Abstract
A ‘Down Syndrome critical region’ (DSCR) sufficient to induce the most constant phenotypes of Down syndrome (DS) had been identified by studying partial (segmental) trisomy 21 (PT21) as an interval of 0.6–8.3 Mb within human chromosome 21 (Hsa21), although its existence was later questioned. We propose an innovative, systematic reanalysis of all described PT21 cases (from 1973 to 2015). In particular, we built an integrated, comparative map from 125 cases with or without DS fulfilling stringent cytogenetic and clinical criteria. The map allowed to define or exclude as candidates for DS fine Hsa21 sequence intervals, also integrating duplication copy number variants (CNVs) data. A highly restricted DSCR (HR-DSCR) of only 34 kb on distal 21q22.13 has been identified as the minimal region whose duplication is shared by all DS subjects and is absent in all non-DS subjects. Also being spared by any duplication CNV in healthy subjects, HR-DSCR is proposed as a candidate for the typical DS features, the intellectual disability and some facial phenotypes. HR-DSCR contains no known gene and has relevant homology only to the chimpanzee genome. Searching for HR-DSCR functional loci might become a priority for understanding the fundamental genotype-phenotype relationships in DS.
Introduction
The concept that the main symptoms and signs of Down syndrome (DS) may be caused by overexpression of one or a few genes located on a delimited, small region on human chromosome 21 (Hsa21) has had changing fortunes in the last decades. Since the fundamental discovery of Lejeune et al. (1), we know that Hsa21 is present in an extra copy in the cells of subjects with DS (trisomy 21). While it is widely accepted that excess genetic material from Hsa21 is responsible for the typical picture of DS in particular intellectual disability (ID), cardiovascular defects and craniofacial dysmorphisms (2), to date there is no pathogenetic model linking specific structural and functional aspects of Hsa21 to DS picture. For most of the Hsa21 loci that were studied, the overexpression model, that is the excess of gene products in a 3:2 ratio with respect to the normal cells expected by the additional Hsa21 template, has proven to be true (2–5). In addition, several genes on Hsa21 are known to have functions biologically consistent with pathways known to be altered in DS, in particular brain development and function, oxidative metabolism and one-carbon pathway (6). However, the actual correlation of individual Hsa21 genes to DS still remains hypothetical in humans and has currently not led to therapeutic success by rationally targeting functions encoded by Hsa21 genes (7).
Natural occurrence of partial (segmental) trisomy 21 (PT21), the duplication of only a delimited segment of Hsa21 associated or not to DS, was first reported by Ilbery et al. (8) as ‘incomplete trisomy’ and is considered to be exceptional or extremely rare (9,10). Approximately 200 cases have actually been reported in literature in comparison to an estimated 5.8 million people with DS in the world (6). However, this condition is invaluable in providing strong evidence that not all Hsa21 loci are required for the manifestation of DS and at the same time offers a powerful way to ‘detect the culprits among so many innocents’ (11). Indeed, from a general biological and pathogenic point of view, it is reasonable that overexpression of a certain fraction of genes may be tolerated: ‘Surely, most of the genes do not produce harm when in triplicate, because trisomic children would not survive at all. Few of the accelerated reactions are dangerous’ (11). As a recent remarkable example (12), a terminal duplication of at least 5 Mb on distal 21q22.3 including 77 protein-coding genes (30% of all protein-coding Hsa21 genes) was not related to an abnormal phenotype (Hsa21 interval 41.6–46.6 Mb on GenBank sequence NC_000021.9; all chromosomal coordinates are adjusted in this work to the current Genome Reference Consortium (GRC) human genome assembly GRCh38, or hg38, December 2013). Similarly, while this work was in revision, a new case of a 4.4 Mb interstitial duplication on 21q22.2q22.3 (39 612 165–43 981 027 interval in hg38), including 51 genes of which 7 are in the Online Mendelian Inheritance in Man (OMIM) Genetic Morbid Map, was reported both in a mother with a completely normal clinical picture and her female fetus with no dysmorphism and normal psycho-motor development at two years of age (13). In addition, overexpression of one gene might compensate overexpression of another gene present in the duplicated chromosome (e.g. overexpression of RCAN1, also known as DSCR1, may compensate damage induced by overexpression of APP, both genes being located on Hsa21) (14). Therefore, no effort should be spared to undertake a systematical investigation of partial trisomies in order to focus on the possibility that a low number of genes conserved in the segments shared by DS subjects may have a significant role in the DS pathogenesis. It is necessary at this point to briefly review the rise and fall of the concept of a ‘critical’ region for DS on Hsa21 to justify this study.
In the first reports of PT21 (8,15,16), it was impossible to identify which subregion of Hsa21 was lacking. Actually, at that time, it was the phenotype to guide the interpretation of the cases, due to difficulties in identifying the small G-group supernumerary chromosome seen in DS as Hsa21 itself (17,18). Since the introduction of a worldwide accepted banding method in the human cytogenetic analysis in the early 1970s (19,20), it became possible to demonstrate that children with a phenotype indistinguishable from DS appeared to have only a specific portion of Hsa21 rather than a complete Hsa21 long arm (21q). Hsa21 short arm (21p) is considered genetically empty in practice, as shown by centric fusion (leading to robertsonian translocation) in which the loss of 21p is consistent with a DS phenotype indistinguishable from the one due to free trisomy 21 if the 21q dose is unbalanced (16,21) and with no clinical consequences if 21q dose is balanced (18,22,23). The main mechanisms leading to this so-called ‘partial’ or ‘segmental’ trisomy were described as discordant segregation of interstitially deleted chromosomes 21; translocations involving segments of 21q; tandem translocations with an incomplete long arm of replicated Hsa21 (24).
On these bases, in 1974 Niebuhr was the first to put forward the hypothesis (25), by reviewing 14 previously described cases with tandem translocations of G-group chromosomes and reporting a new one, that ‘trisomy of a rather delimited segment on chromosome No. 21 is essential for the development of typical features in Down's syndrome’, suggesting that ‘the very distal segment (21q22)’ (17.4 Mb) ‘may be pathogenetic in Down's syndrome’, thus already excluding that 65% of Hsa21 (total length 46.7 Mb) may give a fundamental contribution to the very basic features of DS.
In the subsequent 20 years, several single cases as well as case series of PT21 were reported. The development of fluorescence in situ hybridization (FISH) techniques (26) allowed a more detailed description of the duplicated 21q segments associated to DS culminating in the early 1990s in the suggestion that a delimited sequence interval on Hsa21 overlapping among different cases contributes significantly to the basic phenotype characteristic of trisomy 21. In particular, the data of Rahmani et al. (27) (two cases, D21S17-ETS2 interval, <3 Mb size), McCormick et al. (28) (16 cases, D21S55-COL6A1, 8.3 Mb) as well as Delabar et al. (29) (20 cases, D21S55-proximal to ERG, 0.6 Mb) converged toward a region within 21q22 and restricted up to 0.6 Mb, from 37.7 to 38.3 Mb on Hsa21.
The first time that the term ‘Down Syndrome critical region’ (DSCR) was used appears to have been in Rahmani et al. (30); the terms ‘Minimal Chromosomal Region’ (MCR) (28), ‘Down Syndrome minimum critical region’ (DCR) (OMIM entry #190685) or ‘Down Syndrome minimal region’ (31) have also been used. Subsequently, in the naming of genes believed to be localized in the DSCR, the ‘C’ in the acronym was sometimes meant as ‘critical’ and sometimes ‘candidate’ (32). Currently, although the concept itself of ‘DSCR’ is questioned (see below), the term ‘critical’ has remained prevalent in the gene nomenclature and biomedical literature. This appears to be adequate because, if existing, the DSCR concept should indicate a region causing most critical, shared symptoms/signs of DS, rather than a region candidate for causing the (whole) phenotype of DS. The DSCR concept led to a flourishing of structural and functional studies of its content (33).
Korenberg et al. (34) argued against a single DSCR, because in 3 of the 16 cases, they reported only proximal duplications of 21 were observed; Antonarakis (10) observed in this respect that more cases were necessary to clarify the contributions of different Hsa21 regions to DS phenotypes. It is very important that in several cases of PT21 the karyotype was reported as normal but the observation of a phenotype strongly suggestive of DS prompted further studies revealing subtle duplication of a Hsa21 region [e.g. Scott et al. (35) and so on].
At the end of the 1990s, the availability of the nucleotide sequence of Hsa21 (36) as well as methods with higher resolution such as Array Comparative Genomic Hybridization (CGH) have allowed the obtainment of new valuable data, along with the continuing report of new PT21 cases. Two landmark studies were published in 2009 (37,38), each reporting results for an unprecedented number of subjects presenting with this condition, in particular 16 (37) and then 30 cases (38), including both new (12 and 9, respectively) and previously described cases. Although both studies clearly showed associations of specific Hsa21 regions to distinct DS phenotypes, they were not focused on the search of a ‘core’ DSCR, because of the observation that one single region cannot explain all DS phenotypes in all subjects. As a matter of fact, since 2009, there has not actually been another systematic attempt to identify a restricted DSCR and further analysis on the subject has been explicitly considered unwarranted by some authors (39). However, the subject to date has not yet been thoroughly investigated following basic criteria allowing to reach definite conclusions, in particular by collecting the largest possible number of cases and by computerizing all the data, as it was proposed since 1996 (40).
We propose here a systematic reanalysis of all described cases with PT21 that is innovative under several aspects. First, while previous works have always focused on a limited number of cases, we have systematically searched for and compared any available case published from 1973 (just following the introduction of human chromosome banding) to 2015. An accurate bibliographical search has been performed to include any report of interest. We have also largely exploited the availability of a Hsa21 detailed physical map to which consistently anchor data produced through several decades by many different cytogenetic methods. For this reason, we have applied several computational biology methods to establish a common framework and then used it to build our original integrated, Hsa21 comparative map of PT21 in humans. In addition, we put forward the concept of an ‘exclusion map’, in addition to that of a ‘candidate map’, accurately studying cases with a non-DS phenotype but with duplicated (trisomic) segments of Hsa21 that, therefore, cannot be assumed to have a relevant role in DS; the final map integrates data from both these complementary approaches, also using available copy number variants (CNVs) (41) data to highlight Hsa21 regions whose duplication has been found in variation studies of the human genome in healthy subjects. Moreover, we have not selected for the inclusion in the map, following accurate manual curation, any case whose description of cytogenetic, molecular or clinical features did not allow an unambiguous assessment of genotype–phenotype relationship. Finally, rather than trying to identify subregions responsible for distinct phenotypes, we focus on the diagnosis of DS itself as the phenotype to be mapped, thus implying its most common clinical findings: a recognizable form of ID that is also the most serious problem for people with DS, in presence of a typical DS facies. The fact that DS ID frequency in trisomy 21 is virtually 100% and most typical facial phenotypes are the second most frequent signs of DS allows us to avoid confusion factors such as variability of the presence of all other symptoms and signs, which may be related to additive factors (allelic isoforms, CNVs, epistasis, positional effects in cases with translocations, epigenetics, environment) rather than the pure duplication of critical Hsa21 portions.
The presented approach based on stringent criteria to review cytogenetic/clinical features and on systematic map integration led us to accurately revise data points from past analyses and to propose a more fine delimitation at a structural level of a Hsa21 region constantly duplicated in individuals with diagnosis of DS and thus likely associated to the most typical DS symptoms and signs such as a recognizable form of ID and some facial phenotypes.
Results
Partial trisomy 21 comparative map building
Following systematic bibliographic searches, 110 reports from 1973 to 2015 were identified as containing case descriptions fulfilling cytogenetic and clinical criteria (see the following Results section below) and then selected for the study, as described in Table 1, Supplementary Material Tables S1 and S2 and Supplementary Material References file.
Table 1.
Reports of partial trisomy 21 cases from 1973 to 2015
| Reports matching various PubMed queries | 1322 |
| Reports describing partial trisomy 21 apparently suitable for the study | 110 |
| 1. Reports describing cases included in the map | 96 |
| 2. Reports describing only cases excluded from the map | 14 |
| Reports describing partial trisomy 21 but in mosaicism | 20 |
| Reports describing partial trisomy 21 but with X-translocation t(X;21) | 3 |
| Total of reports describing partial trisomy 21* | 133 |
Derived from PubMed searches plus examination of cross references. References are listed in the Supplementary Material References file.
Apart from the first case listed, published in 1965 (42) but characterized at chromosome banding level later, the first considered report is the one published by Aula et al. (9). These authors themselves state that cases of partial Hsa21 trisomy ‘previously reported in the literature cannot be considered’ in the respect of identifying Hsa21 segments required for the expression of a typical DS ‘because the differential staining techniques of chromosomes were not available’. A comparative map showing the localization of segmental anomalies of Hsa21 was obtained as described in the Materials and Methods section from 125 subjects fulfilling stringent criteria for genotype–phenotype mapping out of a total of 180 reported cases (Supplementary Material Table S2). Among the six morphological signs studied here, the most common in PT21 subjects with DS were upslanted palpebral fissures (91.7%), epichantus (90.8%) and flat nasal bridge (87.5%), while the most frequently absent in PT21 subjects without DS were flat occipit/brachicefaly, broad hands and open mouth (absent in 88.9%, 87.5% and 87.0% of cases, respectively) (Supplementary Material Tables S2 and S3). The frequency of all these signs was significantly different (at P < 0.0001, except for open mouth where P = 0.0002) by Fisher test between the DS and non-DS groups.
Supplementary Material Table S4 summarizes family relationships among the subjects included in the PT21 map. Only 13 subjects out of a total of 125 are relatives of an index case. In two of these families, one relative has an opposite DS/non-DS condition in comparison to the index case, thus enriching information about the role of the involved Hsa21 segments. In the other families, the related cases serve as an additional confirmation (similar to a ‘biological replicate’) of the genotype-phenotype relationship, although not totally independent.
The final picture of the comparison of candidate regions could be derived from 88 subjects with DS, in whom trisomic regions are not excluded (and therefore candidate) as responsible for DS, while disomic regions are excluded as responsible. In the 37 subjects without DS, trisomic regions are excluded as candidates for DS, while disomic regions cannot be excluded as possibly responsible.
Main alterations represented in the map were: 64 (51%) cases with reciprocal translocations involving segments of 21q; 42 (34%) cases with segmental replication of Hsa21; 7 (6%) cases with tandem translocations involving an incomplete long arm of the replicated Hsa21. Case summary is reported in Table 2.
Table 2.
Types of partial trisomy 21 reanalyzed to build the Hsa21 integrated map
| Chromosome alteration | Number of cases |
|---|---|
| t(n;21) | 64 |
| dup(21) | 42 |
| t(21;21) | 7 |
| inv(21) | 4 |
| der(21) | 3 |
| ins(21) | 2 |
| r(21) | 2 |
| rec(21) | 1 |
| Total | 125 |
t, translocation; n, any autosome involved in the translocation except Hsa21; dup, duplication; inv, inversion; der, chromosome derived from an uncharacterized alteration; ins, insertion; r, ring chromosome; rec, recombinant chromosome.
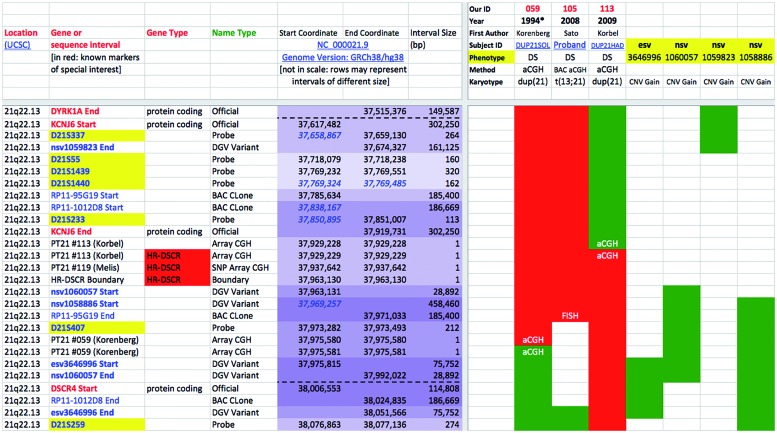
Each case description was independently reviewed and mapped by at least two authors and any discrepancy between them in the results was discussed and reconciled. The complete map with all details (698 sequence intervals color-labeled for all the 125 selected cases: matrix of 87 250 data points) is available as Supplementary Material Table S1. A concise, sample outlook of a portion of the map is depicted in Fig. 1.
Figure 1.
HR-DSCR as highlighted by the trisomy 21-integrated map (simplified view). Only cases (columns) and CNVs strictly defining HR-DSCR limits are shown here. Rows: Hsa21 sequence intervals (only those centered on HR-DSCR are represented here). Red = candidate or not excluded regions; green = excluded regions. Levels of overlapping among intervals are indicated by increasingly darker violet color of the coordinates; blue italics indicate coordinates overlapping (Start or End) or nesting (Start and End) with the just previous interval (row). HR-DSCR coordinates: 37 929 229–37 963 130. Complete map is available as Supplementary Material Table S1.
Partial trisomy 21 comparative map analysis
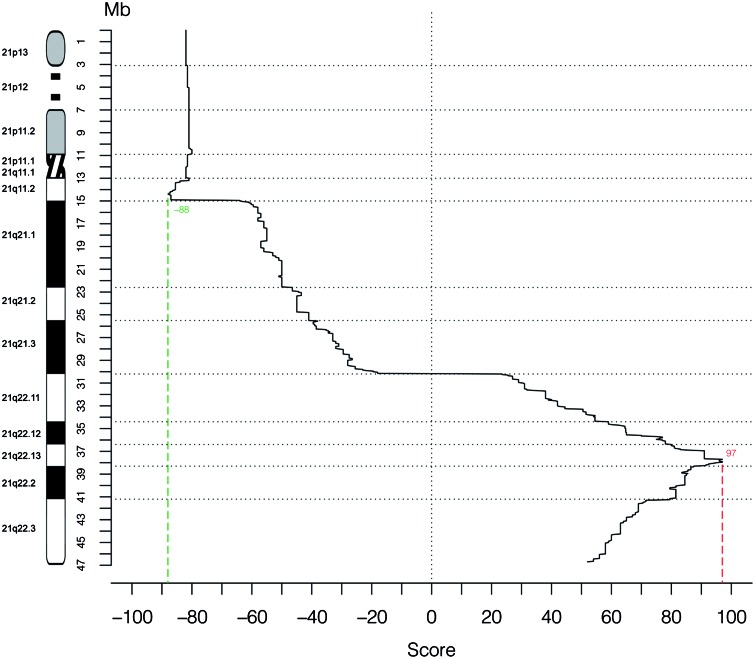
Scores for association with DS for each sequence interval are graphed in Fig. 2. Complete data for each distinct sequence interval placed in the map are given in Supplementary Material Table S1.
Figure 2.
Genotype–phenotype correlation in 125 cases of partial trisomy 21. The X-axis displays the score for association with DS for each sequence interval of 50 kb, shown as median of the values assigned to each map row (Supplementary Material Table S1) that is comprised in each interval. The Y-axis represents the position along Hsa21 (scale in Mb).
First, analysis of the PT21 comparative map allowed the exclusion of several regions of Hsa21 as associated to DS, in particular 21p, 21q1 and 21q21. Highest scores were found in the 21q22.13 and 21q22.2 subbands (Fig. 2). Further inspection of these regions identifies a minimal region of approximately 46 kb, located on 21q22.13 (from 37 929 229 to 37 975 580), as having the highest scores, even when considering pure positively candidating scores not integrated with penalization/exclusion scores (Supplementary Material Table S1). In addition, although its precise boundaries are derived from a few different informative cases (Fig. 1), the duplication of this region is shared by all 80 DS subjects with available data about it and is absent in all 36 non-DS subjects with available data about it, thus defining a new, highly restricted DSCR (HR-DSCR) (Supplementary Material Table S1, Fig. 1). In detail, the most informative data for the HR-DSCR proximal boundary came from a subject (our case #113) analyzed by high-density oligonucleotide tiling array platform, covering all regions of Hsa21 to which probes could be uniquely mapped, with a median probe distance of 90 bp and enabling breakpoint-mapping at 200–300 bp resolution (38), while distal boundary was refined using CNV data as detailed below.
There are no known genes located in the HR-DSCR.
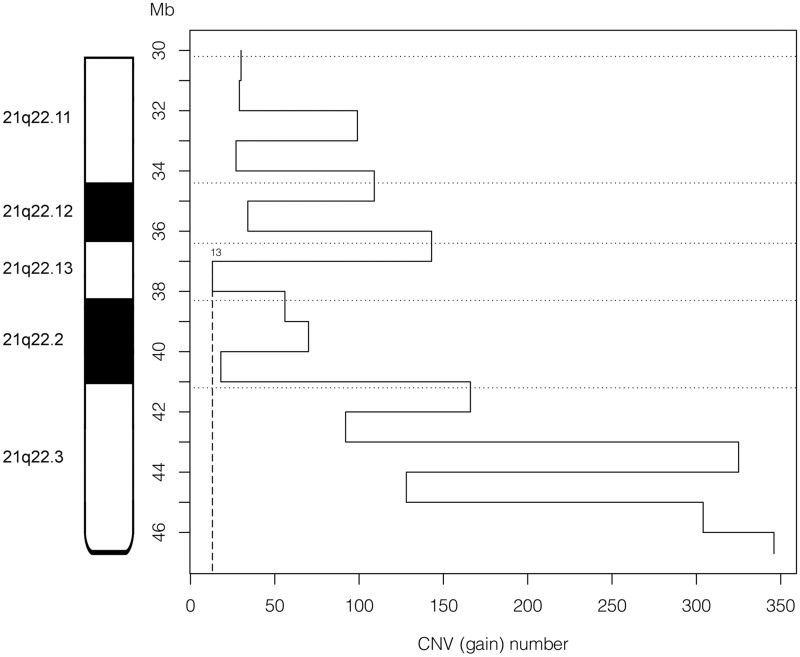
21q22 CNV analysis
CNV analysis shows that partial duplications of Hsa21 have been repeatedly reported in healthy subjects for regions including whole genes, whose gain appears to therefore not be associated to DS phenotypes (Fig. 3). The 1 Mb 21q22 interval with the least number of reported gains coincides with the one including HR-DSCR. A more detailed analysis shows that no duplication in the 37 674 328–37 963 130 Hsa21 interval has been described in healthy subjects, therefore, allowing further distally narrowing of the HR-DSCR, in association to PT21 data, to the 34 kb interval from 37 929 229 to 37 963 130 (Supplementary Material Table S1, Fig. 1).
Figure 3.
CNV frequency in human 21q22. The X-axis displays the number of duplication CNVs found in normal human genomes registered in the Database of Genomic Variants (DGV) for each sequence interval of 1 Mb in the band 21q22. The Y-axis represents the position along Hsa21 (scale in Mb).
In detail, the most informative data for the HR-DSCR distal boundary came from the duplication CNV coded as structural variant nsv1060057, supported by nssv3733448 (source: http://dgv.tcag.ca/, explanation in the Materials and Methods section). The experimental platform used was Affymetrix® Genome-Wide Human SNP Array 6.0, featuring 1.8 million genetic markers, including more than 906 600 single nucleotide polymorphisms and more than 946 000 probes for the detection of copy number variation, with an average/median intermarker spacing of 1600 bp/680 bp, respectively (source, Affymetrix: http://www.affymetrix.com/), that could possibly increase the distal boundary coordinate by some hundreds of nucleotides.
Sequence analysis
BLAST analysis of the 34 kb HR-DSCR region (1000 bp consecutive stretches, BLASTN tool, searches against nr/nt database by default parameters without filters), allowed us to find regions with similarity typically covering 99–100% of the query with 99% of identity in Pan troglodytes (chimpanzee) chromosome 22. Similarity with Pongo abelii (orangutan) or other primates was also found in some of the segments although dropped to below 50% of the query cover and below 90% of identity. No other relevant similarity was observed. Attention was brought to specifically search for similarity with mouse genome, in particular with the mouse chromosome 16, which contains the greatest part of regions syntenic with Hsa21. Only when forcing human–mouse selective comparison for 1000 bp consecutive stretches and changing BLAST default parameters by adding filter for human repeats and using the more sensitive blastn algorithm rather than default megablast, 8 small interspersed segments of homology (from 64 bp to 552 bp, with an identity of 66–78%) were found with mouse chromosome 16, covering in total less than 6.1% of the HR-DSCR, or with other mouse chromosomes (3 small segments of 68, 90 and 117 bp on chromosomes 1, 17 and 18, respectively), thus confirming the lack of relevant homology between the 33 902 bp of HR-DSCR and mouse genome.
Discussion
It is indisputable that DS is constantly associated to the presence of an excess of Hsa21 genetic material (with the very extremely rare exceptions of a few described ‘phenocopies’, discussed below). In some subjects, the presence in excess of only a part of Hsa21 has been demonstrated, often indeed to explain manifestation of DS in subjects with an apparently normal karyotype when studied with more simple methods. These cases represent an invaluable natural source of knowledge about the rational association of delimited regions/functions of Hsa21 and DS phenotypes.
We present here the first systematic reanalysis and comparison of all known and reported cases of PT21 with usable mapping data (published during the last 43 years, from 1973 to 2015), enriched with analysis of Hsa21 CNVs in healthy subjects which has become possible in the last years. It has not been a trivial task to collect and study this article series, mainly because of intricacies in terminology hampering straightforward PubMed/MeSH (Medical Subject Heading)-based searches, publication in only paper form and decades ago, writing in languages different from English. We propose that our systematic strategy to explore all the nodes of the network made by citations in each article retrieved by PubMed searches may have bridged the gaps remaining following the on line queries. Our result of 125 identified cases with PT21 fulfilling stringent criteria for genotype-phenotype mapping outperforms by far any attempt made in the past to review previously described cases. These cases represent a highly curated subset excluding cases with ambiguous assessment at cytogenetic, molecular or clinical level. In addition, the thorough reanalysis and comparison of these data allowed correction of multiple mapping errors made in the past thanks to the update of band boundaries as well as relative order and exact position of many genomic markers by exploiting the latest version of Hsa21 genomic coordinates and updated human gene databases; integration of multiple analyses performed subsequently at different level of detail on the same subjects and reported by different authors in the course of years; correction of incongruences in clinical classification or cytogenetic characterization and removal of cases when resolution of uncertainties or ambiguities was not possible even after having contacted when appropriate the corresponding authors of the original reports.
The minimal region whose duplication is shared by all the subjects with DS and is not present in all the subjects without DS revealed to be in fact a short stretch of DNA, the HR-DSCR (Fig. 1), also showing the highest score indicating association to DS, embedded within the traditional DSCR, confirming it and further narrowing it, when also including CNV data in the map (obtained from a database with >2.5 million entries identified in >22 300 human genomes) to a smaller size (34 kb, i.e. 0.07% of Hsa21) that might contain a small number of unknown genetic determinants associated to the key manifestations of DS.
Regarding the proposal of a minimal DSCR associated to the condition of DS, it should be noted that the scoring approach possibly has a systematic bias depending on the level and the detail of the type of analysis available in the reported data. A region could have a high score (Fig. 2) simply because it was often investigated, but it could be duplicated in some subjects without DS thus excluding its association to the phenotype. In this respect, the coherence for all reported cases of the candidature of a region in absence of any reason for its exclusion is actually the most important fact to define its relevance (Supplementary Material Table S1, Fig. 1). In our case, however, the two lines of demonstration (quantity and quality) are convergent and support and strengthen each other.
Remarkably, analysis of gain (duplication) CNVs reported through 21q22 confirmed the peculiarity of this trait, because it appears to be spared by duplication in healthy subjects (Supplementary Material Table S1), and it is included in a wider zone presenting with the minimal number of gain CNVs among all proximal and distal surrounding zones (Fig. 3). The fact that many PT21 cases are not well characterized with respect to CNVs should be underlined. However, when CNVs have been studied in detail in PT21 cases, it has been found that most of the detected CNVs overlapped with those previously found in unaffected individuals (38). Variability originating from CNVs might contribute to both normal and DS phenotypes (e.g. through silencing), both in Hsa21 (43) or in other chromosomes (44), possibly accounting for discordant genotype–phenotype relationships for certain features.
A limit of our study is that a single subject carrying a pure HR-DSCR duplication has not been reported to date, providing positive evidence of a critical region ‘stricto sensu’. However, in a great number of genetic conditions, it has been widely shown that integration of data from even a few subjects with different cytogenetic alterations and similar phenotypes has been fundamental to define a ‘critical’, ‘common minimal’, ‘minimal overlapping’, ‘minimally shared’ region, leading to the narrowing of the location of the underlying critical genetic determinants and in many cases to their following identification (e.g. searching for ‘critical region’ or ‘minimal region’ in OMIM at http://www.omim.org/ retrieved 313 or 47 ‘phenotype description, molecular basis known’ entries, respectively, and the two terms were used interchangeably).
Another clue supporting the association of HR-DSCR to the most constant DS phenotypes, somewhat independently from the coexistence of duplication of other specific Hsa21 regions, is the fact that HR-DSCR is shared by subjects with DS that in the rest of Hsa21 show absence of duplication indifferently proximally or distally to it (Fig. 1, Supplementary Material Table S1).
As a consequence of the identification of a HR-DSCR, a relevant causative role in DS for genes located outside it should be rediscussed. There are no known genes located in the HR-DSCR, while KCNJ6 and DSCR4 are the adjacent characterized genes, proximally and distally, respectively. Actually, the map shows that regions both proximal and distal to the HR-DSCR may be disomic in subjects manifesting DS. Lack of support for the synergistic roles of RCAN1 and DYRK1A, or APP, as main contributors to many DS phenotypes had been already underlined (38). While a relevant causative role of DYRK1A for DS has been widely discussed (45), and while it certainly affects the development and function of the nervous system, we have not found evidence that it is located within the HR-DSCR, although it lies close to it. Interestingly, a subject reported by Cetin et al. (46) manifests DS in the absence of duplication of DYRK1A as shown by both FISH and Array CGH (although this case has been excluded from our integrated map due to discrepancy between the results of the two methods in another small region, between KCNJ6 and DSCR8).
The proposed limits of the HR-DSCR add support to the likely exclusion of a critical role, for DS as such, exerted by other known genes whose role in DS has been often discussed, although they may be involved in individual, non-constant DS phenotypes, such as APP, SOD1, KCNJ6, ETS2 and DSCAM (33).
Currently available annotation of human genome maps does not give any clue that HR-DSCR contains sequences associated to known protein families, and its very small 34 kb size is lower than the 67 kb mean size of a human protein-coding gene (as determined by searching the recent GeneBase database (47): mean of gene size from 17 958 ‘reviewed’ or ‘validated’ entries in the National Center for Biotechnology Information—NCBI—Gene April 2015 annotation release). An unknown microRNA (miRNA) gene might represent an excellent candidate for being a single major contributor to DS because of its pleiotropy of action. MiR-155 miRNA overexpression has been associated to DS through nexin 27 (48); however, no discussion was presented about its chromosomal localization proximally on 21q21.3 at 25.5 Mb, which is far from any described DSCR.
Further detailed analysis at bioinformatic and molecular genetic levels of the DSCR is needed to define its mechanism of action in causing DS, and the task will be facilitated by the restriction that we have presented here. If a causative role of functional elements in this region will be shown, this will also be of high interest in checking that they are actually silenced in the alternative therapeutic molecular approaches aimed at Hsa21 inactivation by XIST (49).
Our results are not inconsistent with the view that there is not a single chromosomal region on Hsa21 responsible for most of the DS phenotypic features (34). There are certainly different regions mostly associated to one or the other of the DS specific symptoms and signs (37,38). However, while none of the clinical characteristic can be considered pathognomonic of DS, the combination of the overall external appearance with relatively typical ID is specific (50), so we focused on presence or absence of DS as such. Due to the fact that ID also represents the main real problem for persons with DS, and the most relevant for the search of a cure which is still lacking in comparison with the availability of treatments for heart, thyroid and other disorders (51), our approach focused on the search for a very specific and restricted region associated with the manifestation of the basic, common features of DS and in particular ID appears to be well justified. The possibility that genes outside this candidate region are also contributing to ID is not problematic: it is likely for any imbalance of Hsa21 as well as indeed of any other chromosome to affect cognitive functions, considering that it is well known that the human brain is characterized by a very high number of active genes (52) as well as by a high prevalence of alternative splicing (53).
Our approach also solves the puzzle of the apparent involvement of a more proximal Hsa21 region in DS if ID is mapped as a single phenotype: indeed, when considering DS itself as the phenotype, the proximal region is no longer found because it may also be associated to ID in subjects without DS (17), in agreement with the fact that duplication of only proximal part of 21q never results in DS phenotype (Supplementary Material Table S1). Two subjects with ID contributed to the identification of a proximal region for ID because they were clustered with other cases with DS ID, but they were clearly identified as non-DS by Park et al. (54) (cases #039 and #139 in Supplementary Material Tables S1 and S2). It should be underlined here that typical DS cognitive deficits, that is delayed psychomotor development, disturbance in abstract thinking and inhibited speech development in presence of good learning, memory, imitation and socialization abilities (50) are well recognizable both by ‘gestalt’ impression gained through clinical experience as well as by natural history of developmental trajectory (55), neuropsychological testing (56) and brain imaging (57).
Genomics has contributed to present the concept of a ‘personalized medicine’ (each patient is different), somewhat fragmenting the traditional unity of nosographic entities (each disease is equal to itself). However, the ‘splitter’ and ‘lumper’ approaches are indeed complementary and each adds knowledge on its side (58). In DS, we have a well-defined cytogenetic marker, Hsa21, so, while we see obvious phenotype differences among each person with DS, a basic, defined common mechanism for the consistent, reproducible DS ‘core’ phenotype must exists. In several studies about DSCR, this has been ruled out by observing that a certain single duplicated region, for instance, is not ‘being responsible for all aspects of the phenotype’ (37), is not ‘responsible for all or most severe DS features’ (38), and ‘is not sufficient to cause the full DS phenotype’ (59). This view appears to be quantitative (‘all’, ‘most’ features) and does not account for the well-confirmed clinical observation that there are some features constituting a small group but present at the highest frequency in subjects with DS, so that a DSCR should rather be defined as the region which ‘suffices to induce the main phenotypic symptoms of the classic syndrome of trisomy 21’ (17) (where this region was indicated in the subbands 21q22.1 and 21q22.2, at whose boundary we find now the HR-DSCR).
Actually, clinical observations reported in the literature (60–62) as well as our experience in the follow-up of children with DS (63) clearly indicate that ID and some of the craniofacial dysmorphisms are the most frequent and constant features of DS, so that ‘the face of a person with DS is peculiar and typical from birth’ (50), while other symptoms and signs are less or much less frequent and may be reasonably associated to other Hsa21 regions contributing to the complex DS phenotype (along with nucleotide variation, CNVs and other individual genetic and possibly epigenetic features). These regions appear to be dispensable if the crucial DS stigma are concerned, that is fundamentally the same that allowed the historical recognition of the nosographic entity to Dr. John Langdon Down: a ‘close resemblance to one another in mental power’ in individuals with typical facies he called of ‘Mongolian’ type (64) before this inappropriate term was replaced by ‘Down syndrome’ or better ‘trisomy 21’ (65). It is not a case that, of the two most discriminating dysmorphic signs of DS found by statistical analysis since 1976 (61), oblique eyes and flat nasal bridge, the former was chosen by Down to initially name the syndrome itself, and the latter has become widely used to suspect DS since prenatal age by ultrasound scanning. The incipit of a fundamental 1990 article by Lejeune about DS pathogenesis (11) also started by describing these signs: ‘With upward-slanting eyelids, a little nose in a round face, and incompletely chiseled features, DS patients look more like children than the usual child does’, then adding that their character is ‘cheerful and affectionate, they have a special charm easier to cherish than to describe’, but that the condition deprives ‘the children of the most precious quality afforded by our genetic patrimony, the full power of rational thinking’. In fact, these two facial signs typically allow to anticipate the full manifestation of ID, the most constant feature in absolute, along with which they can be legitimately defined ‘critical’ without necessarily being associated to a ‘full’ symptomatological spectrum. It should be also noted that the whole Hsa21 itself is not able to cause the full range of symptoms and signs of DS which are never all displayed by the same subject, are present with a great variability even among subjects with complete trisomy 21, and may involve virtually any organ. Rather, the nearly constant presence of a few specific features (ID and facies) has been well recognized clinically as shared by subjects with complete as well as PT21.
With regard to the presence in our series of numerous cases of PT21 arising from reciprocal translocation, potential positional effects should be discussed. We observe that, due to the wide variety of translocation type or the autosome involved along with Hsa21, the consistency of the core DS phenotype and the coherence of the genotype–phenotype relationship with regard to DS/non-DS status in both non-translocated or translocated PT21 cases (Supplementary Material Tables S1 and S2) suggests that positional effects are of little relevance for the problem considered in this work, while they could affect severity of the core phenotypes as well as manifestation or not of less frequent symptoms and signs.
Some of the subjects described in the works that we have examined present with a DS phenotype and no evidence of cytogenetic/molecular alteration; these cases are known as phenocopies, 4 were described by McCormick et al. (28), and one by Anneren et al. (66) and later by Ahlbom et al. (67). In these cases, no marker was analyzed within the HR-DSCR. Significantly, all reports of phenocopies predate the availability of methods with high resolution at molecular level. In many cases, as reviewed in the Introduction section, a PT21 was indeed discovered following a reanalysis at higher resolution of the apparently normal chromosomes in a subject with DS. In addition, no case of DS with a specific chromosomal/genic alteration involving a chromosome different from Hsa21 has been reported to date. Taken together, these findings are consistent with the possibility that a very small Hsa21 region might be duplicated in the subjects described as ‘phenocopies’ and might be sufficient for the expression of basic DS features. We have been unsuccessful in the attempt to obtain samples from these subjects to reanalyze their DNA by current high-resolution methods.
While this work was in revision, two additional PT21 cases were reported (13) in two Taiwanese fetuses without DS, who both inherited from their respective healthy mothers a proximal (14.3 Mb at 21q11.2q21.3) and a distal (4.4 Mb on 21q22.2q22.3) duplication, respectively. These two cases alone allow the confirmation that duplication of regions covering a total of 55% of 21q, and not including 21q22.13 where we locate HR-DSCR, is not associated to DS. Our conclusions can be in a similar way subjected to prospective validation by accurately following the future literature about PT21, as well as by stimulating experimental reanalysis at higher resolution of the already described cases when possible and useful.
Naturally occurring autosomal trisomy with resemblance to DS in an animal has been described to our knowledge only in a chimpanzee (68). Remarkably, the HR-DSCR we have identified here has no homology as such to the mouse genome, while it is conserved only in humans and chimpanzee. This is not inconsistent with a role in superior cognitive functions and with previous observations that gene content in the previously defined larger DSCR is not identical between humans and mouse (69,70). The DSCR4 locus (71), which is expressed in the placenta and is the known gene located nearest to the distal boundary of HR-DSCR, is present only in humans.
Should further analysis demonstrate critical functions for DS in HR-DCSR, this might require a rediscussion of the usefulness of mouse models for confuting the existence of a DSCR in mouse (72) as well as for delimiting it (73). Actually, mouse chromosome band 16C4 presents the same loci that surround human HR-DSCR, and in the same order (Dyrk1a, Kcnj6, Kcnj15, Erg, Ets2), but it lacks any relevant homology with the HR-DSCR sequence itself that is located between KCNJ6 and KCNJ15 in humans, and such homology cannot be found in the rest of mouse genome. Although mouse models of DS are now available containing rearranged chromosomes 10, 16 and 17 in order to possibly include the full spectrum of genes localized on Hsa21 (74) and although formally it cannot be excluded that a similar function may be exerted in different vertebrates by diverged sequences with no homology between them (75), our finding contributes to bring attention to the known multiple limits of genetic similarities between trisomy of Hsa21 and mouse models (76). More generally, there is also the open problem of the legitimacy of comparing the impairment of superior functions most typical of human intelligence (77) and selectively damaged in DS, such as abstraction and language, to alterations of mouse behavior (78).
Finally, we note that while we have focused here on ID and facies as the very basic and more constant DS features, the framework we have built could constitute a valuable resource to record and map in the future other particular phenotypes described in the 125 cases from whom we have obtained the integrated map, to validate in a much larger number of cases different specific regions described as associated to, for example, congenital heart disease (37,38) or other phenotypes with a not constant presentation in DS. A relevant issue in this regard is the association between the duplication of APP Hsa21 gene, encoding amyloid beta precursor protein involved in Alzheimer disease (AD) and early onset of an AD-like dementia in a progressively larger proportion of subjects with DS with increasing age. While we have pointed out here that the ID evident since the first years is a universal sign of DS, formerly known as mental retardation and diagnosed before the age of 18 years, a form of dementia similar to AD affects only a proportion of subjects with trisomy 21 (79,80), although the risk may be upward of 70% when reaching 70 years of age, and is seen as progressive deterioration of cognitive and functional abilities. No convincing relationship between severity of ID (or Intelligence Quotient—IQ—score) and risk of AD has been found in people with DS (79). As these facts suggest, the intimate mechanisms at the base of these cognitive alterations must be different. An interesting finding that emerges from our map is that 42 subjects diagnosed with DS and with a demonstrated DS ID present with no duplication of APP (Supplementary Material Tables S1 and S2), further underlining that APP duplication is dispensable for DS ID and therefore that at least some core molecular mechanisms for DS ID are distinct from those for AD. It would be very interesting, on the other hand, to do an analogue mapping study correlating presence or absence of APP duplication with development or not of DS AD. However, only a few cases are at present useful in this respect; for example, two subjects with PT21 in absence of APP duplication did not develop AD until at least 65 (38) or 78 (81,82) years of age (our cases #111 and #77, respectively). Articles about PT21 cases are typically not prospectively followed by successive reports about the onset or not of AD, although the potential relevance of a similar observation is clear; for example, the just published report by Su et al. (13) states that ‘whether these two probands have CBS- or APP-associated disorders is an issue that remains to be closely followed up in their later lives’.
In conclusion, our results support the view that a single region associated with the manifestation of core DS features lies on 21q22.13, appearing to be much smaller than previously suspected and possibly containing currently undescribed genes whose identification might become a priority in searching for highly relevant targets for a cure of DS ID.
Materials and Methods
Bibliographic search
In order to retrieve all published articles related to PT21 we first performed systematic queries on PubMed at the NCBI site (http://www.ncbi.nlm.nih.gov/pubmed/), using a combination of wildcards (*), Boolean operators (AND/OR) and MeSH terms. Details of the bibliographic search are given in the Supplementary Material Methods file.
In summary, a non-redundant set of 1322 PubMed records (from the total of 1414 PubMed results found with all the queries) was examined (Table 1).
All items with certain or suspect matching the aim of the study, based on title and/or abstract, were obtained as full text. For many articles, there was not a downloadable electronic version, so we used library services of the University of Bologna through ‘NILDE—Network Inter Library Document Exchange’ to obtain scanned copies of the old paper reports. Language translation was sought, when necessary, from French or Polish.
The references cited in all the selected articles were examined, and when pointing to articles that were not identified by PubMed searches, those articles were obtained and their references were examined in turn. The process was repeated until convergence, that is, as long as no further article pertinent to the study subject was identified.
Three further articles describing PT21 cases were retrieved by a PubMed search in 2016, following the completion of our map and while this work was in revision. The first article, although actually published in 2015 (83), was not retrieved in our previous search performed through December 9, 2015 (Supplementary Material Methods file) because it did not mention ‘Down syndrome’ (a concept that was added only after the later MeSH terms indexing by PubMed) nor the word root ‘21q*’, nor the expression ‘Chromosome 21’. The same subject was first described in a 1965 report (84), which was not retrieved because of using the term ‘mongoloid’ without containing the ‘trisom*’ word root, although all terms related to the ‘mongol’ root were officially banned from 1961 (65). This report was not further cited by other articles related to the matter. The subject showed a DS phenotype although attenuated and a derivative chromosome 15 with a duplication of 21q spanning from 21q21.2 to the telomer, thus consistent with our findings reported above.
The second article (85) describes a de novo duplication of ∼0.3 Mb in 21q22.3 associated with duplication of ∼0.3 Mb in 12p13.33 in a subject with ID and distinctive phenotypic features, deriving from both of the two genetic alterations, therefore, it would not have been considered for our study because of possibly confounding effects on the DS phenotype of the 12p duplication, according to the criteria outlined in the ‘Case selection’ subsection below. The third article (13) has been cited and discussed in the Introduction and Discussion sections above.
Case selection
An accurate selection of cases was made by reevaluation of each report describing cases of PT21 in order to only include cases with sufficient and unambiguous description at cytogenetic (and possibly molecular) and clinical level for the purpose of creation of the integrated map.
Cytogenetic inclusion criteria were duplication of a partial portion of 21q in absence of complex cytogenetic anomaly: trisomy 21 with one interstitial deletion or segmentally replicated Hsa21; reciprocal translocations involving segments of 21q; tandem translocations with an incomplete long arm of the replicated Hsa21. Exclusion criteria were: reciprocal or tandem translocations and ring Hsa21 with a complete 21q; tetrasomies of Hsa21 that have a different gene dosage in comparison with trisomy 21; mosaic trisomy 21, less informative for determining the phenotypic effects caused by an excess of Hsa21 segments (10) due to effects of the cell mosaicism on the phenotype possibly confounding those derived from the incomplete duplication of 21q (16); chromosomal rearrangements involving Hsa21 and the X chromosome, for the effects due to variable inactivation of Hsa21 regions translocated to X (17); chromosomal alterations described in leukemic cell clones.
Molecular analysis criteria were description in sufficient detail of the duplicated segment boundaries: availability of at least the banding pattern; availability of sequence data allowing placement of sequence-tagged sites (STSs) and FISH probes on the map; coherence among different methods when they were used to study the same subject. Cases with incomplete description of the limits of the duplicated segments or with discrepancies among results obtained by different methods of analysis were not further considered.
Clinical criteria for selection of cases were: all subjects with or without DS clinical features, as long as they present with a PT21. Subjects were classified as ‘DS’ or ‘non-DS’ according to: explicit statements found in the study, when authors judged DS recognizable as present or absent, irrespectively of other symptoms or signs associated to possibly concurrent aneuploidies of non-Hsa21 chromosomal segments, or assessment of detailed phenotype description when present in the article. Subjects with uncertain diagnosis of DS or non-DS status were excluded. This led, for example, to exclusion of 13 cases analyzed by Lyle et al. (37) because while the description made the study of individual phenotypes possible, which was the object of their work, it did not allow the certain assignment of these subjects to DS or non-DS group. Fetuses were excluded due to the impossibility of ascertaining phenotype in detail (60). Subjects with DS phenotype but with no visible chromosomal abnormality as well as no described molecular alteration (possible phenocopies of DS, for example (28,66,67), where the last two references describe the same case) could not be considered for the goal of PT21 map building.
Map framework building
First, we used the software Transcriptome Mapper (TRAM) (86) to process human genome data downloaded from the ‘NCBI Gene’ (http://www.ncbi.nlm.nih.gov/gene) databank and to build an updated framework of the structure of Hsa21 based on known Hsa21 genes. The framework was imported in a spreadsheet table and then enriched with coordinates for single nucleotide polymorphisms (SNPs), STSs, bacterial artificial chromosome (BAC) clones, nucleotide positions determined by Array CGH as limits of altered regions in individual subjects, cytogenetic band limits and key CNVs. Full details are given in the Supplementary Material Methods file.
All the genomic coordinates related to previous versions of the human genome sequence were converted in the matching current coordinates on hg38 using the on line tool LiftOver (https://genome.ucsc.edu/cgi-bin/hgLiftOver). When this was not possible due to lack of information for the STS-related primer sequences or the location of the two primer sequences was inconsistent, the STS was not placed on our comparative map. When mapping of FISH data required placement of BAC clone intervals on the map, the related coordinates were derived by ‘NCBI Clone’ (http://www.ncbi.nlm.nih.gov/clone) database. Finally, CGH data were also converted to hg38 coordinates using the LiftOver tool when appropriate.
In summary, spreadsheet rows represented any specific and relevant sequence feature on Hsa21 for a total of 698 sequence intervals (rows), providing anchor points useful to homogeneously map each cytogenetic feature described in the reports of PT21 from 1973 to 2015.
Comparative map building
For each subject studied, a column on the file built as explained above was added, representing the structure of his/her Hsa21. Each row represented a specific sequence interval on Hsa21, and for each subject with DS the corresponding cell was colored following this code: red = trisomic, therefore, possible candidate as causing DS; green = disomic, therefore, excluded as causing DS; blue = monosomic, considered as ‘not duplicated’, therefore, excluded as candidate; white = information not available with certainty. A complementary reasoning was used to color the cells representing sequence intervals when the subject presented cytogenetically with a segmental trisomy 21 in absence of a typical DS picture. In particular: red = disomic in non-DS, therefore, not excluded as causing DS; green = trisomic in non-DS, therefore, excluded as causing DS; blue = monosomic, considered as ‘not duplicated’, therefore, indirectly not excluded as candidate; white = information not available with certainty. Headings of non-DS cases are highlighted in yellow.
Each usable detail provided in the analyzed reports has been exploited to build the comparative map. When only cytogenetic banding was available, the whole band has been labeled according to the report, using the most updated correlation between Hsa21 bands/subbands and human genome coordinates available in University of California Santa Cruz (UCSC) Genome Browser (https://genome.ucsc.edu). In more recent works, STS, polymerase chain reaction (PCR), FISH and CGH data were used. All efforts were made to position each feature on the map, converting the reported coordinates, usually preceding 2013, in the matching current coordinates on hg38 (December 2013) using the on line tool LiftOver.
Subjects repeatedly reported by different authors in the course of years were identified, when it was possible to do so, based on the cross references in the case description and the data produced by different authors and methods for the same subject were integrated on the same column of the map.
Scoring system
A score was assigned to each interval sequence substantially following the scoring system applied by Lyle et al. (37), but attributing a lower score to not excluded regions in non-DS subjects due to the fact that these regions would be candidate regions only indirectly.
A score of +1 was assigned to each trisomic (candidate) sequence interval in DS subjects, while +0.5 was assigned to disomic (not excluded) intervals in non-DS subjects. A score of −1 was assigned to each sequence interval that was excluded as candidate for DS, being disomic in DS or trisomic in non-DS subjects. Monosomic regions are considered as ‘not duplicated’, therefore, they should be excluded as candidates in DS subjects (score = −1) and indirectly not excluded in non-DS (score = +0.5). For each sequence interval, the algebraic sum of the scores is calculated, generating the final score for the interval. The Excel macro and the Python scripts implementing the described algorithms for the calculations of the scores and for summarizing scores along Hsa21 regular intervals (Fig. 2), respectively, are available upon request.
Higher scores indicate increased probability of association to DS. Detailed partial and final scores for each interval are reported in Supplementary Material Table S1 at the right of the columns representing mapping for the 125 analyzed cases.
CNV analysis
The Database of Genomic Variants (DGV), a curated catalogue of human genomic structural variation (87) (http://dgv.tcag.ca/), was searched to identify any reported gain/duplication CNV in the region identified as DSCR, using hg38 map. In addition, subsequent intervals of 1 Mb were searched to count for described gain/duplication CNVs along 21q22 sequence. For the searches, DGV data were downloaded and imported in a FileMaker Pro (FileMaker, Santa Clara, CA, USA) database. Structural variants were also checked on the just released map by Sudmant et al. (88).
Phenotype recording
In recording phenotype, we focused on ID and on the most typical signs characteristic of the facies of subjects with DS, being the particular facies, the most frequent sign associated to ID in DS (see Introduction section). Hypotonia was not considered because it has to be observed in particular at birth and in many reports, it was not recorded; in addition, its evaluation suffers of some grade of subjectivity.
Oblique eye fissure (upslanted palpebral fissures) and flat nasal bridge have been recognized in the classic work by Jackson et al. (61) as the most discriminating signs. Korenberg et al. (34) reported that in their series of PT21 cases with DS ID, upslanted palpebral fissures and broad hands were the most constant signs. We recorded presence or absence, when known, of these three mentioned signs, along with epichantus, flat occipit and mouth permanently open for each case analyzed to build the present map. This choice is in agreement with other reports (50,62).
The six chosen individual signs were recorded as present or absent when this was explicitly stated, and as not available (N/A) when no explicit statement was available. In some cases where the picture of the subject was presented and it was clear, it was possible to deduce the presence or absence of a phenotype not explicitly described or excluded in the text. Diagnosis of presence or absence of DS was accepted as such when clearly stated in the original article describing the case.
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
We wish to sincerely thank the Fondazione Umano Progresso, Milano, Italy for their fundamental support to our research on trisomy 21 and to this study.
We thank all the other people that very kindly contributed by individual donations to support part of the fellowships as well as computer hardware that allowed to conduct this research. In particular, we are profoundly grateful to Matteo and Elisa Mele, to the ‘Gruppo Arzdore’, ‘Parrocchia di Dozza’ and ‘Associazione Turistica Pro Loco di Dozza’ (Dozza, Bologna, Italy), and to Illumia, Bologna, as well as to the Costa, Dal Monte, Ghignone, Morini and Valetti families, the architects of the Jérôme Lejeune exhibition at the Rimini Meeting in 2012, Rina Bini, Lara Bosa, Toni Brandi, Centro Bresciano Down—Onlus, Centro Culturale Piergiorgio Frassati—Ravenna, Anna Fusina, Roberta Galassi, Giovanni Guida, Tiziana Martinelli, Carla Molinari Samarani and Guido Samarani, Marco Sfirra and Giuditta Samarani.
We are sincerely grateful to Dr. Giuseppe Colpani (Director General) and Dr. Elisabetta Zanette (Rector and General Director Staff Coordinator), University of Bologna, for starting the web site for donations to Bologna University including our research project, and to Dr. Julia Hoffmann and Dr. Daniela Tibaldi for their valuable support with the web site, as well as to Dr. Claudio Negro, Elisa Bettini, Monica Fiori and Sabina Meneghetti (Research Office, Department of Experimental, Diagnostic and Specialty Medicine—DIMES at University of Bologna) for their kind and continuous support in managing the research funding. We are really pleased to thank Vittoria Aiello and Massimiliano Albanese for starting a new international initiative in support of our research.
Special thanks to Maria Cristina Labanti (NILDE service, University of Bologna) for her kind and valuable assistance in the document delivery, to Pamela Magini for her kind and expert consultation about Array CGH, and to Alessandro Ghezzo (University of Bologna) for his kind and expert consultation about neuropsychology of DS.
A very special thanks to Dr. Ombretta Salvucci (NIH, Bethesda, USA) for her incomparable and friendly support in discussing Lejeune's life and ideas and encouragement and scientific advice in pursuing the present project, along with Drs. Mark Basik (McGill University, Montreal, Canada), Anna C. Berardi (Santo Spirito's Hospital, Pescara, Italy), Maria Chiara Mimmi (University of Udine, Italy), Maria Chiara Monaco (NIH, Bethesda, USA), Annalisa Radeghieri and Doris Ricotta (University of Brescia, Italy), and Marcello Villanova (Nigrisoli Hospital, Bologna).
We are genuinely grateful to the parents and families of the Bologna associations CEPS (Centro Emiliano Problemi Sociali per la trisomia 21) and GRD (Genitori Ragazzi Down), in particular to Alfredo Donati, Vittorio Lazzari, Marina Magagnoli, Antonella Misuraca, Pier Luigi Sforza and Patrizia Torchi; and to Annalisa Sereni for their testimony and their continuing interest and confident support to our research, along with other families of children with DS.
We are grateful to Kirsten Welter for her kind and expert revision of the manuscript.
This work is dedicated to Professor Jérôme Lejeune (1926–1994), who has taught us ‘to fight against disease and to love the disabled’ and whose scientific thought has inspired this research, and to his fellow and teacher to some of us Professor Maria Zannotti, who brought back research on DS in Bologna in the late ‘60’s and is still advising us on the subject.
Conflict of Interest statement. The authors have declared that no conflict of interest exists.
Funding
Fellowships for MCP, AP and MC for the duration of the project have been mainly funded by the Fondazione Umano Progresso, Milano, Italy; the Foundation is a non-profit entity. A donation from the company Illumia, Bologna, Italy, supported one year of the MC fellowship. GP's fellowship has been funded by Lions Club Bologna Colli Augusto Murri, Bologna.
Donations from Fondazione Umano Progresso and from other donors acknowledged above supported the purchase of the hardware and software that were necessary to conduct the research.
Part of the computational biology analysis was executed on the Apple Mac Pro ‘Multiprocessor Server’ available at the ‘Center for Research in Molecular Genetics Fondazione CARISBO’, Bologna, and funded by Fondazione CARISBO. Funding to pay the Open Access publication charges for this article was provided by donations to the Laboratory of Genomics, DIMES, University of Bologna, for the study of trisomy 21.
References
- 1.Lejeune J., Gauthier M., Turpin R. (1959) Human chromosomes in tissue cultures. C. R. Hebd Seances Acad. Sci., 248, 602–603. [PubMed] [Google Scholar]
- 2.Gardiner K., Herault Y., Lott I.T., Antonarakis S.E., Reeves R.H., Dierssen M. (2010) Down syndrome: from understanding the neurobiology to therapy. J. Neurosci., 30, 14943–14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinet P.M., Allard D., Lejeune J., Jerome H. (1975) Letter: gene dosage effect in trisomy 21. Lancet, 1, 276.. [DOI] [PubMed] [Google Scholar]
- 4.Giannone S., Strippoli P., Vitale L., Casadei R., Canaider S., Lenzi L., D'Addabbo P., Frabetti F., Facchin F., Farina A, et al. (2004) Gene expression profile analysis in human T lymphocytes from patients with Down Syndrome. Ann. Hum. Genet., 68, 546–554. [DOI] [PubMed] [Google Scholar]
- 5.Patterson D. (2009) Molecular genetic analysis of Down syndrome. Hum. Genet., 126, 195–214. [DOI] [PubMed] [Google Scholar]
- 6.Roizen N.J., Patterson D. (2003) Down's syndrome. Lancet, 361, 1281–1289. [DOI] [PubMed] [Google Scholar]
- 7.Strippoli P., Pelleri M.C., Caracausi M., Vitale L., Piovesan A., Locatelli C., Mimmi M.C., Berardi A.C., Ricotta D., Radeghieri A, et al. (2013) An integrated route to identifying new pathogenesis-based therapeutic approaches for trisomy 21 (Down Syndrome) following the thought of Jérôme Lejeune. Sci. Postprint, 1, e00010. [Google Scholar]
- 8.Ilbery P.L., Lee C.W., Winn S.M. (1961) Incomplete trisomy in a mongoloid child exhibiting minimal stigmata. Med. J. Aust., 48, 182–184. [DOI] [PubMed] [Google Scholar]
- 9.Aula P., Leisti J., von Koskull H. (1973) Partial trisomy 21. Clin. Genet., 4, 241–251. [DOI] [PubMed] [Google Scholar]
- 10.Antonarakis S.E. (1998) 10 years of Genomics, chromosome 21, and Down syndrome. Genomics, 51, 1–16. [DOI] [PubMed] [Google Scholar]
- 11.Lejeune J. (1990) Pathogenesis of mental deficiency in trisomy 21. Am. J. Med. Genet., 7 (suppl.), 20–30. [DOI] [PubMed] [Google Scholar]
- 12.Gijsbers A.C., van Haeringen A., Bosch C.A., Hansson K., Verschuren M., Bakker E., Breuning M.H., Ruivenkamp C.A. (2010) A subtle familial translocation t(3;21)(p26.3;q22.3): an apparently healthy boy with a 3p deletion and 21q duplication. Cytogenet. Genome Res., 128, 245–249. [DOI] [PubMed] [Google Scholar]
- 13.Su M.T., Kuan L.C., Chou Y.Y., Tan S.Y., Kuo T.C., Kuo P.L. (2016) Partial trisomy of chromosome 21 without the Down syndrome phenotype. Prenat. Diagn, doi: 10.1002/pd.4796. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 14.Shaw J.L., Chang K.T. (2013) Nebula/DSCR1 upregulation delays neurodegeneration and protects against APP-induced axonal transport defects by restoring calcineurin and GSK-3beta signaling. PloS Genet., 9, e1003792.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Migeon B.R., Kaufmann B.N., Young W.J. (1962) A chromosome abnormality with fragment in a paramongol child. Bull. Johns Hopkins Hosp., 111, 221–229. [PubMed] [Google Scholar]
- 16.Dent T., Edwards J.H., Delhanty J.D. (1963) A partial mongol. Lancet, 2, 484–487. [DOI] [PubMed] [Google Scholar]
- 17.Subrt I., Prchlikova H. (1970) An extra chromosomal centric fragment in an infant with stigmata of Down's syndrome. J. Med. Genet., 7, 407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rethore M.O. (1981) In Burgio G.R., Fraccaro M., Tiepolo L., Wolf U. (eds.), Trisomy 21. Springer-Verlag, Berlin, pp. 173–182. [Google Scholar]
- 19.Caspersson T., Zech L., Johansson C., Modest E.J. (1970) Identification of human chromosomes by DNA-binding fluorescent agents. Chromosoma, 30, 215–227. [DOI] [PubMed] [Google Scholar]
- 20.Seabright M. (1971) A rapid banding technique for human chromosomes. Lancet, 2, 971–972. [DOI] [PubMed] [Google Scholar]
- 21.Gustavson K.H. (1964) Down's Syndrome: a clinical and cytogenetic investigation. Almqvist & Wiksell, Uppsala. [Google Scholar]
- 22.Hamerton J.L. (1971) Banding patterns of metaphase chromosomes in Down's syndrome. Lancet, 2, 709.. [DOI] [PubMed] [Google Scholar]
- 23.Eggermann T, Schwanitz G. (2011) In Dey S. (ed.), Genetics and Etiology of Down Syndrome. InTech, Rijeka, Croatia, pp. 3–22. [Google Scholar]
- 24.Daniel A. (1979) Normal phenotype and partial trisomy for the G positive region of chromosome 21. J. Med. Genet., 16, 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niebuhr E. (1974) Down's syndrome. The possibility of a pathogenetic segment on chromosome no. 21. Humangenetik, 21, 99–101. [DOI] [PubMed] [Google Scholar]
- 26.Pinkel D., Straume T., Gray J.W. (1986) Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc. Natl Acad. Sci. USA, 83, 2934–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rahmani Z., Blouin J.L., Creau-Goldberg N., Watkins P.C., Mattei J.F., Poissonnier M., Prieur M., Chettouh Z., Nicole A., Aurias A, et al. (1989) Critical role of the D21S55 region on chromosome 21 in the pathogenesis of Down syndrome. Proc. Natl Acad. Sci. USA, 86, 5958–5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCormick M.K., Schinzel A., Petersen M.B., Stetten G., Driscoll D.J., Cantu E.S., Tranebjaerg L., Mikkelsen M., Watkins P.C., Antonarakis S.E. (1989) Molecular genetic approach to the characterization of the “Down syndrome region” of chromosome 21. Genomics, 5, 325–331. [DOI] [PubMed] [Google Scholar]
- 29.Delabar J.M., Theophile D., Rahmani Z., Chettouh Z., Blouin J.L., Prieur M., Noel B., Sinet P.M. (1993) Molecular mapping of twenty-four features of Down syndrome on chromosome 21. Eur. J. Hum. Genet., 1, 114–124. [DOI] [PubMed] [Google Scholar]
- 30.Rahmani Z., Blouin J.L., Creau-Goldberg N., Watkins P.C., Mattei J.F., Poissonnier M., Prieur M., Chettouh Z., Nicole A., Aurias A, et al. (1990) Down syndrome critical region around D21S55 on proximal 21q22.3. Am. J. Med. Genet., 7 (suppl.), 98–103. [DOI] [PubMed] [Google Scholar]
- 31.Shapiro B.L. (1999) The Down syndrome critical region. J. Neural. Transm., 57 (suppl.), 41–60. [DOI] [PubMed] [Google Scholar]
- 32.Fuentes J.J., Pritchard M.A., Estivill X. (1997) Genomic organization, alternative splicing, and expression patterns of the DSCR1 (Down syndrome candidate region 1) gene. Genomics, 44, 358–361. [DOI] [PubMed] [Google Scholar]
- 33.Deitz S.L., Blazek J.D., Solzak J.P, Roper R.J. (2011) In Dey S. (ed.), Genetics and Etiology of Down Syndrome. InTech, Rijeka, Croatia, pp. 65–96. [Google Scholar]
- 34.Korenberg J.R., Chen X.N., Schipper R., Sun Z., Gonsky R., Gerwehr S., Carpenter N., Daumer C., Dignan P., Disteche C, et al. (1994) Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc. Natl Acad. Sci. USA, 91, 4997–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott J.A., Wenger S.L., Steele M.W., Chakravarti A. (1995) Down syndrome consequent to a cryptic maternal 12p;21q chromosome translocation. Am. J. Med. Genet., 56, 67–71. [DOI] [PubMed] [Google Scholar]
- 36.Hattori M., Fujiyama A., Taylor T.D., Watanabe H., Yada T., Park H.S., Toyoda A., Ishii K., Totoki Y., Choi D.K, et al. (2000) The DNA sequence of human chromosome 21. Nature, 405, 311–319. [DOI] [PubMed] [Google Scholar]
- 37.Lyle R., Bena F., Gagos S., Gehrig C., Lopez G., Schinzel A., Lespinasse J., Bottani A., Dahoun S., Taine L, et al. (2009) Genotype-phenotype correlations in Down syndrome identified by array CGH in 30 cases of partial trisomy and partial monosomy chromosome 21. Eur. J. Hum. Genet., 17, 454–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korbel J.O., Tirosh-Wagner T., Urban A.E., Chen X.N., Kasowski M., Dai L., Grubert F., Erdman C., Gao M.C., Lange K, et al. (2009) The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proc. Natl Acad. Sci. USA, 106, 12031–12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sturgeon X., Le T., Ahmed M.M., Gardiner K.J. (2012) Pathways to cognitive deficits in Down syndrome. Prog. Brain Res., 197, 73–100. [DOI] [PubMed] [Google Scholar]
- 40.Barnicoat A.J., Bonneau J.L., Boyd E., Docherty Z., Fennell S.J., Huret J.L., King M., Maltby E.L., McManus S., Pilz D.T, et al. (1996) Down syndrome with partial duplication and del (21) syndrome: study protocol and call for collaboration. Study I: Clinical assessment. Clin. Genet., 49, 20–27. [DOI] [PubMed] [Google Scholar]
- 41.Beckmann J.S., Estivill X., Antonarakis S.E. (2007) Copy number variants and genetic traits: closer to the resolution of phenotypic to genotypic variability. Nat. Rev. Genet., 8, 639–646. [DOI] [PubMed] [Google Scholar]
- 42.Lejeune J., Berger R., Vidal O.R., Rethore M.O. (1965) [A case of G-G translocation in tandem]. Ann. Genet., 8, 60–62. [PubMed] [Google Scholar]
- 43.Sailani M.R., Makrythanasis P., Valsesia A., Santoni F.A., Deutsch S., Popadin K., Borel C., Migliavacca E., Sharp A.J., Duriaux Sail G, et al. (2013) The complex SNP and CNV genetic architecture of the increased risk of congenital heart defects in Down syndrome. Genome Res., 23, 1410–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Priest J.R., Girirajan S., Vu T.H., Olson A., Eichler E.E., Portman M.A. (2012) Rare copy number variants in isolated sporadic and syndromic atrioventricular septal defects. Am. J. Med. Genet. A, 158A, 1279–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de la Torre R., Dierssen M. (2012) Therapeutic approaches in the improvement of cognitive performance in Down syndrome: past, present, and future. Prog. Brain Res., 197, 1–14. [DOI] [PubMed] [Google Scholar]
- 46.Cetin Z., Yakut S., Mihci E., Manguoglu A.E., Berker S., Keser I., Luleci G. (2012) A patient with Down syndrome with a de novo derivative chromosome 21. Gene, 507, 159–164. [DOI] [PubMed] [Google Scholar]
- 47.Piovesan A., Caracausi M., Ricci M., Strippoli P., Vitale L., Pelleri M.C. (2015) Identification of minimal eukaryotic introns through GeneBase, a user-friendly tool for parsing the NCBI Gene databank. DNA Res., 22, 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X., Zhao Y., Zhang X., Badie H., Zhou Y., Mu Y., Loo L.S., Cai L., Thompson R.C., Yang B, et al. (2013) Loss of sorting nexin 27 contributes to excitatory synaptic dysfunction by modulating glutamate receptor recycling in Down's syndrome. Nat. Med., 19, 473–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang J., Jing Y., Cost G.J., Chiang J.C., Kolpa H.J., Cotton A.M., Carone D.M., Carone B.R., Shivak D.A., Guschin D.Y, et al. (2013) Translating dosage compensation to trisomy 21. Nature, 500, 296–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tolksdorf M, Wiedemann H.R. (1981) In Burgio G.R., Fraccaro M., Tiepolo L., Wolf U. (eds.), Trisomy 21 . Springer-Verlag, Berlin, pp. 3–32. [Google Scholar]
- 51.Weijerman M.E., de Winter J.P. (2010) Clinical practice. The care of children with Down syndrome. Eur. J. Pediatr., 169, 1445–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramskold D., Wang E.T., Burge C.B., Sandberg R. (2009) An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol., 5, e1000598.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de la Grange P., Gratadou L., Delord M., Dutertre M., Auboeuf D. (2010) Splicing factor and exon profiling across human tissues. Nucleic Acids Res., 38, 2825–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park J.P., Wurster-Hill D.H., Andrews P.A., Cooley W.C., Graham J.M., Jr. (1987) Free proximal trisomy 21 without the Down syndrome. Clin. Genet., 32, 342–348. [DOI] [PubMed] [Google Scholar]
- 55.Fidler D.J. (2005) The emerging Down syndrome behavioral phenotype in early childhood – implications for practice. Infants Young Children, 18, 86–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghezzo A., Salvioli S., Solimando M.C., Palmieri A., Chiostergi C., Scurti M., Lomartire L., Bedetti F., Cocchi G., Follo D, et al. (2014) Age-related changes of adaptive and neuropsychological features in persons with Down syndrome. PLoS One, 9, e113111.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Menghini D., Costanzo F., Vicari S. (2011) Relationship between brain and cognitive processes in Down syndrome. Behav. Genet., 41, 381–393. [DOI] [PubMed] [Google Scholar]
- 58.Watson R.A. (2013) Searching for the answer to cancer: new perspective may help to resolve an ageless conundrum. Urol. Oncol., 31, 721–726. [DOI] [PubMed] [Google Scholar]
- 59.Papoulidis I., Papageorgiou E., Siomou E., Oikonomidou E., Thomaidis L., Vetro A., Zuffardi O., Liehr T., Manolakos E., Vassilis P. (2014) A patient with partial trisomy 21 and 7q deletion expresses mild Down syndrome phenotype. Gene, 536, 441–443. [DOI] [PubMed] [Google Scholar]
- 60.Hall B. (1966) Mongolism in newborn infants. An examination of the criteria for recognition and some speculations on the pathogenic activity of the chromosomal abnormality. Clin. Pediatr. (Phila), 5, 4–12. [DOI] [PubMed] [Google Scholar]
- 61.Jackson J.F., North E.R, 3rd., Thomas J.G. (1976) Clinical diagnosis of Down's syndrome. Clin. Genet., 9, 483–487. [DOI] [PubMed] [Google Scholar]
- 62.Devlin L., Morrison P.J. (2004) Accuracy of the clinical diagnosis of Down syndrome. Ulster Med. J., 73, 4–12. [PMC free article] [PubMed] [Google Scholar]
- 63.Cocchi G., Gualdi S., Bower C., Halliday J., Jonsson B., Myrelid A., Mastroiacovo P., Amar E., Bakker M.K., Correa A, et al. (2010) International trends of Down syndrome 1993–2004: Births in relation to maternal age and terminations of pregnancies. Birth Defects Res. A: Clin. Mol. Teratol., 88, 474–479. [DOI] [PubMed] [Google Scholar]
- 64.Down J.L. (1995) Observations on an ethnic classification of idiots. 1866. Ment. Retard., 33, 54–56. [PubMed] [Google Scholar]
- 65.Allen G., Benda C.E., Book J.A., Carter C.O., Ford C.E., Chu E.H., Hanhart E., Jervis G., Langdon-Down W., Lejeune J, et al. (1961) Mongolism. Am. J. Hum. Genet., 13, 426.. [PMC free article] [PubMed] [Google Scholar]
- 66.Anneren G., Edman B. (1993) Down syndrome–a gene dosage disease caused by trisomy of genes within a small segment of the long arm of chromosome 21, exemplified by the study of effects from the superoxide-dismutase type 1 (SOD-1) gene. APMIS, 40 (Suppl), 71–79. [PubMed] [Google Scholar]
- 67.Ahlbom B.E., Goetz P., Korenberg J.R., Pettersson U., Seemanova E., Wadelius C., Zech L., Anneren G. (1996) Molecular analysis of chromosome 21 in a patient with a phenotype of Down syndrome and apparently normal karyotype. Am. J. Med. Genet., 63, 566–572. [DOI] [PubMed] [Google Scholar]
- 68.McClure H.M., Belden K.H., Pieper W.A., Jacobson C.B. (1969) Autosomal trisomy in a chimpanzee: resemblance to Down's syndrome. Science, 165, 1010–1012. [DOI] [PubMed] [Google Scholar]
- 69.Luke S., Gandhi S., Verma R.S. (1995) Conservation of the Down syndrome critical region in humans and great apes. Gene, 161, 283–285. [DOI] [PubMed] [Google Scholar]
- 70.Takamatsu K., Maekawa K., Togashi T., Choi D.K., Suzuki Y., Taylor T.D., Toyoda A., Sugano S., Fujiyama A., Hattori M, et al. (2002) Identification of two novel primate-specific genes in DSCR. DNA Res., 9, 89–97. [DOI] [PubMed] [Google Scholar]
- 71.Nakamura A., Hattori M., Sakaki Y. (1997) A novel gene isolated from human placenta located in Down syndrome critical region on chromosome 21. DNA Res., 4, 321–324. [DOI] [PubMed] [Google Scholar]
- 72.Olson L.E., Richtsmeier J.T., Leszl J., Reeves R.H. (2004) A chromosome 21 critical region does not cause specific Down syndrome phenotypes. Science, 306, 687–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang X., Liu C., Yu T., Zhang L., Meng K., Xing Z., Belichenko P.V., Kleschevnikov A.M., Pao A., Peresie J, et al. (2015) Genetic dissection of the Down syndrome critical region. Hum. Mol. Genet., 24, 6540–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu T., Li Z., Jia Z., Clapcote S.J., Liu C., Li S., Asrar S., Pao A., Chen R., Fan N, et al. (2010) A mouse model of Down syndrome trisomic for all human chromosome 21 syntenic regions. Hum. Mol. Genet., 19, 2780–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Doglio L., Goode D.K., Pelleri M.C., Pauls S., Frabetti F., Shimeld S.M., Vavouri T., Elgar G. (2013) Parallel evolution of chordate cis-regulatory code for development. PLoS Genet., 9, e1003904.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nelson D.L., Gibbs R.A. (2004) Genetics. The critical region in trisomy 21. Science, 306, 619–621. [DOI] [PubMed] [Google Scholar]
- 77.Lejeune J. (1966) Chromosomal studies in psychiatry. Recent Adv. Biol. Psychiatry, 9, 13–20. [DOI] [PubMed] [Google Scholar]
- 78.Nuffield Council on Bioethics. (2002) Research in behavioural genetics involving animals. In: Genetics and Human Behaviour: The Ethical Context. Nuffield Council on Bioethics, London. [Google Scholar]
- 79.Zigman W.B., Devenny D.A., Krinsky-McHale S.J., Jenkins E.C., Urv T.K., Wegiel J., Schupf N., Silverman W. (2008) Alzheimer's disease in adults with Down syndrome. Int. Rev. Res. Ment. Retard., 36, 103–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wiseman F.K., Al-Janabi T., Hardy J., Karmiloff-Smith A., Nizetic D., Tybulewicz V.L., Fisher E.M., Strydom A. (2015) A genetic cause of Alzheimer disease: mechanistic insights from Down syndrome. Nat. Rev. Neurosci., 16, 564–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prasher V.P., Farrer M.J., Kessling A.M., Fisher E.M., West R.J., Barber P.C., Butler A.C. (1998) Molecular mapping of Alzheimer-type dementia in Down's syndrome. Ann. Neurol., 43, 380–383. [DOI] [PubMed] [Google Scholar]
- 82.Zigman W.B. (2013) Atypical aging in Down syndrome. Dev. Disabil. Res. Rev., 18, 51–67. [DOI] [PubMed] [Google Scholar]
- 83.Hamm J.A., Carroll A.J., Mikhail F.M., Korf B.R., Finley W.H. (2015) Partial trisomy 21: a fifty-year follow-up visit. Am. J. Med. Genet. A, 167, 1610–1613. [DOI] [PubMed] [Google Scholar]
- 84.Finley S.C., Finley W.H., Rosecrans C.J., Phillips C. (1965) Exceptional intelligence in a mongoloid child of a family with a 13-15/partial 21 (d-partial g) translocation. N. Engl. J. Med, 272, 1089–1092. [DOI] [PubMed] [Google Scholar]
- 85.Mekkawy M.K., Mazen I.M., Kamel A.K., Vater I., Zaki M.S. (2016) Genotype/phenotype correlation in a female patient with 21q22.3 and 12p13.33 duplications. Am. J. Med. Genet. A, 170, 1050–1058. [DOI] [PubMed] [Google Scholar]
- 86.Lenzi L., Facchin F., Piva F., Giulietti M., Pelleri M.C., Frabetti F., Vitale L., Casadei R., Canaider S., Bortoluzzi S, et al. (2011) TRAM (Transcriptome Mapper): database-driven creation and analysis of transcriptome maps from multiple sources. BMC Genomics, 12, 121.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.MacDonald J.R., Ziman R., Yuen R.K., Feuk L., Scherer S.W. (2014) The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res., 42, D986–D992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Hsi-Yang Fritz M, et al. (2015) An integrated map of structural variation in 2,504 human genomes. Nature, 526, 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.