Nishida N., Gafa R., Song J., Guo Z. et al. (2013) CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res., 23, 1446–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nikpay M., Goel A., Won H.H., Hall L.M., Willenborg C., Kanoni S., Saleheen D., Kyriakou T., Nelson C.P., Hopewell J.C. et al. (2015) A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet., 47, 1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grarup N., Rose C.S., Andersson E.A., Andersen G., Nielsen A.L., Albrechtsen A., Clausen J.O., Rasmussen S.S., Jorgensen T., Sandbaek A. et al. (2007) Studies of association of variants near the HHEX, CDKN2A/B, and IGF2BP2 genes with type 2 diabetes and impaired insulin release in 10,705 Danish subjects: validation and extension of genome-wide association studies. Diabetes, 56, 3105–3111. [DOI] [PubMed] [Google Scholar]
- 44.Shea J., Agarwala V., Philippakis A.A., Maguire J., Banks E., Depristo M., Thomson B., Guiducci C., Onofrio R.C., Kathiresan S. et al. (2011) Comparing strategies to fine-map the association of common SNPs at chromosome 9p21 with type 2 diabetes and myocardial infarction. Nat. Genet., 43, 801–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Helgeland O., Hertel J.K., Molven A., Raeder H., Platou C.G., Midthjell K., Hveem K., Nygard O., Njolstad P.R., Johansson S. (2015) The Chromosome 9p21 CVD- and T2D-Associated Regions in a Norwegian Population (The HUNT2 Survey). Int. J. Endocrinol., 2015, 164652.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klattenhoff C.A., Scheuermann J.C., Surface L.E., Bradley R.K., Fields P.A., Steinhauser M.L., Ding H., Butty V.L., Torrey L., Haas S. et al. (2013) Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell, 152, 570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li P., Ruan X., Yang L., Kiesewetter K., Zhao Y., Luo H., Chen Y., Gucek M., Zhu J., Cao H. (2015) A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab., 21, 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu G., Tang Q., Sharma S., Yu F., Escobar T.M., Muljo S.A., Zhu J., Zhao K. (2013) Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat. Immunol, 14, 1190–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang P., Xue Y., Han Y., Lin L., Wu C., Xu S., Jiang Z., Xu J., Liu Q., Cao X. (2014) The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science, 344, 310–313. [DOI] [PubMed] [Google Scholar]
- 50.Michalik K.M., You X., Manavski Y., Doddaballapur A., Zornig M., Braun T., John D., Ponomareva Y., Chen W., Uchida S. et al. (2014) Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ. Res, 114, 1389–1397. [DOI] [PubMed] [Google Scholar]
- 51.Bell R.D., Long X., Lin M., Bergmann J.H., Nanda V., Cowan S.L., Zhou Q., Han Y., Spector D.L., Zheng D. et al. (2014) Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler. Thromb. Vasc. Biol., 34, 1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carpenter S., Aiello D., Atianand M.K., Ricci E.P., Gandhi P., Hall L.L., Byron M., Monks B., Henry-Bezy M., Lawrence J.B. et al. (2013) A long noncoding RNA mediates both activation and repression of immune response genes. Science, 341, 789–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ilott N.E., Heward J.A., Roux B., Tsitsiou E., Fenwick P.S., Lenzi L., Goodhead I., Hertz-Fowler C., Heger A., Hall N. et al. (2015) Corrigendum: Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat. Commun., 6, 6814.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun L., Goff L.A., Trapnell C., Alexander R., Lo K.A., Hacisuleyman E., Sauvageau M., Tazon-Vega B., Kelley D.R., Hendrickson D.G. et al. (2013) Long noncoding RNAs regulate adipogenesis. Proc. Natl. Acad. Sci. USA, 110, 3387–3392., [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao T., Liu L., Li H., Sun Y., Luo H., Li T., Wang S., Dalton S., Zhao R.C., Chen R. (2015) Long Noncoding RNA ADINR Regulates Adipogenesis by Transcriptionally Activating C/EBPalpha. Stem Cell Rep., 5, 856–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao X.Y., Li S., Wang G.X., Yu Q., Lin J.D. (2014) A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol. Cell, 55, 372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alvarez-Dominguez J.R., Bai Z., Xu D., Yuan B., Lo K.A., Yoon M.J., Lim Y.C., Knoll M., Slavov N., Chen S. et al. (2015) De Novo Reconstruction of Adipose Tissue Transcriptomes Reveals Long Non-coding RNA Regulators of Brown Adipocyte Development. Cell Metab., 21, 764–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guil S., Esteller M. (2012) Cis-acting noncoding RNAs: friends and foes. Nat. Struct. Mol. Biol., 19, 1068–1075. [DOI] [PubMed] [Google Scholar]
- 59.Pal A., Barber T.M., Van de Bunt M., Rudge S.A., Zhang Q., Lachlan K.L., Cooper N.S., Linden H., Levy J.C., Wakelam M.J. et al. (2012) PTEN mutations as a cause of constitutive insulin sensitivity and obesity. N. Engl. J. Med, 367, 1002–1011., [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qian J., Nunez S., Reed E., Reilly M.P., Foulkes A.S. (2016) A Simple Test of Class-Level Genetic Association Can Reveal Novel Cardiometabolic Trait Loci. PLoS One, 11, e0148218.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu J.Z., McRae A.F., Nyholt D.R., Medland S.E., Wray N.R., Brown K.M., Investigators A., Hayward N.K., Montgomery G.W., Visscher P.M. et al. (2010) A versatile gene-based test for genome-wide association studies. Am. J. Hum. Genet., 87, 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luo L., Peng G., Zhu Y., Dong H., Amos C.I., Xiong M. (2010) Genome-wide gene and pathway analysis. Eur. J. Hum. Genet., 18, 1045–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karro J.E., Yan Y., Zheng D., Zhang Z., Carriero N., Cayting P., Harrrison P., Gerstein M. (2007) Pseudogene.org: a comprehensive database and comparison platform for pseudogene annotation. Nucleic Acids Res., 35, D55–D60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A.U., Wheeler E., Glazer N.L., Bouatia-Naji N., Gloyn A.L. et al. (2010) New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet., 42, 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segre A.V., Steinthorsdottir V., Strawbridge R.J., Khan H., Grallert H., Mahajan A. et al. (2012) Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet., 44, 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schunkert H., Konig I.R., Kathiresan S., Reilly M.P., Assimes T.L., Holm H., Preuss M., Stewart A.F., Barbalic M., Gieger C. et al. (2011) Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet., 43, 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reilly M.P., Li M., He J., Ferguson J.F., Stylianou I.M., Mehta N.N., Burnett M.S., Devaney J.M., Knouff C.W., Thompson J.R. et al. (2011) Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet, 377, 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saleheen D., Zaidi M., Rasheed A., Ahmad U., Hakeem A., Murtaza M., Kayani W., Faruqui A., Kundi A., Zaman K.S. et al. (2009) The Pakistan Risk of Myocardial Infarction Study: a resource for the study of genetic, lifestyle and other determinants of myocardial infarction in South Asia. Eur. J. Epidemiol., 24, 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J. et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Genomes Project, C., Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature, 491, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ulitsky I., Shkumatava A., Jan C.H., Sive H., Bartel D.P. (2011) Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell, 147, 1537–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu Y., Ferguson J.F., Xue C., Ballantyne R.L., Silverman I.M., Gosai S.J., Serfecz J., Morley M.P., Gregory B.D., Li M. et al. (2014) Tissue-specific RNA-Seq in human evoked inflammation identifies blood and adipose LincRNA signatures of cardiometabolic diseases. Arterioscler. Thromb. Vasc. Biol., 34, 902–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.