Abstract
Orofacial clefts (OFCs), which include non-syndromic cleft lip with or without cleft palate (CL/P), are among the most common birth defects in humans, affecting approximately 1 in 700 newborns. CL/P is phenotypically heterogeneous and has a complex etiology caused by genetic and environmental factors. Previous genome-wide association studies (GWASs) have identified at least 15 risk loci for CL/P. As these loci do not account for all of the genetic variance of CL/P, we hypothesized the existence of additional risk loci. We conducted a multiethnic GWAS in 6480 participants (823 unrelated cases, 1700 unrelated controls and 1319 case–parent trios) with European, Asian, African and Central and South American ancestry. Our GWAS revealed novel associations on 2p24 near FAM49A, a gene of unknown function (P = 4.22 × 10−8), and 19q13 near RHPN2, a gene involved in organizing the actin cytoskeleton (P = 4.17 × 10−8). Other regions reaching genome-wide significance were 1p36 (PAX7), 1p22 (ARHGAP29), 1q32 (IRF6), 8q24 and 17p13 (NTN1), all reported in previous GWASs. Stratification by ancestry group revealed a novel association with a region on 17q23 (P = 2.92 × 10−8) among individuals with European ancestry. This region included several promising candidates including TANC2, an oncogene required for development, and DCAF7, a scaffolding protein required for craniofacial development. In the Central and South American ancestry group, significant associations with loci previously identified in Asian or European ancestry groups reflected their admixed ancestry. In summary, we have identified novel CL/P risk loci and suggest new genes involved in craniofacial development, confirming the highly heterogeneous etiology of OFCs.
Introduction
Orofacial clefts (OFCs) are a heterogeneous group of craniofacial malformations that comprise a significant fraction of all human birth defects, occurring in approximately 1 in 700 individuals worldwide (1). The prevalence of OFCs varies widely across populations as Native American and Asian populations have the highest rates (∼1/500 live births), whereas African and African-American populations have the lowest rates (∼1/2500 live births). European-derived populations have intermediate rates at approximately 1/1000 live births. Populations from Central and South America also have intermediate prevalence rates that vary regionally, possibly reflecting their mixed ancestries.
Non-syndromic OFCs, which occur in the absence of other cognitive or structural abnormalities, have a complex etiology that reflects the combined actions of multiple genetic and environmental risk factors. The focus of much of the genetics research has been on the common forms of OFCs: cleft lip alone (CL), CL with palate (CLP) and cleft palate alone (CP) (2). As is true for many complex traits, substantial progress in gene identification has occurred for OFCs, owing to multiple successful genome-wide association studies (GWASs). To date, there have been six OFC GWASs (3–8), a meta-analysis (9) and several replication studies (10–12). A major finding from this work is that OFCs have significant genetic heterogeneity; collectively, these studies have identified at least 15 genetic loci with compelling statistical and biological support. However, these loci account only for a modest portion of the genetic variance of CL/P, suggesting that additional genetic risk factors are likely involved.
In addition, some of these loci appear to have far stronger effect sizes in some populations (3,9), which may contribute to the variability in prevalence rates. For example, the 8q24 locus shows strong evidence of association in populations with European ancestry, whereas the 1q32 (IRF6) and 20q12 (MAFB) loci show stronger effects among Asian populations (3,13). However, the majority of GWASs to date have focused on populations with European ancestry; only two included Asian populations (3,8,10), and none focused on Hispanic/Latino or African populations. In order to identify additional risk loci in diverse and understudied populations, we conducted a large multi-ethnic GWAS comprising unrelated cases, unrelated controls and trios of European, Asian and African ancestry, as well as admixed populations from Central and South America.
Results
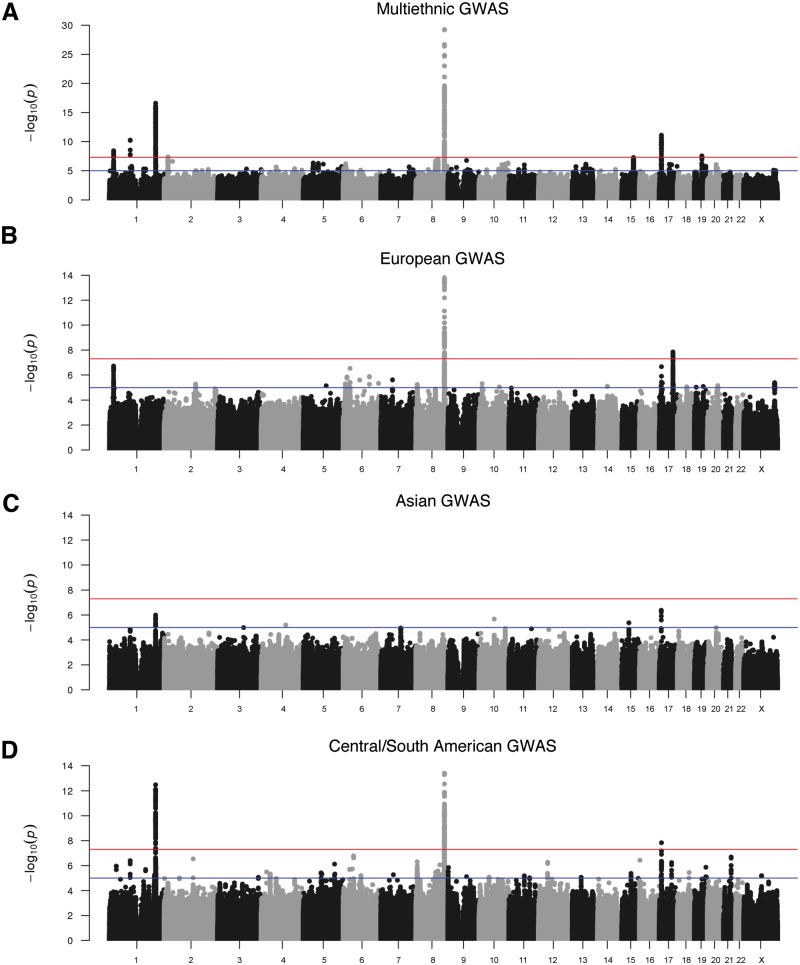
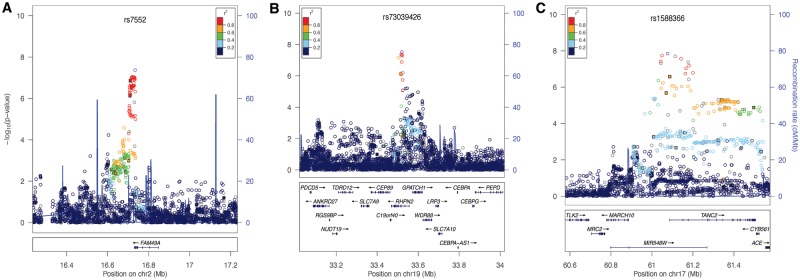
We performed tests of association in the two subsets of our multi-ethnic sample. The first was a set of 1319 case–parent trios in which the probands had CL/P, and the second was an independent case–control set consisting of 823 unrelated cases with CL/P and 1700 unrelated controls (Supplementary Material, Fig. S3). The 293,633 genotyped single nucleotide polymorphisms (SNPs) and 33,669,354 imputed SNPs were analyzed separately in each subset and combined by meta-analysis. Here, we focus on the results of the meta-analysis, but we note that similar results were obtained in both the case–control and case–parent trio analyses, indicating that the meta-analysis results are not driven by only one of the analyses. In the combined multi-ethnic sample, 316 (imputed) and 113 (genotyped) SNPs from seven loci were associated with CL/P at genome-wide significance (Fig. 1A, Table 1). Five of these loci were previously reported in GWASs of CL/P (1p36, near PAX7, 1p22 near ARHGAP29, 1q32 near IRF6, 8q24 at a gene desert and 17p13.1 near NTN1). Associations for two novel loci were observed on chromosome 2q24.2 near FAM49A (lead SNP rs7552, P = 4.22 × 10−8; Fig. 2A) and 19q13.11 near RHPN2 (lead SNP rs73039426, P = 2.92 × 10−8, Fig. 2B).
Figure 1.
Manhattan plots of –log10(P-values) from meta-analysis of case–control and TDT results in a multiethnic CL/P cohort. Results are shown for the combined multiethnic sample (A) and groups with (B) European ancestry, (C) Asian ancestry and (D) Central and South American ancestry. Colored lines denote suggestive (blue) and genome-wide (red) thresholds for significance. The genomic inflation factors, λ, are 1.088, 1.01, 0.979 and 1.045, for the multiethnic, European, Asian and Central/South American scans, respectively, indicating minor inflation in p-values consistent with the observation of multiple strongly associated loci.
Table 1.
Summary of GWAS meta-analysis results for loci with P < 5 × 10−7
| GWAS analysis population | Locus | SNP | Risk allele | TDT OR | CC OR | Meta OR | 95% CI | P-value |
|---|---|---|---|---|---|---|---|---|
| Multi-ethnic | 1p36.13 | rs4920524 | G | 1.33 | 1.34 | 1.34 | 1.21-1.47 | 3.72E−09 |
| 1p22.1 | rs4147882 | T | 1.49 | 1.33 | 1.42 | 1.28-1.58 | 5.59E−11 | |
| 1q32 | rs11119345 | G | 1.78 | 1.85 | 1.81 | 1.57-2.07 | 2.52E−17 | |
| 2p22 | rs372148350 | AT | 1.37 | 1.80 | 1.49 | 1.28-1.74 | 2.50E−07 | |
| 2p24.2 | rs7552 | G | 1.36 | 1.18 | 1.28 | 1.17-1.40 | 4.22E−08 | |
| 5p12 | rs59569344 | G | 1.31 | 1.39 | 1.34 | 1.19-1.50 | 4.75E−07 | |
| 5q13.1 | rs831227 | T | 1.24 | 1.26 | 1.25 | 1.15-1.36 | 5.34E−07 | |
| 8q21.3 | rs12543318 | C | 1.31 | 1.16 | 1.25 | 1.15-1.36 | 2.11E−07 | |
| 8q22.2 | rs1788160 | G | 1.35 | 1.20 | 1.29 | 1.17-1.41 | 9.52E−08 | |
| 8q24 | rs55658222 | A | 2.08 | 1.91 | 2.00 | 1.78-2.26 | 5.63E−30 | |
| 9q21.31 | rs6559624 | T | 1.45 | 1.19 | 1.34 | 1.20-1.50 | 1.72E−07 | |
| 10q26.3 | rs7922405 | A | 1.24 | 1.25 | 1.25 | 1.14-1.36 | 4.60E−07 | |
| 15q24 | rs28689146 | A | 1.24 | 1.35 | 1.28 | 1.17-1.40 | 5.40E−08 | |
| 17p13.1 | rs11273201 | AACCCAAAACCCAC | 1.44 | 1.43 | 1.44 | 1.29-1.59 | 7.84E−12 | |
| 19q13.11 | rs73039428 | A | 1.65 | 1.31 | 1.50 | 1.30-1.73 | 2.92E−08 | |
| European | 1p36.13 | rs9439714 | C | 1.62 | 1.38 | 1.52 | 1.30-1.89 | 1.91E−07 |
| 6p21.33 | rs79411602 | C | 1.67 | 1.31 | 1.52 | 1.29-1.78 | 2.92E−07 | |
| 8q24 | rs72728734 | G | 2.22 | 1.84 | 2.04 | 1.70-2.44 | 7.33E−15 | |
| 17p13.1 | rs7406226 | A | 1.50 | 1.73 | 1.59 | 1.33-1.89 | 2.16E−07 | |
| 17q23.2 | rs1588366 | A | 1.63 | 2.09 | 1.78 | 1.46-2.17 | 1.41E−08 | |
| Asian | 1q32 | rs1044516 | G | 1.63 | 2.04 | 1.71 | 1.38-2.11 | 1.01E−06 |
| 17p13.1 | rs11078735 | C | 2.62 | 1.52 | 2.27 | 1.65-3.12 | 4.20E−07 | |
| Central/ South American | 1p22.1 | rs4147882 | T | 1.55 | 1.41 | 1.48 | 1.27-1.73 | 3.99E−07 |
| 1p32 | rs11119345 | G | 1.85 | 1.77 | 1.81 | 1.55-2.13 | 3.40E−13 | |
| 2q21 | rs4850293 | G | 2.56 | 3.55 | 2.94 | 1.95-4.44 | 2.87E−07 | |
| 6p12.3 | rs6918746 | A | 1.27 | 1.59 | 1.40 | 1.23-1.59 | 1.65E−07 | |
| 8p23.1 | rs10780175 | T | 1.38 | 1.41 | 1.39 | 1.22-1.59 | 4.90E−07 | |
| 8q24 | rs55658222 | A | 1.92 | 1.92 | 1.92 | 1.62-2.27 | 3.98E−14 | |
| 16p13 | rs111698437 | T | 2.33 | 3.39 | 2.74 | 1.86-4.03 | 3.67E−07 | |
| 17p13.1 | rs7406226 | A | 1.60 | 1.28 | 1.45 | 1.28-1.65 | 1.46E−08 | |
| 17q22 | rs227727 | T | 1.41 | 1.37 | 1.39 | 1.22-1.58 | 5.73E−07 | |
| 21q22 | rs4920143 | T | 1.52 | 1.38 | 1.46 | 1.26-1.68 | 2.04E−07 |
Figure 2.
Regional association plots showing –log10(P-values) for genotyped (filled squares) and imputed (open circles) SNPs for novel genome-wide significant peaks. (A) Results at the 2p24 locus from the meta-analysis in the multi-ethnic cohort. (B) Results at the 19q13 locus from the meta-analysis in the multiethnic cohort. (C) Results at the 17q23 locus from meta-analysis in the European ancestry group. Plots were generated using LocusZoom (52). The recombination overlay (blue line, right y-axis) indicates the boundaries of the LD block. Points are color coded according to pairwise LD (r2) with the index SNP.
Seven additional regions approached genome-wide significance with P-values <5 × 10−7. All genotyped SNPs with P-values <1 × 10−5 are available in Supplementary Material, Tables S4–S7. One of these was a previously reported locus on 8q21 near DCAF4L2 and MMP16. Four novel loci had multiple SNPs approaching genome-wide significance: 5p12 near FGF10 (lead SNP rs59569344, P = 4.75 × 10−7), 8q22 in VPS13B (lead SNP rs1788160, P = 9.52 × 10−8), 10q26.3 in MGMT (lead SNP rs7922405, P = 4.60 × 10−7) and 15q24.1 in ARID3B (lead SNP rs28689146, P = 5.4 × 10−8). Regional plots for these four novel loci are in Supplementary Material, Figure S4. At the two remaining novel loci (2p22 and 9q21.3), single SNPs approached genome-wide significance.
To explore population-specific associations, we stratified our sample into genetically defined continental ancestry groups and used the same analysis approach (i.e., meta-analysis of results from the trio and case–control subsets) in European, Asian and Central/South American groups. Stratified analysis was not performed separately in the African group due to small sample size. Manhattan plots for case–control and transmission disequilibrium test (TDT) analyses in the subpopulations are presented in Supplementary Material, Figures S5–S7.
In the European group, two loci on 8q24 and 17q23 showed genome-wide significant associations (Fig. 1B, Supplementary Material, Table S5). The most significant SNP at the 8q24 gene desert was rs72728734 (P = 7.33 × 10−15). At the novel 17q23 locus, the most significant SNP was rs1588366 (P = 1.41 × 10−8) near TANC2 (Fig. 2C). Three loci approached genome-wide significance: 1p36 (PAX7), 17p13.1 (NTN1) and a novel locus on 6p21 at the HLA-B locus (lead SNP rs79411602, P = 2.92 × 10−7).
In the Asian group, no significant associations were observed (Fig. 1C). However, the previously reported region on 1q32 near IRF6 (lead SNP rs1044516; P = 1.01 × 10−6) and several SNPs on 17p13 (lead SNP rs12451139; P = 4.20 × 10−7) showed ‘suggestive’ evidence of association, represented by multiple SNPs with P-values in the order of magnitude range 10−7 to 10−6 (Supplementary Material, Table S6). SNPs on 1q32 and 17p13 have shown association with CL/P in Asian populations in prior studies and were observed in our multi-ethnic analysis. However, the most significant SNPs on chromosome 17 in the present study are located nearly 800 kb centromeric to the previously reported 17p13 region overlapping NTN1 and may represent a novel region unrelated to NTN1.
In the Central and South American group, three loci showed genome-wide significant evidence of association (1q32, 8q24 and 17p13) (Fig. 1D, Supplementary Material, Table S7). Five loci approached genome-wide significance on chromosomes 1p22, 6p12, 8p23, 16p13 and 21q22.3 (Table 1). We further scrutinized the peaks with suggestive evidence of association (P < 1.0 × 10−5), among which were a combination of signals originally observed in both European and Asian groups including 8q21 near MMP16 (lead SNP rs111608481, P = 3.43 × 10−6), and 17q22 near NOG (lead SNP rs227727, P = 5.73 × 10−7). The combination of signals originally observed in both European and Asian groups likely reflects the admixture and population history of the region.
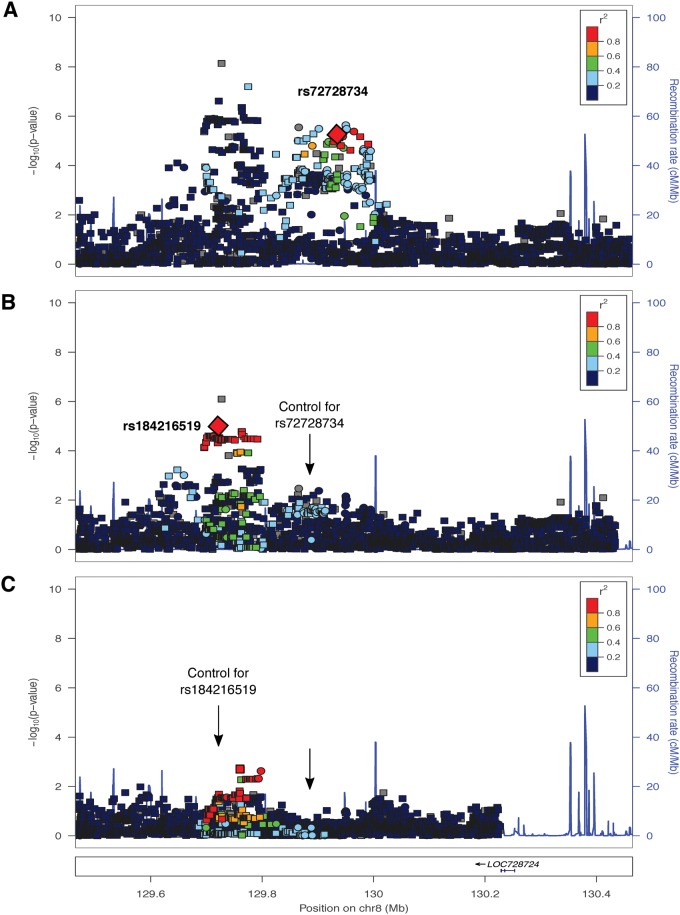
We performed conditional analyses on each of the genome-wide significant peaks within the case–control subset to determine whether these peaks represent multiple independent association signals. We identified two independent signals at the 8q24 locus in Europeans (Fig. 3). One signal was observed in previous studies (3–5); our lead SNP, rs72728734 (P = 6.60 × 10−6) was in high linkage disequilibrium (LD) with rs987525 (r2 = 0.81), the lead SNP identified previously. The second emerged after conditioning on our lead SNP (rs184216519, P = 9.57 × 10−6). We similarly observed two independent signals in the multi-ethnic meta-analysis but did not find evidence of two signals at this locus in the Central/South American analyses (data not shown). Conditioning on lead SNPs at 1q32 (IRF6) and 17p13 (NTN1) in the multiethnic analysis (Supplementary Material, Fig. S8) and 1q32 in the Central/South American analysis (Supplementary Material, Fig. S9) suggested the presence of multiple independent signals.
Figure 3.
Conditional analysis results for the 8q24 locus in European cases and controls. (A) Regional association plot showing –log10(P-values) for genotyped (squares) and imputed (circles) SNPs. The lead SNP in the European meta-analysis, rs72728734, is indicated by the large red diamond. (B) Regional association plot after conditioning on rs72728734. The lead SNP for the residual signal, rs184216519, is indicated by the large red diamond. The arrow indicated the located of the conditioned SNP. (C) Regional association plot after conditioning on rs184216519 and rs72728734. Arrows indicate the positions of the conditioned SNPs.
Our replication efforts focused on 10 SNPs in five loci: 2p24 (2 SNPs), 10q26.3 (2 SNPs) and 19q13 (3 SNPs) from the multi-ethnic analysis and 6p21 (1 SNP) and 17q23 (2 SNPs) from the European analysis. We genotyped 607 cases with CL/P and 1685 controls from three population-based case–control cohorts of European ancestry. The strongest evidence of association was with SNPs on 2p24 (rs7552, P = 2.68 × 10−7; Table 2). Nominal P-values were observed for SNPs at the 17q23 locus (P = 0.02). Although none of the three 19q13 SNPs were significant for CL/P, nominal P-values were observed for the CLP subgroup when stratified by cleft type (P < 0.01). A limitation of this study is that the replication cohort is European, reducing our ability to replicate multi-ethnic signals like 19q13.
Table 2.
Association results for replication study in independent European CL/P cases and controls
| Region | SNP | Minor allele | P-value | OR | 95% CI |
|---|---|---|---|---|---|
| 2p24.2 | rs7552 | G | 2.68E−07 | 1.47 | 1.27–1.71 |
| rs1465614 | C | 9.69E−06 | 1.41 | 1.21–1.64 | |
| 6p21.33 | rs2523554 | C | 0.21 | 1.09 | 0.95–1.26 |
| 10q26.3 | rs4751113 | C | 0.91 | 1.01 | 0.88–1.16 |
| rs7922405 | G | 0.89 | 0.99 | 0.86–1.14 | |
| 17q23.2 | rs16946713 | G | 0.03 | 0.82 | 0.68–0.98 |
| rs1588366 | G | 0.02 | 0.82 | 0.69–0.97 | |
| 19q13.11 | rs11881514 | A | 0.16 | 1.13 | 0.95–1.33 |
| rs73039426 | T | 0.13 | 0.78 | 0.56–1.08 | |
| rs8112217 | G | 0.23 | 0.81 | 0.57–1.15 |
Overall, the results of our multi-ethnic study corroborate findings from previous GWAS. Fifteen loci had reached genome-wide significance in the previous six GWASs or meta-analysis (3–6,8,9). Supplementary Material, Table S8 lists the P-values from the present study for the previously published SNPs. As discussed above, five of these loci reached genome-wide significance in this study. Five additional loci showed suggestive evidence of association (8q21 near MMP16, 10q25 near VAX1, 13q31 near SPRY2, 17q22 near NOG and 20q12 near MAFB). Weaker evidence for association was observed at three loci: 2p21 near THADA, 3p11 near EPHA3 and 16p13 near CRBBP. In aggregate, our results support a majority of the previously reported risk loci and implicate additional novel loci, indicating that the etiology of CL/P is highly heterogeneous.
Discussion
GWASs have allowed for the identification of risk loci for complex diseases and traits. For CL/P, six GWASs and a meta-analysis have identified 15 loci with genome-wide significance, confirming the multifactorial nature of this disease (3–6,8,14). In addition, these efforts have revealed population specificity of several loci (3,9). The current study adds to this body of literature by identifying new risk loci (some with population specificity), by confirming the previously nominated loci in European and Asian populations, and by expanding the GWAS approach to African and Central/South American populations.
Insights into population differences
As has been seen in other OFC GWASs, there are several peaks that are much more statistically significant in certain populations. Two such regions that have been consistently replicated, 1q32 (IRF6) and 8q24 (gene desert), were associated with CL/P in the current study’s Asian and European groups, respectively, as expected. Interestingly, both regions were also genome-wide significant in our Central/South American group. As summarized in Supplementary Material, Table S1, the Central/South American group comes from a variety of locations, with the majority of study subjects in this group from Colombia (276/449 cases and 405/601 trios). The Colombian families were ascertained from Paisa region with Medellin as the largest city (15). Paisas who speak a ‘pure’ dialect of Spanish comprise the majority of the population; previous genetic studies have shown that the Paisas are admixed including Caucasian (made up of Spaniards, Basques and to a small degree Sephardic Jews), Amerindians and African (15,16). Notably, data from mitochondrial and Y chromosome markers suggest that the population is an admixture of immigrant men and native women (17,18). The principal components of ancestry (PCA) analyses for ancestry were consistent with this admixture history for the Colombians and also for the other Central/South American study subjects. Therefore, it is particularly notable that the Central/South American group showed evidence of association with both OFC GWAS peaks previously identified with either European or Asian ancestry.
Insights into new risk loci
19q13. SNPs at the new 19q13 risk locus spanned RHPN2. This gene encodes the ubiquitously expressed RhoA binding protein, Rhophilin 2 (19). Although there are few studies devoted to elucidating the function of RHPN2, biochemical assays suggest that its normal function is to decrease stress fibers, a component of the actin cytoskeleton (19). RhoA belongs to a family of precisely regulated molecular switches that modulate a variety of cellular processes such as cytoskeletal reorganization, cell migration and cell–cell interactions. Several lines of evidence point to the critical role of Rho signaling and cell migration in craniofacial development. First, Irf6-deficient keratinocytes exhibit delayed migration, prominent stress fibers and increased RhoA activity (20). In contrast, Grhl3-deficient keratinocytes have decreased stress fibers and RhoA activity (21). Both IRF6 and GRHL3 are mutated in Van der Woude syndrome, the most common OFC syndrome (22,23). Activation of RhoA is required for transforming growth factor beta 3 signaling during palate formation (24). We previously investigated the 1p22 GWAS locus with expression studies and sequencing that implicated ARHGAP29 (a RhoA GTPase activating protein) as the etiologic OFC gene (25). Although additional studies into the role of RHPN2 in craniofacial development are needed, these association results are additional evidence pointing to abnormal regulation of Rho activity as a pathophysiological mechanism for OFCs.
17q23. SNPs in the 415 kb associated region on 17q23 span several genes including TANC2, CYB561 and a hypothetical micro-RNA. A definitive role for TANC2 in craniofacial development is not immediately apparent; however, mice homozygous for a gene trap allele die prior to embryonic day 12, suggesting that this gene is required for development (26). In addition, TANC2 is an oncogene that regulates epithelial cell growth and survival (27). If future studies confirmed a role in craniofacial development, TANC2 would join of the growing list of genes in which allelic variants increase the risk for certain cancers and also confer risk for OFCs including IRF6 (28), CDH1 (29) and FOXE1 (30). Another compelling candidate in the 17q23 region is DCAF7, located ∼500 kb from the peak signal. DCAF7, also known as WDR68, encodes a conserved Trp-Asp repeat protein that is thought to serve as a scaffold to facilitate protein–protein interactions. In zebrafish, wdr68 is required for craniofacial development (31,32). The dys mutant, which harbors an insertion in this gene, has reduced Meckel's and palatoquadrate cartilages, which serve as the embryonic lower and upper jaws, respectively (31). This phenotype resembled that of endothelin-1 (edn1) mutants leading to the conclusion that wdr68 is required for all edn1-dependent cartilages of the first arch. Although a Dcaf7−/− mouse has not been generated, Edn1−/− mice have many craniofacial anomalies including auricular and mandibular hypoplasia and CP (33). Moreover, micrognathia, bifid uvula and other oral anomalies have been described in humans with homozygous EDN1 mutations (34). Together, these data suggest that DCAF7/WDR68 and endothelin signaling have conserved roles in craniofacial development in vertebrates. Although this gene is located far from the peak signal, common etiologic variants could be located in regulatory elements of DCAF7, as has been shown for other OFC risk loci (13,25,30).
2p24. The associated SNPs at the 2p24 locus span the 3′-untranslated region of FAM49A and its downstream region. Little is known about FAM49A, which is expressed in the brain in mouse and zebrafish (35,36) and encodes a conserved protein of unknown function. Another compelling gene in the region is MYCN, the gene mutated in Feingold syndrome. This syndrome is characterized by digital anomalies, microcephaly and facial dysmorphism that can include OFCs.
Among the second tier hits, two loci (5p12 and 15q24) included compelling putative OFC risk genes. The 5p12 locus contains FGF9 and FGF10, both of which have been implicated in palatogenesis in humans and mice (37,38). Our peak signal at the 15q24 locus is within the 1.1-Mb critical region for 15q24 microdeletion syndrome, which includes facial dysmorphism and CP among the described features (39).
Conclusion
The goals of this study were to identify novel risk genes for CL/P and to extend the GWAS approach to understudied populations, such as admixed populations from Central and South America. The large sample sizes allowed us to accomplish both of these goals. In addition to confirming previous findings for nine risk loci, we identified three new risk loci with genome-wide significance and many other loci with compelling statistical support. We also confirmed the population specificity of some risk loci (e.g. 8q24 in Europeans) and showed that risk loci from both European and Asian groups were associated in the Central/South American group although no genome-wide significant novel risk loci specific to this admixed group were observed. Taken together, these results advance our understanding of the genetic architecture of OFCs, highlight the importance of a multi-ethnic approach to this common birth defect and may ultimately prove useful for predicting recurrence and prognosis.
Materials and Methods
Cohort description
The cohort for this study comes from a worldwide sample totaling 11,727 participants recruited from 18 sites across 13 countries from North America, Central or South America, Asia, Europe and Africa. Most of the sites were a part of ongoing genetic and phenotyping studies led by the University of Pittsburgh Center for Craniofacial and Dental Genetics; some sites were also a part of ongoing genetic studies led by the University of Iowa. In general, recruitment and study assessments were done at regional centers for OFC treatment. All sites had Institutional Review Board approval both locally and at the University of Pittsburgh or University of Iowa as appropriate for participation after informed consent and for large-scale genomic studies including data sharing.
The overall study cohort includes OFC-affected probands, their unaffected family members and controls with no known history of OFC or of other craniofacial anomalies. The specific affection status included for the current study was CL with or without CP (CL/P). We partitioned the total sample into two mutually exclusive analysis sets for the current study: (1) a subset of 1319 case–parent trios (i.e. 3957 individuals; note, from each multiplex family only one trio was chosen), and (2) a subset of 823 unrelated CL/P cases and 1700 unrelated controls (Supplementary Material, Table S1). Note that these unrelated cases did not have DNA available for one or both parents, and further that there is no overlap between the trios– and the case–control group. See the flow chart depicted in Supplementary Material, Figure S2 showing the process for assigning participants into analysis subset. A total of 317 individuals (35 cases, 282 individuals in case–parent trios) were previously genotyped and included in the CL/P GWAS by Beaty et al. (3). All Guatemalan individuals were previously genotyped and included in the GWAS by Wolf et al. (14).
Genotyping, quality control and imputation
Samples were genotyped for ∼580K SNPs using an Illumina platform by the Center for Inherited Disease Research (CIDR) at Johns Hopkins University, and data quality assurance and data cleaning were performed in collaboration with the CIDR Genetics Coordinating Center (GCC) at the University of Washington using analysis pipelines developed by the GCC (40). A total of 539,473 genotyped SNPs (comprising 96.74% of those attempted) passed quality filters recommended by the GCC (Supplementary Material, Table S2). Of these, 293,633 SNPs had a minor allele frequency (MAF) of ≥1%.
Imputation of 34,985,077 unobserved genetic polymorphisms was conducted using the software IMPUTE2 (41) and phase 3 of the 1000 Genomes Project (comprising 2504 individuals from 26 populations worldwide) as the reference panel. SNPs passing quality filters were ‘pre-phased’ to create haplotypes using SHAPEIT2 software (42) prior to imputation. Imputation accuracy was assessed by masked variant analysis, demonstrating high-quality imputation, with mean concordance of 0.995 for SNPs with MAF < 0.05 and 0.960 for SNPs with MAF ≥ 0.05. The ‘most-likely’ genotypes (i.e. genotypes with the highest probability (Q)) were selected for statistical analysis if and only if the highest probability >0.5. Imputed SNPs with deviations from Hardy–Weinberg equilibrium in European controls were filtered out of downstream analyses. Further details regarding the quality assurance, ancestry analyses, imputation, and data cleaning efforts are available in the Quality Control Report issued by the University of Washington GCC for this project and are available online (http://www.ccdg.pitt.edu/docs/Marazita_ofc_QC_report_feb2015.pdf, last accessed April 25, 2016).
PCA
PCA analysis was used, incorporating identity-by-descent estimates calculated for all pairs of participants (43) to extract the maximal subset of unrelated participants who were then included in PCA (using a pruned set of 67K SNPs chosen to be in low LD). The resulting eigenvectors were projected onto the remaining set of relatives. Supplementary Material, Figure S1 shows plots of the PCs, which strongly tracked global recruitment site and self-reported race/ethnicity. Further, although PCs of ancestry represent orthogonal information across the total multi-ethnic sample, individual PCs may be highly collinear in specific strata. Therefore, in addition to the cosmopolitan PCs generated across all participants, PCs of ancestry were also generated within each continental group (European, Central/South American and Asian, as defined by cosmopolitan PCs) for use in stratified analyses.
Statistical analyses
The TDT (44) was used to perform a GWAS in the trio set. In brief, this test detects evidence of over-transmission of a particular allele from parents to affected offspring and is robust to any population stratification. The GWAS in the unrelated case–control analysis set was performed using logistic regression while adjusting for the first 18 PCA in order to protect against genomic inflation due to population structure. Both the TDT test and the logistic regression analyses were performed as implemented in PLINK (45). The weighted OR method of meta-analysis (46) was used to combine TDT and case–control results. We performed a GWAS in the total sample, and also stratified by continental ancestry group (European, Central/South American and Asian, but not African due to insufficient sample size). For stratified analyses, we adjusted for the first five PCs in European and Central/South American samples and first three PCs in the Asian sample.
We used the conventional threshold for declaring genome-wide significance, P-value of 5 × 10−8, corresponding to the Bonferroni correction for 1 million tests. Furthermore, as an exploratory effort we scrutinized ‘suggestive’ associations with P-values <5 × 10−7 for nearby (i.e. physically proximal) genes with known biological functions that may be involved in craniofacial development. Calculations of the genomic inflation factor, ʎ, showed no significant inflation. Results of these analyses will be available for browsing as a part of the Facebase Consortium’s Human Genomics Analysis Interface Tool (www.facebase.sdmgenetics.pitt.edu, last accessed April 25, 2016).
Replication cohort and genotyping
Samples utilized for replication genotyping came from five OFC case–control population-based studies (two American and three European): the Danish National Birth Cohort (47), the Iowa Case–Control Study (Iowa) (48), the Norway Facial Clefts Study (49), the Norwegian Mother and Child Cohort (MoBa) (50), and the Utah Child and Family Health Study (Utah) (51) (Supplementary Material, Table S3). Cases and controls were identified primarily through national or state-based birth registries. Ten SNPs were selected for replication. If the best genotyped or imputed SNP was not available, a surrogate SNP was chosen. Genotyping was performed using TaqMan SNP Genotyping Assays (Life Technologies, Carlsbad, CA) on the Fluidigm microfluidic EP1 SNP Genotyping System and GT192.24 Dynamic Array Integrated Fluidic Circuits (Fluidigm, San Francisco, CA). We considered replication P-values <0.005 to be statistically significant (0.05/10 SNPs).
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
Authors thank the participating families who made the studies summarized here possible, and who provided on-going inspiration to the investigators. The tireless efforts by the dedicated field staff and collaborators was similarly essential to the success of this study. Many thanks to Kathy Bardi, Rebecca DeSensi, Margaret Cooper, Joel Anderton, Wendy Carricato, Kathleen Deeley, Keri Simonette, Matthew Ford, and Dr. Joseph Losee (University of Pittsburgh); Jennifer Standley, Nichole Nidey, Camden Bay and Erin Brothers-Smith (University of Iowa); Marcia Adams, Marie Hurley and Mary Jewell (Center for Inherited Disease Research, Johns Hopkins University); András Kiss and Eszter Nyulászi (Hungary); Rosemary Vance and the Children of the Americas (www.childrenoftheamericas.org, Guatemala); Elena Serna, Rosa Martinez, and Syed S. Hashmi (University of Texas, Houston); Dr. Ajit Ray (India); Dr. You-e Liu (China); Dr. Rick Martin (Missouri); Sondra Valdez (University of Colorado); the Smile Train and Transforming Faces Worldwide (Africa).
Conflict of Interest statement. None declared.
Funding
National Institutes of Health (X01-HG007485 to MLM, R01-DE016148 to M.L.M., U01-DE024425 to M.L.M, R37-DE008559 to J.C.M. and MLM, R01-DE009886 to M.L.M., R21-DE016930 to M.L.M., R01-DE014667 to A.C.L. and L.M.M., R21-DE016930 to M.L.M., R01-DE012472 to M.L.M., R01-DE011931 to J.T.H., R01-DE011948 to K.C., R01-DD000295 to G.L.W., U54-MD007587 to C.J.B., R25-MD007607 to C.J.B., K99-DE024571 to C.J.B., K99/R00-DE022378 to A.B., K99-DE025060 to E.J.L., and R01-DE020895 to G.L.W.). Genotyping and data cleaning were provided via an National Institutes of Health contract HHSN268201200008I to the Johns Hopkins Center for Inherited Disease Research. Additional support provided by: the Robert Wood Johnson Foundation, AMFDP Grant (72429 to A.B.); an intramural grant from the Research Institute of the Children's Hospital of Colorado (FWD); operating costs support in the Philippines was provided by the Institute of Human Genetics, National Institutes of Health, University of the Philippines, Manila (C.P.); grants through Fundação de Amparo á Pesquisa do Estado do Rio de Janeiro, Brazil (I.M.O.): grant numbers (E-26/102.797/2012, E-26/110.140/2013); grants through CNPq, Brazil (I.M.O.): grant numbers: (481069/2012-7, 306396/2013-0, 400427/2013-3). Some replication samples were from the Utah Child and Family Health study (R01-DE016877 [R.G.M.]. Some replication samples were from the Danish National Birth Cohort (PIs Mads Melbye and Jørn Olsen) funded by a major grant from the Danish National Research Foundation and initiated by the Danish Epidemiology Science Centre. Additional support for the Danish National Birth Cohort was obtained from the Pharmacy Foundation, the Egmont Foundation, the March of Dimes Birth Defects Foundation, the Augustinus Foundation, and the Health Foundation. Some replication samples were from the Norwegian Cleft Study and from the Norwegian MoBa Study, both supported by the National Institute of Environmental Health Sciences/National Institutes of Health Intramural Research Program and the Norwegian Research Council.
References
- 1.Leslie E.J., Marazita M.L. (2013) Genetics of cleft lip and cleft palate. Am. J. Med. Genet. C Semin. Med. Genet., 163C, 246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marazita M.L. (2012) The evolution of human genetic studies of cleft lip and cleft palate. Annu. Rev. Genomics Hum. Genet., 13, 263–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaty T.H., Murray J.C., Marazita M.L., Munger R.G., Ruczinski I., Hetmanski J.B., Liang K.Y., Wu T., Murray T., Fallin M.D. et al. (2010) A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat. Genet., 42, 525–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnbaum S., Ludwig K.U., Reutter H., Herms S., Steffens M., Rubini M., Baluardo C., Ferrian M., Almeida de Assis N., Alblas M.A. et al. (2009) Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat. Genet., 41, 473–477. [DOI] [PubMed] [Google Scholar]
- 5.Grant S.F., Wang K., Zhang H., Glaberson W., Annaiah K., Kim C.E., Bradfield J.P., Glessner J.T., Thomas K.A., Garris M. et al. (2009) A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24. J. Pediatr., 155, 909–913. [DOI] [PubMed] [Google Scholar]
- 6.Mangold E., Ludwig K.U., Birnbaum S., Baluardo C., Ferrian M., Herms S., Reutter H., de Assis N.A., Chawa T.A., Mattheisen M. et al. (2010) Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat. Genet., 42, 24–26. [DOI] [PubMed] [Google Scholar]
- 7.Wolf Z.T., Brand H.A., Shaffer J.R., Leslie E.J., Arzi B., Willet C.E., Cox T.C., McHenry T., Narayan N., Feingold E. et al. (2015) Genome-wide association studies in dogs and humans identify ADAMTS20 as a risk variant for cleft lip and palate. PLoS Genet., 11, e1005059.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun Y., Huang Y., Yin A., Pan Y., Wang Y., Wang C., Du Y., Wang M., Lan F., Hu Z. et al. (2015) Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate. Nat. Commun., 6, 6414.. [DOI] [PubMed] [Google Scholar]
- 9.Ludwig K.U., Mangold E., Herms S., Nowak S., Reutter H., Paul A., Becker J., Herberz R., AlChawa T., Nasser E. et al. (2012) Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat. Genet., 44, 968–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beaty T.H., Taub M.A., Scott A.F., Murray J.C., Marazita M.L., Schwender H., Parker M.M., Hetmanski J.B., Balakrishnan P., Mansilla M.A. et al. (2013) Confirming genes influencing risk to cleft lip with/without cleft palate in a case-parent trio study. Hum. Genet., 132, 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia Z., Leslie E.J., Cooper M.E., Butali A., Standley J., Rigdon J., Suzuki S., Gongorjav A., Shonkhuuz T.E., Natsume N. et al. (2015) Replication of 13q31.1 association in nonsyndromic cleft lip with cleft palate in Europeans. Am. J. Med. Genet. A., 167, 1054–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butali A., Suzuki S., Cooper M.E., Mansilla A.M., Cuenco K., Leslie E.J., Suzuki Y., Niimi T., Yamamoto M., Ayanga G. et al. (2013) Replication of genome wide association identified candidate genes confirm the role of common and rare variants in PAX7 and VAX1 in the etiology of nonsyndromic CL(P). Am. J. Med. Genet. A., 161A, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leslie E.J., Taub M.A., Liu H., Steinberg K.M., Koboldt D.C., Zhang Q., Carlson J.C., Hetmanski J.B., Wang H., Larson D.E. et al. (2015) Identification of functional variants for cleft lip with or without cleft palate in or near PAX7, FGFR2, and NOG by targeted sequencing of GWAS loci.Am. J. Hum. Genet. 96(3), 397–411.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolf Z.T., Leslie E.J., Arzi B., Jayashankar K., Karmi N., Jia Z., Rowland D.J., Young A., Safra N., Sliskovic S. et al. (2014) Genome-wide association studies in dogs and humans identify ADAMTS20 as a risk variant for cleft and palate. PLoS Genet., 10, e1004257.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bravo M.L., Valenzuela C.Y., Arcos-Burgos O.M. (1996) Polymorphisms and phyletic relationships of the Paisa community from Antioquia (Colombia). Gene Geogr, 10, 11–17. [PubMed] [Google Scholar]
- 16.Jimenez I., Mora O., Lopez G., Jimenez M.E., Zuluga L., Isaza R., Sanchez J.L., Uribe C.S., Valenzuela C.Y., Blanco R. et al. (1996) Idiopathic epilepsy with generalized tonic clonic seizures in Antioquia, Colombia: is the joint Amerindian and Negroid racial admixture the cause of its high prevalence? Biol. Res., 29, 297–304. [PubMed] [Google Scholar]
- 17.Carvajal-Carmona L.G., Soto I.D., Pineda N., Ortiz-Barrientos D., Duque C., Ospina-Duque J., McCarthy M., Montoya P., Alvarez V.M., Bedoya G. et al. (2000) Strong Amerind/white sex bias and a possible Sephardic contribution among the founders of a population in northwest Colombia. Am. J. Hum. Genet., 67, 1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mesa N.R., Mondragon M.C., Soto I.D., Parra M.V., Duque C., Ortiz-Barrientos D., Garcia L.F., Velez I.D., Bravo M.L., Munera J.G. et al. (2000) Autosomal, mtDNA, and Y-chromosome diversity in Amerinds: pre- and post-Columbian patterns of gene flow in South America. Am. J. Hum. Genet., 67, 1277–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peck J.W., Oberst M., Bouker K.B., Bowden E., Burbelo P.D. (2002) The RhoA-binding protein, rhophilin-2, regulates actin cytoskeleton organization. J. Biol. Chem., 277, 43924–43932. [DOI] [PubMed] [Google Scholar]
- 20.Biggs L.C., Naridze R.L., DeMali K.A., Lusche D.F., Kuhl S., Soll D.R., Schutte B.C., Dunnwald M. (2014) Interferon regulatory factor 6 regulates keratinocyte migration. J. Cell Sci., 127, 2840–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caddy J., Wilanowski T., Darido C., Dworkin S., Ting S.B., Zhao Q., Rank G., Auden A., Srivastava S., Papenfuss T.A. et al. (2010) Epidermal wound repair is regulated by the planar cell polarity signaling pathway. Dev. Cell, 19, 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondo S., Schutte B.C., Richardson R.J., Bjork B.C., Knight A.S., Watanabe Y., Howard E., de Lima R.L., Daack-Hirsch S., Sander A. et al. (2002) Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat. Genet., 32, 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peyrard-Janvid M., Leslie E.J., Kousa Y.A., Smith T.L., Dunnwald M., Magnusson M., Lentz B.A., Unneberg P., Fransson I., Koillinen H.K. et al. (2014) Dominant mutations in GRHL3 cause Van der Woude Syndrome and disrupt oral periderm development. Am. J. Hum. Genet., 94, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaartinen V., Haataja L., Nagy A., Heisterkamp N., Groffen J. (2002) TGFbeta3-induced activation of RhoA/Rho-kinase pathway is necessary but not sufficient for epithelio-mesenchymal transdifferentiation: implications for palatogenesis. Int. J. Mol. Med., 9, 563–570. [PubMed] [Google Scholar]
- 25.Leslie E.J., Mansilla M.A., Biggs L.C., Schuette K., Bullard S., Cooper M., Dunnwald M., Lidral A.C., Marazita M.L., Beaty T.H. et al. (2012) Expression and mutation analyses implicate ARHGAP29 as the etiologic gene for the cleft lip with or without cleft palate locus identified by genome-wide association on chromosome 1p22. Birth Defects Res. A Clin. Mol. Teratol., 94, 934–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han S., Nam J., Li Y., Kim S., Cho S.H., Cho Y.S., Choi S.Y., Choi J., Han K., Kim Y. et al. (2010) Regulation of dendritic spines, spatial memory, and embryonic development by the TANC family of PSD-95-interacting proteins. J. Neurosci., 30, 15102–15112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahmood S.F., Gruel N., Chapeaublanc E., Lescure A., Jones T., Reyal F., Vincent-Salomon A., Raynal V., Pierron G., Perez F. et al. (2014) A siRNA screen identifies RAD21, EIF3H, CHRAC1 and TANC2 as driver genes within the 8q23, 8q24.3 and 17q23 amplicons in breast cancer with effects on cell growth, survival and transformation. Carcinogenesis, 35, 670–682. [DOI] [PubMed] [Google Scholar]
- 28.Botti E., Spallone G., Moretti F., Marinari B., Pinetti V., Galanti S., De Meo P.D., De Nicola F., Ganci F., Castrignano T. et al. (2011) Developmental factor IRF6 exhibits tumor suppressor activity in squamous cell carcinomas. Proc. Natl. Acad. Sci. U. S. A., 108, 13710–13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frebourg T., Oliveira C., Hochain P., Karam R., Manouvrier S., Graziadio C., Vekemans M., Hartmann A., Baert-Desurmont S., Alexandre C. et al. (2006) Cleft lip/palate and CDH1/E-cadherin mutations in families with hereditary diffuse gastric cancer. J. Med. Genet, 43, 138–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lidral A.C., Liu H., Bullard S.A., Bonde G., Machida J., Visel A., Uribe L.M., Li X., Amendt B., Cornell R.A. (2015) A single nucleotide polymorphism associated with isolated cleft lip and palate, thyroid cancer and hypothyroidism alters the activity of an oral epithelium and thyroid enhancer near FOXE1. Hum. Mol. Genet., 24, 3895–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nissen R.M., Amsterdam A., Hopkins N. (2006) A zebrafish screen for craniofacial mutants identifies wdr68 as a highly conserved gene required for endothelin-1 expression. BMC Dev. Biol., 6, 28.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang B., Doan D., Roman Petersen Y., Alvarado E., Alvarado G., Bhandari A., Mohanty A., Mohanty S., Nissen R.M. (2013) Wdr68 requires nuclear access for craniofacial development. PLoS ONE, 8, e54363.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurihara Y., Kurihara H., Suzuki H., Kodama T., Maemura K., Nagai R., Oda H., Kuwaki T., Cao W.H., Kamada N. et al. (1994) Elevated blood pressure and craniofacial abnormalities in mice deficient in endothelin-1. Nature, 368, 703–710. [DOI] [PubMed] [Google Scholar]
- 34.Gordon C.T., Petit F., Kroisel P.M., Jakobsen L., Zechi-Ceide R.M., Oufadem M., Bole-Feysot C., Pruvost S., Masson C., Tores F. et al. (2013) Mutations in endothelin 1 cause recessive auriculocondylar syndrome and dominant isolated question-mark ears. Am. J. Hum. Genet., 93, 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richardson L., Venkataraman S., Stevenson P., Yang Y., Burton N., Rao J., Fisher M., Baldock R.A., Davidson D.R., Christiansen J.H. (2010) EMAGE mouse embryo spatial gene expression database: 2010 update. Nucleic Acids Res., 38, D703–D709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Howe D.G., Bradford Y.M., Conlin T., Eagle A.E., Fashena D., Frazer K., Knight J., Mani P., Martin R., Moxon S.A. et al. (2013) ZFIN, the Zebrafish Model Organism Database: increased support for mutants and transgenics. Nucleic Acids Res., 41, D854–D860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H., Zhang T., Wu T., Hetmanski J.B., Ruczinski I., Schwender H., Liang K.Y., Murray T., Fallin M.D., Redett R.J. et al. (2013) The FGF and FGFR gene family and risk of cleft lip with or without cleft palate. Cleft Palate. Craniofac. J., 50, 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nie X., Luukko K., Kettunen P. (2006) FGF signalling in craniofacial development and developmental disorders. Oral Dis., 12, 102–111. [DOI] [PubMed] [Google Scholar]
- 39.Mefford H., Shur N, Rosenfeld J. (1993). 15q24 microdeletion syndrome. In Pagon R.A., Adam M.P., Ardinger H.H., Wallace S.E., Amemiya A., Bean L.J.H., Bird T.D., Fong C.T., Mefford H.C., Smith R.J.H., Stephens K. (eds), In GeneReviews(R), University of Washington, Seattle, WA. [PubMed] [Google Scholar]
- 40.Laurie C.C., Doheny K.F., Mirel D.B., Pugh E.W., Bierut L.J., Bhangale T., Boehm F., Caporaso N.E., Cornelis M.C., Edenberg H.J. et al. (2010) Quality control and quality assurance in genotypic data for genome-wide association studies. Genet. Epidemiol., 34, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howie B.N., Donnelly P., Marchini J. (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet., 5, e1000529.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delaneau O., Zagury J.F., Marchini J. (2013) Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods, 10, 5–6. [DOI] [PubMed] [Google Scholar]
- 43.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.M. (2010) Robust relationship inference in genome-wide association studies. Bioinformatics, 26, 2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spielman R.S., McGinnis R.E., Ewens W.J. (1993) Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet, 52, 506–516. [PMC free article] [PubMed] [Google Scholar]
- 45.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J. et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kazeem G.R., Farrall M. (2005) Integrating case-control and TDT studies. Ann. Hum. Genet., 69, 329–335. [DOI] [PubMed] [Google Scholar]
- 47.Olsen J., Melbye M., Olsen S.F., Sorensen T.I., Aaby P., Andersen A.M., Taxbol D., Hansen K.D., Juhl M., Schow T.B. et al. (2001) The Danish National Birth Cohort–its background, structure and aim. Scand. J. Public Health, 29, 300–307. [DOI] [PubMed] [Google Scholar]
- 48.Munger R.G., Romitti P.A., Daack-Hirsch S., Burns T.L., Murray J.C., Hanson J. (1996) Maternal alcohol use and risk of orofacial cleft birth defects. Teratology, 54, 27–33. [DOI] [PubMed] [Google Scholar]
- 49.Wilcox A.J., Lie R.T., Solvoll K., Taylor J., McConnaughey D.R., Abyholm F., Vindenes H., Vollset S.E., Drevon C.A. (2007) Folic acid supplements and risk of facial clefts: national population based case-control study. BMJ (Clinical Research Ed, 334, 464.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Magnus P., Irgens L.M., Haug K., Nystad W., Skjaerven R., Stoltenberg C., MoBa Study G. (2006) Cohort profile: the Norwegian Mother and Child Cohort Study (MoBa). Int. J. Epidemiol, 35, 1146–1150. [DOI] [PubMed] [Google Scholar]
- 51.Munger R.G., Tamura T., Johnston K.E., Feldkamp M.L., Pfister R., Cutler R., Murtaugh M.A., Carey J.C. (2011) Plasma zinc concentrations of mothers and the risk of oral clefts in their children in Utah. Birt. Defects Res. A. Clin. Mol. Teratol., 91, 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics, 26, 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.