Abstract
Alzheimer’s disease (AD) is a chronic neurodegenerative disorder characterized by progressive deterioration of cognitive function. Pathogenesis of AD is incompletely understood; evidence suggests a role for epigenetic regulation, in particular the cytosine modifications 5-methylcytosine and 5-hydroxymethylcytosine (5hmC). 5hmC is enriched in the nervous system and displays neurodevelopment and age-related changes. To determine the role of 5hmC in AD, we performed genome-wide analyses of 5hmC in DNA from prefrontal cortex of post-mortem AD patients, and RNA-Seq to correlate changes in 5hmC with transcriptional changes. We identified 325 genes containing differentially hydroxymethylated loci (DhMLs) in both discovery and replication datasets. These are enriched for pathways involved in neuron projection development and neurogenesis. Of these, 140 showed changes in gene expression. Proteins encoded by these genes form direct protein–protein interactions with AD-associated genes, expanding the network of genes implicated in AD. We identified AD-associated single nucleotide polymorphisms (SNPs) located within or near DhMLs, suggesting these SNPs may identify regions of epigenetic gene regulation that play a role in AD pathogenesis. Finally, using an existing AD fly model, we showed some of these genes modulate AD-associated toxicity. Our data implicate neuronal projection development and neurogenesis pathways as potential targets in AD. By incorporating epigenomic and transcriptomic data with genome-wide association studies data, with verification in the Drosophila model, we can expand the known network of genes involved in disease pathogenesis and identify epigenetic modifiers of Alzheimer’s disease.
Introduction
Alzheimer’s disease (AD) is the most common neurodegenerative disease and the leading cause of dementia (1). The essential clinical feature of AD is a progressive decline in memory and other cognitive abilities (2). The neuropathological hallmarks of AD are extracellular amyloid plaques, intracellular neurofibrillary tangles and selective neuronal loss in vulnerable regions of the brain (2). Neurons located in medial temporal lobe and areas of the temporal, parietal and frontal neocortex are particularly vulnerable. Genetic, biochemical, and neuropathological studies implicate the aggregation of beta-amyloid (Aβ, the main component of amyloid plaques) as a central process of AD pathogenesis (3). The majority of AD cases begin after the age of 65 and are known as late-onset or sporadic AD (4). While the risk of sporadic AD has been associated with Apolipoprotein E (APOE) and a growing number of single nucleotide polymorphisms (SNPs) in more than 20 loci identified by genome-wide association studies (GWAS), the exact causes of sporadic AD remain unknown (5). While late-onset AD is largely (∼70%) heritable, with the best-characterized risk allele for AD, APOE4, accounting for a small proportion (∼4%) of heritability, it has been suggested that the remaining risk may be due to alterations in epigenetic processes (6–9).
Cytosine modifications are one of the three widely recognized epigenetic marks (8,9). Methylation of the fifth position of cytosine (5-methylcytosine; 5mC) is the best characterized and is known to be involved in the regulation of gene expression (10). Recently, the importance of 5-hydroxymethylcytosine (5hmC) was recognized when two independent research teams demonstrated the presence of 5hmC in mouse Purkinje neurons and embryonic stem cells (11,12). 5hmC is generated by conversion of 5mC by the ten-eleven translocation (TET) protein family and is recognized by specific 5hmC-binding proteins (13,14)
While 5mC levels are similar among cell types, the overall level of 5hmC varies between different tissues and cell types (15,16). Specifically, 5hmC is highly enriched in the central neural system (CNS), with levels ∼10-fold higher than those in embryonic stem cells (17). Our group and others have characterized specific distributions of 5hmC across the genome in the CNS and shown developmental- and age-related changes (16,18–23). Taken together, the high levels of 5hmC in the CNS, the identification of specific 5hmC binding proteins, and the specific distribution of 5hmC within the genome suggest that the patterns of 5hmC acquired throughout neuronal development are critical for proper neurodevelopment and neurological function in the adult brain. Thus, dysregulation of 5hmC due to aging or environmental stressors may play a role in age-related neurodegenerative diseases, such as AD (7,24–26). While many studies have examined alterations in 5mC in post-mortem AD brain tissue, few have explored the role of 5hmC (9,25,27–32). Thus, we sought to map genome-wide changes in 5hmC in late-onset AD and compare these changes to gene expression measured in these same samples. The analyses reported here demonstrate that combining epigenomic, transcriptomic and GWAS data can reveal valuable information about disease pathways. This study implicates pathways involved in neuron projection development and neurogenesis pathways as potential targets in AD.
Results
Identification and characterization of differentially hydroxymethylated loci
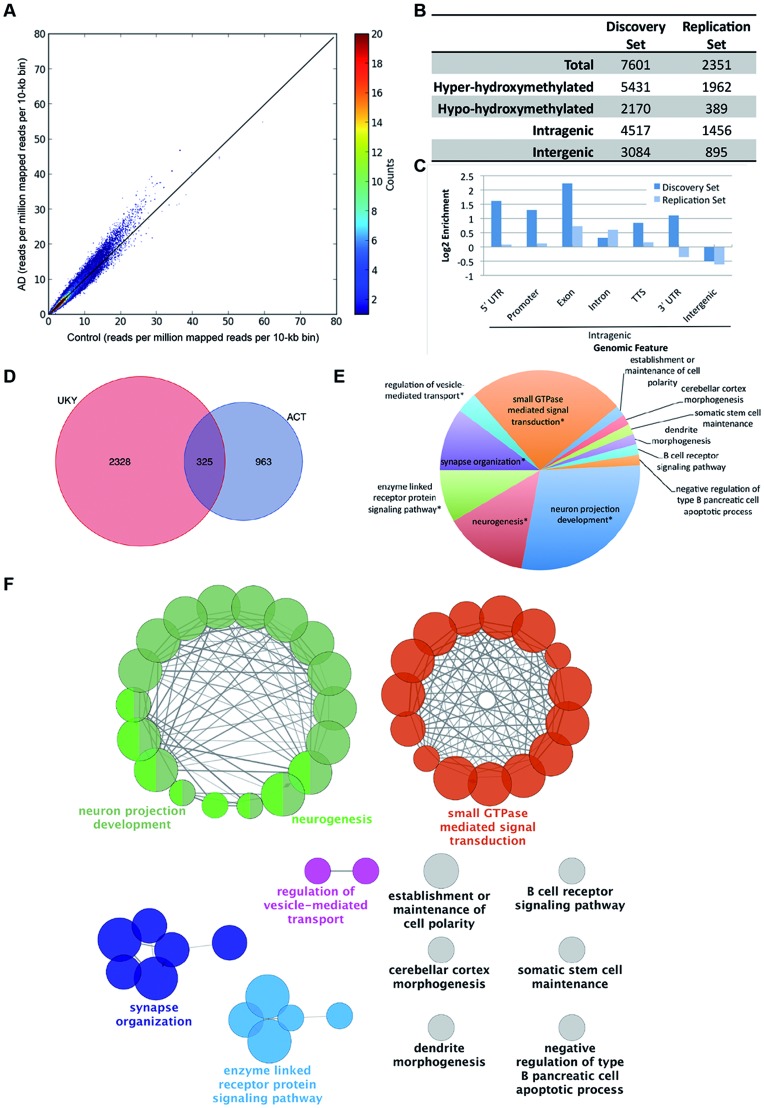
To identify differentially hydroxymethylated regions, we isolated genomic DNA from prefrontal cortical tissues from patients with significant Alzheimer’s pathology according to established criteria, and control patients of similar age with no history of neurological illness and no significant neuropathology from a discovery set and a replication set (Supplementary Material, Table S1) (33). We generated genome-wide 5hmC profiles using a previously established chemical labeling and affinity purification method, followed by high-throughput sequencing (18). High-throughput sequencing resulted in a range from 8.2 to 25.6 million non-duplicated reads. Unique, non-duplicated reads with no more than two mismatches in the first 25 bp were aligned to the human genome (hg19) (34). Genome-wide patterns of 5hmC were evaluated by counting mapped reads per 10-kb bin, normalized to sequencing coverage, demonstrating a global increase in 5hmC in AD compared to control (Fig. 1A). Differentially hydroxymethylated loci (DhMLs) were identified separately within each dataset with the software diffReps, using the specified settings (window = 1000, step = 100, fragment = 300) (35). Unlike peak callers, diffReps uses a sliding window-based approach to detect differential modification sites, natively handles multiple samples, and provides quantitative and statistical information about the size and significance of these differences (35). In the discovery set, 7601 DhMLs were identified (5431 showing hyper-hydroxymethylation in AD and 2170 showing hypo-hydroxymethylation in AD) (Fig. 1B, Supplementary Material, Table S2). Fewer DhMLs were identified in the replication set, likely due to the smaller number of samples; 2351 DhMLs were identified in this set (1962 hyper-hydroxymethylated and 389 hypo-hydroxymethylation in AD) (Fig. 1B, Supplementary Material, Table S3).
Figure 1.
Identification and characterization of DhMLs. (A) Scatterplot of normalized genome-wide 5hmC reads within each 10-kb bin. Reads were normalized to coverage. Color of each point shows the number of bins with the given number of reads. (B) Characteristics of DhMLs from each dataset identified with diffReps. (C) Annotation of DhMLs by Homer shows enrichment and depletion of DhMLs by genomic feature. (D) Genes containing DhMLs were identified by Homer annotation for each dataset, and 325 genes were identified in both datasets. (E) The number of gene ontology terms assigned to significantly enriched gene ontology term groups for DhML-containing genes identified by ClueGO. Asterisks indicate that the displayed term is the most significant gene ontology term in a related group of gene ontology terms. (F) Gene ontology network of shared genes. For simplicity, only the most significant gene ontology term for each group is shown.
Next, we annotated each set of DhMLs to the genome (hg19) using Homer (36). Homer assigns each specified region to genomic features. This analysis showed that, in both datasets, ∼60% of the DhMLs are located within genes (Fig. 1B, Supplementary Material, Tables S4 and S5). A more detailed breakdown of these features is shown in Figure 1C. DhMLs are enriched in intragenic regions and depleted in intergenic regions. Thus, we focused the remainder of our analysis on DhMLs located in intragenic regions. Homer annotated these intragenic DhMLs to 2653 genes in the discovery set and 1288 genes in the replication set. Of these genes, 325 were identified in both datasets, and we used these for further analysis (Fig. 1D, Supplementary Material, Table S6).
False discovery rates are provided in diffReps as a Benjamini–Hochberg correction (37). For the default P-value cutoff (P < 1E-4) using the negative binomial test, the false discovery rate for diffReps is 0.2 Applying more stringent cutoff of 0.1 reduced the number of identified DhMLs by 10 for the discovery set and 4 for the replication set. A further reduction to 0.05, reduce the number of identified DhMLs a further 12 and 18 DhMLs from the discovery and replication sets, respectively. In addition, by focusing the analysis on genes identified in two independent sets, we increase the likelihood that the regions identified for further analysis are not false positives.
Gene ontology analysis
We performed a gene ontology enrichment analysis with ClueGO, a gene ontology analysis plug-in for Cytoscape, a software platform for network analysis, that visualizes non-redundant gene ontology terms for large gene clusters and presents the data as a functionally grouped network that groups enriched gene ontology terms based on the similarity of associated genes within those terms (38–40). We used the gene ontology terms in the Biological Process ontology, with the total number of genes associated with all terms in this source used as reference. This analysis revealed that the most highly enriched network of gene ontology terms included biological processes related to neuron projection development and neurogenesis (Fig. 1E and F). Other enriched groups include enzyme-linked receptor protein signaling, synapse organization and regulation of vesicle-mediated transport. For clarity, only the group leading term (most significant term in each group) is indicated on the figure. The individual terms and genes within these groups are listed in Supplementary Material, Table S7 and genes associated with the most enriched terms are shown in Table 1. Enriched terms that were not grouped with related terms include: the establishment of cell polarity, cerebellar cortex morphogenesis and dendrite morphogenesis (Fig. 1F). Together, these terms represent pathways involved in neuronal morphology and synaptic function.
Table 1.
Genes associated with most significantly enriched GO terms
| GO Terms | Genes |
|---|---|
| Group leading term | |
| Neurogenesis | ABLIM1, AGRN, ANK1, AP2A2, ARHGAP35, ARHGAP39, ARHGEF12, ASIC2, ATP8A2, ATXN2, BIN1, BMP7, CACNA1A, CACNA1S, CACNB2, CACNB4, CAMK1D, CDH4, CLASP1, COL4A2, COL5A1, COL9A3, CTNNA1, CTNNA2, CUX1, DCLK1, DSCAML1, EPHA8, EPHB2, FZR1, GLI2, GPR56, KIRREL3, KNDC1, LDB2, LLGL1, LYN, MAGI2, MAPK1, MOB2, MYO10, NCK2, NFIB, NGF, NRXN3, PAK2, PARD3, PCNT, PRDM16, PTPRG, PTPRM, RAPGEF1, RBFOX2, SLIT1, SLIT3, SPTAN1, SPTBN1, SYNDIG1, TCF12, TCF7L2, TIAM1, TRAF3, VAV2, WNT9A, YWHAE, ZHX2 |
| Enzyme-linked receptor protein signaling | ADRBK1, ADRBK1, AFAP1L2, AGRN, AP2A2, ARHGEF12, ARHGEF18, BIN1, BMP7, CAMK1D, CDH13, COL4A2, DUSP22, EPHA8, EPHB2, FGFRL1, GATA5, GNG7, INPP5D, INSR, ITPR2, ITSN1, KALRN, LYN, MAGI2, MAPK1, MVB12B, MYOF, NCK2, NEDD4L, NGF, NOS1, NQO1, OBSCN, PARD3, PCSK6, PDZD2, PRDM16, PRKAG2, PRKCB, PRKCZ, PTPRG, PTPRT, RAPGEF1, RPTOR, SOGA1, SPTBN1, TCF7L2, TIAM1, TNFRSF8, VAV2, YWHAE, ZBTB16 |
| Synapse organization | AGRN, ASIC2, AP2A2, APBA2, ARF1, EPHB2, CACNA1A, CACNA1S, CACNB2, CACNB4, CTNNA2, LDB2A2, CTNND2, DLGAP2, NRXN3, FZR1, GNG7, ITSN1, KCNAB1, KCNIP1, LLGL1, MAPK1, NGF, NOS1, NQO1, SYNDIG1, PRKCB, PRKCZ, SEPT9, SHANK2, SLIT1, SLIT3, SYN3 |
| Regulation of vesicle-mediated transport | ARF1, ATXN2, BIN1, CACNA1A, CAMK1D, CD300A, CDH13, HIP1, LLGL1, LYN, MAGI2, MAPK1, NEDD4L, RPH3AL, SEPT9 |
| Small GTPase-mediated signal transduction | ABHD12, AGRN, ARF1, ARHGAP35, ARHGAP39, ARHGEF12, ARHGEF18, ASAP2, ATP8B1, BCAR3, BIN1, CACNB4, CDH13, DNMT3A, GNG7, GPR56, ICAM1, IMPA2, INSR, IQSEC3, ITSN1, KALRN, KNDC1, LLGL1, MACF1, MACROD1, MAPK1, MTHFR, MYO10, NGF, NOS1, OBSCN, PARK2, PDE10A, PDE6B, PDZD2, PGAM5, PLCG2, PRKAG2, PTPRN2, RAB31, RAB40B, RAB6B, RAD51B, RAP1GAP2, RAPGEF1, RAPGEF5, RASA3, RASGEF1A, RGS10, RREB1, SEPT9, SHMT1, SIPA1L3, SLC25A25, SOGA1, SPATA13, TBC1D14, TIAM1, TRAF3, USP8, UVRAG, VAV2, WRN |
| Ungrouped GO terms | |
| Establishment or maintenance of cell polarity | ANK1, CLASP1, CTNNA1, FRMD4B, LLGL1, MACF1, MAP4, MAP7, PARD3, PRKCZ, TRAF3 |
| Cerebellar cortex morphogenesis | ATXN2, CACNA1A, GLI2, KNDC1, LDB2, PCNT |
| Somatic stem cell maintenance | BMP7, LDB2, LRP5, PRDM16, TCF7L2, ZHX2 |
| Dendrite morphogenesis | CACNA1A, CTNNA2, CUX1, DCLK1, EPHB2, KNDC1, LLGL1, RBFOX2 |
| B cell receptor signaling pathway | CD300A, LYN, MAPK1, NFATC2, PLCG2, PRKCB |
| Negative regulation of type B pancreatic cell apoptotic process | ERC2, NGF, TCF7L2 |
AD-associated SNPs located in or near DhMLs
In addition to identifying DhMLs associated with changes in gene expression, we also determined whether any disease-associated SNPs were located in or near DhMLs. We used SNPs from the International Genomics of Alzheimer’s Project (IGAP) to identify AD-associated SNPs that are located within 50 kb of regions of altered hydroxymethylation (41). We chose 50 kb upstream and downstream of each SNPs to cover a range likely to be on the same haplotype block. We performed this analysis for the discovery and replication datasets separately, and then we identified genes that were contained in both datasets. We considered all DhMLs for this analysis, both intergenic and intragenic. In the University of Kentucky dataset, there were 277 AD-associated SNPs, representing 33 genes, within 50 kb of DhMLs (Supplementary Material, Table S8); in the Emory University dataset, there were 85 SNPs representing 16 genes (Supplementary Material, Table S9). Of these, 49 SNPs and 9 genes were shared (Table 1). These genes are listed in Table 2, along with the number of disease-associated SNPs identified in this analysis and the most significant of those SNPs as reported by IGAP. To visualize this, we used the WashU Epigenome Browser to map the DhMLs and that the identified DhMLs are located in areas of known epigenetic modifiers that are also associated with AD by GWAS (Fig. 2) (42). The diagrams of each identified genomic region in Figure 2 show the location of each DhML and each SNP in relation to block of linkage disequilibrium in the relevant genomic interval, as well as known areas of epigenetic regulation (ChromHMM and RRBS tracks). This analysis suggests that disease associated SNPs may tag regions of epigenetic regulation that may be important for disease pathogenesis and may help uncover the functional consequences of these disease-associated variants.
Table 2.
Genes and AD-associated SNPs within 50 kb of DhMLs
| Gene symbol | Gene name | Chr | No. of SNPs | SNP | P-value | Function |
|---|---|---|---|---|---|---|
| BIN1 | Bridging integrator 1 | 2 | 20 | rs35114168 | 3.72E-16 | Adapter protein involved with synaptic vesicle endocytosis |
| INPP5D | Inositol polyphosphate-5-phosphatase | 2 | 1 | rs10933431 | 6.62E-06 | Nuclear inositol phosphate signaling processes |
| CELF1 | CUGBP, elav-like family member 1 | 11 | 8 | rs66749409 | 3.24E-06 | mRNA alternative splicing |
| MADD | MAP-kinase activating death domain | 11 | 2 | rs7944584 | 3.05E-06 | Apoptosis |
| MYBPC3 | Myosin binding protein C, cardiac | 11 | 3 | rs2071305 | 2.24E-06 | Myosin-associated protein |
| NDUFS3 | NADH dehydrogenase (ubiquinone) Fe-S protein 3 | 11 | 3 | rs71475924 | 1.44E-06 | Core subunit of mitochondrial complex I |
| PTPMT1 | Protein tyrosine phosphatase, mitochondrial 1 | 11 | 5 | rs12798346 | 5.20E-06 | Dephosphorylates mitochondrial lipids |
| SPI1 | Spi-1 proto-oncogene | 11 | 5 | rs3740686 | 3.20E-06 | Transcription factor involved in myeloid and B-lymphoid cell development |
| FCF1 | FCF1 RRNA-processing protein | 14 | 2 | rs12883118 | 3.58E-07 | pre-rRNA processing and 40S ribosomal subunit assembly |
Genes annotated to AD-associated SNPs were within 50 kb of DhMLs from both the Discovery set and Replication set are shown. The number of AD-associated SNPs identified within these regions associated with each gene is indicated. The SNP with highest significance according to IGAP is reported.
Figure 2.
Genomic regions containing DhMLs and AD-associated SNPs. (A) Genomic loci containing (A) BIN1; (B) MADD, MYBPC3, SPI1, CELF1, PTPMT1 and NDUFS3; (C) INPP5D and (D) FCF1. Shown in each panel are, from top to bottom: 1) ChromHMM analysis and 2) RRBS results from a Roadmap Epigenomics Project reference sample (neuronal nuclei of frontal cortex from post-mortem brain tissue of an 81-year-old male). The legend for colors of the ChromHMM track is shown at the bottom of the figure. 3) The DhMLs identified in this analysis. Blue represents loss of 5hmC in AD; red represents a gain. Shading represents the fold change (with darker colors representing larger changes). 4) AD-associated SNPs located within 50 kb of the identified DhMLs. 4) Blocks of linkage disequilibrium for the CEU population generated by the HapMap project. 5) The RefSeq gene or genes located in the visualized regions.
Proteins encoded by genes with altered 5-hydroxymethylation form a protein–protein interaction network with an AD-disease network
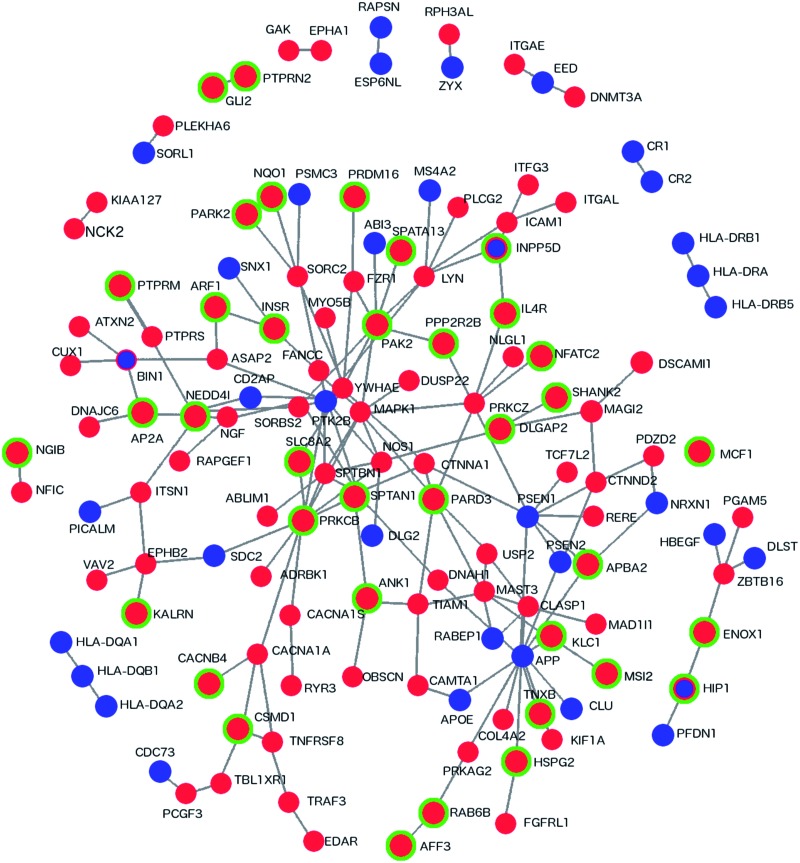
To determine whether the 325 DhML-containing genes are functionally related to previously discovered AD-associated loci, we generated a protein–protein interaction (PPI) network with DAPPLE (43). DAPPLE uses PPI information from the database InWeb to identify direct and indirect (up to 1 non-specified protein) interactions between proteins in the supplied dataset (44). First, we assembled a list of monogenic AD-related genes (APP (amyloid precursor protein), Presenilin 1 and Presenilin 2), as well as AD-susceptibility loci (APOE and genes identified by the IGAP), and generated a PPI network with DAPPLE. We then added the 325 DhML-containing genes and performed PPI analysis with DAPPLE (Fig. 3). This network includes 35 of 114 AD genes and 106 of 325 DhML-containing genes. A network with this degree of connectivity with this number of genes is unlikely to occur by chance (P = 0.001). Adding DhML-containing genes to this network provides a new degree of interconnectivity in the AD-susceptibility network, providing connections between unconnected groups of proteins. The network module most expanded by incorporating 5hmC data is the Aβ production-related module surrounding APP. Thus, this analysis demonstrates that incorporating genome-wide 5hmC data can identify new genes in the network of genes involved in AD pathogenesis.
Figure 3.
Direct PPIs between DhML-containing genes and an AD susceptibility network. Direct PPI network between genes identified as containing intragenic DhMLs in the current analysis and AD susceptibility genes generated by DAPPLE (P = 0.001). Red indicates that a gene contains a DhML. Green indicates a significant result by RNA-Seq. Blue indicates an AD susceptibility gene.
Genes with altered 5hmC show changes in gene expression
To assess whether these changes in hydroxymethylation affected gene expression, we also analyzed the RNA-Seq data that we published previously (45). After alignment to the transcriptome by TopHat and the genome by Bowtie, differential expression was determined by CuffLinks (34,46,47). Of the 325 genes, 140 also showed significant changes in expression (Supplementary Material, Tables S10 and S11). Analysis with GeneOverlap, an R package that tests the significance of overlap between two sets of genes, indicates that genes with alterations in 5hmC are 3 times more likely to also show changes in expression (P = 4.2 × 10−21) (48). There is no consistent pattern in our data of increased or decreased 5hmC influencing the direction of gene expression changes. Many of the identified genes contain multiple DhMLs in different locations, sometimes in opposing directions (Supplementary Material, Table S13). Further studies in human cells would be required to elucidate mechanism by which these epigenetic modifiers regulate gene expression. In addition, we overlaid the RNA-Seq results on the PPI network in Figure 3 and found that 38 of these 140 genes are a part of this network.
5hmC has been proposed to play a role in RNA splicing, since there are tissue-specific distributions of 5hmC at exon–intron boundaries (23). Thus, we identified genes containing DhMLs that also show significant changes in splicing as detected by RNA-Seq. Three genes showed significant differences in splicing: intersectin 2 (ITSN2), ecto-NOX disulfide-thiol exchanger 1 and protein tyrosine phosphatase receptor mu (PTPRM). ITSN2 is an SH3 domain-containing protein involved in clathrin-mediated endocytosis. Members of the ecto-NOX family are involved in plasma membrane electron transport. PTPRM is involved in cell migration, and more specifically with neurite outgrowth in neurons (Supplementary Material, Table S10).
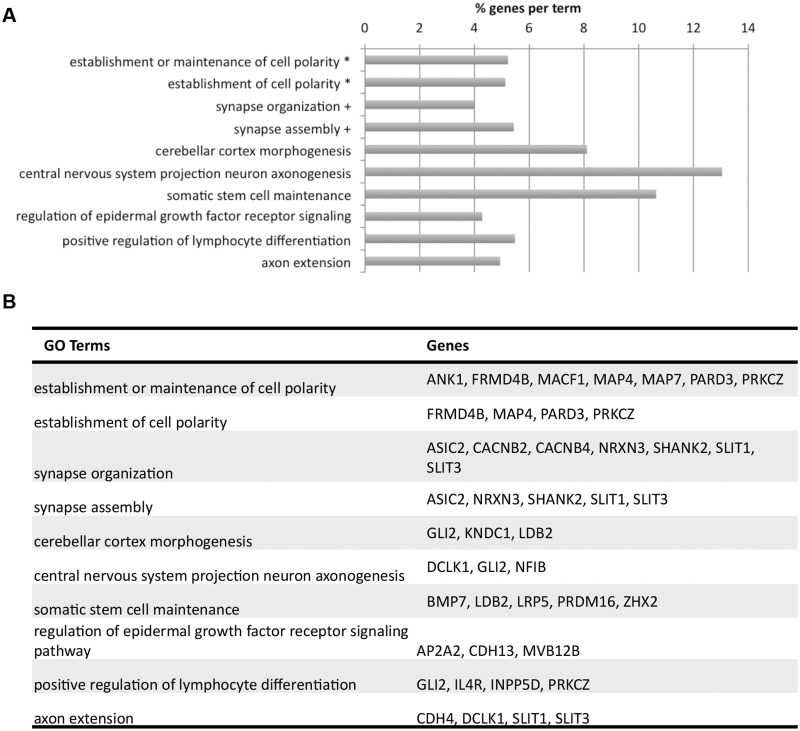
To determine if these 140 genes are involved in common pathways, we again performed gene ontology analysis with ClueGO on this subset of genes (38) (Fig. 4, Supplementary Material, Table S11). As expected, the significantly enriched gene ontology terms represent a subset of terms from the analysis of the previous gene set (Figs 1E and 4A). The most significantly enriched gene ontology term groups are the establishment or maintenance of cell polarity and synapse organization, with other enriched terms including axonogenesis, axon extension, somatic stem cell maintenance and cerebellar cortex morphogenesis. The specific genes in each of these groups is shown in Figure 4B.
Figure 4.
Enriched gene ontology terms for 140 DhML-containing genes with significant RNA-Seq results. (A) Symbols indicate terms that have been grouped. Bar length indicates the % of genes within each gene ontology term that are in the dataset. (B) Specific genes assigned to each GO term.
Drosophila orthologs of DhML-containing genes modify Tau-induced neurotoxicity
To verify that DhML-containing genes are functionally relevant to AD pathogenesis, we tested Drosophila orthologs of these genes for their ability to modify Tau-induced neurotoxicity. There are several Drosophila experimental models relevant to AD, including transgenic systems based on the neurotoxicity of both Aβ and Tau (49,50). For functional screening, we selected the Tau transgenic model because it has been successfully employed for rapid genetic screening (51), and there is growing consensus that Tau is a downstream mediator of Aβ toxicity in AD (52,53). Expression of human Tau in the fly nervous system recapitulates several features of AD, including age-dependent neurodegeneration, decreased lifespan and abnormally phosphorylated and misfolded Tau (49). Importantly, wild-type and mutant forms of human Tau demonstrate similar mechanisms of toxicity when expressed in the Drosophila nervous system and show consistent interactions with known genetic modifiers (49,54). Therefore, the fly model selected for our study is relevant to understanding the mechanisms of Tau toxicity in AD.
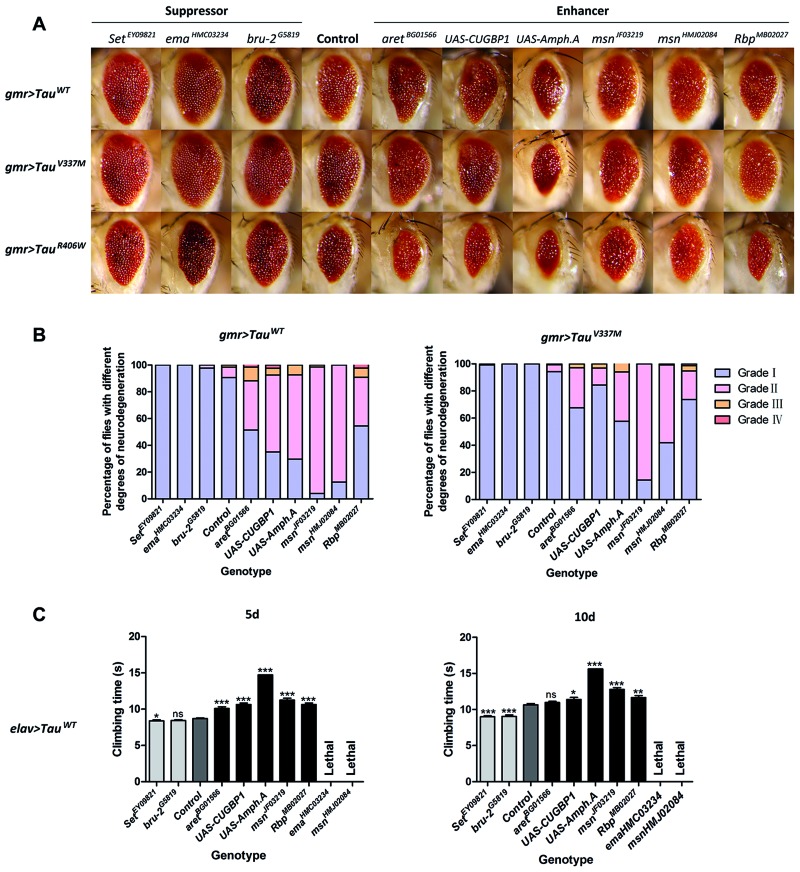
Expression of human wild-type or mutant Tau in the fly eye using gmr-GAL4 drivers causes a moderately reduced eye size and roughened surface, a phenotype that is amenable to rapid screening for second-site genetic modifiers. Specifically, by scoring for lines that either exacerbate or rescue the eye phenotype, genes can be characterized as enhancers or suppressors of Tau toxicity, respectively. Drosophila orthologs of 15 candidate genes that associated with the DhMLs close to AD-associated SNPs were investigated in the fly model of Tau-mediated neurotoxicity. Three suppressors and six enhancers were identified by evaluating eye phenotype. Overexpression of bru-2 (CELF1) and Set (TSPYL5) and a loss-of-function allele of ema (CLEC16A) rescued reduced eye size from 50% to 70–80% of normal size in flies expressing wild-type Tau. Reciprocally, overexpression of human CUGBP1 (CELF1), Amph (BIN1) and Rbp (BZRAP1) and reduced expression of aret (another Drosophila ortholog of CELF1) or msn (MINK1) further disrupted eye morphology, with smaller eye size and more frequent appearance of loss-of-pigmentation, retinal collapse and necrosis. On its own, dysregulation of these genes showed no effect on eye morphology, except for human CUGBP1, with an induced rough eye phenotype. Parallel experiments in flies expressing mutant Tau (V337M and R406W) showed consistent results (Table 3; Fig. 5A and B) while the mis-expression of Tau (R406W), one of the most toxic forms of Tau, in the fly eye decreased viability with much less progeny flies. We next tested whether these genes could modulate the Tau-induced motor deficits reported previously (51). Similarly, the modulation of tested alleles on the climbing deficits caused by the expression of Tau is consistent with our analyses of eye phenotype, except for two alleles that led to the lethality of both Tau and control flies, without the emergence of adult progeny flies (Table 3 and Fig. 5C).
Table 3.
Modifiers of Tau toxicity in Drosophila
| Human gene | Drosophila ortholog | Loss-of-function allele | Overexpression allele | Modification on Tau toxicity | |
|---|---|---|---|---|---|
| Eye evaluation | Climbing assay | ||||
| CELF1 (CUGBP1) | aret | aretBG01566 | Enhancer | Enhancer | |
| bru-2 | bru-2G5819 | Suppressor | Suppressor | ||
| UAS-CUGBP1 | Enhancer | Enhancer | |||
| BIN1 | Amph | UAS-Amph.A | Enhancer | Enhancer (reduced viability) | |
| MINK1 | msn | msnJF03219 | Enhancer | Enhancer | |
| msnHMJ02084 | Enhancer | Lethal | |||
| BZRAP1 | Rbp | RbpMB02027 | Enhancer | Enhancer | |
| CLEC16A | ema | emaHMC03234 | Suppressor | Lethal | |
| TSPYL5 | Set | SetEY09821 | Suppressor | Suppressor | |
ANK1, Ankryin 1; DIP2A, Disco interacting protein 2 homolog A.
Figure 5.
Identification of epigenetic modifiers of Tau-mediated neurotoxicity using an AD fly model. (A) Eye phenotype of 5-day-old flies expressing both Tau (wild-type or mutant) and modifiers. (B) Percentage bar chart of eye phenotypes. Data show percentage of flies divided into four grades (I, II, III and IV) on the basis of eye phenotypes. (C) Comparison of climbing ability of 5-day-old and 10-day-old Tau WT flies with the presence or absence of modifiers. Data show mean climbing time ± SEM (Student’s t-test or Mann–Whitney test (*** = P < 0.0001, ** = P < 0.01, * = P < 0.05, ns = no significant difference). Details of all the indicated genotypes in this figure are described in Supplementary Material.
Among these modifiers, we found that the loss-of-function allele of aret enhanced the Tau-induced rough eye phenotype and deficits in climbing ability, which is consistent with previous findings (51). Furthermore, the overexpression of Amph enhanced Tau-dependent neurodegeneration, and overexpression of both Tau and Amph was lethal in most flies. Sixteen crosses were conducted, and only 21 female and 11 male progeny flies were collected. In the climbing assay, overexpression of Amph causes defects in its own climbing ability and exacerbates the Tau-induced deficit (Fig. 5). This is consistent with a previous report that reduced expression of Amph suppresses Tau-mediated neurotoxicity in the eye phenotype, notal bristle number and mushroom body morphology (55).
Discussion
In our current analysis, we identified AD DhMLs in two independent datasets. These DhMLs are enriched in intragenic regions, with 325 genes that contain DhMLs in both datasets. Most of these 325 DhML-containing genes have never before been implicated in AD pathogenesis. Only 11 of these intragenic DhMLs are located in or near genes at AD susceptibility loci identified by GWAS. The most highly enriched gene ontology terms for these 325 genes are for biological processes involved in neuron projection development and neurogenesis (Fig. 1F, Supplementary Material, Table S7). Additional enriched groups include enzyme-linked receptor protein signaling, synapse organization and regulation of vesicle-mediated transport; ungrouped enriched terms include the establishment of cell polarity, cerebellar cortex morphogenesis and dendrite morphogenesis (Fig. 1E and Table 1). The 140 genes of these 325 that also show changes in gene expression are enriched for a subset of these gene ontology terms: the establishment or maintenance of cell polarity, synapse organization, axonogenesis, axon extension, somatic stem cell maintenance and cerebellar cortex morphogenesis (Fig. 4). Together, these enriched gene ontology terms represent pathways involved in the development and maintenance of neuronal morphology and synaptic function, highlighting the potential importance of these pathways in AD and suggesting a need for further study of the roles in AD pathogenesis of specific DhML-containing genes with changes in expression in these gene ontology groups (Fig. 4B).
Of the proteins encoded by the 325 DhML-containing genes, 106 interact with a PPI network of 35 proteins encoded by known AD genes and expand this AD susceptibility network; 43 of these also show changes in expression (Fig. 3). Specifically, the Aβ production-related module is the most expanded part of the network. Thus, our analysis has identified genes not previously implicated in AD pathogenesis that interact with a network of genes known to be involved in AD, demonstrating that these 106 genes, with priority on the 43 that showed changes in expression, are candidates for further in-depth study of their role in AD pathogenesis. Interestingly, two recent analyses of differential methylation in AD identified genes with altered methylation that are correlated with neuropathology in AD (31,32). Our analysis identified two genes (Ankryin 1 and Disco interacting protein 2 homolog A) that were also uncovered by these methylation analyses. That we identified overlapping but distinct sets of genes indicates it is important to explore both 5mC and 5hmC changes in AD. The success of this analysis of 5hmC and the analyses of 5mC by other groups in AD suggests that the incorporation of genome-wide epigenetic data is a useful addition to identify mechanisms of disease pathology (31,32). This analysis also identified 49 AD susceptibility SNPs identified by GWAS in nine loci within 50 kb upstream and downstream of DhMLs (Table 2) (41). For most of these loci, the functional variants are not known (5,56). Our analysis suggests that these SNPs are tagging regions of epigenetic regulation importance in AD. Together, these results suggest that the functional variants within these risk loci may affect gene expression and disease risk by altering cytosine modifications within that locus.
Disease-related changes can occur at multiple levels, including DNA, RNA and protein. DNA methylation or hydroxymethylation may contribute to disease risk through pathways other than gene expression, for example, by affecting DNA and/or chromatin stability, or by interacting with other as yet unknown pathways. As such, a model system is ideal to investigate the overall functional consequence of molecular dysfunction at multiple levels. Drosophila has emerged as a useful model in genetic studies of AD due to the striking functional and structural similarities (known as orthologs) between humans and Drosophila (57–59). For instance, nearly 70% of disease-causing genes in humans have orthologs in Drosophila, including the APP ortholog (59,60). Moreover, fly orthologs associated with known AD genes exhibit functional conservation (61). In this study, we used a Drosophila AD model to rapidly test the roles of fly orthologs of DhML-containing genes also associated with AD by GWAS to determine if altered expression of these genes can modify processes involved in AD pathogenesis. We confirmed that these AD-associated genes with altered 5-hydroxymethylation modify Tau-mediated neurotoxicity in Drosophila (Fig. 5). These results support the power of combining genome-wide profiling and the fly model to identify additional factors contributing to human diseases.
Other studies have explored global changes in 5hmC, making this is the first study to our knowledge to map genome-wide changes in 5hmC in AD (25,29,62–64). Although the sample size is small and we did not use a single base resolution method, we identified a large amount of overlap of genes with differential 5-hydroxymethylation between our discovery and replication sets, suggesting that these genes represent some shared AD pathology. In addition, the inclusion of one male in the replication set raises some concerns due to the gender differences in cytosine modification patterns and in AD pathogenesis. However, we repeated the analysis while excluding the male sample and identified 136 of the 325 DhML-containing genes and 62 of 140 DhML-containing genes. Therefore, we included the male in the analysis to maintain sample numbers (Supplementary Material, Table S4). Importantly, our findings also overlap with recent epigenome-wide association studies of 5mC and GWAS-associated SNPs, supporting that this method identifies regions that are relevant for disease and justifies incorporating 5hmC into studies of single-base resolution analysis of cytosine modifications in larger cohorts (31,32). This analysis of alterations of 5hmC in AD has expanded the network of known genes involved in AD. The gene ontology enrichment results highlight the importance of pathways involved in neuronal morphology and synaptic function in AD pathogenesis, identifying these genes as candidates for further study in cellular and animal models for their role in AD pathogenesis. These genes can be prioritized based on our results, with highest priority given to the 43 genes that contain DhMLs, show changes in expression, and encode proteins involved in the AD susceptibility PPI network. We also found 11 previously identified AD susceptibility loci in or near DhMLs, suggesting that variants within these loci may affect disease risk via epigenetic regulation of gene expression. Finally, this report also demonstrates that incorporating an analysis of 5-hmC is a useful strategy for future studies to yield a more complete picture of pathways that contribute to AD pathogenesis and suggest possible new targets for AD treatments.
Materials and Methods
Case materials
Human post-mortem frozen tissue from frontal cortex were provided from clinically and pathologically well-characterized cases at the University of Kentucky Alzheimer's Disease Research Center and from the Emory University Alzheimer’s Disease Research Center. Subjects and neuropathologic evaluations were conducted as described previously (65,66). All procedures (including informed consent for each patient) was performed in accordance with the Institutional Review Boards for the respective institutions. For details on the characteristics of the samples and the individuals from whom the samples were obtained, see Supplementary Material. AD diagnoses were made in accordance with established criteria (NIA-AA) (67). Controls had no history of neurological illness.
Genomic DNA preparation
Genomic DNA was isolated from brain samples with standard protocols. Tissues were homogenized on ice and then treated with proteinase K (0.667 µg/µl) in digestion buffer (100 mM Tris-HCl (pH 8.5), 5 mM EDTA, 0.2% SDS (vol/vol), 200 mM NaCl) overnight at 55 °C. On the second day, an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1, saturated with 10 mM Tris (pH 8.0) and 1 mM EDTA; P-3803, Sigma) was added, mixed completely, centrifuged for 5 min at 20 817g, and precipitated with an equal volume of isopropanol. Genomic DNA was recovered in 10 mM Tris-HCl (pH 8.0) and sonicated to ∼500 bp by Misonix 3000 (microtip, 4 pulses of 27 s each, 1-min rest on ice, output 2).
5hmC–specific enrichment
5-hmC enrichment was performed as previously described (18). 5-hmC labeling reactions were performed in a 100-µl solution containing 50 mM HEPES buffer (pH 7.9), 25 mM MgCl2, 300 ng/µl sonicated genomic DNA (100–500 bp), 250 µM UDP-6-N3-Glu and 2.25 µM wild-type β-glucosyltransferase. Reactions were incubated for 1 h at 37 °C. DNA substrates were purified via Qiagen DNA purification kit or by phenol–chloroform precipitation and reconstituted in H2O. Click chemistry was performed by adding 150 µM dibenzocyclooctyne-modified biotin into the DNA solution and incubating for 2 h at 37 °C. Samples were purified by Pierce Monomeric Avidin Kit (Thermo) following the manufacturer’s recommendations. After elution, biotin-5-N3-gmC–containing DNA was concentrated with 10 K Amicon Ultra 0.5-ml Centrifugal Filters (Millipore) and purified using a Qiagen DNA purification kit.
Sequencing of 5hmC–enriched DNA
Libraries were generated following the Illumina protocol for ‘Preparing Samples for ChIP Sequencing of DNA’ (Part 111257047 Rev. A). We used 25 ng of input genomic DNA or 5hmC–captured DNA to initiate the protocol. DNA fragments of ∼150–300 bp were gel-purified after the adaptor ligation step. Polymerase chain reaction-amplified DNA libraries were quantified on an Agilent 2100 Bioanalyzer and diluted to 6–8 pM for cluster generation and sequencing. We performed 38-cycle single-end sequencing Generation using Version 4 Cluster Generation and Sequencing Kits (Part 15002739 and 15005236, respectively) and Version 7.0 recipes. Image processing and sequence extraction were done using the standard Illumina pipeline.
Sequence alignment and binning
FASTQ files were aligned to the human genome (hg19) with Bowtie, retaining only unique non-duplicate genomic matches with no more than two mismatches in the first 25 bp (34). Unique, non-duplicate reads were counted in 10 000-bp bins and normalized to the total number of non-duplicate reads.
Identification and annotation of DhMLs
DhMLs were identified with the software diffReps using unique non-duplicate reads with the specified settings (window = 1000, step = 100, fragment = 300) (35). Annotation of these regions was carried out with Homer (36). Cytoscape and the ClueGO plugin were used to carry out gene ontology analysis (38–40). DAPPLE was used to generate PPI networks (43). GeneOverlap was used to test the overlap between sets of genes (48).
RNA-Seq data acquisition
The RNA-Seq data were reported previously (45). The samples with high-quality RNA samples (RIN 7.8-8.8) were used for further analyses.
RNA-Seq sequencing alignment and analysis
Paired-end reads were mapped to University of California Santa Cruz human reference genome hg19 using Illumina iGenomes pre-built indexes (https://ccb.jhu.edu/software/tophat/igenomes.shtml, last accessed April 13, 2016), TopHat (v2.0.3), and Bowtie (34,46). The Cufflinks suite of tools was used to assemble transcripts and determine differential expression of transcripts (47).
IGAP is a large two-stage study based upon (GWAS) on individuals of European ancestry. In stage 1, IGAP used genotyped and imputed data on 7 055 881 (SNPs) to meta-analyze four previously published GWAS datasets consisting of 17 008 AD cases and 37 154 controls (The European Alzheimer's Disease Initiative, EADI, the Alzheimer Disease Genetics Consortium, ADGC, The Cohorts for Heart and Aging Research in Genomic Epidemiology consortium, CHARGE and the Genetic and Environmental Risk in AD consortium, GERAD). In stage 2 11 632 SNPs were genotyped and tested for association in an independent set of 8572 AD cases and 11 312 controls. Finally, a meta-analysis was performed combining results from stages 1 and 2.
Drosophila strains and culture conditions
The transgenic Drosophila lines expressing human wild-type and mutant Tau (V337M and R406W) were described previously (49). UAS-CUGBP1 was constructed and reported previously (68). The gmr-GAL4, elav-GAL4 (C155), wild-type w1118 strain and Drosophila lines of the orthologs of candidate modifiers were obtained from the Bloomington Drosophila Stock Center. Fly cultures and crosses were carried out using standard fly medium under controlled temperature conditions at 25 °C.
Scoring eye phenotype
Targeted expression of either human wild-type or mutant Tau (V337M and R406) in the fly eye presents highly uniform eye degeneration of reduced eye size, accompanied by rough eye surface. Crosses between Tau and candidate genes were performed to investigate modifiers with enhancement or suppression effects in view of eye phenotype under light microscopy. In no cases could the modifier rescue the disrupted eye morphology to normal; thus, we defined reduced eye size with rough surface as the basic eye phenotype. Upon the basic phenotype, eyes were examined and given 1 point for the presence of each of the additional phenotypes: loss of pigmentation, retinal collapse and necrosis. A higher point stands for a more severe phenotype. The observed eye phenotypes are categorized into different phenotypic groups according to points of severity: Grade I (mild, basic phenotype), II (moderate, 1 point), III (severe, 2 points) and IV (extremely severe, 3 points). For each genotype, over 200 5-day-old male flies were examined. For LM images, whole flies were analyzed using an OLYMPUS DP72 microscope.
Climbing assay
Groups of ten 5-day-old or 10-day-old male flies were transferred into 1.25-cm-diameter and 28-cm-height plastic tubes with 1 h incubation at room temperature to wake from anesthesia and acclimatize to the new environment. The arrival time of the fifth fly at the 15-cm finish line was collected and analyzed. Three trials were repeated for each group. For each genotype, over 15 groups of male flies were examined, except for co-expression of wild-type Tau and Amph with reduced viability. For statistical analyses, comparisons were made using Student’s t-test or Mann–Whitney test (*** = P < 0.0001, ** = P < 0.01, * = P < 0.05, ns = no significant difference).
Accession Numbers
The Gene Expression Omnibus accession number for 5hmC data reported in this paper is GSE72782. Raw RNA-Seq files have been deposited in the National Center for Biotechnology Information Sequence Read Archive database, www.ncbi.nlm.nih.gov/sra (accession no. SRA060572).
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
The authors would like to thank C. Strauss for critical reading of the manuscript.
Conflict of Interest statement. None declared.
Funding
This work was supported in part by National Institutes of Health (NS079625 to P.J., AG025688 to A.I.L./J.J.L./M.G./P.J., HG006699 to Q.D., ES024570 to A.I.B.), and National Natural Science Foundation of China (81571253, 81172513, 81071028 to R.D.).
References
- 1.Barker W.W., Luis C.A., Kashuba A., Luis M., Harwood D.G., Loewenstein D., Waters C., Jimison P., Shepherd E., Sevush S. et al. (2002) Relative frequencies of Alzheimer disease, Lewy body, vascular and frontotemporal dementia, and Hippocampal sclerosis in the State of Florida Brain Bank. Alzheimer Dis. Assoc. Disord., 16, 203–212. [DOI] [PubMed] [Google Scholar]
- 2.Holtzman D.M., Morris J.C., Goate A.M. (2011) Alzheimer's disease: the challenge of the second century. Sci. Transl. Med., 3, 77sr1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardy J., Selkoe D.J. (2002) The amyloid hypothesis of Alzheimer's disease, progress and problems on the road to therapeutics. Science, 297, 353–356. [DOI] [PubMed] [Google Scholar]
- 4.Blennow K., de Leon M.J., Zetterberg H. (2006) Alzheimer's disease. Lancet, 368, 387–403. [DOI] [PubMed] [Google Scholar]
- 5.Karch C.M., Cruchaga C., Goate A.M. (2014) Alzheimer's disease genetics, from the bench to the clinic. Neuron, 83, 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wingo T.S. (2012) Autosomal recessive causes likely in early-onset Alzheimer disease. Arch. Neurol., 69, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bihaqi S.W., Schumacher A., Maloney B., Lahiri D.K., Zawia N.H. (2012) Do epigenetic pathways initiate late onset Alzheimer disease (LOAD), towards a new paradigm. Curr. Alzheimer Res., 9, 574–588. [DOI] [PubMed] [Google Scholar]
- 8.Irier H.A., Jin P. (2012) Dynamics of DNA methylation in aging and Alzheimer's disease. DNA Cell Biol., 31, S42–S48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng Y., Bernstein A.I., Chen D., Jin P. (2014) 5-Hydroxymethylcytosine: a new player in brain disorders? Exp. Neurol., 268, 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith Z.D., Chan M.M., Mikkelsen T.S., Gu H., Gnirke A., Regev A., Meissner A. (2012) A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature, 484, 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kriaucionis S., Heintz N. (2009) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science, 324, 929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L. et al. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science, 324, 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y. (2011) TET proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science, 333, 1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spruijt C.G., Gnerlich F., Smits A.H., Pfaffeneder T., Jansen P.W.T.C., Bauer C., Münzel M., Wagner M., Müller M., Khan F. et al. (2013) Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell, 152, 1146–1159. [DOI] [PubMed] [Google Scholar]
- 15.Nestor C.E., Ottaviano R., Reddington J., Sproul D., Reinhardt D., Dunican D., Katz E., Dixon J.M., Harrison D.J., Meehan R.R. (2012) Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res., 22, 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pastor W.A., Pape U.J., Huang Y., Henderson H.R., Lister R., Ko M., McLoughlin E.M., Brudno Y., Mahapatra S., Kapranov P. et al. (2011) Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature, 473, 394–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Globisch D., Münzel M., Müller M., Michalakis S., Wagner M., Koch S., Brückl T., Biel M., Carell T. (2010) Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One, 5, e15367.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song C.X., Szulwach K.E., Fu Y., Dai Q., Yi C., Li X., Li Y., Chen C.H., Zhang W., Jian X. et al. (2011) Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol., 29, 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szulwach K.E., Li X., Li Y., Song C.X., Wu H., Dai Q., Irier H., Upadhyay A.K., Gearing M., Levey A.I. et al. (2011) 5-hmC–mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci., 14, 1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y., Damayanti N.P., Irudayaraj J., Dunn K., Zhou F.C. (2014) Diversity of two forms of DNA methylation in the brain. Front. Genet., 5, 46.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ficz G., Branco M.R., Seisenberger S., Santos F., Krueger F., Hore T.A., Marques C.J., Andrews S., Reik W. (2011) Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature, 473, 398–402. [DOI] [PubMed] [Google Scholar]
- 22.Wu H., D'Alessio A.C., Ito S., Wang Z., Cui K., Zhao K., Sun Y.E., Zhang Y. (2011) Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev., 25, 679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khare T., Pai S., Koncevicius K., Pal M., Kriukiene E., Liutkeviciute Z., Irimia M., Jia P., Ptak C., Xia M. et al. (2012) 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nat. Struct. Mol. Biol., 19, 1037–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang F., Yang Y., Lin X., Wang J.Q., Wu Y.S., Xie W., Wang D., Zhu S., Liao Y.Q., Sun Q. et al. (2013) Genome-wide loss of 5-hmC is a novel epigenetic feature of Huntington's disease. Hum. Mol. Genet., 22, 3641–3653. [DOI] [PubMed] [Google Scholar]
- 25.Coppieters N., Dieriks B.V., Lill C., Faull R.L.M., Curtis M.A., Dragunow M. (2014) Global changes in DNA methylation and hydroxymethylation in Alzheimer's disease human brain. Neurobiol. Aging, 35, 1334–1344. [DOI] [PubMed] [Google Scholar]
- 26.Coppieters N., Dragunow M. (2011) Epigenetics in Alzheimer's disease, a focus on DNA modifications. Curr. Pharm. Des., 17, 3398–3412. [DOI] [PubMed] [Google Scholar]
- 27.Mastroeni D., Grover A., Delvaux E., Whiteside C., Coleman P.D., Rogers J. (2010) Epigenetic changes in Alzheimer's disease, decrements in DNA methylation. Neurobiol. Aging, 31, 2025–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao J.S., Keleshian V.L., Klein S., Rapoport S.I. (2012) Epigenetic modifications in frontal cortex from Alzheimer's disease and bipolar disorder patients. Transl. Psychiatry, 2, e132.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bradley-Whitman M.A., Lovell M.A. (2013) Epigenetic changes in the progression of Alzheimer's disease. Mech. Ageing Dev., 134, 486–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Mahdawi S., Virmouni S.A., Pook M.A., Evans-Galea M. (2014) The emerging role of 5-hydroxymethylcytosine in neurodegenerative diseases. Front. Neurosci., 8, 397.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lunnon K., Smith R., Hannon E., De Jager P.L., Srivastava G., Volta M., Troakes C., Al-Sarraj S., Burrage J., Macdonald R. et al. (2014) Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer's disease. Nat. Neurosci., 17, 1164–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Jager P.L., Srivastava G., Lunnon K., Burgess J., Schalkwyk L.C., Yu L., Eaton M.L., Keenan B.T., Ernst J., McCabe C. et al. (2014) Alzheimer's disease, early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat. Neurosci., 17, 1156–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mirra S.S., Heyman A., McKeel D., Sumi S.M., Crain B.J., Brownlee L.M., Vogel F.S., Hughes J.P., Belle G.V., Berg L. (1991) The Consortium to Establish a Registry for Alzheimer's Disease (CERAD), Part II. Standardization of the neuropathologic assessment of Alzheimer's disease. Neurology, 41, 479–486. [DOI] [PubMed] [Google Scholar]
- 34.Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol., 10, R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen L., Shao N.Y., Liu X., Maze I., Feng J., Nestler E.J. (2013) diffReps, detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One, 8, e65598.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell, 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol., 57, 289–300. [Google Scholar]
- 38.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pagès F., Trajanoski Z., Galon J. (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics, 25, 1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopes C.T., Franz M., Kazi F., Donaldson S.L., Morris Q., Bader G.D. (2010) Cytoscape Web: an interactive web-based network browser. Bioinformatics, 26, 2347–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smoot M.E., Ono K., Ruscheinski J., Wang P.L., Ideker T. (2011) Cytoscape 2.8, new features for data integration and network visualization. Bioinformatics, 27, 431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B. et al. (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet., 45, 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou X., Maricque B., Xie M., Li D., Sundaram V., Martin E.A., Koebbe B.C., Nielsen C., Hirst M., Farnham P. et al. (2011) The Human Epigenome Browser at Washington University. Nat. Methods, 8, 989–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rossin E.J., Lage K., Raychaudhuri S., Xavier R.J., Tatar D., Benita Y., Constortium I.I.B.D.G., Cotsapas C., Daly M.J. (2011) Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet., 7, e1001273.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lage K., Karlberg E.O., Størling Z.M., Olason P.I., Pedersen A.G., Rigina O., Hinsby A.M., Tümer Z., Pociot F., Tommerup N. et al. (2007) A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol., 25, 309–316. [DOI] [PubMed] [Google Scholar]
- 45.Bai B., Hales C.M., Chen P.C., Gozal Y., Dammer E.B., Fritz J.J., Wang X., Xia Q., Duong D.M., Street C. et al. (2013) U1 small nuclear ribonucleoprotein complex and RNA splicing alterations in Alzheimer's disease. Proc. NatI Acad. Sci. U. S. A., 110, 16562–16567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trapnell C., Pachter L., Salzberg S.L. (2009) TopHat, discovering splice junctions with RNA-Seq. Bioinformatics, 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol., 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen L. (2013) GeneOverlap: Test and visualize gene overlaps R package version 1.6.0, http://shenlab-sinai.github.io/shenlab-sinai/.
- 49.Wittmann C.W., Wszolek M.F., Shulman J.M., Salvaterra P.M., Lewis J., Hutton M., Feany M.B. (2001) Tauopathy in Drosophila, neurodegeneration without neurofibrillary tangles. Science, 293, 711–714. [DOI] [PubMed] [Google Scholar]
- 50.Finelli A., Kelkar A., Song H.J., Yang H., Konsolaki M. (2004) A model for studying Alzheimer's Abeta42-induced toxicity in Drosophila melanogaster. Mol. Cell. Neurosci., 26, 365–375. [DOI] [PubMed] [Google Scholar]
- 51.Shulman J.M., Chipendo P., Chibnik L.B., Aubin C., Tran D., Keenan B.T., Kramer P.L., Schneider J., Bennett D.A., Feany M.B. et al. (2011) Functional screening of Alzheimer pathology genome-wide association signals in Drosophila. Am. J. Hum. Genet., 88, 232–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vossel K.A., Zhang K., Brodbeck J., Daub A.C., Sharma P., Finkbeiner S., Cui B., Mucke L. (2010) Tau reduction prevents Abeta-induced defects in axonal transport. Science, 330, 198.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roberson E.D., Scearce-Levie K., Palop J.J., Yan F., Cheng I.H., Wu T., Gerstein H., Yu G.Q., Mucke L. (2007) Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer's disease mouse model. Science, 316, 750–754. [DOI] [PubMed] [Google Scholar]
- 54.Fulga T.A., Elson-Schwab I., Khurana V., Steinhilb M.L., Spires T.L., Hyman B.T., Feany M.B. (2007) Abnormal bundling and accumulation of F-actin mediates tau-induced neuronal degeneration in vivo. Nat. Cell Biol., 9, 139–148. [DOI] [PubMed] [Google Scholar]
- 55.Chapuis J., Hansmannel F., Gistelinck M., Mounier A., Van Cauwenberghe C., Kolen K.V., Geller F., Sottejeau Y., Harold D., Dourlen P. et al. (2013) Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol. Psychiatry, 18, 1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bettens K., Sleegers K., Van Broeckhoven C. (2013) Genetic insights in Alzheimer's disease. Lancet Neurol., 12, 92–104. [DOI] [PubMed] [Google Scholar]
- 57.Jeibmann A., Paulus W. (2009) Drosophila melanogaster as a model organism of brain diseases. Int. J. Mol. Sci., 10, 407–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moloney A., Sattelle D.B., Lomas D.A., Crowther D.C. (2010) Alzheimer's disease, insights from Drosophila melanogaster models. Trends Biochem. Sci., 35, 228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rubin G.M., Yandell M.D., Wortman J.R., Gabor Miklos G.L., Nelson C.R., Hariharan I.K., Fortini M.E., Li P.W., Apweiler R., Fleischmann W. et al. (2000) Comparative genomics of the eukaryotes. Science, 287, 2204–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fortini M.E., Skupski M.P., Boguski M.S., Hariharan I.K. (2000) A survey of human disease gene counterparts in the Drosophila genome. J. Cell Biol., 150, F23–F30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prüßing K., Voigt A., Schulz J.B. (2013) Drosophila melanogaster as a model organism for Alzheimer’s disease. Mol. Neurodegener., 8, 35.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chouliaras L., Mastroeni D., Delvaux E., Grover A., Kenis G., Hof P.R., Steinbusch H.W., Coleman P.D., Rutten B.P., van den Hove D.L. (2013) Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer's disease patients. Neurobiol. Aging, 34, 2091–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mastroeni D., McKee A., Grover A., Rogers J., Coleman P.D. (2009) Epigenetic differences in cortical neurons from a pair of monozygotic twins discordant for Alzheimer's disease. PLoS One, 4, e6617.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Condliffe D., Wong A., Troakes C., Proitsi P., Patel Y., Chouliaras L., Fernandes C., Cooper J., Lovestone S., Schalkwyk L. et al. (2014) Cross-region reduction in 5-hydroxymethylcytosine in Alzheimer's disease brain. Neurobiol. Aging, 35, 1850–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmitt F.A., Nelson P.T., Abner E., Scheff S., Jicha G.A., Smith C., Cooper G., Mendiondo M., Danner D.D., Van Eldik L.J. et al. (2012) University of Kentucky Sanders-Brown healthy brain aging volunteers, donor characteristics, procedures and neuropathology. Curr. Alzheimer Res., 9, 724–733., [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nelson P.T., Jicha G.A., Schmitt F.A., Liu H., Davis D.G., Mendiondo M.S., Abner E.L., Markesbery W.R. (2007) Clinicopathologic correlations in a large Alzheimer disease center autopsy cohort, neuritic plaques and neurofibrillary tangles “do count” when staging disease severity. J. Neuropathol. Exp. Neurol., 66, 1136–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Montine T.J., Phelps C.H., Beach T.G., Bigio E.H., Cairns N.J., Dickson D.W., Duyckaerts C., Frosch M.P., Masliah E., Mirra S.S. et al. (2012) National Institute on Aging-Alzheimer's Association guidelines for the neuropathologic assessment of Alzheimer's disease: a practical approach. Acta. Neuropathol., 123, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sofola O.A., Jin P., Qin Y., Duan R., Liu H., de Haro M., Nelson D.L., Botas J. (2007) RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron, 55, 565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.